Jády et al. identify a novel class of Alu RNAs that are expressed only in the nucleoplasm of human or primate cells. They analyze Wdr79-associated RNAs and show that the right arms of nascent intronic Alu RNAs can be processed and packaged into metabolically stable box H/ACA RNPs. Their results have functional and evolutionary implications.
Keywords: Alu elements, AluACA RNAs, box H/ACA RNAs, intron-encoded RNAs, RNA processing
Abstract
Alu repetitive sequences are the most abundant short interspersed DNA elements in the human genome. Full-length Alu elements are composed of two tandem sequence monomers, the left and right Alu arms, both derived from the 7SL signal recognition particle RNA. Since Alu elements are common in protein-coding genes, they are frequently transcribed into pre-mRNAs. Here, we demonstrate that the right arms of nascent Alu transcripts synthesized within pre-mRNA introns are processed into metabolically stable small RNAs. The intron-encoded Alu RNAs, termed AluACA RNAs, are structurally highly reminiscent of box H/ACA small Cajal body (CB) RNAs (scaRNAs). They are composed of two hairpin units followed by the essential H (AnAnnA) and ACA box motifs. The mature AluACA RNAs associate with the four H/ACA core proteins: dyskerin, Nop10, Nhp2, and Gar1. Moreover, the 3′ hairpin of AluACA RNAs carries two closely spaced CB localization motifs, CAB boxes (UGAG), which bind Wdr79 in a cumulative fashion. In contrast to canonical H/ACA scaRNPs, which concentrate in CBs, the AluACA RNPs accumulate in the nucleoplasm. Identification of 348 human AluACA RNAs demonstrates that intron-encoded AluACA RNAs represent a novel, large subgroup of H/ACA RNAs, which are apparently confined to human or primate cells.
Mammalian cells express thousands of non-protein-coding RNAs (ncRNAs), which, in the form of ribonucleoproteins (ncRNPs), function in all steps of gene expression (Mattick and Makunin 2006; Wilusz et al. 2009; Taft et al. 2010). Functional ncRNPs are produced through diverse and complex biosynthetic processes, which usually include the synthesis and processing of precursor ncRNAs, the assembly of mature ncRNAs with RNP proteins, and finally, the targeting of nascent ncRNPs to the appropriate subcellular compartments where they function.
Box H/ACA RNAs represent an abundant and evolutionarily conserved group of ncRNAs (for reviews, see Meier 2005; Terns and Terns 2006; Matera et al. 2007; Kiss et al. 2010; Watkins and Bohnsack 2012). Most box H/ACA RNAs function as guide RNAs in site-specific pseudouridylation of rRNAs and spliceosomal snRNAs (Ganot et al. 1997a; Ni et al. 1997; Darzacq et al. 2002) but also participate in pre-rRNA processing, telomere synthesis, and probably other processes (Morrissey and Tollervey 1993; Mitchell et al. 1999; Chen et al. 2000; Atzorn et al. 2004; Fayet-Lebaron et al. 2009). The majority of human H/ACA RNAs are processed from pre-mRNA introns, and the mature RNAs are typically composed of two 55- to 60-nucleotide (nt)-long hairpins connected and followed by short single-stranded hinge and tail sequences (Fig. 1A; Kiss et al. 1996; Ganot et al. 1997b). While the hinge region contains the conserved H box (consensus AgAnnA), the 3′-terminal tail carries the ACA or, less frequently, the AUA box motif located always 3 nt before the RNA end (Balakin et al. 1996; Ganot et al. 1997b). The bipartite guide sequences occupy the rising and descending strands of the internal pseudouridylation loop in the 5′ and/or 3′ hairpin (Ganot et al. 1997a). Each hairpin unit associates with four H/ACA core proteins: Nhp2, Nop10, Gar1, and the pseudouridine synthase dyskerin (for reviews, see Reichow et al. 2007; Ye 2007; Li 2008; Kiss et al. 2010; Watkins and Bohnsack 2012). In the H/ACA RNP, base-pairing of the H/ACA guide sequences with the complementary target RNA places the unpaired substrate uridine at the active site of dyskerin (Li and Ye 2006; Duan et al. 2009; Liang et al. 2009). Optimally, the so-called “pseudouridylation pocket” of the guide RNA is located 14 nt upstream of the H or ACA box that is a fundamental coordinator of correct dyskerin binding. Box H/ACA RNPs mediating rRNA pseudouridylation accumulate in the nucleolus and are termed the small nucleolar RNPs (snoRNPs), while those H/ACA RNPs that direct snRNA pseudouridylation reside in the nucleoplasmic Cajal bodies (CBs) and are called the small CB RNPs (scaRNPs) (Darzacq et al. 2002; Kiss et al. 2006). The terminal loop of the 5′ and/or 3′ hairpin of scaRNAs carries the CAB box (consensus, ugAG) that binds Wdr79 (also known as Wrap53) and targets scaRNPs into CBs (Richard et al. 2003; Tycowski et al. 2009; Venteicher et al. 2009).
Figure 1.
Human box H/ACA and Alu RNAs. (A) Structure and function of human box H/ACA pseudouridylation guide RNAs. The highly and moderately conserved nucleotides in the H, ACA, and CAB box motifs are in capital and lowercase letters, respectively. (N) Any nucleotide; (Ψ) uridines selected for pseudouridylation in the substrate RNA. The distance between the selected uridine and the H or ACA box of the guide RNA is ∼14 nt. (B) Organization and expression of Pol III-specific Alu RNA genes. The upstream enhancer, the internal A and B promoter, and the downstream terminator elements are indicated. The internal and terminal A-rich regions (An) are shown. Schematic structures of the full-length and 3′-terminally processed (scAlu) Alu RNAs are shown. The internal A-rich (A) and L regions required for Pol II inhibition are in shaded boxes.
Alu elements are abundant primate-specific repetitive sequences that are present in >1 million similar but not identical copies in the human genome (Lander et al. 2001). Full-length Alu elements are ∼300 nt long and are composed of two tandem monomer units, called the left and right arms, both followed by short A-rich sequences (Fig. 1B; for reviews, see Batzer and Deininger 2002; Kramerov and Vassetzky 2005; Berger and Strub 2011). Alu elements arose from the RNA polymerase III (Pol III)-specific signal recognition particle (SRP) 7SL RNA gene and were amplified throughout the genome by RNA retrotransposition (Batzer and Deininger 2002; Dewannieux et al. 2003; Hasler et al. 2007; Kriegs et al. 2007). Due to their preferential interspersion within gene-rich regions, Alu elements are coincidently transcribed by Pol II into pre-mRNAs (Lander et al. 2001; Chen et al. 2002; Versteeg et al. 2003). While Alu sequences imbedded in the untranslated regions of mRNAs may control mRNA stability and translation, the intronic Alu RNAs are believed to be rapidly degraded after pre-mRNA splicing (Cordaux and Batzer 2009; Ponicsan et al. 2010; Berger and Strub 2011).
Using the internal A and B Pol III-binding motifs inherited from the 7SL gene, some Alu elements are independently transcribed, albeit with low efficiency, into 300- to 400-nt-long Alu RNAs that terminate in at least four consecutive U residues encoded by downstream DNAs (Fig. 1B). The Pol III-specific primary Alu transcripts can be further processed into more stable small cytoplasmic Alu (scAlu) RNAs encompassing only the left monomer (Chang et al. 1996; Sarrowa et al. 1997). Both full-length and processed Alu RNAs interact with the heterodimer of the Spr9 and Srp14 7SL SRP proteins (Bovia et al. 1995; Hsu et al. 1995). Alu RNAs accumulate at very low levels (100–1000 copies per cell), but upon stress induction, cellular Alu concentrations show 10-fold to 20-fold increases, indicating that Alu RNAs play a role in stress response (Hasler et al. 2007; Ponicsan et al. 2010; Berger and Strub 2011). Nuclear full-length Alu RNAs have been reported to repress the transcription of protein-coding genes during heat shock because synthetic Alu RNAs were found to bind Pol II to prevent its interaction with promoter DNA in in vitro assembled preinitiation complexes (Mariner et al. 2008; Yakovchuk et al. 2009). Although both Alu arms seem to bind Pol II, the transcriptional inhibitory activity is limited to the right arm. More recently, aberrant overaccumulation of Alu RNAs in retinal pigment epithelium (RPE) cells has been reported to induce cytotoxicity, leading to the development of geographic atrophy, a severe form of age-related macular degeneration (Kaneko et al. 2011).
