Mroczek et al. propose a model in which the yeast protein Usb1 and its human homolog act as RNases that remove terminal nucleotides from the U6 transcript, leading to mature snRNA formation. These findings suggest that C16orf57 encodes the long-sought U6 snRNA phosphodiesterase, an essential mRNA splicing regulator, and provide a molecular basis for human diseases such as poikilothermia, Rothmund-Thomson syndrome, and dyskeratosis congenita.
Keywords: C16orf57, U6 snRNA, USB1, hUSB1, phosphodiesterase, poikiloderma with neutropenia
Abstract
C16orf57 encodes a human protein of unknown function, and mutations in the gene occur in poikiloderma with neutropenia (PN), which is a rare, autosomal recessive disease. Interestingly, mutations in C16orf57 were also observed among patients diagnosed with Rothmund-Thomson syndrome (RTS) and dyskeratosis congenita (DC), which are caused by mutations in genes involved in DNA repair and telomere maintenance. A genetic screen in Saccharomyces cerevisiae revealed that the yeast ortholog of C16orf57, USB1 (YLR132C), is essential for U6 small nuclear RNA (snRNA) biogenesis and cell viability. Usb1 depletion destabilized U6 snRNA, leading to splicing defects and cell growth defects, which was suppressed by the presence of multiple copies of the U6 snRNA gene SNR6. Moreover, Usb1 is essential for the generation of a unique feature of U6 snRNA; namely, the 3′-terminal phosphate. RNAi experiments in human cells followed by biochemical and functional analyses confirmed that, similar to yeast, C16orf57 encodes a protein involved in the 2′,3′-cyclic phosphate formation at the 3′ end of U6 snRNA. Advanced bioinformatics predicted that C16orf57 encodes a phosphodiesterase whose putative catalytic activity is essential for its function in vivo. Our results predict an unexpected molecular basis for PN, DC, and RTS and provide insight into U6 snRNA 3′ end formation.
Poikiloderma with neutropenia (PN) is a rare, autosomal recessive skin condition (OMIM 604173) in which mutations in the C16orf57 gene were found (Volpi et al. 2010). C16orf57 encodes a protein of unknown molecular function. To date, 19 distinct C16orf57 mutations have been identified in 31 patients with PN (for details, see Supplemental Table S1; Arnold et al. 2010; Tanaka et al. 2010; Volpi et al. 2010; Clericuzio et al. 2011; Colombo et al. 2012), all of which lead to the generation of truncated, and most likely nonfunctional, proteins. C16orf57 is also mutated in a subset of patients diagnosed with dyskeratosis congenita (DC) and Rothmund-Thomson syndrome (RTS) (Walne et al. 2010; Piard et al. 2012). Interestingly, DC and RTS share many similar clinical phenotypes with PN; however, mutations in C16orf57 can be used as molecular markers that allow for correct diagnosis (Piard et al. 2012). DC is often caused by mutations in factors involved in telomere maintenance (Armanios 2009), while RTS is primarily caused by mutations in the RECQL4 DNA helicase involved in DNA repair and replication (Larizza et al. 2010). The correlation between these diseases and mutations in C16orf57 suggests that it may encode a protein involved in ensuring genome integrity. However, no changes in telomere length were observed in RTS patients with C16orf57 mutations (Walne et al. 2010), which raised the question of what the physiological function of C16orf57 is.
In this study, we took advantage of an unbiased yeast genetic screen to investigate the function of C16orf57. Our results, coupled with functional experiments in yeast and human cells as well as bioinformatic analyses, indicated that C16orf57 and its yeast ortholog, USB1 (for U Six Biogenesis 1), may encode the long-sought U6 small nuclear RNA (snRNA) phosphodiesterase, which is an essential enzyme in the biogenesis of the splicing apparatus. Splicing is the process of removing introns and is an essential step in mRNA maturation (Wahl et al. 2009). In eukaryotes, this process is mediated by a dynamic macromolecular assembly composed of five snRNA particles and associated proteins, which are together referred to as small nuclear ribonucleoproteins (snRNPs; U1, U2, U4, U5, U6). The U1, U2, U4, and U5 snRNAs are transcribed by RNA polymerase II and subsequently capped, similar to other transcripts. The U6 snRNA, however, is transcribed by RNA polymerase III and never leaves the nucleus. Mature U6 is involved in catalysis of the splicing transesterification reactions and 5′ splice site selection (Kandels-Lewis and Seraphin 1993; Lesser and Guthrie 1993; Valadkhan 2010). The U6 snRNA undergoes large conformational changes during splicing. At the beginning of the splicing cycle, the U1 and U2 snRNPs base-pair with the pre-mRNA. The U6 snRNA, together with the U4 and U5 snRNAs, as a part of assembly called the U4/U6.U5 tri-snRNP, then enters the prespliceosome. Prior to the splicing of the pre-mRNA, the U1 and U4 snRNAs dissociate from the complex, which allows the U6 snRNA to base-pair with the U2 snRNA and the pre-mRNA to form an active site of the spliceosome (Valadkhan 2010). After the catalytic splicing steps, the U6 snRNA dissociates from the spliceosome with the U6-associated LSM proteins. The proper recycling of the U6 snRNA requires base-pairing with the U4 snRNA, which is promoted by Prp24, another U6 snRNP protein. The cycle is completed when the U5 snRNP is recruited to form the U4/U6.U5 tri-snRNP.
In addition to the general mechanism of mRNA splicing, there is substantial research regarding the regulation and processing of the U6 snRNA itself (Patel and Bellini 2008). Transcription of the U6 snRNA terminates at a uridine tract, which is the typical RNA polymerase III termination signal, similar to other transcripts, such as tRNAs and 5S rRNA. In humans, the U6 snRNA is post-transcriptionally oligouridynylated by a specific poly(U) polymerase (TUTase) (Trippe et al. 2003, 2006). Interestingly, the last nucleotide of ∼90% of human U6 snRNAs in the cell is modified to form a terminal 2′,3′-cyclic phosphate (U>P) (Lund and Dahlberg 1992; Schutz et al. 2010). On the other hand, in yeast, uridines at the 3′ end of the U6 snRNA are generated by transcription, and the RNA molecule is terminated with a 2′ or 3′ phosphate moiety or hydroxyl groups (Lund and Dahlberg 1992). It has been suggested that this U6 snRNA modification is required for the proper assembly of the mature snRNP (Gu et al. 1997). Similar to most polymerase III transcripts, the nascent U6 snRNA is bound by La antigen, and the modification of its 3′ end promotes the interaction between the RNA and the heptameric ring of LSM proteins (Pannone et al. 2001). It has also been suggested that the modification of the U6 snRNA 3′ end takes place dynamically during every spliceosome assembly and disassembly; however, the function of such a mechanism in splicing remains obscure (Tazi et al. 1993).
Despite our understanding of RNA splicing and the role of the U6 snRNA in this essential process, the enzymes responsible for modifying the U6 snRNA 3′ end in yeast and humans remain unknown. In this study, we identified C16orf57 (hUsb1) as a putative phosphodiesterase essential for the generation of the cyclic phosphate at the 3′ end of the U6 snRNA. The C16orf57 gene product is therefore a factor involved in the biogenesis of the pre-mRNA splicing apparatus, which would have been extremely difficult to identify had it not been for its yeast ortholog. We propose a model in which Usb1/hUsb1 acts as an RNase responsible for the removal of the terminal nucleotide from the U6 transcript, leading to formation of the mature snRNA.
Results
Usb1 depletion causes in vivo splicing defects and loss of U6 snRNA
The C16orf57 gene is evolutionarily conserved in all major eukaryotic phyla, with an ortholog in yeast, YLR132C, encoding yeast Usb1 (Supplemental Fig. S1), which, similar to its human counterpart, has no annotated function. In order to determine the role of Usb1, we first studied its role in yeast growth. We constructed a GAL#x2237;USB1 strain in which the expression of USB1 is under the galactose-inducible GAL1 promoter. In the presence of galactose, Usb1 is produced, whereas in glucose-rich medium, the GAL1 promoter is repressed, leading to a depletion of Usb1 protein. Using this strain, we confirmed that Usb1 was essential for yeast viability, and its metabolic depletion led to a growth defect (Fig. 1A). To gain insight into the function of Usb1, we next performed a multicopy suppressor screen using the GAL#x2237;USB1 strain. The GAL#x2237;USB1 strain was transformed with a yeast multicopy library, plated on selective medium with galactose as the sole carbon source, and then transferred to restrictive, glucose-rich medim to suppress USB1 expression. We screened 10,000 clones and isolated 30 colonies that were able to restore growth on glucose medium. Sequencing the plasmids isolated from these 30 colonies identified the wild-type USB1 or SNR6 gene, encoding the U6 snRNA, as the suppressors of the growth defect caused by Usb1 depletion (Fig. 1B). This result strongly suggested that Usb1 might be involved in U6 snRNA regulation and play a role in splicing.
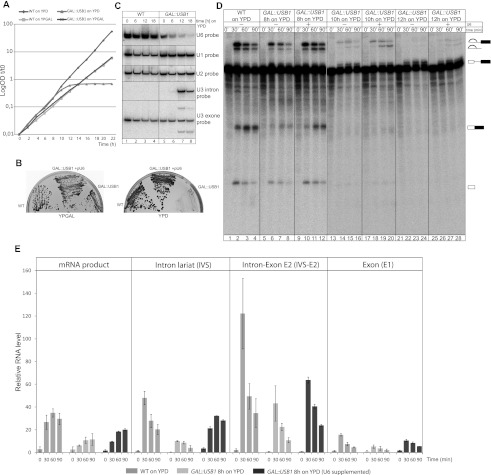
Figure 1.