In this study, analysis of human Wdr79-associated RNAs followed by in vivo processing studies demonstrates that the right arms of nascent intronic Alu RNAs, instead of being degraded after pre-mRNA splicing, can be processed and packaged into metabolically stable box H/ACA RNPs. We also demonstrate that the newly discovered Alu-derived H/ACA RNPs, contrary to their association with the CB localization protein Wdr79, accumulate in the nucleoplasm of human cells. Since AluACA RNAs are processed from the primate-specific right Alu monomers, they represent a large group of ncRNAs that are expressed exclusively in human or primate cells.
Results
Human Wdr79 copurifies with novel Alu-derived H/ACA RNAs
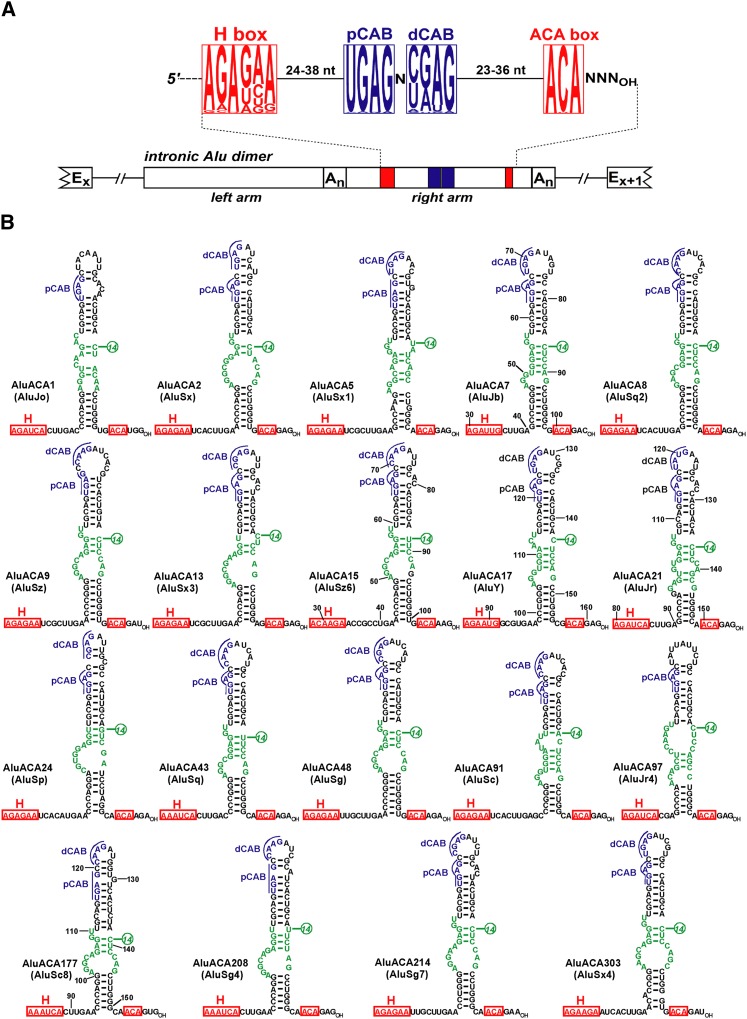
To identify novel Wdr79-associated RNAs, a Flag-tagged version of human Wdr79 (FL-Wdr79) was transiently expressed in human HeLa cells and purified by immunoprecipitation (IP) with an anti-Flag antibody. RNAs coprecipitated with FL-Wdr79 were converted into cDNAs, and after PCR amplification, their 3′-terminal nucleotide sequences were determined by strand-specific high-throughput sequencing with an Illumina Genome Analyzer (Fasteris AG). The majority (60%) of the 5.75 million total reads that aligned to the human genome sequence represented contaminating RNAs, mainly spliceosomal snRNAs, box C/D and H/ACA snoRNAs, and rRNAs. The abundant snRNAs and snoRNAs transiently accumulate in CBs and might associate, most probably nonspecifically, with the Wdr79 CB protein. Nevertheless, 36% of the positive reads corresponded to known or predicted H/ACA and C/D scaRNAs (data not shown). To our surprise, the remaining 235,000 reads, ∼4% of the total reads, represented Alu repetitive sequences. A detailed analysis of the obtained Alu sequences resulted in the identification of 348 novel putative Alu RNAs (Supplemental Table 1). To unambiguously distinguish between closely related Alu sequences, we considered only those Alu reads that were at least 75 nt long and perfectly aligned with a unique region of the reported human genome sequence (University of California at Santa Cruz [UCSC] Genome Browser). Likewise, Alu RNAs represented by <20 reads were also excluded from the list of Alu RNAs. The 3′-terminal sequences of the new Alu RNAs were present within the right arms of dimeric or, in a few cases, monomeric Alu elements, which fell into 19 different subgroups of the three major Alu subfamilies (AluS, AluJ, and AluY), indicating that expression and association of Alu RNAs with Wdr79 is not an Alu family-specific affair (Fig. 2A; Supplemental Table 2).
Figure 2.
Human AluACA RNAs. (A) Organization of human intronic AluACA RNA genes. Exons (Ex and Ex+1) of the host gene and the A-rich sequences (An) following the left and right arms of the intronic full-length Alu element are indicated. Frequencies of the four ribonucleotides in the predicted H, pCAB, dCAB, and ACA motifs of 348 AluACA RNAs are indicated by the heights of the corresponding letters. The 5′-terminal hairpin regions of AluACA RNAs are not included. (B) Proposed two-dimensional structures of the 3′ end regions of human AluACA RNAs. For each Alu subgroup, one sample RNA represented by the highest number of sequence reads is shown. The conserved H and ACA box motifs are in red, and the proximal and distal CAB boxes (pCAB and dCAB) are highlighted in blue. Potential pseudouridylation guide sequences are in green. Nucleotides located 14 nt upstream of the ACA box are indicated. Numbering of AluACA7, AluACA15, AluACA17, AluACA21, and AluACA177 RNAs was based on mappings of transiently expressed RNAs (see Fig. 4).
We noticed that the newly discovered Wdr79-associated Alu RNAs carried an ACA or, in two instances, an AUA motif located invariantly 3 nt before the RNA 3′ end (Fig. 2A; Supplemental Table 1). Inspection of the genomic coding sequences of the new Alu RNAs revealed that they always contained a potential H box motif (consensus, AGAgaA) situated 67–76 nt upstream of the 3′ end of the RNA. Computer-mediated structure predictions revealed that the putative H and ACA motifs of all Alu RNAs were folded together by 50- to 60-nt-long hairpins that are highly reminiscent of the 3′ hairpins of canonical box H/ACA RNAs. The predicted 3′-terminal structures of 19 Alu RNAs representing different Alu subgroups are shown in Figure 2B. Finally, we also noticed that the 3′-terminal hairpin of the great majority of Alu RNAs carried two closely spaced potential CB localization signals: the highly conserved proximal CAB box (pCAB) located in the raising strand of the hairpin and the less conserved distal CAB box (dCAB) in the terminal loop (Fig. 2A,B; Supplemental Table 1). These observations together strongly suggested that the newly detected Wdr79-associated Alu RNAs, hereafter called AluACA RNAs, represent a novel, large subclass of human H/ACA RNAs.
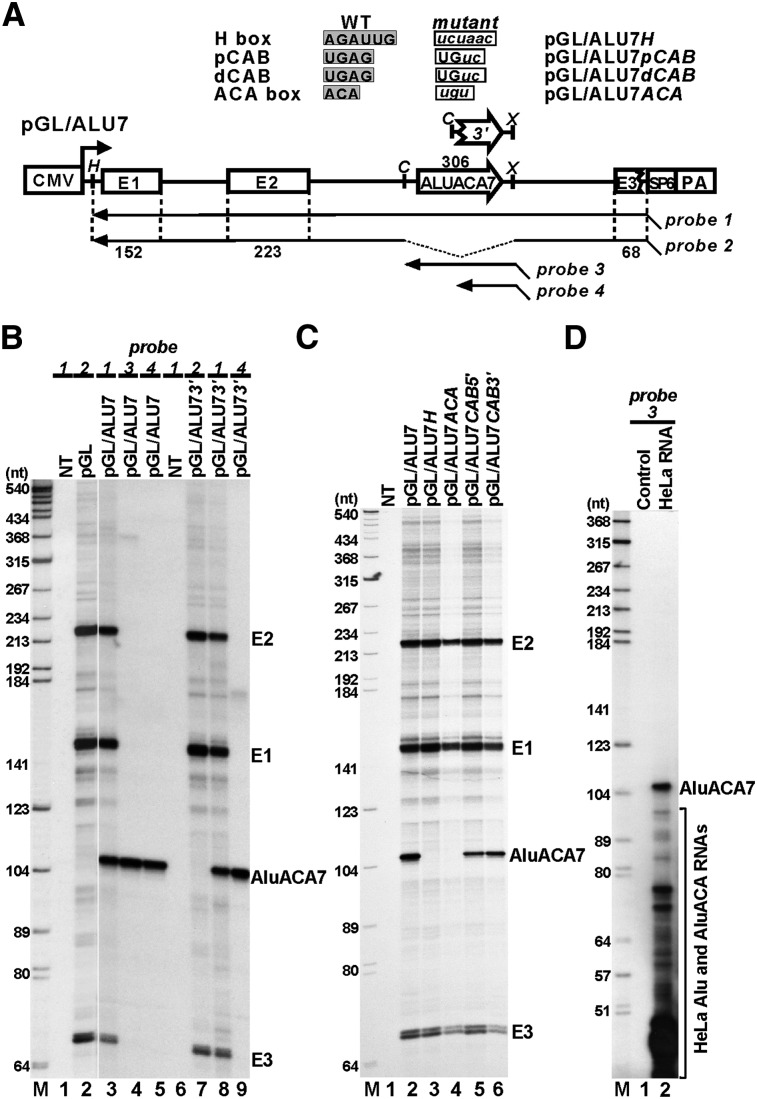
AluACA7 is processed from the second intron of the β-globin pre-mRNA reporter in an H and ACA box-dependent manner
Apart from a few exceptions (see the Discussion), the genomic Alu DNAs encoding the new AluACA RNAs were found within introns of established or predicted protein-coding genes (Fig. 2A; Supplemental Table 3). Importantly, the expressed RNA sequences lay invariantly on the sense intronic DNA strand, suggesting that they were synthesized within and processed from their host pre-mRNAs. To test this notion, the full-length human ALUACA7 element that encodes the AluACA7 RNA and resides within the first intron of the PARP1 (poly-ADP-ribose polymerase family, member 1) gene was PCR-amplified and inserted into the second intron of the human β-globin gene in the pGL expression vector (Fig. 3A). The resulting pGL/ALU7 and the empty pGL expression plasmids were transfected into HeLa cells, and accumulation of the processed AluACA7 and β-globin RNAs was monitored by RNase A/T1 protection using internally labeled sequence-specific antisense RNA probes (Fig. 3A). Mapping of the RNAs derived from pGL-transfected cells with a β-globin-specific probe (probe 2) confirmed the accumulation of the three processed β-globin mRNA exons: E1, E2, and E3 (Fig. 3B, lane 2). In pGL/ALU7-transfected cells, besides the spliced globin exons, a novel, ∼105-nt-long RNA was detected with the GL/ALU7-specific probe 1 (Fig. 3B, lane 3). Mapping of the same RNA with probes complementary to the full-length ALUACA7 DNA (probe 3) or to the 3′-terminal right arm of ALUACA7 (probe 4) demonstrated that the accumulating new RNA was processed from the right arm of the expressed intronic ALUACA7 RNA (Fig. 3B, lanes 4,5). This conclusion was further corroborated by the observation that upon transfection of HeLa cells with the pGL/ALU73′ expression plasmid that carried only the right arm of ALUACA7, the 105-nt-long AluACA7 RNA efficiently accumulated (Fig. 3B, lanes 8,9). This also demonstrated that the accumulating AluACA7 RNA, instead of being synthesized from putative internal Pol III promoter elements located in the left arm of ALUACA7 DNA, was processed from the globin pre-mRNA synthesized by Pol II from the CMV promoter.
Figure 3.
Processing of AluACA7 RNA from the second intron of transiently expressed β-globin pre-mRNA. (A) Schematic structure of the pGL/ALU7 expression vector. The cytomegalovirus promoter (CMV), the exon regions of the β-globin gene (E1–E3), the polyadenylation site (PA), and the SP6 promoter used for synthesis of antisense RNA probes are indicated. The open arrow indicates the full-length dimeric ALUACA7 DNA, inserted into the second intron of the human β-globin gene. The broken arrow represents the 3′-terminal right arm of the ALUACA7 Alu element. Relevant restriction sites are shown. (H) HindIII; (C) ClaI; (X) XhoI. The antisense RNA probes (probes 1–4), together with the lengths of globin exons, are shown. Nucleotide alterations introduced into the wild-type (WT) box H, pCAB, dCAB, and ACA sequence motifs of ALUACA7 are indicated. (B) In vivo processing of AluACA7 from the globin pre-mRNA. Cellular RNAs extracted from HeLa cells nontranfected (NT) or transfected with the pGL, pGL/ALU7, or pGL/ALU73′ expression plasmids were mapped by RNase protection performed with antisense RNAs indicated above the lanes. Probe RNAs protected by the overexpressed AluACA7 RNA and the processed exons (E1–E3) of the globin mRNA were separated on a 6% sequencing gel and visualized by autoradiography. (Lane M) Size markers in nucleotides. (C) Expression of AluACA7 depends on the H and ACA boxes. RNAs from HeLa cells transfected with the indicated expression plasmids were mapped with sequence-specific RNA probes. (D) Detection of HeLa endogenous AluACA7 RNA. HeLa cellular RNA (30 μg) was mapped with a high-specific-activity RNA probe complementary to the ALUACA7 DNA. Probe RNA fragments protected by AluACA7 or partially protected by other AluACA and Pol III-transcribed Alu RNAs were separated on a sequencing gel. Control mapping with Escherichia coli tRNA is shown.
To test whether the predicted H, ACA, pCAB, and dCAB boxes are essential for the in vivo processing of AluACA7 from the transiently expressed globin pre-mRNA, these sequence motifs were altered in the pGL/ALU7 expression construct (Fig. 3A). The resulting mutant pGL/ALU7H, pGL/ALU7ACA, pGL/ALU7pCAB, and pGL/ALU7dCAB expression plasmids were transfected into HeLa cells. RNase mappings with specific probes demonstrated that disruption of the H and ACA motifs fully abolished accumulation of the mutant AluACA7 RNAs but had no effect on globin mRNA expression (Fig. 3C, lanes 3,4). Alteration of the putative pCAB and dCAB motifs only slightly reduced the accumulation of the mutant AluACA7 RNAs (Fig. 3C, lanes 5,6).
To confirm that the transiently expressed AluACA7 RNA was faithfully processed from the β-globin pre-mRNA, we determined the length of endogenous HeLa AluACA7 RNA. Mapping of HeLa RNAs from nontransfected cells failed to detect AluACA7, indicating that this RNA accumulates at a very low level in HeLa cells (Fig. 3B [lanes 1,6], C [lane 1]). However, when the amount of HeLa RNA used for mapping, the specific activity of the ALUACA7-specific probe (probe 3), and the time of autoradiography were increased ∼10-fold each, the endogenous HeLa AluACA7 RNA was readily detected and, similarly to the transiently expressed RNA, protected an ∼105-nt-long fragment of the RNA probe (Fig. 3D, lane 2). Thus, we concluded that the AluACA7 RNA was faithfully processed from the second intron of the transiently expressed globin pre-mRNA.
The 5′-terminal hairpin of AluACA RNAs shows strong structural variation
The correct 5′ and 3′ termini of transiently overexpressed AluACA7 RNA were determined by primer extension and 3′ end race analysis, respectively (Fig. 4A,B). These experiments confirmed that the human AluACA7 RNA is composed of 105 nt and therefore represents the shortest human H/ACA RNA reported thus far. Notably, AluACA7 carries a 29-nt-long 5′-terminal hairpin that is unusually short for H/ACA RNAs (Fig. 4C). To test whether a short 5′ hairpin is a common hallmark of AluACA RNAs, four other RNAs (AluACA15, AluACA17, AluACA21, and AluACA177), representing different Alu subfamilies (Fig. 2B), were transiently overexpressed in HeLa cells using the pGL expression vector (Fig. 4D). RNase mappings demonstrated that all Alu RNAs were processed from the transiently expressed globin pre-mRNA, albeit with different efficiency (Fig. 4D, lanes 3,4,7,8,11,12,15,16). The accumulating AluACA RNAs showed strong size variations. While the apparent length of AluACA15 was similar to that of AluACA7, the other three RNAs (AluACA17, AluACA21, and AluACA177) migrated above the 152-nt-long first exon (E1) of the globin mRNA. Importantly, mapping of endogenous HeLa AluACA15, AluACA17, AluACA21, and AluACA177 using increased amounts of cellular RNAs and high specific activity probes confirmed that the transiently expressed AluACA15, AluACA17, AluACA21, and AluACA177 RNAs were faithfully processed from the globin pre-mRNA (Fig. 5A; data not shown). Computer folding of the 5′-terminal genomic sequences predicted by the measured lengths of the mature RNAs revealed that the human AluACA15, AluACA17, AluACA21, and AluACA177 RNAs carry 27-, 85-, 79-, and 82-nt-long 5′-terminal hairpins, respectively (Fig. 4C). While the short AluACA7 (105-nt) and AluACA15 (107-nt) RNAs were processed exclusively from the right arms of intronic Alu dimers, the 5′-terminal nucleotides of the long AluACA17 (162-nt) and AluACA21 (155-nt) RNAs derived from the terminal regions of the upstream left Alu arms. Interestingly, the 5′-terminal sequences of the 158-nt-long AluACA177 RNA that is encoded by a right Alu monomer originated from upstream non-Alu-flanking sequences. Thus, we propose that in contrast to the highly conserved 3′-terminal hairpin unit, the 5′-terminal hairpins of human intron-encoded AluACA RNAs show strong sequence and structural variations.