Usb1 depletion leads to splicing defects caused by a reduction in U6 snRNA levels. (A) Usb1 depletion causes growth arrest in yeast. Growth curves of wild type (WT) and the GAL#x2237;USB1 strain (with the USB1 gene under the control of a galactose-inducible promoter) cultured on permissive YPGAL and repressive YPD medium. The presented values were corrected for dilution and are shown as log OD600, where t is the time in hours. (B) U6 snRNA overexpression suppresses the growth arrest caused by Usb1 depletion. The expression of additional copies of U6 snRNA or USB1 fully restores growth of the GAL#x2237;USB1 strain on glucose-rich repressive medium (YPD). (C) Depletion of Usb1 leads to in vivo splicing defects and the accumulation of precursor RNAs, which is correlated with reduced levels of the U6 snRNA. Northern blot analysis for the U1, U2, and U6 snRNAs, and precursor and mature U3 snRNA is shown. Total RNA was isolated from wild-type and GAL#x2237;USB1 strains grown in repressive YPD medium for up to 18 h. All hybridizations were performed with the same blot. (D) Extracts lacking Usb1 have reduced splicing activity in vitro, a defect that is partially restored by the addition of U6 snRNA isolated from a TAP-purified Prp24 fraction to the splicing reaction. Splicing reactions were performed using extracts prepared from wild-type (lanes 1–4) or GAL#x2237;USB1 cells after 8 h (lanes 5–12), 10 h (lanes 13–20), and 12 h (lanes 21–28) of Usb1 depletion. Reactions were carried out for 0, 30, 60, and 90 min. Reaction products were fractionated in 15% polyacrylamide gels and detected by autoradiography. The cell extracts were supplemented with the U6 snRNA in order to resume splicing activity. (E) Quantifications of the in vitro splicing reaction product and its intermediates. The graph shows relative levels of mature RP51a mRNA and its splicing intermediates, intron–lariat (IVS), intron–exon2 (IVS-E2), and exon1 (E1), in splicing reactions performed using extracts from wild type and the GAL#x2237;USB1 strain after 8 h of Usb1 depletion. All values are averages from three independent splicing reactions. Autoradiograms were quantified using MultiGauge software.
To test the idea that Usb1 might play a role in splicing, we assayed for splicing defects caused by Usb1 depletion. RNA isolated from wild-type and GAL#x2237;USB1 strains at various time points after the induction of Usb1 depletion was analyzed by Northern blot (Fig. 1C). We first analyzed the U3 snoRNA, which contains spliceosomal introns as pre-mRNAs. We observed an accumulation of U3 snoRNA precursors upon Usb1 depletion, confirming the involvement of Usb1 in splicing. We next investigated the steady-state levels of snRNAs by Northern blots and observed that the levels of U6 snRNA decreased as Usb1 depleted. Importantly, the levels of the U1, U2, U4, and U5 snRNAs remained unaffected, indicating that Usb1 had a specific role in U6 snRNA regulation (Fig. 1C; Supplemental Fig. S2). In order to further analyze the splicing defects caused by Usb1 depletion, we performed splicing reactions using extracts from wild-type and Usb1-depleted cells to splice RP51a pre-mRNA in vitro. Splicing activity of the Usb1-depleted extracts grown in repressive YPD medium for 8 h or longer was significantly lower than that of the wild-type extracts (Fig. 1D,E). Importantly, low splicing efficiency of the Usb1-depleted extracts was partially rescued by the addition of U6 snRNA isolated from a TAP-purified Prp24 fraction to the splicing reactions (Fig. 1D,E). At the 10- and 12-h time points, the rescue efficiency was low; however, the excised introns were clearly visible. This indicated that the splicing reaction took place and that the amount of spliced introns was higher after supplementation with purified U6 snRNA. Our data suggest that the main function of Usb1 is to regulate U6 snRNA biogenesis, rather than the splicing reaction itself (Fig. 1D [lanes 9–12,17–20,25–28], E).
Usb1 is essential for U6 snRNA stability
Since Usb1 depletion causes splicing defects and reduction in U6 snRNA levels, we investigated whether Usb1 is a part of the U6 snRNP or its assembly intermediates. We performed IgG affinity purification using a Usb1 TAP strain followed by gel filtration and mass spectrometry. Our results clearly showed that Usb1 behaved as a monomer (Supplemental Fig. S3A,B) and was not a part of any stable macromolecular complex under our purification conditions. We then tested whether Usb1 was involved in snRNP formation. Northern blot analyses of extracts from wild-type and Usb1-depleted cells separated in native gels indicated that Usb1 was not involved in U4/U6 di-snRNP and U4/U6.U5 tri-snRNP formation, even though their levels were limited by the amount of U6 (Supplemental Fig. S4A,B).
We then tested the possibility that Usb1 might be involved in regulation of U6 snRNA gene transcription. This hypothesis was examined using chromatin immunoprecipitation (ChIP) experiments, which revealed that RNA polymerase occupancy over the U6 snRNA in comparison with the control 5S rRNA gene did not change significantly upon Usb1 depletion (Supplemental Fig. S5). We next examined U6 snRNA stability in Usb1-depleted cells. Half-life analysis via transcription inhibition by thiolutin treatment followed by quantitative Northern blot hybridization revealed that the U6 snRNA half-life decreased from ∼10 h to 2.5 h in the absence of Usb1 (Fig. 2A1–B3; Supplemental Fig. S6A,B). In addition, U6 snRNA degradation products were present upon Usb1 depletion with high-resolution Northern blot analysis (Fig. 2C, lanes 5,6). However, primer extension analysis showed no difference at the 5′ end of the U6 snRNA (Supplemental Fig. S6C), suggesting that the observed intermediate products resulted from 3′ end degradation. These results led us to conclude that Usb1 is a U6 snRNA stability factor in yeast.
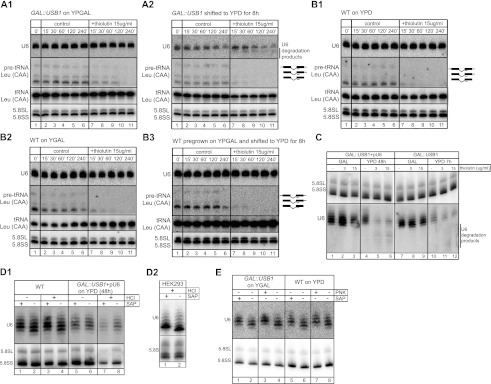
Figure 2.
Depletion of Usb1 reduces U6 snRNA half-life and leads to accumulation of decay intermediates. (A,B) The GAL#x2237;USB1 strain was grown in permissive YPGAL medium (A1) or repressive YPD medium (A2) for 8 h. The wild-type (WT) strain was grown on YPD (B1) or YPGAL (B2) or pregrown on YPGAL medium and shifted to YPD for 8 h (B3). Cells were treated with the RNA polymerase inhibitor thiolutin, then total RNA was extracted at the indicated times for Northern blot hybridizations against U6 snRNA, 5.8S rRNA as the loading control, and tRNALeu (CAA) precursor as the thiolutin treatment indicator. Depletion of Usb1 led to U6 snRNA instability and decreased half-life from 10 h to 2.5 h (cf. A2, lanes 7–11 and A1, lanes 7–11). For additional quantification analyses, see Supplemental Fig. S6A,B. (C) High-resolution Northern blot analysis of RNA from GAL#x2237;USB1 + pU6 and GAL#x2237;USB1 growing in permissive YPGAL (lanes 1–3,7–9, respectively) or repressive YPD (lanes 4–6,10–12, respectively) medium and treated with thiolutin for 4 h. (Lanes 5,6) The degradation products of U6 snRNA were detectable in the samples after 48 h of depletion. (Lanes 1,4) Long periods of Usb1 depletion caused changes in the migration of U6 snRNA. (D1) Analysis of U6 snRNA 3′ termini after Usb1 depletion. Total RNA from wild type (lanes 1–4) and GAL#x2237;USB1 + pU6 (lanes 5–8) growing in repressive YPD medium for 48 h was treated with HCl and/or shrimp alkaline phosphatase (SAP) and analyzed by Northern blotting. Only the U6 snRNA isolated from wild-type cells changed its migration toward the longer species upon phosphate removal by SAP, whereas HCl treatment, which can open the cyclic phosphate ring, had no effect on migration. (D2) Total RNA from HEK293 cells was treated with SAP and HCl similar to that for yeast, and U6 snRNA was detected by high-resolution Northern blot. (E) The U6 snRNA is terminated with 3′ phosphate at the 3′ end. Total RNA from GAL#x2237;USB1 grown on YPGAL (lanes 1–4) and wild type cultured on YPD (lanes 5–8) were treated with T4 PNK (lanes 3,7) or SAP (lanes 1,5) as control.
USB1 is essential for the presence of the phosphate moiety at the 3′ end of the U6 snRNA
Previous studies showed that the yeast U6 snRNA is terminated with a single phosphate moiety in position 2′ or 3′ or with hydroxyl groups (Lund and Dahlberg 1992). The presence of a phosphate at the 3′ (U-P) would be a feature specific to this snRNA, which we wanted to analyze in greater detail. In order to determine the properties of the U6 snRNA 3′ end, the RNA extracted from wild-type and Usb1-depleted cells was treated with HCl. If there was indeed a cyclic phosphate group at this end, HCl treatment would open the cyclic phosphate, which could then be removed by shrimp alkaline phosphatase (SAP). RNA extracted from human HEK293 cells was used as positive controls of SAP and polynucleotide kinase (PNK) treatment (Fig. 2D). Treated RNA was separated with high-resolution sequencing gels and analyzed with Northern blots for the U6 snRNA. As expected, in wild-type RNA extracts, the majority of the U6 snRNA molecules contained a phosphate moiety at the 2′ or 3′ end, evident from the slower mobility upon treatment with SAP (Fig. 2D1, lanes 1,2). As expected, HCl treatment had no effect on yeast U6 snRNA RNA mobility (Fig. 2D1, lanes 3,4) but resulted in SAP sensitivity of the human U6 snRNA (Fig. 2D2). In contrast to wild type, in RNA extracted from USB1-depleted cells, SAP/HCl treatment had no effect on U6 snRNA mobility, and in general, the U6 snRNA molecules migrated similar to those from wild-type cells after SAP treatment (Fig. 2D1, cf. lanes 1 and 6). In order to clarify whether the U6 snRNA in yeast contains a 2′ or 3′ phosphate at the 3′ end, we performed PNK treatment, which is known to have 3′ but not 2′ phosphatase activity (Cameron and Uhlenbeck 1977). Our results clearly demonstrate that there is a 3′ phosphate moiety at the end of the U6 snRNA (Fig. 4E), which is critical for U6 snRNA stability, and most importantly, that Usb1 is essential for the presence of this modification.
Figure 4.
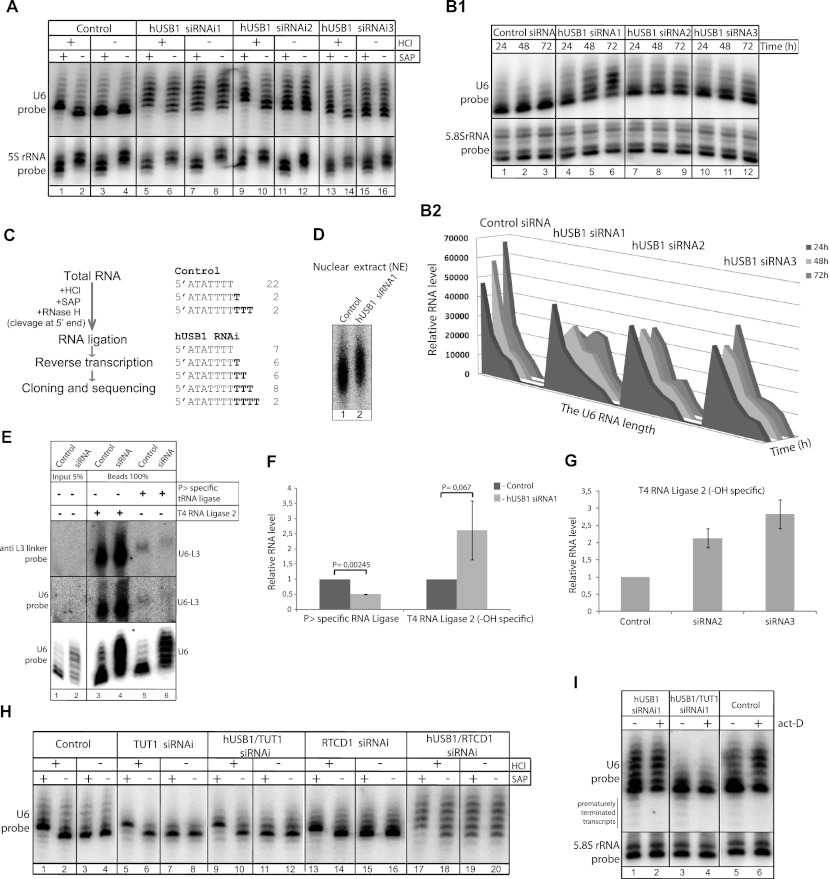
hUSB1 is essential for the presence of the cyclic phosphate at the U6 snRNA 3′ end. (A) U6 snRNA molecules are more heterogeneous and elongated after hUSB1 depletion by RNAi. High-resolution Northern blot analysis of RNA from control (lanes 1–4) and siRNA-treated (lanes 5–16) HeLa cells is shown. In order to examine the phosphorylation status, RNA was treated with HCl and/or SAP phosphatase, which in control cells led to a shift in U6 snRNA on the gel caused by cyclic phosphate removal. (B) Kinetics of the effect of three siRNAs against hUsb1 on the U6 snRNA. The HEK293 cells were treated with three siRNAs against hUSB1 and then harvested at the indicated time points. (B1) Total RNA was analyzed with high-resolution Northern blots. (B2) The U6 snRNA molecules were quantified with MultiGauge software and are shown as a 3D plot. (C) U6 snRNA molecules are more heterogeneous at the 3′ end upon hUSB1 depletion. CR-RT–PCR analysis of U6 snRNA from control and siRNA-treated HeLa cells is shown. Before ligation, the RNA was treated with HCl and SAP in order to remove the phosphate moiety from the 3′ end. RNase H cleavage was performed in order to remove the γ-monomethylguanosine triphosphate (meGTP) structure from the U6 5′ end. After reverse transcription and PCR, the reaction products were cloned and sequenced. This confirmed increased heterogeneity at the 3′ ends of U6 snRNA molecules after siRNA treatment. The numbers indicate the amount of independent clones that were sequenced. (D) RNA labeling in nuclear extracts confirms the changes in U6 snRNA mobility in high-resolution gels. Native U6 snRNA was labeled by incubating nuclear extracts from control (lane 1) or siRNA-treated (lane 2) HeLa cells with α-32PUTP. Isolated RNA was separated on 6% sequencing polyacrylamide gels and detected by autoradiography. (E–G) The formation of the U6 snRNA cyclic phosphate moiety is hUSB1-dependent. (E) U6 snRNA from control (lanes 1,3,5) and siRNA-treated (lanes 2,4,6) cells was captured with streptavidin magnetic beads with biotinylated antisense oligonucleotide complementary to its 5′ end. Subsequently, RNA was ligated with the preadenylated L3 linker using T4 RNA ligase 2 or cyclic phosphate-specific tRNA ligase from Arabidopsis thaliana. Reaction products were separated on 6% sequencing polyacrylamide gels and analyzed by Northern blotting using probes against the L3 linker and U6 snRNA. Larger blots with unligated U6 snRNA are presented in Supplemental Figure S8. (F,G) After hUSB depletion, the relative number of U6 snRNA molecules terminated with 2′,3′-cyclic phosphate decreases, and those terminated with OH groups increases, as quantified by MultiGauge software. All values are averages from three independent experiments, and the respective P-values calculated with an unpaired two-tailed Student's t-test are presented. Please note that the low signal intensity for the tRNA ligase reaction products compared with the T4 reaction was due to a lower reaction efficiency. (H) TUTase or RTCD1 activity is not required for the formation of the U6 snRNA 3′ end cyclic phosphate. RNA from cells treated with siRNAs against TUTase (lanes 5–8) or RTCD1 (lanes 13–16) and from control HeLa cells (lanes 1–4) was analyzed using high-resolution Northern blots. The RNA phosphorylation status was determined as described in A. (I) Depletion of hUSB1 or TUTase does not change the stability of mature U6 snRNA. High-resolution Northern blot analysis of RNA from cells treated with siRNAs against hUSB1 (lanes 1,2) and both hUSB1 and TUTase (lanes 3,4) and from control cells (lanes 5,6) after 2 h of actinomycin-D treatment is shown. (Lanes 2,6) The oligouridylated transcripts are more stable compared with mature forms and hence accumulate. (Lanes 2,4,6) Note that shorter precursor transcripts are eliminated after actinomycin-D treatment.
C16orf57/hUSB1 is essential for the presence of the cyclic phosphate at the 3′ end of the human U6 snRNA
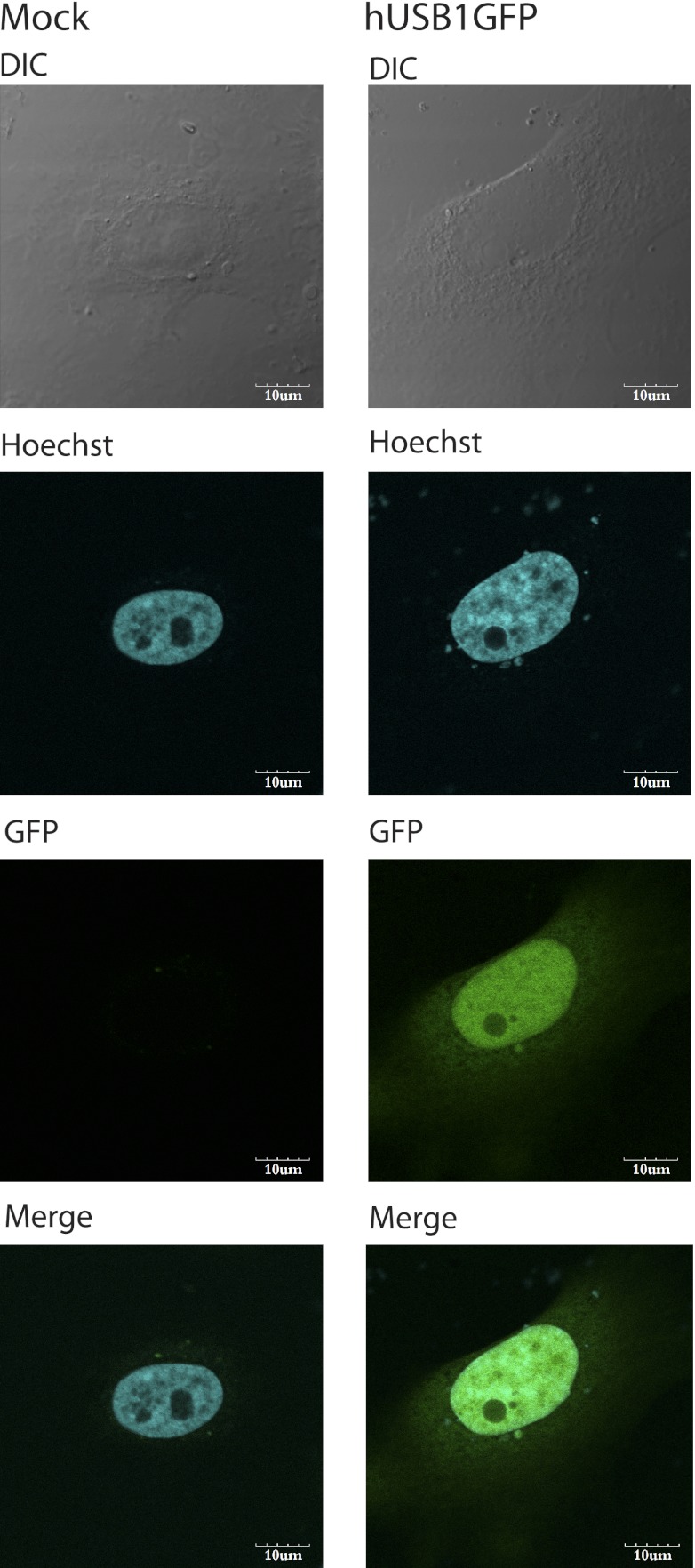
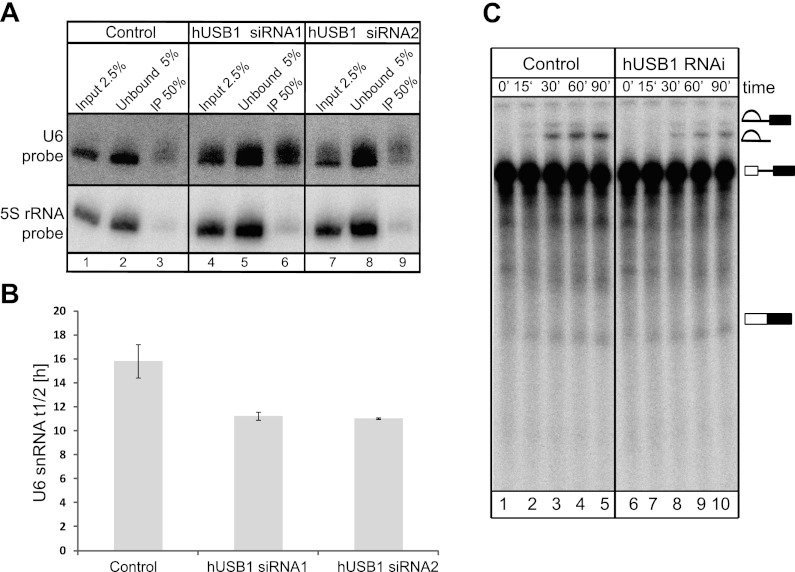
Having established a role for Usb1 in yeast, we then focused our attention on the protein encoded by the human C16orf57, which we named hUSB1. We first asked whether hUSB1 is also involved in U6 snRNA 3′ end processing in human cells. Using HeLa cells transiently transfected with a construct expressing a hUSB1-GFP C-terminal fusion protein, we confirmed that hUSB1 was also localized to the nucleus (Fig. 3). We next performed RNAi experiments in HeLa cells using three different siRNAs to examine the effect of knocking down hUSB1 on the U6 snRNA. The siRNAs exhibited similar silencing efficiencies, as verified by reverse transcription and quantitative PCR (Supplemental Fig. S7B). We used high-resolution sequencing gels and Northern blot analysis combined with HCl/SAP treatments to analyze the effect of siRNA-mediated knockdown on the U6 snRNA. Upon hUSB1 knockdown, U6 snRNA levels did not change; however, the U6 snRNA molecules became extended and more heterogeneous in length compared with control cells (Fig. 4A). The effect of all three different siRNAs has similar kinetics (Fig. 4B1,B2), but the effect of siRNA1 on the U6 snRNA was the most pronounced. Our experiment also confirmed the presence of 2′,3′-cyclic phosphate at the 3′ end of the human U6 snRNA (Fig. 4A); however, due to the U6 snRNA molecule heterogeneity in hUSB1-depleted cells, it was difficult to draw a clear conclusion about the effect of depletion on the U6 snRNA 3′ end phosphorylation. Heterogeneity of the U6 snRNA after hUSB1 depletion was also confirmed by circularized RNA RT–PCR (CR-RT–PCR) analysis and sequencing the 3′ ends of the U6 snRNA molecules (Fig. 4C). In addition, in vitro labeling of endogenous U6 snRNA by incubation of HeLa nuclear extracts with α-P32-UTP under conditions in which the U6 snRNA would be the major labeled species revealed that native U6 snRNA molecules were longer upon hUSB1 depletion (Fig. 4D).