Figure 4.
Mapping of transiently overexpressed AluACA RNAs. (A) Determination of the 5′ terminus of transiently overexpressed AluACA7 with primer extension analysis. The 5′-terminal nucleotides of AluACA7 are indicated. (B) Mapping of the 3′ terminus of AluACA7 with RNA 3′ end race. The 3′-terminal AluACA7 and the 5′-terminal oligonucleotide tag sequences are shown. (C) Proposed secondary structures of the 5′ hairpins of AluACA RNAs. The 5′-terminal sequences of AluACA15, AluACA17, AluACA21, and AluACA117 were inferred from the lengths of transiently expressed RNAs measured by RNase mappings. The predicted H boxes are indicated. Nucleotides derived from left Alu arms or from upstream non-Alu-flanking sequences are shown. (D) Transient expression of AluACA RNAs in HeLa cells. RNAs from HeLa cells nontransfected (NT) or transfected with the indicated expression plasmids were analyzed by RNase mappings with sequence-specific probes indicated above the lanes. For other details, see the legend for Figure 3B.
Figure 5.
Protein composition of AluACA RNPs. (A) Association of HeLa endogenous AluACA7 and AluACA15 RNAs with H/ACA core proteins and Wdr79. Extracts prepared from HeLa cells nontransfected (NT) or transfected with pFL-DYS, pNHP2-GFP, pGAR1-GFP, or pNOP10-GFP were subjected to IP with the indicated antibodies. IP of transiently expressed FL-Dys, Nhp2-GFP, Gar1-GFP, and Nop10-GFP proteins and endogenous HeLa Wdr79 and Srp14 was monitored by Western blot analysis. RNAs prepared from cell extracts (Ext) and the pellets of IP reactions were analyzed by RNase mapping with sequence-specific RNA probes indicated on the right. (Lane M) DNA and protein size markers. (B) Association of transiently overexpressed AluACA RNAs with scaRNP proteins. HeLa cells transfected with pGL/ALU7 or pGL/ALU15 were cotransfected with the pFL-DYS or pFL-WDR expression plasmids as indicated. Extracts prepared from nontransfected (NT) or transfected cells were subjected to IP with the indicated antibodies. RNAs prepared from cell extracts and pellets of IP reactions were analyzed by RNase protection.
AluACA RNAs associate with box H/ACA core proteins and Wdr79
We next assayed whether the newly discovered human AluACA RNAs are associated with the four box H/ACA RNP proteins: dyskerin, Nhp2, Gar1, and Nop10. Flag-tagged dyskerin (FL-Dys) and green fluorescent protein (GFP)-tagged Nhp2-GFP, Gar1-GFP, and Nop10-GFP proteins were transiently expressed in HeLa cells and, after extract preparation, immunoprecipitated with anti-Flag and anti-GFP antibodies (Fig. 5A). IP of the epitope-tagged H/ACA proteins was confirmed by Western blot analysis (Fig. 5A, lanes 4,8,12,16). RNase protection analysis was used to monitor the association of the recovered H/ACA proteins with HeLa endogenous AluACA7 and AluACA15 RNAs as well as with the U85 H/ACA-C/D composite scaRNA and the 7SL SRP RNA, which were used as positive and negative controls, respectively (Darzacq et al. 2002). Similar to the U85 scaRNA, HeLa AluACA7 and AluACA15 RNAs copurified with epitope-tagged dyskerin, Nhp2, Gar1, and Nop10. Likewise, IP of the Wdr79 scaRNP protein from a HeLa cell extract with a specific antibody recovered U85, AluACA7, and AluACA15, confirming the specificity of the observed interaction of transiently expressed Flag-tagged Wdr79 with HeLa AluACA RNAs (Fig. 5A, lane 20). As expected, none of the tested H/ACA scaRNP proteins interacted with HeLa 7SL RNA (Fig. 5A, lanes 4,8,12,16,20). On the other hand, IP of HeLa Srp14, a component of the Srp9/Srp14 heterodimer that binds to Pol III-specific Alu RNAs and the “Alu domain” of 7SL RNA (Bovia et al. 1995; Hsu et al. 1995), efficiently recovered the 7SL RNA but failed to pull down U85, AluACA7, and AluACA15 (Fig. 5A, lane 24).
Finally, we wanted to confirm that similar to the endogenous HeLa AluACA RNAs, transiently overexpressed AluACA RNAs processed from the globin pre-mRNA are accurately packaged into H/ACA scaRNPs. The AluACA7 and AluACA15 RNAs, together with FL-Dys and FL-Wdr79, were transiently expressed in HeLa cells (Fig. 5B). Upon IP of the accumulating FL-Dys and FL-Wdr79 proteins, RNase mapping demonstrated that similar to the control U85 scaRNA, both AluACA7 and AluACA15 were present in the pellets of the IP reactions, which, on the other hand, lacked the 7SL RNA (Fig. 5B, lanes 3,5,10,12). As predicted, IP of the Srp14 SRP protein, which recovered the 7SL RNA, failed to precipitate the AluACA7, AluACA15, and U85 RNAs (Fig. 5B, lanes 7,14). These observations demonstrated that human AluACA RNAs associate with the four H/ACA core proteins dyskerin, Nhp2, Nop10, and Gar1 as well as with the proposed CB localization protein Wdr79.
The CAB boxes of AluACA7 are essential for both Wdr79 binding and efficient RNA accumulation
Binding of the Wdr79 scaRNP protein is directed by a short sequence motif, the CB localization signal (CAB box), located in the terminal loop of the 5′ and/or 3′ hairpin of box H/ACA scaRNAs. Surprisingly, the terminal stem–loop region of the 3′ hairpin of AluACA RNAs carries two closely spaced potential Wdr79-binding motifs: the distal and proximal CAB boxes (Fig. 2). To test the functional significance of the two potential CAB boxes of AluACA RNAs, mutant AluACA7 RNAs with altered proximal (AluACA7pCAB) or distal (AluACA7dCAB) CAB motifs were transiently expressed in HeLa cells (Fig. 6A). Association of the accumulating mutant AluACA7 RNAs with HeLa Wdr79 was measured by co-IP experiments using an anti-Wdr79 antibody. Compared with the control wild-type AluACA7 (Fig. 6A, lane 2), the mutant AluACA7pCAB and AluACA7dCAB RNAs showed reduced Wdr79-binding capacities (Fig. 6A, lanes 4,6). Simultaneous disruption of both CAB boxes fully abolished the Wdr79-binding ability of the double mutant AluACA7pdCAB RNA (Fig. 6A, lane 8). Moreover, as compared with the wild-type RNA, the double mutant AluACA7pdCAB RNA lacking functional CAB boxes accumulated at a highly reduced level, contrary to the fact that accumulation of the host globin mRNA showed no alteration (Fig. 6A, cf. lanes 1 and 7). This suggested that besides binding of Wdr79, the CAB boxes are also critical for efficient accumulation of AluACA RNAs.
Figure 6.

The CAB boxes of AluACA RNAs function in Wdr79 binding. (A) The proximal and distal CAB boxes of AluACA7 bind Wdr79 in a cumulative manner. Nucleotide alterations introduced into the proximal and distal CAB box of AluACA7 are indicated. Extracts prepared from HeLa cells transfected with the pGL/ALU7 (WT), pGL/ALU7pCAB, pGL/ALU7dCAB, or pGL/ALU7pdCAB expression plasmids were reacted with an anti-Wdr79 antibody. RNAs isolated from cell extracts and the pellets of IP reactions were analyzed by RNase mappings. (B) The CAB boxes of AluACA RNAs are essential for both Wdr79 binding and efficient RNA accumulation. Schematic structure of the U64-AluACA7 composite RNA is shown. Extracts prepared from HeLa cells nontransfected (NT) or transfected with pGL/U64-ALU7 (WT) or pGL/U64-ALU7pdCAB were reacted with an anti-Wdr79 antibody. RNAs isolated from cell extracts and the pellets of IP reactions were analyzed by RNase protection. The first exon (E1) of globin mRNA is indicated.