Figure 3.
hUSB1 is a nuclear protein. HeLa cells were transiently transfected with a plasmid bearing a hUSB1-GFP fusion gene. Twenty-four hours after transfection, cells were examined by confocal microscopy. The fluorescence images for GFP and Hoechst nucleic acid stain, a differential interference contrast (DIC) image, and a merged image are presented for hUSB1GFP and the negative control.
Subsequently, we determined the phosphorylation status of the 3′ ends of the U6 snRNAs after hUSB1 depletion using Northern blot experiments. The U6 snRNA was purified and ligated to preadenylated oligonucleotides using either 3′-OH-dependent T4 RNA ligase 2 or 2′,3′-cyclic phosphate-dependent tRNA ligase to determine the ratio of the U6 snRNA containing a 3′-cyclic phosphate. In control HeLa cells, the U6 snRNA molecules contained U>P at their 3′ ends. In contrast, the level of U>P-containing molecules was reduced in the siRNA-treated cells. This finding correlated with the accumulation of U6 snRNA molecules extended with a 3′-OH group in Figure 4 (E and F) and Supplemental Figure S8. Accumulation of U6 snRNA molecules with 3′-OH groups was confirmed for HeLa cell treatment with two additional siRNAs against hUSB1 (Fig. 4G). Our results indicate that the presence of 2′,3′-cyclic phosphate at the U6 snRNA 3′ end is dependent on hUSB1, suggesting that, as in yeast, this protein is involved in U6 snRNA biogenesis in human cells.
In order to further understand the role of hUSB1 in U6 snRNA regulation, we analyzed the relationship between hUSB1 and other enzymes that are thought to participate in U6 snRNA 3′ end processing (Fig. 4H; Supplemental Fig. S7A). To this end, we first performed RNA-3′-phosphate cyclase (RTCD1) knockdowns in HeLa cells. RTCD1 was suggested to be involved in U6 3′-phosphate cyclization (Genschik et al. 1997); however, we did not observe any effects on the U6 3′ end upon siRNA-mediated RTCD1 knockdown as judged by Northern blot analyses of HCl/SAP-treated cell extracts (Fig. 4H, lanes 13–16). We next knocked down U6 snRNA-specific poly(U) polymerase (TUTase), which significantly reduced the accumulation of extended U6 snRNA molecules, especially in cells with concurrent hUSB1 knockdown (Fig. 4H, cf. lanes 5 and 1,17).
In the Northern blot analyses, we always observed U6 snRNA molecules that were shorter than the major mature species (e.g., see Fig 4H, lane 1). To determine whether these shorter species are decay intermediates or precursors prior to TUTase-mediated extension, we combined hUsb1 and double hUSB1/TUT1 siRNA-mediated knockdown with actinomycin-D transcription inhibitor treatment. Northern blot analysis revealed that the shorter species disappeared after 2 h of actinomycin-D treatment, strongly suggesting that these shorter molecules were precursors (Fig. 4I, lanes 2,4,6). Moreover, actinomycin-D treatment led to accumulation of the extended species in hUSB1-depleted cells, which were reduced in the hUSB1/TUTase double knockdown (Fig. 4I, cf. lanes 2 and 4). Here we present the first siRNA-based evidence that TUT1 is indeed responsible for U6 snRNA 3′ uridylation. However, TUTase does not seem to be directly involved in the formation of the cyclic phosphate at the end of the U6 snRNA (Fig. 4H, lanes 5–8).
To further characterize the effect of hUSB1 knockdown on U6 snRNA, we measured the half-life of the U6 snRNA in control and siRNA-treated cells. Newly synthesized RNA was metabolically labeled by 4-thiouridine (4sU) incorporation (Dolken et al. 2008). RNA isolated from the cells was biotinylated in a 4sU-specific reaction and then purified using streptavidin beads to separate newly synthesized RNAs from pre-existing RNA fractions (Fig. 5A,B). The ratio between pre-existing and newly synthesized U6 snRNA was used to calculate the half-life of the transcript. In contrast to what we observed in yeast, the stability of the U6 snRNA was only mildly affected by the lack of 3′ modifications, its half-life reducing from 15 h to 11 h. This may reflect differences in U6 biogenesis in humans and yeast, for in humans, U6 TUTase can add uridines to the U6 snRNA 3′ ends to repair molecules and prevent their rapid degradation. Furthermore, the lack of hUSB1 only slightly affected the splicing activity of nuclear extracts, which is in agreement with our findings in Figure 5, A and B, confirming a difference between U6 snRNA biogenesis in yeast and human cells (Fig. 5C).
Figure 5.
hUSB1 knockdown does not significantly change the half-life of human U6 snRNA or its in vitro splicing activity. (A) Northern blot analysis of RNA transcripts after thiouridine labeling of HeLa cells in vivo. Total RNA from control cells (lanes 1–3) and cells treated with siRNA1 (lanes 4–6) and siRNA2 (lanes 7–9) was biotinylated and captured with streptavidin beads. Captured RNA was separated on 6% polyacrylamide gels, blotted, and hybridized against the U6 snRNA and 5S rRNA. Quantitative analyses were performed with MultiGauge software, as described in the Materials and Methods. (B) The graph shows measurements of the U6 snRNA half-life in control and siRNA-treated HeLa cells. The values are averages from three independent experiments. (C) hUSB1 depletion slightly affects the in vitro splicing activity. Nuclear extracts from HeLa cells after hUsb1 depletion (lanes 6–10) exhibited a slightly decreased splicing activity compared with extracts from control cells (lanes 1–5). The reactions were carried out for 90 min, after which RNA was purified, fractionated on 15% polyacrylamide gels, and detected by autoradiography.
USB1 encodes a putative phosphodiesterase whose potential catalytic activity is essential for its function
Our results prompted us to carefully examine the Usb1 protein sequence as well as its orthologs. PSI-Blast (Altschul et al. 1997) searches initiated with the Usb1 and hUSB1 sequences identified nearly 200 uncharacterized homologs from all major eukaryotic phyla. These searches, however, did not reveal any detectable sequence similarity to any other proteins of known structure or function. Using highly sensitive methods for distant homology detection (Ginalski et al. 2004) and fold recognition (Ginalski et al. 2003), we mapped Usb1 and hUSB1 to various 2H phosphodiesterase structures, including that of a cyclic nucleotide phosphodiesterase, 1FSI (Hofmann et al. 2000), and a 2′–5′ RNA ligase, 2FYH (Kanai et al. 2009). Interestingly, hUSB1 was previously classified as a 2H phosphodiesterase superfamily member (CG16790-like family of eukaryotic ligT ligases) (Mazumder et al. 2002). In addition, a recent theoretical publication also identified hUSB1 as a putative phosphodiesterase, but since its molecular function was not determined, hUSB1 was instead suggested to be an RNA ligase (Colombo et al. 2012).