To further assay the role of the two CAB boxes of AluACA RNAs in RNA accumulation, we constructed and transiently expressed a hybrid AluACA RNA encompassing the 3′ hairpin unit of AluACA7 and the 5′-terminal hairpin region of the U64 box H/ACA snoRNA (Fig. 6B). The U64-AluACA7 composite RNA accumulated with 15–20 times higher efficiency than the wild-type AluACA7, indicating that the inherently weak expression of AluACA7 is at least partially due to its unusually short 5′ hairpin (Fig. 6B, lane 2). Upon IP of HeLa Wdr79, the U64-AluACA7 composite RNA coprecipitated with Wdr79, demonstrating that the 5′ hairpin unit of AluACA7 is dispensable for Wdr79 binding (Fig. 6B, lane 3). Alteration of both the distal and proximal CAB boxes of U64-AluACA7, besides fully disrupting Wdr79 binding, largely reduced the accumulation of the double mutant U64-AluACA7pdCAB RNA (Fig. 6B, lanes 4,5). Based on these observations, we concluded that the two CAB box motifs of AluACA7, besides recruiting Wdr79 in a cumulative fashion, contribute to the efficient accumulation of AluACA7.
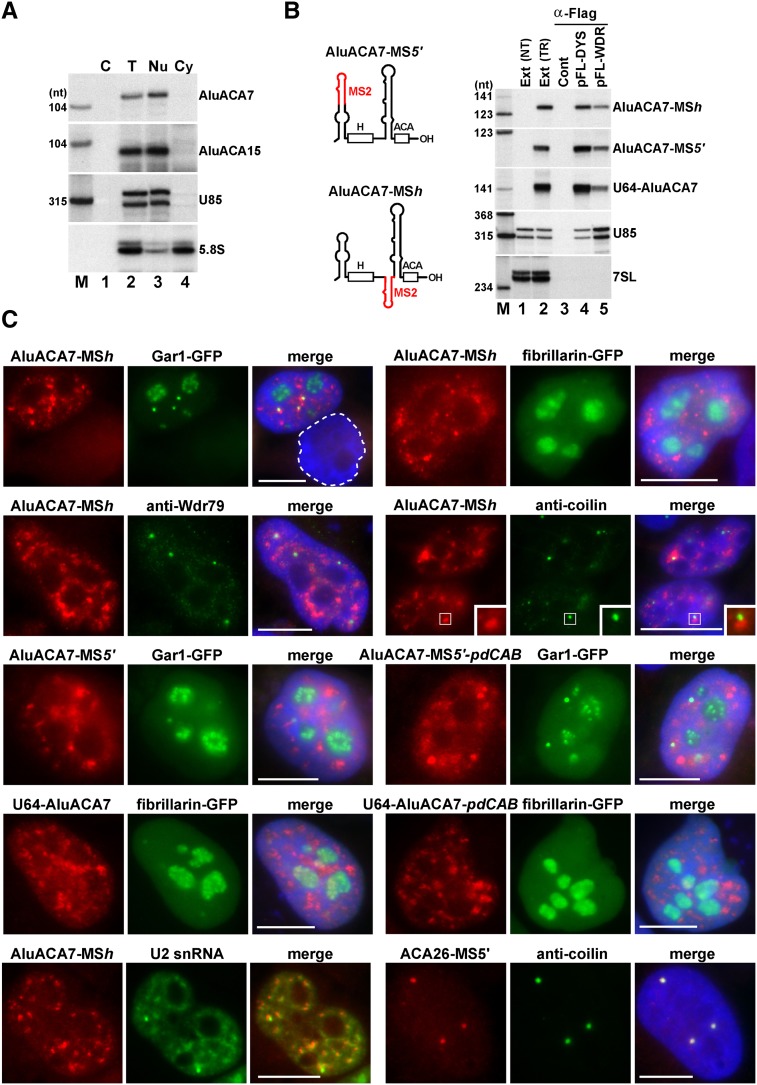
Transiently expressed tagged AluACA7 RNAs accumulate in the nucleoplasm
While all box H/ACA RNAs tested so far accumulated within the nucleus, Alu RNAs have been detected in both the nucleus and cytoplasm (for reviews, see Kiss et al. 2006; Berger and Strub 2011). To learn about the subcellular localization of human AluACA RNAs, HeLa cells were fractionated into nuclear and cytoplasmic fractions, and the distribution of the endogenous AluACA7 and AluACA15 RNAs was monitored by RNase mapping (Fig. 7A). Together with the control U85 scaRNA that accumulates in the nucleoplasmic CBs (Darzacq et al. 2002), both AluACA7 and AluACA15 RNAs were detected in the nuclear fraction and were missing from the cytoplasm, where the majority of the 5.8S rRNA accumulated (Fig. 7A, lanes 3,4).
Figure 7.
Subcellular localization of AluACA RNAs. (A) HeLa AluACA7 and AluACA15 RNAs accumulate in the nucleus. RNA samples isolated from either HeLa cells (T) or the nuclear (Nu) and cytoplasmic (Cy) fractions of HeLa cells were mapped by RNase A/T1 protection with antisense RNA probes as indicated on the right. (Lane C) Control mapping with E. coli tRNA. (B) Expression of internally tagged AluACA7 RNAs in HeLa cells. Schematic structures of transiently expressed AluACA7-MSh and AluACA7-MS5′ RNAs with the inserted MS2 coat protein-binding motifs (in red) are shown. For the structure of U64-AluACA7, see Figure 6B. Extracts (Ext) prepared from HeLa cells nontransfected (NT) or transfected (TR) with the indicated combination of the pGL/ALU7-MSh, pGL/ALU7-MS5′ pGL/U64-ALU7, pFL-DYS, and pFLWDR79 expression plasmids were reacted with an anti-Flag antibody. RNAs prepared from cell extracts or the pellets of IP reactions were analyzed by RNase mapping. Lane Cont shows mapping with E. coli tRNA. (C) Fluorescent in situ hybridization. HeLa cells transiently expressing the indicated tagged or chimeric version of AluACA7 RNA were stained with sequence-specific fluorescent oligonucleotide probes. Nucleoli were visualized by expression of fibrillarin-GFP or Gar1-GFP. CBs were immunostained with antibodies against Wdr79 or coilin. DAPI staining of nuclear DNA was omitted at U2 localization. Bar, 10 μm.
We used fluorescent in situ hybridization microscopy to determine the precise subnuclear localization of AluACA RNAs. To avoid undesired cross-hybridization of AluACA-specific oligonucleotide probes with Pol III-synthesized full-length or 3′-terminally processed Alu RNAs or with pre-mRNAs containing Alu sequences, modified AluACA7 RNAs carrying an MS2 coat protein-binding motif in either the hinge (AluACA7-MSh) or the 5′-terminal hairpin (AluACA7-MS5′) were transiently expressed in HeLa cells using the pGL expression vector (Fig. 7B). RNase mappings confirmed that both MS2-tagged AluACA RNAs were correctly and efficiently processed from the β-globin host pre-mRNA (Fig. 7B, lane 2). Coexpression and IP of FL-Dys and FL-Wdr79 proteins revealed that the accumulating AluACA7-MSh and AluACA7-MS5′ RNAs were packaged into Wdr79-associated H/ACA RNPs (Fig. 7B, lanes 4,5). Moreover, cell fractionation experiments showed that the transiently expressed AluACA7-MS RNPs, similar to the endogenous AluACA7 and AluACA15 RNPs, accumulated within the nucleus of transfected cells (data not shown).
Probing of cells expressing AluACA7-MSh or AluACA7-MS5′ RNAs with a fluorescently labeled MS2-specific oligonucleotide probe revealed that both RNAs accumulated in the nucleoplasm with predominant exclusion from the nucleolar territories, which were stained by coexpression of GFP-tagged nucleolar marker proteins Gar1 and fibrillarin (Fig. 7C). Importantly, our MS2 probe failed to specifically stain nontransfected cells (Fig. 7C, circled) or highlight any RNA upon Northern blotting of HeLa total RNAs (data not shown). Moreover, insertion of the MS2 tag into the 5′ hairpin of the ACA26 H/ACA scaRNA failed to alter the CB-specific accumulation of the transiently expressed ACA26-MS RNA, excluding the formal possibility that the MS2 tag sequences targeted AluACA7 into the nucleoplasm. Since all Wdr79-associated RNAs, including box H/ACA and C/D scaRNAs, concentrate in nucleoplasmic CBs, HeLa cells expressing AluACA7-MSh were costained with the MS2 probe and antibodies against the coilin and Wdr79 CB marker proteins. While coilin and Wdr79 were enriched in CBs, the AluACA7-MSh RNA showed no significant concentration within CBs, although it frequently accumulated in the close vicinity of CBs. Similarly to the MS2-tagged AluACA7 RNAs, the transiently expressed U64-AluACA7 chimeric RNA showed a predominant nucleoplasmic distribution. A weak accumulation of U64-AluACA7 within the nucleolar territories was probably due to its high level of expression. Nevertheless, these results confirm that the cis-acting elements supporting the nucleoplasmic accumulation of AluACA7 are located in the 3′-terminal hairpin of the RNA. Simultaneous disruption of the distal and proximal CAB box motifs failed to alter the nucleoplasmic localization of the mutant U64-AluACA7-pdCAB and the AluACA7-MS5′-pdCAB RNAs, indicating that Wdr79 binding is not required for the nucleoplasmic localization of AluACA RNAs.
Instead of being evenly dispersed in the nucleoplasm, the MS2-tagged AluACA7 RNAs showed a speckled localization pattern highly reminiscent of the nucleoplasmic distribution of spliceosomal snRNAs (Carmo-Fonseca et al. 1991a,b). Upon costaining of HeLa cells with fluorescent oligonucleotide probes specific for the transiently expressed AluACA7-MSh RNA and the endogenous U2 snRNA, the AluACA7 and U2 signals failed to perfectly overlap in the nucleoplasm. Moreover, in contrast to U2 that concentrated in the nucleoplasmic speckles, the AluACA7-MSh RNA showed no significant enrichment in speckles and paraspeckles (data not shown). In conclusion, in situ localization experiments confirmed that the newly discovered AluACA RNPs represent a novel subgroup of human box H/ACA RNPs that accumulate in the nucleoplasm of human cells.
Discussion
Despite the fact that Alu repetitive elements constitute >10% of the human genome, expression of Alu RNAs is remarkably low (Berger and Strub 2011). Genomic Alu elements can be transcribed by either Pol III using the weak internal promoters of Alu DNAs or Pol II as parts of long pre-mRNA transcripts. In this study, we demonstrated that intron-embedded nascent Alu RNAs can be processed into metabolically stable RNAs. The novel intron-encoded Alu RNAs, termed AluACA RNAs, feature the characteristic hallmarks of box H/ACA scaRNAs: They fold into the common hairpin–hinge–hairpin–tail structure and carry the conserved H, ACA, and CAB boxes of H/ACA scaRNAs. Consequently, the mature AluACA RNAs associate with the five H/ACA scaRNP proteins dyskerin, Nop10, Nhp2, Gar1, and Wdr79.
Our in vivo processing studies confirmed that the intronic AluACA RNAs, instead of being transcribed independently by Pol III, are post-transcriptionally processed from their host pre-mRNAs in a box H- and ACA-dependent fashion (Fig. 3). Consistent with this biosynthetic pathway, the genomic copies of 337 new AluACA RNAs, out of 348, were found within introns of known or predicted protein-coding genes invariantly in the sense orientation (Supplemental Table 3). Although the remaining 11 new RNAs (AluACA11, AluACA63, AluACA82, AluACA83, AluACA100, AluACA111, AluACA134, AluACA324, AluACA328, AluACA342, and AluACA348) were mapped to intergenic regions lacking established Pol II transcription units, it is unlikely that these RNAs are synthesized independently by Pol III because primary H/ACA RNA transcripts generated by Pol III are not recognized as substrates for in vivo H/ACA RNA processing (Richard et al. 2006). Therefore, we favor the idea that these intergenic Alu elements are transcribed within yet-undetected Pol II transcripts or are misplaced in the human genome sequence.
We identified 348 novel AluACA RNAs (Supplemental Table 1), but the final number of human AluACA RNAs is apparently much higher. During the processing of our sequence data, we followed a stringent filtering strategy to definitively distinguish between highly similar genomic Alu elements. We ignored almost 200 potential AluACA RNA sequences that were either <75 nt or absent from the human genome reference sequence (UCSC Genome Browser). It is important to mention that dozens of our Alu reads, although missing from the UCSC genome sequence, were detected in other databases, indicating that they likely represent bona fide AluACA RNAs. We can also predict that human cells in different tissues and at different stages of development express many additional intronic AluACA RNAs. Thus, we propose that human cells express several hundreds or maybe thousands of AluACA RNAs. However, contrary to their high number, AluACA RNAs show a low level of global accumulation in HeLa cells. As predicted by the distribution of AluACA and scaRNA reads in our deep sequencing data set, AluACA RNAs accumulate at a 10 times lower level than do canonical scaRNAs.
Human Alu elements fall into three major subfamilies of different evolutionary ages: the young AluY and the more ancient AluS and AluJ subfamilies (Batzer and Deininger 2002). Pol III preferentially transcribes AluY elements, which constitute the smallest Alu subfamily (Shaikh et al. 1997). In contrast, the subfamily distribution of the 348 new intronic AluACA RNAs—specifically, 37 AluY, 78 AluJ, and 233 AluS RNAs—shows a strong correlation with the genomic abundance of AluY, AluJ, and AluS elements (Supplemental Table 2). In primates, fusion of the more ancient free left Alu monomer with the primate-specific free right Alu monomer led to the prevailing dimeric Alu organization (Quentin 1992; Kriegs et al. 2007). The new AluACA RNAs, with no exception, derived from right Alu monomers. As both originated from the 7SL RNA, the left and right Alu monomers are highly similar, except that the right monomer contains a 31-nt-long insertion missing from the left monomer (Supplemental Fig. 2). In fact, this inserted extra sequence accounts for the AluACA-coding capacity of the right Alu monomer because it constitutes the terminal stem–loop region and carries the proximal and distal CAB boxes of the 3′ hairpins of AluACA RNAs. Given that right Alu arm sequences are confined to primate genomes, the AluACA RNAs represent a human-specific or maybe primate-specific subgroup of box H/ACA RNAs (Quentin 1992). Therefore, it seems that gaining AluACA RNA-coding capacity by Alu DNAs is a recent event of primate genome evolution, although we cannot exclude the possibility that primordial SRP RNA originated from ancient H/ACA RNAs.
Besides their apparent similarity to canonical H/ACA scaRNPs, the AluACA RNPs also possess distinctive structural features. The AluACA RNAs may carry unusually short 5′ hairpins that are not anticipated to support stable H/ACA RNP assembly (Fig. 4). The archaeal homolog of Nhp2, called L7Ae, tethers Nop10 and the catalytic domain of dyskerin to the upper stem of H/ACA RNAs by forming a specific interaction with the conserved Kink turn motif (Rozhdestvensky et al. 2003; Baker et al. 2005; Charpentier et al. 2005; Liang et al. 2007). Eukaryotic H/ACA RNAs lack Kink turns, suggesting that Nhp2, contrary to its structural similarity to L7Ae (Koo et al. 2011), forms a more flexible interaction with the structurally variable upper stems of H/ACA RNAs (Li et al. 2012). Accumulation of AluACA7 RNA requires the H box motif (Fig. 3), indicating that the short 5′ hairpin of AluACA7 still binds dyskerin that recognizes the H box and the preceding basal stem of H/ACA RNAs (Li and Ye 2006). Eukaryotic dyskerin can form a stable complex with Nop10, Nhp2, and Gar1 even in the absence of H/ACA RNAs (Henras et al. 2004; Wang and Meier 2004; Li et al. 2012). This suggests that the short 5′ hairpins of AluACA RNAs can bind the complete set of H/ACA core proteins, contrary to the apparent absence of a docking surface for Nhp2. Nevertheless, it seems that suboptimal structural organization of the 5′ hairpin units is the major reason of the remarkable weak cellular accumulation of AluACA RNAs. Supporting this idea, replacement of the short and long 5′ hairpin units of AluACA7 and AluACA17, respectively, for the 5′ hairpin of the U64 box H/ACA snoRNA dramatically increased the accumulation levels of both RNAs (Fig. 6B; data not shown).
The 3′-terminal or 5′-terminal hairpin of canonical H/ACA scaRNAs contains one CAB box motif that is essential for Wdr79 binding and CB localization but dispensable for RNA accumulation (Richard et al. 2003; Jády et al. 2004). In contrast, the 3′ hairpin of AluACA RNAs carries two closely spaced CAB motifs that bind Wdr79 in a cumulative fashion and are crucial for efficient RNA expression (Fig. 6). It is unclear whether the two CAB boxes promote RNA accumulation directly through binding Wdr79 or indirectly through recruiting putative trans-acting RNA processing and/or RNP assembly factors (Theimer et al. 2007). Wdr79 has been proposed to function as a localization signal protein that targets box H/ACA and C/D scaRNPs into CBs (Tycowski et al. 2009; Venteicher et al. 2009). However, the demonstration that Wdr79-associated AluACA RNPs accumulate in the nucleoplasm suggests that the molecular mechanism supporting the subnuclear localization of human box H/ACA RNPs is more complex than anticipated before (Fig. 7).