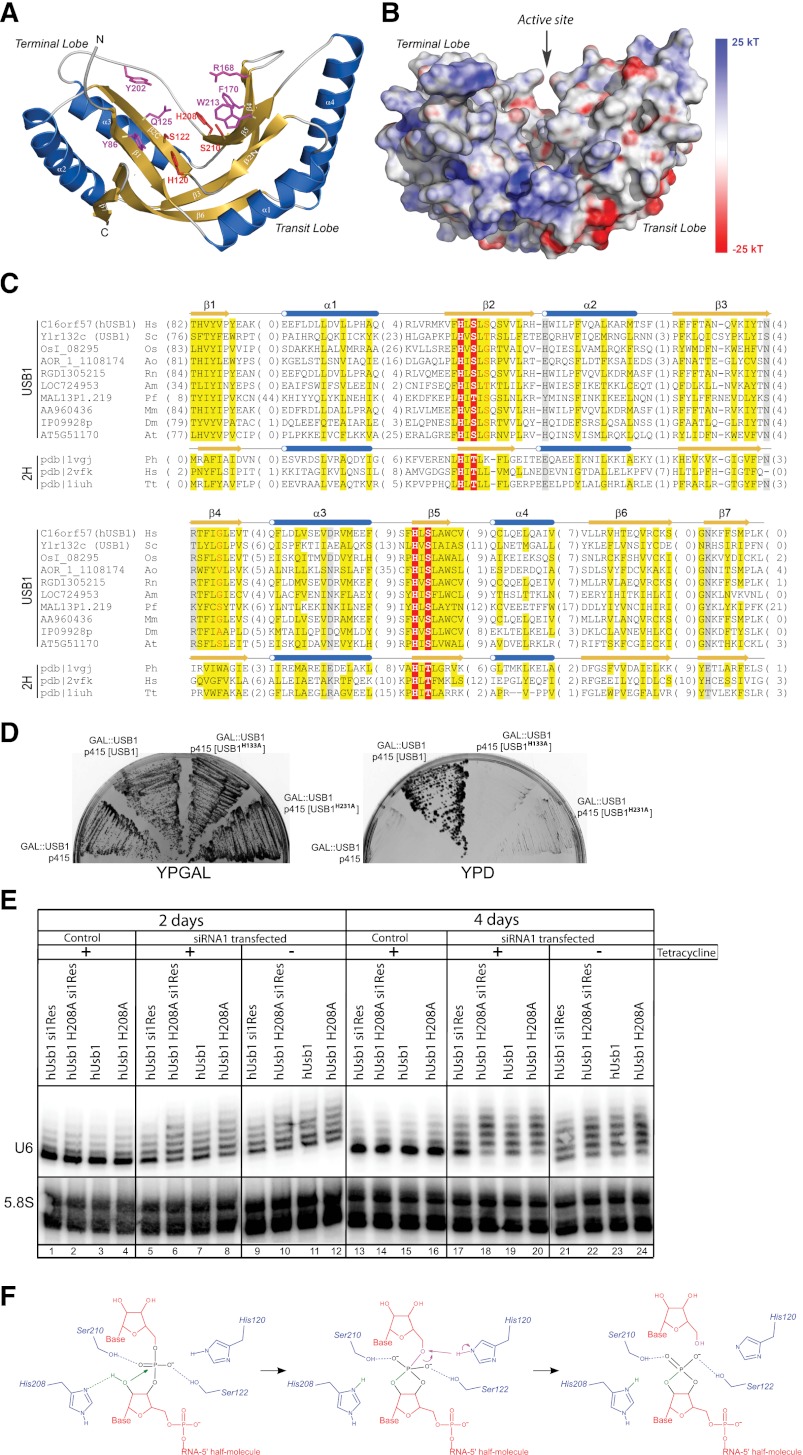
To analyze the sequence and structure features of Usb1/hUSB1, we generated multiple sequence alignments of USB1 family proteins and selected structures of 2H superfamily members and constructed their three-dimensional (3D) homology models of Usb1 and hUSB1 (Fig. 6A,B). Although one of the hallmarks of the 2H superfamily is its extreme sequence diversity (Mazumder et al. 2002), we were able to identify structural and sequence motifs that are common to all superfamily members in Usb1 and hUSB1. Similar to other 2H phosphodiesterases, our 3D models had two closely interacting and topologically equivalent repeats (lobes) with a pseudo-twofold rotational symmetry (Sakamoto et al. 2005). Each lobe consists of a four-stranded, anti-parallel β sheet flanked by two helices on one side that provides a conserved HXT/S catalytic motif within a V-shaped active site cleft (Fig. 6A). According to the model, the hUSB1 family members conserved all amino acids critical for the enzymatic activity of 2H phosphodiesterases, including the invariant catalytic residues from two HXT/S motifs: H120, S122, H208, and S210 (numbered according to hUSB1) (Fig. 6C). In order to check whether putative phosphodiesterase activity is essential for Usb1 function, we introduced a mutant with putative catalytic residues replaced by alanines (H133A and H231A) into the GAL#x2237;USB1 strain. We found that the mutant Usb1 could not restore yeast growth in repressive glucose-rich medium, indicating that the putative catalytic activity of Usb1 is necessary for its essential function in yeast (Fig. 6D). We then preformed rescue experiments in human HEK293 stable cell lines where we complemented the effect of RNAi by expression of siRNA-insensitive wild-type and H208A mutant hUSB1 constructs. High-resolution Northern blot analysis clearly showed that expression of the wild-type RNAi-insensitive construct largely rescued the phenotype (Fig. 6E, lanes 5,17), while expression of the catalytic mutant did not and, if anything, had a slight negative effect (Fig. 6E, cf. lanes 5 and 6, and lanes 17 and 18). These results indicated that the effect of siRNA treatment on U6 snRNA is dependent on depletion of hUSB1 and that putative catalytic activity is essential for its function.
Figure 6.
USB1 encodes a putative phosphodiesterase whose potential catalytic activity is essential for its function. (A) A 3D model of the hUSB1 protein. Invariant catalytic histidines and serines from HXS/T motifs are shown in red, whereas other conserved residues potentially responsible for substrate binding are shown in magenta. (B) The electrostatic potential surface for hUSB1 suggests that the terminal lobe has the ability to bind nucleic acids. Negatively charged regions are colored in red, and positively charged regions are colored in blue (ranging from −25 kT up to 25 kT). The protein is in the same orientation as in A. Please see Supplemental Figure S10 for more details. (C) Usb1 belongs to the 2H phosphodiesterase superfamily. Multiple sequence alignments of USB1 family representatives and selected distantly related 2H phosphodiesterase structures. Numbers of the residues that are not shown are specified in parentheses. Residue conservation is denoted by the following scheme: uncharged are highlighted in yellow; charged or polar are highlighted in gray; small are letters in red; and invariant catalytic residues of HXT/S motifs are highlighted in red. Locations of predicted (hUSB1) and observed (Protein Data Bank [PDB], 1VGJ) secondary structure elements are marked above the corresponding sequences. (Hs) Homo sapiens; (Sc) Saccharomyces cerevisiae; (Os) Oryza sativa; (Ao) Aspergillus oryzae; (Rn) Rattus norvegicus; (Am) Apis mellifera; (Pf) Plasmodium falciparum; (Mm) Mus musculus; (Dm) Drosophila melanogaster; (At) Arabidopsis thaliana; (Ph) Pyrococcus horikoshii; (Tt) Thermus thermophilus. (D) A complementation assay reveals that the Usb1 catalytic mutant cannot restore cell growth. Plasmids containing either wild-type (WT) or H133A, H231A mutant versions of the USB1 gene were transformed into the GAL#x2237;USB1 strain. The resulting strains were plated on permissive (YPGAL) and restrictive (YPD) media. (E) Expression of siRNA-insensitive wild-type hUSB1 mRNA, but not H208A hUSB1 mutant mRNA, leads to partial rescue of the molecular phenotype caused by hUSB1 depletion. HEK293 stable cell lines expressing constructs under the control of a tetracycline-regulated promoter, including hUsb1 mRNA resistant to siRNA (si1Res hUsb1) or its corresponding H208A mutant mRNA (H208A si1Res) as well as hUsb1 mRNA sensitive to siRNA (hUsb1) or its corresponding H208A mutant mRNA (hUsb1 H208A), were transfected with siRNA against hUSB1 and then induced with tetracycline. (Lines 5–8,17–20) Cells were collected 2 and 4 d after transfection, and U6 snRNA was detected by high-resolution Northern blots. Untransfected cells (lines 1–4,13–16) and uninduced cells (lines 9–12,21–24) were used as controls. (F) Proposed reaction mechanism catalyzed by hUSB1. The nucleophilic attack on the phosphate group results in the formation of a cyclic phosphate moiety. Groups involved in the first and second steps of the reaction are shown in green and magenta, respectively, whereas the catalytic residues are denoted in blue.
Electrostatic surface potential is one of the main factors that may help to distinguish between different substrates of 2H phosphodiesterase enzymes (Sakamoto et al. 2005). As seen in Supplemental Figure S10, various 2H phosphodiesterases retain specific features of their electrostatic surface potential in order to bind molecules of different physicochemical properties. As observed by Kato et al. (2003), an extensive positively charged region is essential for the recognition of large negatively charged molecules like RNA. For example, the 2′–5′ tRNA ligase has positively charged regions on both sides of the active site cleft, encompassing its terminal and transit lobes, which allows it to correctly bind both 3′ and 5′ tRNA half-molecules. In contrast, plant CPDase and cyclic CMP phosphodiesterase lack positive charges around their active site regions, as they are involved in metabolizing small molecules, such as ADP-ribose 1″,2″-cyclic phosphate (Appr>p) (Supplemental Fig. S10; Hofmann et al. 2000). As shown in Figure 6B, hUSB1 has a positively charged region only on the terminal lobe. This suggests that hUSB1 is involved in metabolizing a terminal fragment of the RNA molecule, such as the poly(U) chain of U6 snRNA.
In the reaction mechanism proposed for 2H phosphodiesterases (Supplemental Fig. S9A,B), serines or threonines are necessary to stabilize the position of a cyclic phosphate moiety within the active site, while histidines are responsible for activating a nucleophile and the formation of reaction intermediates. Interestingly, a similar overall active site shape with two catalytic histidines is observed in the evolutionarily unrelated RNase A (Aravind and Koonin 2001). Furthermore, RNase A not only cleaves the RNA P–O5′ bond (leading to the formation of a cyclic phosphate), it also hydrolyzes a cyclic phosphate moiety using a mechanism strikingly similar to that of 2H phosphodiesterases (Supplemental Fig. S9C,D; Raines 1998; Cuchillo et al. 2011). Consequently, we propose that hUSB1 catalyzes the cleavage of the P–O5′ bond at the 3′ end of poly(U), resulting in the formation of a 2′,3′-cyclic phosphate and the removal of the terminal uridine via a mechanism similar to that of RNase A (Fig. 6F; Supplemental Fig. S9D).
Discussion
In this study, we identified a putative phosphodiesterase that is essential for the U6 snRNA 3′ end processing. We propose that the Usb1 protein is a phosphodiesterase acting as an RNase responsible for trimming the poly(U) tract of the last nucleotides in the pre-U6 snRNA molecule. This leads to the formation of mature U6 snRNA 3′ end-terminated with a 3′ phosphate in yeast or a 2′,3′-cyclic phosphate in humans. However, since we did not show the activity directly, we cannot formally rule out the possibility that Usb1/hUSB1 is involved not in the generation, but rather the maintenance of 3/2′,3′-cyclic phosphate or biogenesis of the actual enzyme responsible for generation of U6 snRNA 3/2′,3′-cyclic phosphate. However, such possibilities seem unlikely.