The function of AluACA RNPs remains elusive. The great majority of box H/ACA RNAs direct site-specific RNA pseudouridylation (see above). Intriguingly, the 3′-terminal hairpins of AluACA RNAs contain perfect pseudouridylation guide loops that, in principle, could position the putative target uridine 14 nt upstream of the ACA box, which is an optimal configuration for efficient pseudouridylation (Fig. 2B). Compared with the bracketing basal and upper stems, the putative pseudouridylation loops of AluACA RNAs show increased sequence variability. Thus, AluACA RNAs could direct pseudouridylation of many different target RNAs. We are currently testing whether in vitro and in vivo assembled AluACA RNPs possess pseudouridine synthase activity. The AluACA RNAs might also function as transcriptional regulators, as the right arms of human Alu RNAs have been reported to inhibit Pol II transcription through binding to core Pol II at promoters of transcriptionally repressed genes (Mariner et al. 2008; Yakovchuk et al. 2009). However, mature AluACA RNAs partially lack the internal L region and frequently miss the entire A-rich motif, which are essential for Alu-mediated Pol II repression (Fig. 1). Understanding the nucleoplasmic function of human AluACA snRNPs remains an exciting task for the future.
Playing important roles in all steps of gene expression, ncRNAs contribute to the biological complexity of eukaryotic organisms. In this study, besides uncovering a novel, unpredicted role for human Alu repetitive elements, we identified and partially characterized a novel, large subgroup of box H/ACA RNPs that are apparently confined to human or primate cells. Although the nucleoplasmic role of the newly discovered AluACA RNPs remains unknown, they likely contribute to the genetic and biochemical complexity of human cells.
Materials and methods
Unless stated otherwise, all manipulations were performed according to standard laboratory protocols (Sambrook et al. 1989). Modified and unmodified oligodeoxynucleotides were purchased from Eurofins MWG Operon.
Expression constructs
Full-length human Alu DNA elements encoding the AluACA7, AluACA15, AluACA17, AluACA21, and AluACA177 RNAs, together with their 69-/45-, 58-/80-, 65-/74-, 71-/75-, and 159-/124-base-pair (bp) 5′- and 3′-flanking sequences, respectively, were PCR-amplified using HeLa nuclear DNA as a template. To obtain the pBST/ALU7, pGL/ALU7, pBST/ALU15, pGL/ALU15, pBST/ALU17, pGL/ALU17, pBST/ALU21, pGL/ALU21, pBST/ALU177, and pGL/ALU117, the amplified Alu fragments were inserted into the ClaI and XhoI sites of the pBluescript (pBST) and pGL plasmids (Darzacq et al. 2002). Construction of the pGL/ALU7H, pGL/ALU7pCAB, pGL/ALU7dCAB, pGLALU7pdCAB pGL/ALU7ACA, pGL/ALU73′, pGL/ALU7MSh, pGL/ALU7MS5′, and pGL/ALU7MS5′pdCAB expression vectors carrying mutants versions of the ALUACA7 gene was performed by PCR amplification using the appropriate mutagenic primers. The MS2 coat protein-binding sequence 5′-AAACAUGAGGAUCACCCAUGUC-3′ was inserted between the A9 and U20 (AluACA-MS5′) or U38 and A40 (AluACA-MSh) residues of AluACA7. To generate pGL/U64-ALUACA7, the 5′ hairpin and the adjacent H box of the human U64 snoRNA gene (1–72 nt) were PCR-amplified and fused to the coding DNA of the second hairpin of AluACA7 (36–105 nt) using a two-step PCR amplification approach. The resulting U64-ALUACA7 DNA was inserted into the ClaI and XhoI sites of pGL and pBST. The pGL/U64-ALUACA7pdCAB expression plasmid was constructed by PCR amplification using an appropriate mutagenic oligonucleotide primer and pGL/U64-ALUACA7 as a template. To obtain pGL/ACA26-MS5′, the MS2 coat protein-binding motif was inserted between the A31 and A32 residues of the ACA26 gene in the pGL/ACA26 expression plasmid (Kiss et al. 2004). The pFL-WDR79 expression vector was purchased for GeneCopoeia (EX-V0379-M12). To obtain FL-DYS, the human dyskerin cDNA was PCR-amplified with an upstream oligonucleotide that contained a Flag motif and was inserted into the KpnI and BamHI sites of pcDNA3. The pNHP2-GFP, pGAR1-GFP, pNOP10-GFP, and pFIB-GFP expression plasmids have been reported (Dundr et al. 2000; Darzacq et al. 2006). The identity of all of the constructs was verified by sequence analysis.
RNA analyses
RNAs from HeLa cells and from the nuclear and cytoplasmic fractions of HeLa cells were extracted by the guanidine thyocyanate/phenol-chloroform extraction method (Goodall et al. 1990). Primer extension analysis was performed using a 5′ end-labeled oligonucleotide primer complementary to AluACA7 from G59 to C79. RNA 3′-terminal sequence race and RNase A/T1 protection analysis have been described (Goodall et al. 1990; Kiss and Filipowicz 1993). To generate low and high specific activity (sp. act.) antisense RNA probes, the pGL/ALU and pBST/ALU recombinant plasmids were digested with HindIII and ClaI and used as a template for in vitro transcription with SP6 and T7 RNA polymerases, respectively, in the presence of [α-P32]CTP (sp. act. 220 or 2700 Ci/mmol). All probes were purified on denaturing polyacrylamide gels. About 2 μg of human HeLa RNA coimmunoprecipitated with transiently expressed Flag-tagged Wdr79 was used as a template for generation of a cDNA library that was analyzed by high-throughput sequencing using Solexa technology (Fasteris SA).
Protein analyses
Preparation of HeLa cell extracts, cell fractionation, IP of snRNP complexes, and Western blot analysis of proteins have been described (Tyc and Steitz 1989). For Western blot analysis, we used the following antibodies: mouse monoclonal anti-Flag M2 (1:10,000 dilution; Sigma), anti-GFP (1:1000 dilution; Roche), rabbit polyclonal anti-WDR (1:200 dilution; Abcam ab99376), and anti-SRP14 antibodies (1:1000 dilution; ProteinTech Group, Inc.).
Fluorescent in situ hybridization
Fluorescent in situ hybridization has been described (Darzacq et al. 2002). Specific oligodeoxynucleotides containing aminoallyl nucleotides (*) were labeled with FluoroLink Cy3 monofunctional reactive dye (GE Healthcare) to detect the MS2 tag (5′-AT*TTTGTACTCCT*AGTGGGTACAGT*A-3′) or the U64-AluACA7 and U64-AluACA7pdCAB RNAs (5′-C*TCCCAGGCTCAAGT*CTTTTCTCTCTCTCCC*-3′). Nuclear DNA was visualized with 1 μg/mL DAPI. Wdr79 and coilin were detected by rabbit polyclonal anti-Wdr79 (1:50 dilution; Abcam ab99376) and a monoclonal mouse anti-coilin antibody (1:100 dilution; a kind gift of Dr. M. Carmo-Fonseca).
European nucleotide archive accession numbers
The accession numbers for the 348 AluACA RNA sequences, from AluACA1–AluACA348, reported in this study are HE85591917–HE856264, respectively.
Acknowledgments
We thank X. Darzacq (Ecole Normale Supérieure, Paris, France) and M. Dundr (Rosalind Franklin University of Medicine and Science, Chicago) for providing us with pGAR-GFP, pNHP2-GFP, pNOP10-GFP, and pFIB-GFP expression plasmids. We are indebted to M. Carmo-Fonseca (Universidade de Lisboa, Portugal) for providing us with anti-coilin antibody. We are grateful to H. Seitz for his help in initial data analysis. Our work was supported by la Fondation pour la Recherche Médicale grant to T.K.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.197467.112.