Moreover, we showed that, in yeast, the presence of a phosphate at the 3′ end of the U6 snRNA is essential for its stability, but not for its successful incorporation into snRNPs. In contrast to yeast, the knockdown of hUSB1 in human cells does not lead to significant changes in the U6 snRNA half-life. We propose that this difference is due to activity of the U6-specific poly(U) polymerase, which can constantly extend shortened U6 snRNA molecules. Therefore, the major phenotype of hUSB1 dysfunction is the appearance of extended U6 snRNA species in human cells. The sensing mechanism that ensures the proper length of the oligouridine tract at the 3′ end of the human U6 snRNA must exist. Our results do not allow us to provide a molecular description of such a phenomenon, but formation of cyclic phosphate is crucial for its occurrence, since depletion of hUSB1 leads to accumulation of extended species. The majority of U6 snRNA molecules in humans have four uridines that may correspond to the region protected by LSM proteins.
Previously, it was suggested that the U6 snRNA 3′ end modification is essential for the exchange between the La RNA chaperone and doughnut-shaped heteromer of seven human Sm-like proteins (Lsm2–8) as well as for the incorporation of the U6 snRNA into larger snRNP assemblies (Seraphin 1995; Achsel et al. 1999). However, our results suggest otherwise; we showed that splicing efficiency did not change significantly upon hUSB1 knockdown in human cells, while the overexpression of the U6 snRNA restored growth in Usb1-depleted yeast cells. Moreover, it seems very unlikely that a dysfunctional exchange between La and LSM proteins would lead to yeast U6 snRNA destabilization, since La protein recognizes and, as a result, protects the 3′ end of target RNAs. Therefore, we propose that the 3′ end modification itself has a protective role for the U6 snRNA. In fact, the lack of hUSB1 leads to human diseases that are severe, yet not lethal, arguing that the U6 snRNA modification is not essential for splicing. Moreover, our preliminary analysis of deep RNA sequencing of RNA samples isolated from hUSB1-depleted HeLa cells does not show a significant accumulation of unspliced introns (data not shown). This suggests that the lack of the U6 snRNA cyclic phosphate does not affect global splicing efficiency and that the diseases caused by hUSB1 mutations may result from subtle differences in splicing.
It is intriguing why the U6 snRNA in yeast is terminated with a 3′ phosphate, while in humans, it is terminated with a 2′,3′-cyclic phosphate. One of the possible explanations is a difference in the catalytic properties of Usb1 and hUSB1. Unfortunately, our intensive efforts to recapitulate the reactions catalyzed by these enzymes in vitro have been unsuccessful to date. Another explanation for the difference is that a terminal phosphate cyclase catalyzes cyclization in human cells, while the Usb1 leaves a 3′-terminal phosphate in yeast cells. However, our results showed that the depletion of an obvious candidate for U6 snRNA cyclase RTCD1 did not influence the U6 snRNA 3′ end formation (Tanaka and Shuman 2009). Conversely, it is possible that there is a yeast-specific enzyme that resolves the cyclic phosphate bond, but there are no known candidates with such an activity.
Interestingly, although hUSB1 is localized to the nucleus, which is compatible with its proposed enzymatic activity, the yeast ortholog exhibits nuclear and mitochondrial localization (Glatigny et al. 2011). Furthermore, Usb1 overexpression was shown to be able to compensate for specific respiratory defects of an oxa1 yeast mutant, which suggests that Usb1 may be involved in mitochondrial metabolism. However, Usb1's respiratory function was not required for yeast viability, indicating that the growth defects that we observed in Usb1-depleted yeast cells were not due to its function in the mitochondria (Glatigny et al. 2011). Therefore, U6 snRNA 3′ end formation might be the main and essential cellular function of Usb1. It is important to note that hUsb1 does not localize to the nucleolus, suggesting that the 3′ end processing takes place in the nucleoplasm before the RNA transiently passes into the nucleolus, where it is subjected to other post-transcriptional modifications, such as snoRNA-guided 2′-O-methylation and pseudouridydylation (Kiss 2004).
Our studies show that Usb1 is a monomer; however, previous studies have reported that Usb1 may possibly interact with some proteins involved in the formation of catalytically active spliceosome complexes, such as Prp19, Syf1, and Cus2 (Brow 2002; Gavin et al. 2006). These high-throughput studies have certain limitations; however, the proposed interactions further support our hypothesis that Usb1 interacts with the U6 snRNA. U6 snRNA 3′ end formation might be synchronized with the early steps of U6 snRNP assembly before it enters the nucleolus. Additionally, analysis of the electrostatic surface potential of Usb1 and hUsb1 suggests that its terminal lobe has the ability to bind negatively charged RNA molecules. Indeed, Usb1 was detected in a high-throughput screen as an RNA-binding protein (Scherrer et al. 2010).
Finally, our results strongly suggest that U6 TUTtase plays a role in the uridylation of U6 snRNA precursors. The human U6 TUTase was independently identified as a poly(A) polymerase (Star-PAP), which polyadenylates a subset of mRNAs in the nucleus and is regulated by phosphatidylinositol 4,5-bisphosphate (Mellman et al. 2008). The investigators in this study also questioned the function of the U6 TUTase in U6 snRNA metabolism. It would be very intriguing if U6 TUTtase is indeed a dual-function protein, also serving as a poly(A) polymerase.
In summary, we identified a putative U6 snRNA phosphodiesterase as a first step toward understanding the biological mechanism of PN, DC, and RTS, in which hUSB1 is mutated. However, further research is needed to understand the biological and medical consequences of the lack of cyclic phosphate at the 3′ end of the U6 snRNA.
Materials and methods
Strain and plasmid construction
The yeast strains, plasmids, oligonucleotides, and siRNAs used in this study are listed in Supplemental Tables 2–5. The yeast strains were transformed as previously described (Gietz et al. 1992). The GAL#x2237;USB1 and HA-C160/GAL#x2237;USB1 strains were constructed by homologous recombination of a cassette amplified by PCR using the USB1GALF and USB1GALR primers from pFA6-kanMX6-PGAL1 to the wild-type or MW671-HA strain genomes, respectively. Clones were verified by PCR with USB1verF and USB1verR primers. Other strains were prepared by transformation with the appropriate plasmids. Standard yeast genetic methods and selective growth media were used as previously described (Guthrie and Fink 1991).
The plasmids pRS415-USB1H133A and pRS415-USB1H231A were obtained by site-directed mutagenesis using the USB133F/USB133R and USB231F/USB231R primers, respectively, with the pRS415-USB1 plasmid serving as a template. Mutations were confirmed by sequencing.
The tRNA ligase (AT1G07910.1) construct for recombinant protein production in Escherichia coli was prepared by PCR amplification from Arabidopsis thaliana cDNA using pET28AtLigF and pET28AtLigR primers. The PCR product was subsequently cloned into a modified pET28 vector using the SLIC method (Li and Elledge 2007).
The pcDNA-hUSB1GFP construct was prepared by PCR amplification from human cDNA using pcDNAhUSB1F and pcDNAhUSB1R primers. The PCR product was subsequently cloned into a modified version of the pcDNA5/FRT/TO vector (Invitrogen). The details of this method will be published in a forthcoming publication (RJ Szczesny, K Kowalska, LS Borowski, EP Owczarek, PP Stepien, and A Dziembowski, in prep.). Plasmids encoding hUSB1 carrying silent mutations in sequences complementary to siRNA and H208A mutation were obtained by site-directed mutagenesis of the pcDNA-hUSB1GFP plasmid, resulting in pcDNA-hUSB1si1Rec (hUSB1 wild-type siRNA1-insensitive) (oligonucleotides: hUSB1si1RecF and hUSB1si1RecR), pcDNA-hUSB1si1Rec H208A (hUSB1 H208A mutant siRNA1-insensitive) (oligonucleotides: hUSB1si1RecF, hUSB1si1RecR, hUSB1H208AF, and hUSB1H208AR), and pcDNA-hUSB1GFP H208A (hUSB1 H208A siRNA1-sensitive) (oligonucleotides: hUSB1H208AF and hUSB1H208AR).
Cultures
Yeast strains were pregrown in YPGAL medium (1% yeast extract, 2% peptone, 2% galactose) at 30°C and switched to YPD medium (1% yeast extract, 2% peptone, 2% glucose) during the logarithmic growth phase. For the depletions performed over a long time period, growth was monitored throughout the entire experiment, and the cultures were maintained in logarithmic phase. Thiolutin (T3450, Sigma) was added to the yeast culture at a final concentration of 3 or 15 μg/mL. The cells were treated for up to 4 h, and samples were taken at different times over the course of the culture.
HeLa and HEK293 T-Rex FLP-in (Invitrogen) cells were cultured in monolayers in DMEM (Gibco) supplemented with 10% FBS (Gibco) or TET System Approved FBS (Clontech) in the case of stable, inducible cell lines at 37°C in a 5% CO2 humidified atmosphere. Actinomycin-D (Sigma) was added at a final concentration of 5 μg/mL for 2 h. Stably transfected HEK293 T-Rex FLP-in cell lines were selected according to the manufacturer's suggestions (Invitrogen). Expression of exogenous genes was induced by the addition of tetracycline to the culture medium at a concentration of 25 ng/mL.
Yeast genetic screen
The GAL#x2237;USB1 strain was transformed with the Saccharomyces cerevisiae AB320 pYEp13 genomic library (37323, American Type Culture Collection) and plated onto synthetic complete medium (SC; 0.67% yeast nitrogen base, 2% glucose) without leucine to maintain the plasmid. After 3 d, the colonies were replica-plated onto YPGAL and YPD plates and grown for an additional 3 d at 30°C. Plasmids from the 30 positive clones growing on restrictive medium were isolated and sequenced. In order to verify the results of the suppressor screen, the GAL:USB1 strain was transformed with pYX172-U6 and grown on galactose and glucose media.
RNA isolation and Northern blotting
Yeast total RNA was isolated using the GTC/phenol method (Alexander et al. 2010). Total RNA from HeLa cells was purified with TRIzol according to the manufacturer's instructions (Invitrogen). Northern analyses and primer extension reactions were performed as previously described (Mroczek and Kufel 2008). Standard 6% or 8% acrylamide gels were used to separate low-molecular-weight RNAs. High-resolution electrophoresis was performed in 6% sequencing gels in 0.5× TBE buffer.
RNA treatment with HCl, SAP, and PNK
RNA was incubated in the presence of 100 mM HCl for 4 h on ice and then precipitated. The samples were then treated with SAP (Fermentas) or T4 PNK (New England Biolabs) according to the manufacturers' instructions. Finally, RNA was purified with phenol/chloroform extraction.
In vitro splicing assay with yeast extracts
Yeast whole-cell extracts were prepared as previously described (Seraphin and Rosbash 1989). pre-mRNA was generated by in vitro transcription of the pBS195 plasmid (digested with the DdeI). Splicing reactions were carried out for 30–90 min at 30°C, and the resulting RNA was purified with phenol/chloroform. Reaction products were analyzed in 15% polyacrylamide gels. When necessary, purified native U6 snRNA was added at a final concentration of 100 nM.
Purification of native U6 snRNA from yeast
Yeast snRNPs were purified from 20 L of yeast culture by affinity chromatography of TAP-tagged Prp24 as described in “Tap Purification.” RNA was purified from protein fractions by phenol/chloroform, and the U6 snRNA was gel-purified.
RNP and RNA analyses in native gels
Analyses were performed as previously described (Verdone et al. 2004). The samples were fractionated in native gels, then blotted and hybridized with oligonucleotide probes against the U4, U5, and U6 snRNAs.
TAP purification
The TAP purifications were performed from 20 L of yeast culture as previously described (Rigaut et al. 1999) using an Akta FPLC purification system (GE Healthcare). The fractions collected were concentrated by pyrogallol red precipitation, separated by SDS-PAGE, and stained with Coomassie Blue.
ChIP
ChIP was performed as previously described (El Hage et al. 2008) using Dynabeads Protein G (100-03D, Invitrogen) and a α-HA mAb (12CA5, Roche). The precipitated chromatin was amplified in triplicate with Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) using a LightCycler LC480 PCR machine (Roche). The oligonucleotides used for the quantitative PCRs are listed in Supplemental Table 4. The ChIP values for RNA polymerase III occupancy were determined using the formula
where Ct IP is the cycle number for the immunoprecipitate, and Ct background is the cycle number for the control beads without antibody.
hUSB1 localization
HeLa Kyoto cells were transfected with 2 μg of DNA using the TransIT 2020 reagent (Mirus). Four hours after transfection, the cells were replated in dishes with poly-L-lysine-coated cover glasses. Twenty-four hours after transfection, the cells were stained with Hoechst 33342 (Invitrogen) and fixed with formaldehyde. The cover glasses were then mounted in Prolon Gold medium (Invitrogen). The cells were imaged with an FV1000 confocal system (Olympus).
siRNA transfection
For the siRNA-mediated knockdown, 6 × 105 HeLa or 2 × 106 HEK293 cells growing in 100-mm plates were transfected with 20 nM siRNA (Invitrogen) (listed in Supplemental Table 5) using 15 μL of Lipofectamine RNAiMAX (Invitrogen). Cells were harvested at 48, 72, or 96 h after transfection. In the case of 96 h, cells were replated 48 h post-transfection. Appropriate controls were used as recommended by the manufacturer of the siRNAs. For preparation of the nuclear extracts, cells growing in 12 100-mm plates were replated 48 h after transfection in 12 145-mm plates and cultured for an additional 48 h. Control nuclear extracts were prepared from nontransfected cells.
Determination of mRNA levels after siRNA-mediated gene silencing
Ten micrograms of total RNA was treated with Turbo DNase (Ambion) according to the manufacturer's instructions. cDNA was prepared using SuperScript III reverse transcriptase and oligo(dT)18 primer (Invitrogen). Quantitative PCR reactions were performed as described in “ChIP.” Transcript levels were normalized to that of GAPDH.
CR-RT–PCR
The CR-RT–PCR reactions were performed as previously described (Kuhn and Binder 2002) with some modifications. Briefly, to remove γ-monomethylguanosine triphosphate, 10 μg of total RNA was annealed with oligonucleotide hU6RH and then digested with RNase H as described previously (Decker and Parker 1993). Samples were then treated with 10 U of SAP phosphatase (Fermentas), and RNA was purified by phenol/chloroform extraction. RNA was circularized using T4 RNA ligase (New England Biolabs). The cDNA was prepared using SuperScript III reverse transcriptase (Invitrogen) with the oligonucleotide U6CRPRT. The U6 ends were amplified in a PCR reaction with the oligonucleotides U6CR-F1 and U6CR-R1 and cloned using the Zero Blunt TOPO PCR cloning kit (Invitrogen). Sequencing was performed using the M13 Rev primer.
Preparation of nuclear extracts from cell lines, in vitro splicing, and RNA labeling
Small-scale nuclear extracts from siRNA-treated and control HeLa cells were prepared as previously described (Kataoka and Dreyfuss 2008). The AdML pre-mRNA splicing substrate was transcribed in vitro from the HMS81 plasmid digested with BamHI. The splicing reactions were set up as previously described (Le Hir et al. 2000). The reactions were conducted for up to 90 min. RNA was extracted with phenol/chloroform and analyzed in 15% polyacrylamide gels with 7 M urea.
To label the U6 snRNA, 30 μL of nuclear extract was incubated with 50 μCi of α-32PUTP for 60 min at 30°C. RNA was purified using the GTC/phenol method, precipitated, separated on a 6% polyacrylamide gel, and visualized by autoradiography.
Ligation of L3 adapters to U6 snRNA 3′ ends
Twenty micrograms of total RNA was annealed to 500 pM anti-U6BtnTg oligonucleotide (Sigma) in 1× hybridization buffer (250 mM NaCl, 10 mM Tris at pH 7.5, 2 mM EDTA at pH 8.0) and immobilized on Dynabeads M-280 Streptavidin beads (Invitrogen) according to the manufacturer's instructions. Ligations were performed on the beads using 200 μM L3 linker oligonucleotide (IDT DNA) and 200 U of T4 RNA Ligase 2 (New England Biolabs), or cyclic phosphate (P>)-specific A. thaliana tRNA ligase (AT1G07910.1) as previously described (Schutz et al. 2010). The samples were separated in 6% sequencing gels and analyzed by Northern blotting using anti-L3 and hU6 probes.
Labeling and purification of nascent RNA
4sU (Sigma) at a concentration of 200 μM was added to the medium for 2 h, after which the cells were harvested and RNA was purified. RNA biotinylation and capture steps were performed as described previously (Dolken et al. 2008). Purified RNA was separated on 6% polyacrylamide gels, blotted, and hybridized against the U6 snRNA and 5S rRNA. Densitometric analyses were performed using MultiGauge software (Fuji). The U6 snRNA half-life values were calculated according to the following formula:
where R is the newly transcribed RNA:total RNA ratio, tL is the duration of labeling, and t1/2 is the RNA half-life (Dolken et al. 2008).
Recombinant protein expression and purification
A. thaliana tRNA ligase was expressed in the E. coli BL21-CodonPlus-RIL strain (Stratagene) as an N-terminal HIStag-SUMOtag fusion.
Bioinformatic analyses
Proteins belonging to the USB1 family were identified with PSI-Blast (Altschul et al. 1997) searches (E-value threshold = 0.005) performed against the NCBI nonredundant protein sequence database using the hUSB1/USB1 sequence as a query. Multiple sequence alignment was derived using the PCMA program (Pei et al. 2003) followed by manual adjustments. Secondary structures were predicted using PSIPRED (McGuffin et al. 2000). Distantly related proteins of known structure were identified with the metaprofile comparison method Meta-BASIC (Ginalski et al. 2004) and consensus of fold recognition 3D-Jury (Ginalski et al. 2003). Sequence-to-structure alignment between the USB1 family and selected structures was built using a consensus alignment approach and 3D assessment (Ginalski and Rychlewski 2003) based on the results of Meta-BASIC and 3D-Jury mappings as well as conservation of the critical active site residues, hydrophobic patterns, and secondary structure predictions. The 3D models of hUSB1 and USB1 were constructed with MODELLER (Fiser and Sali 2003) using the following structures as templates: RNA ligases from Pyrococcus horikoshii (Protein Data Bank [PDB], 1VGJ) and Thermus thermophilus (PDB, 1IUH) (Kato et al. 2003) and Homo sapiens kinase A anchoring protein (PDB, 2VFK) (Gold et al. 2008). Electrostatic potential was calculated using the APBS tool (Baker et al. 2001).
Acknowledgments
We thank Kasia Kowalska for help with DNA cloning, Krystian Stoduś for recombinant protein purification, Aleksander Chlebowski for help with confocal imaging, Marta Olchowik from the Laboratory of Cytometry at the Nencki Institute for cell sorting, Herve Le Hir for advice on in vitro splicing, and members of the A.D. and K.G. laboratories for stimulating discussions. We thank Joanna Kufel and Bertrand Seraphin for their critical reading of the manuscript. This work was supported by the Ministry of Science and Higher Education, EMBO Installation Programme, and Foundation for Polish Science Team Programme, and cofinanced by the EU European Regional Development Fund and the Operational Program Innovative Economy 2007-2013 (Agreement POIG.02.02.00-14-024/08-00). A.D. and K.G. conceived and directed the studies. S.M. performed all of the experiments on human cell lines and the majority of experiments in yeast. J. Krwawicz performed the initial experiments shown in Figures 1 (A and C) and 6D and Supplemental Figure S2. J. Kutner performed the multicopy suppressor screen shown in Figure 1B and participated in the initial biochemical experiments shown in Supplemental Figure S3. I.K. participated in the initial experiments in yeast. M.L. and K.G. performed bioinformatics analyses. A.D. wrote the manuscript with contributions from S.M., M.L., and K.G.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.193169.112.