References
- Atzorn V, Fragapane P, Kiss T 2004. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol Cell Biol 24: 1769–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DL, Youssef OA, Chastkofsky MI, Dy DA, Terns RM, Terns MP 2005. RNA-guided RNA modification: Functional organization of the archaeal H/ACA RNP. Genes Dev 19: 1238–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakin AG, Smith L, Fournier MJ 1996. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86: 823–834 [DOI] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL 2002. Alu repeats and human genomic diversity. Nat Rev Genet 3: 370–379 [DOI] [PubMed] [Google Scholar]
- Berger A, Strub K 2011. Multiple roles of Alu-related noncoding RNAs. Prog Mol Subcell Biol 51: 119–146 [DOI] [PubMed] [Google Scholar]
- Bovia F, Fornallaz M, Leffers H, Strub K 1995. The SRP9/14 subunit of the signal recognition particle (SRP) is present in more than 20-fold excess over SRP in primate cells and exists primarily free but also in complex with small cytoplasmic Alu RNAs. Mol Biol Cell 6: 471–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Sproat BS, Ansorge W, Swanson MS, Lamond AI 1991a. In vivo detection of snRNP-rich organelles in the nuclei of mammalian cells. EMBO J 10: 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Tollervey D, Pepperkok R, Barabino SM, Merdes A, Brunner C, Zamore PD, Green MR, Hurt E, Lamond AI 1991b. Mammalian nuclei contain foci which are highly enriched in components of the pre-mRNA splicing machinery. EMBO J 10: 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DY, Hsu K, Maraia RJ 1996. Monomeric scAlu and nascent dimeric Alu RNAs induced by adenovirus are assembled into SRP9/14-containing RNPs in HeLa cells. Nucleic Acids Res 24: 4165–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier B, Muller S, Branlant C 2005. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res 33: 3133–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Blasco MA, Greider CW 2000. Secondary structure of vertebrate telomerase RNA. Cell 100: 503–514 [DOI] [PubMed] [Google Scholar]
- Chen C, Gentles AJ, Jurka J, Karlin S 2002. Genes, pseudogenes, and Alu sequence organization across human chromosomes 21 and 22. Proc Natl Acad Sci 99: 2930–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA 2009. The impact of retrotransposons on human genome evolution. Nat Rev Genet 10: 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Jády BE, Verheggen C, Kiss AM, Bertrand E, Kiss T 2002. Cajal body-specific small nuclear RNAs: A novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J 21: 2746–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT 2006. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol 173: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 35: 41–48 [DOI] [PubMed] [Google Scholar]
- Duan J, Li L, Lu J, Wang W, Ye K 2009. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol Cell 34: 427–439 [DOI] [PubMed] [Google Scholar]
- Dundr M, Misteli T, Olson MO 2000. The dynamics of postmitotic reassembly of the nucleolus. J Cell Biol 150: 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayet-Lebaron E, Atzorn V, Henry Y, Kiss T 2009. 18S rRNA processing requires base pairings of snR30 H/ACA snoRNA to eukaryote-specific 18S sequences. EMBO J 28: 1260–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Bortolin ML, Kiss T 1997a. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809 [DOI] [PubMed] [Google Scholar]
- Ganot P, Caizergues-Ferrer M, Kiss T 1997b. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev 11: 941–956 [DOI] [PubMed] [Google Scholar]
- Goodall GJ, Wiebauer K, Filipowicz W 1990. Analysis of pre-mRNA processing in transfected plant protoplasts. In Methods enzymology (ed. JE Dahlberg and JN Abelson), pp. 148–161. Academic Press, New York [DOI] [PubMed] [Google Scholar]
- Hasler J, Samuelsson T, Strub K 2007. Useful ‘junk’: Alu RNAs in the human transcriptome. Cell Mol Life Sci 64: 1793–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras AK, Capeyrou R, Henry Y, Caizergues-Ferrer M 2004. Cbf5p, the putative pseudouridine synthase of H/ACA-type snoRNPs, can form a complex with Gar1p and Nop10p in absence of Nhp2p and box H/ACA snoRNAs. RNA 10: 1704–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K, Chang DY, Maraia RJ 1995. Human signal recognition particle (SRP) Alu-associated protein also binds Alu interspersed repeat sequence RNAs. Characterization of human SRP9. J Biol Chem 270: 10179–10186 [DOI] [PubMed] [Google Scholar]
- Jády BE, Bertrand E, Kiss T 2004. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol 164: 647–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, et al. 2011. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature 471: 325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Filipowicz W 1993. Small nucleolar RNAs encoded by introns of the human cell cycle regulatory gene RCC1. EMBO J 12: 2913–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Bortolin ML, Filipowicz W 1996. Characterization of the intron-encoded U19 RNA, a new mammalian small nucleolar RNA that is not associated with fibrillarin. Mol Cell Biol 16: 1391–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss AM, Jády BE, Bertrand E, Kiss T 2004. Human box H/ACA pseudouridylation guide RNA machinery. Mol Cell Biol 24: 5797–5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Fayet E, Jády BE, Richard P, Weber M 2006. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb Symp Quant Biol 71: 407–417 [DOI] [PubMed] [Google Scholar]
- Kiss T, Fayet-Lebaron E, Jády BE 2010. Box H/ACA small ribonucleoproteins. Mol Cell 37: 597–606 [DOI] [PubMed] [Google Scholar]
- Koo BK, Park CJ, Fernandez CF, Chim N, Ding Y, Chanfreau G, Feigon J 2011. Structure of H/ACA RNP protein Nhp2p reveals cis/trans isomerization of a conserved proline at the RNA and Nop10 binding interface. J Mol Biol 411: 927–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov DA, Vassetzky NS 2005. Short retroposons in eukaryotic genomes. Int Rev Cytol 247: 165–221 [DOI] [PubMed] [Google Scholar]
- Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J 2007. Evolutionary history of 7SL RNA-derived SINEs in supraprimates. Trends Genet 23: 158–161 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Li H 2008. Unveiling substrate RNA binding to H/ACA RNPs: One side fits all. Curr Opin Struct Biol 18: 78–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ye K 2006. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 443: 302–307 [DOI] [PubMed] [Google Scholar]
- Li S, Duan J, Li D, Yang B, Dong M, Ye K 2012. Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase. Genes Dev 25: 2409–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Xue S, Terns RM, Terns MP, Li H 2007. Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex. Nat Struct Mol Biol 14: 1189–1195 [DOI] [PubMed] [Google Scholar]
- Liang B, Zhou J, Kahen E, Terns RM, Terns MP, Li H 2009. Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nat Struct Mol Biol 16: 740–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA 2008. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 29: 499–509 [DOI] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP 2007. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 8: 209–220 [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV 2006. Non-coding RNA. Hum Mol Genet 15: R17–R29 doi: 10.1093/hmg/ddl046 [DOI] [PubMed] [Google Scholar]
- Meier UT 2005. The many facets of H/ACA ribonucleoproteins. Chromosoma 114: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K 1999. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol 19: 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey JP, Tollervey D 1993. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol 13: 2469–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89: 565–573 [DOI] [PubMed] [Google Scholar]
- Ponicsan SL, Kugel JF, Goodrich JA 2010. Genomic gems: SINE RNAs regulate mRNA production. Curr Opin Genet Dev 20: 149–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y 1992. Fusion of a free left Alu monomer and a free right Alu monomer at the origin of the Alu family in the primate genomes. Nucleic Acids Res 20: 487–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichow SL, Hamma T, Ferre-D'Amare AR, Varani G 2007. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res 35: 1452–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Darzacq X, Bertrand E, Jády BE, Verheggen C, Kiss T 2003. A common sequence motif determines the Cajal body-specific localisation of box H/ACA scaRNAs. EMBO J 22: 4283–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Kiss AM, Darzacq X, Kiss T 2006. Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol Cell Biol 26: 630–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A 2003. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: A shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res 31: 869–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T 1989. Molecular cloning: A laboratory manual Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Sarrowa J, Chang DY, Maraia RJ 1997. The decline in human Alu retroposition was accompanied by an asymmetric decrease in SRP9/14 binding to dimeric Alu RNA and increased expression of small cytoplasmic Alu RNA. Mol Cell Biol 17: 1144–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TH, Roy AM, Kim J, Batzer MA, Deininger PL 1997. cDNAs derived from primary and small cytoplasmic Alu (scAlu) transcripts. J Mol Biol 271: 222–234 [DOI] [PubMed] [Google Scholar]
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS 2010. Non-coding RNAs: Regulators of disease. J Pathol 220: 126–139 [DOI] [PubMed] [Google Scholar]
- Terns MP, Terns R 2006. Noncoding RNAs of the H/ACA family. Cold Spring Harb Symp Quant Biol LXXI: 395–405 [DOI] [PubMed] [Google Scholar]
- Theimer CA, Jády BE, Chim N, Richard P, Breece KE, Kiss T, Feigon J 2007. Structural and functional characterization of human telomerase RNA processing and cajal body localization signals. Mol Cell 27: 869–881 [DOI] [PubMed] [Google Scholar]
- Tyc K, Steitz JA 1989. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J 8: 3113–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Kukoyi A, Steitz JA 2009. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell 34: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE 2009. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323: 644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg R, van Schaik BD, van Batenburg MF, Roos M, Monajemi R, Caron H, Bussemaker HJ, van Kampen AH 2003. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res 13: 1998–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Meier UT 2004. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J 23: 1857–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Bohnsack MT 2012. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 3: 397–414 [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL 2009. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev 23: 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovchuk P, Goodrich JA, Kugel JF 2009. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc Natl Acad Sci 106: 5569–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K 2007. H/ACA guide RNAs, proteins and complexes. Curr Opin Struct Biol 17: 287–292 [DOI] [PubMed] [Google Scholar]