References
- Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Luhrmann R 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J 18: 5789–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RD, Barrass JD, Dichtl B, Kos M, Obtulowicz T, Robert MC, Koper M, Karkusiewicz I, Mariconti L, Tollervey D, et al. 2010. RiboSys, a high-resolution, quantitative approach to measure the in vivo kinetics of pre-mRNA splicing and 3′-end processing in Saccharomyces cerevisiae. RNA 16: 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV 2001. A natural classification of ribonucleases. Methods Enzymol 341: 3–28 [DOI] [PubMed] [Google Scholar]
- Armanios M 2009. Syndromes of telomere shortening. Annu Rev Genomics Hum Genet 10: 45–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AW, Itin PH, Pigors M, Kohlhase J, Bruckner-Tuderman L, Has C 2010. Poikiloderma with neutropenia: A novel C16orf57 mutation and clinical diagnostic criteria. Br J Dermatol 163: 866–869 [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA 2001. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow DA 2002. Allosteric cascade of spliceosome activation. Annu Rev Genet 36: 333–360 [DOI] [PubMed] [Google Scholar]
- Cameron V, Uhlenbeck OC 1977. 3′-Phosphatase activity in T4 polynucleotide kinase. Biochemistry 16: 5120–5126 [DOI] [PubMed] [Google Scholar]
- Clericuzio C, Harutyunyan K, Jin W, Erickson RP, Irvine AD, McLean WH, Wen Y, Bagatell R, Griffin TA, Shwayder TA, et al. 2011. Identification of a novel C16orf57 mutation in Athabaskan patients with Poikiloderma with Neutropenia. Am J Med Genet A 155A: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo EA, Bazan FJ, Negri G, Gervasini C, Elcioglu NH, Yucelten D, Altunay I, Cetincelik U, Teti A, Del Fattore A, et al. 2012. Novel C16orf57 mutations in patients with poikiloderma with neutropenia: Bioinformatic analysis of the protein and predicted effects of all reported mutations. Orphanet J Rare Dis 7: 7 doi: 10.1186/1750-1172-7-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuchillo CM, Nogues MV, Raines RT 2011. Bovine pancreatic ribonuclease: Fifty years of the first enzymatic reaction mechanism. Biochemistry 50: 7835–7841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R 1993. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes Dev 7: 1632–1643 [DOI] [PubMed] [Google Scholar]
- Dolken L, Ruzsics Z, Radle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, et al. 2008. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14: 1959–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, Koper M, Kufel J, Tollervey D 2008. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev 22: 1069–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A, Sali A 2003. Modeller: Generation and refinement of homology-based protein structure models. Methods Enzymol 374: 461–491 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636 [DOI] [PubMed] [Google Scholar]
- Genschik P, Billy E, Swianiewicz M, Filipowicz W 1997. The human RNA 3′-terminal phosphate cyclase is a member of a new family of proteins conserved in Eucarya, Bacteria and Archaea. EMBO J 16: 2955–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20: 1425 doi: 10.1093/nar/20.6.1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginalski K, Rychlewski L 2003. Protein structure prediction of CASP5 comparative modeling and fold recognition targets using consensus alignment approach and 3D assessment. Proteins 53: 410–417 [DOI] [PubMed] [Google Scholar]
- Ginalski K, Elofsson A, Fischer D, Rychlewski L 2003. 3D-Jury: A simple approach to improve protein structure predictions. Bioinformatics 19: 1015–1018 [DOI] [PubMed] [Google Scholar]
- Ginalski K, von Grotthuss M, Grishin NV, Rychlewski L 2004. Detecting distant homology with Meta-BASIC. Nucleic Acids Res 32: W576–W581 doi: 10.1093/nar/gkh370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatigny A, Mathieu L, Herbert CJ, Dujardin G, Meunier B, Mucchielli-Giorgi MH 2011. An in silico approach combined with in vivo experiments enables the identification of a new protein whose overexpression can compensate for specific respiratory defects in Saccharomyces cerevisiae. BMC Syst Biol 5: 173 doi: 10.1186/1752-0509-5-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold MG, Smith FD, Scott JD, Barford D 2008. AKAP18 contains a phosphoesterase domain that binds AMP. J Mol Biol 375: 1329–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Shumyatsky G, Makan N, Reddy R 1997. Formation of 2′,3′-cyclic phosphates at the 3′ end of human U6 small nuclear RNA in vitro. Identification of 2′,3′-cyclic phosphates at the 3′ ends of human signal recognition particle and mitochondrial RNA processing RNAs. J Biol Chem 272: 21989–21993 [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR, eds. 1991. Guide to yeast genetics and molecular biology. Methods Enzymol 194: 1–863 [PubMed] [Google Scholar]
- Hofmann A, Zdanov A, Genschik P, Ruvinov S, Filipowicz W, Wlodawer A 2000. Structure and mechanism of activity of the cyclic phosphodiesterase of Appr>p, a product of the tRNA splicing reaction. EMBO J 19: 6207–6217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai A, Sato A, Fukuda Y, Okada K, Matsuda T, Sakamoto T, Muto Y, Yokoyama S, Kawai G, Tomita M 2009. Characterization of a heat-stable enzyme possessing GTP-dependent RNA ligase activity from a hyperthermophilic archaeon, Pyrococcus furiosus. RNA 15: 420–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandels-Lewis S, Seraphin B 1993. Involvement of U6 snRNA in 5′ splice site selection. Science 262: 2035–2039 [DOI] [PubMed] [Google Scholar]
- Kataoka N, Dreyfuss G 2008. Preparation of efficient splicing extracts from whole cells, nuclei, and cytoplasmic fractions. Methods Mol Biol 488: 357–365 [DOI] [PubMed] [Google Scholar]
- Kato M, Shirouzu M, Terada T, Yamaguchi H, Murayama K, Sakai H, Kuramitsu S, Yokoyama S 2003. Crystal structure of the 2′–5′ RNA ligase from Thermus thermophilus HB8. J Mol Biol 329: 903–911 [DOI] [PubMed] [Google Scholar]
- Kiss T 2004. Biogenesis of small nuclear RNPs. J Cell Sci 117: 5949–5951 [DOI] [PubMed] [Google Scholar]
- Kuhn J, Binder S 2002. RT–PCR analysis of 5′ to 3′-end-ligated mRNAs identifies the extremities of cox2 transcripts in pea mitochondria. Nucleic Acids Res 30: 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larizza L, Roversi G, Volpi L 2010. Rothmund-Thomson syndrome. Orphanet J Rare Dis 5: 2 doi: 10.1186/1750-1172-5-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser CF, Guthrie C 1993. Mutations in U6 snRNA that alter splice site specificity: Implications for the active site. Science 262: 1982–1988 [DOI] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ 2007. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods 4: 251–256 [DOI] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE 1992. Cyclic 2′,3′-phosphates and nontemplated nucleotides at the 3′ end of spliceosomal U6 small nuclear RNA's. Science 255: 327–330 [DOI] [PubMed] [Google Scholar]
- Mazumder R, Iyer LM, Vasudevan S, Aravind L 2002. Detection of novel members, structure–function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucleic Acids Res 30: 5229–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT 2000. The PSIPRED protein structure prediction server. Bioinformatics 16: 404–405 [DOI] [PubMed] [Google Scholar]
- Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA 2008. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature 451: 1013–1017 [DOI] [PubMed] [Google Scholar]
- Mroczek S, Kufel J 2008. Apoptotic signals induce specific degradation of ribosomal RNA in yeast. Nucleic Acids Res 36: 2874–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannone BK, Kim SD, Noe DA, Wolin SL 2001. Multiple functional interactions between components of the Lsm2–Lsm8 complex, U6 snRNA, and the yeast La protein. Genetics 158: 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SB, Bellini M 2008. The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res 36: 6482–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Sadreyev R, Grishin NV 2003. PCMA: Fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics 19: 427–428 [DOI] [PubMed] [Google Scholar]
- Piard J, Holder-Espinasse M, Aral B, Gigot N, Rio M, Tardieu M, Puzenat E, Goldenberg A, Toutain A, Franques J, et al. 2012. Systematic search for neutropenia should be part of the first screening in patients with poikiloderma. Eur J Med Genet 55: 8–11 [DOI] [PubMed] [Google Scholar]
- Raines RT 1998. Ribonuclease A. Chem Rev 98: 1045–1066 [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Sakamoto Y, Tanaka N, Ichimiya T, Kurihara T, Nakamura KT 2005. Crystal structure of the catalytic fragment of human brain 2′,3′-cyclic-nucleotide 3′-phosphodiesterase. J Mol Biol 346: 789–800 [DOI] [PubMed] [Google Scholar]
- Scherrer T, Mittal N, Janga SC, Gerber AP 2010. A screen for RNA-binding proteins in yeast indicates dual functions for many enzymes. PLoS ONE 5: e15499 doi: 10.1371/journal.pone.0015499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz K, Hesselberth JR, Fields S 2010. Capture and sequence analysis of RNAs with terminal 2′,3′-cyclic phosphates. RNA 16: 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B 1995. Sm and Sm-like proteins belong to a large family: Identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J 14: 2089–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B, Rosbash M 1989. Identification of functional U1 snRNA–pre-mRNA complexes committed to spliceosome assembly and splicing. Cell 59: 349–358 [DOI] [PubMed] [Google Scholar]
- Tanaka N, Shuman S 2009. Structure–activity relationships in human RNA 3′-phosphate cyclase. RNA 15: 1865–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Morice-Picard F, Lacombe D, Nagy N, Hide M, Taieb A, McGrath J 2010. Identification of a homozygous deletion mutation in C16orf57 in a family with Clericuzio-type poikiloderma with neutropenia. Am J Med Genet A 152A: 1347–1348 [DOI] [PubMed] [Google Scholar]
- Tazi J, Forne T, Jeanteur P, Cathala G, Brunel C 1993. Mammalian U6 small nuclear RNA undergoes 3′ end modifications within the spliceosome. Mol Cell Biol 13: 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe R, Richly H, Benecke BJ 2003. Biochemical characterization of a U6 small nuclear RNA-specific terminal uridylyltransferase. Eur J Biochem 270: 971–980 [DOI] [PubMed] [Google Scholar]
- Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ 2006. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 12: 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S 2010. Role of the snRNAs in spliceosomal active site. RNA Biol 7: 345–353 [DOI] [PubMed] [Google Scholar]
- Verdone L, Galardi S, Page D, Beggs JD 2004. Lsm proteins promote regeneration of pre-mRNA splicing activity. Curr Biol 14: 1487–1491 [DOI] [PubMed] [Google Scholar]
- Volpi L, Roversi G, Colombo EA, Leijsten N, Concolino D, Calabria A, Mencarelli MA, Fimiani M, Macciardi F, Pfundt R, et al. 2010. Targeted next-generation sequencing appoints c16orf57 as clericuzio-type poikiloderma with neutropenia gene. Am J Hum Genet 86: 72–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I 2010. Mutations in C16orf57 and normal-length telomeres unify a subset of patients with dyskeratosis congenita, poikiloderma with neutropenia and Rothmund-Thomson syndrome. Hum Mol Genet 19: 4453–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]