Abstract
We report a convenient new technique for the labeling of filamentous phage capsid proteins. Previous reports have shown that phage coat protein residues can be modified, but the lack of chemically distinct amino acids in the coat protein sequences makes it difficult to attach high levels of synthetic molecules without altering the binding capabilities of the phage. To modify the phage with polymer chains, imaging groups, and other molecules, we have developed chemistry to convert the N-terminal amines of the ~4,200 coat proteins into ketone groups. These sites can then serve as chemospecific handles for the attachment of alkoxyamine groups through oxime formation. Specifically, we demonstrate the attachment of fluorophores and up to 3,000 molecules of 2 kD poly(ethylene glycol) (PEG2k) to each of the phage capsids without significantly affecting the binding of phage-displayed antibody fragments to EGFR and HER2 (two important epidermal growth factor receptors). We also demonstrate the utility of the modified phage for the characterization of breast cancer cells using multicolor fluorescence microscopy. Due to the widespread use of filamentous phage as display platforms for peptide and protein evolution, we envision that the ability to attach large numbers of synthetic functional groups to their coat proteins will be of significant value to the biological and materials communities.
Keywords: phage display, bioorthogonal, bioconjugation, materials science, cancer imaging
Through the use of molecular diversity techniques, filamentous phage can be evolved to bind proteins, polymers, small molecules, and metal ions with high affinity and selectivity.1 The success of this platform is due to the ability of filamentous phage to express a wide variety of peptides and proteins as extensions of the p3 and p8 coat proteins that comprise the capsid (Figure 1). This method has provided useful binders for a variety of research efforts in molecular biology, biotechnology, biomedicine, and materials science.2 In addition, the body of the phage has also proven useful as a robust scaffold for nanoparticle nucleation,3 electrode templating,4 light-collection, 5 cell growth and differentiation,6 and drug delivery.7 To enhance these capabilities, we describe herein a convenient N-terminal-selective modification method that can introduce synthetic functionality on the phage coat proteins without interfering with their binding abilities. We demonstrate the utility of this technique by directly converting evolved phage into targeted imaging agents for in vitro cell targeting experiments. Furthermore, we use this method to attach up to 3,000 polymer chains to these structures without compromising their ability to recognize specific receptors on live cells—a useful capability for reducing background binding and a likely requirement for developing future phage-based agents for in vivo applications.
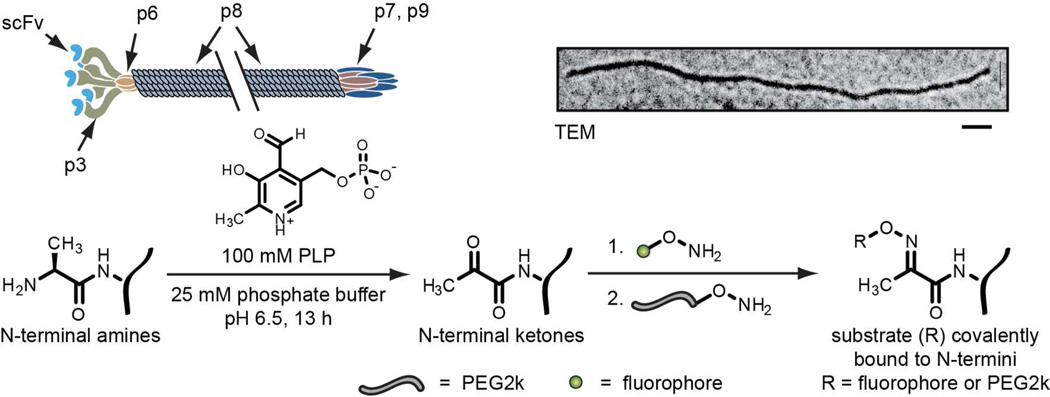
Figure 1.
A cartoon (above) and chemical scheme (below) for the transamination of filamentous phage and the attachment of synthetic molecules. The N-termini are transaminated to yield ketone-bearing proteins, which are then reacted with aminooxy-functionalized fluorophores (green circles), followed by aminooxy-functionalized PEG2k (grey strands). The double slash indicates that the phage is much longer than shown when scaled to the minor coat proteins. The TEM image was stained with uranyl acetate (top right, scale bar represents 100 nm).
Filamentous phage, such as M13 and fd, have approximately five copies of each of their minor coat proteins (p3, p6, p7 and p9, Figure 1). In addition, fd and M13 phage have 4,200 and 2,700 copies of the major coat protein (p8), respectively.2 The p3 sites serve as the principal locations for molecular evolution, especially for large protein inserts such as single chain antibody variable fragments (scFvs) and enzymes. This leaves the p8 sites as abundant locations for the attachment of additional molecules. To introduce synthetic components into these assemblies, the covalent modification of filamentous phage has typically been accomplished through the non-specific modification of amine groups on the capsid surfaces with NHS esters.8,9 However, this approach also leads to extensive acylation of the many lysine residues on the p3 proteins and their associated protein fusions, adding considerable heterogeneity and possible binding interference at high modification levels. Tyrosine residues have also been targeted on the phage surface through the use of diazonium coupling reactions,10 but this approach is also expected to lead to significant modification of critical residues in the evolved proteins. Methods requiring the genetic modification of phage DNA have been attempted to increase specificity. In one case, serine or threonine was genetically introduced at p3 N-termini and oxidized with sodium periodate to produce an aldehyde for chemical labeling.11 This was not demonstrated for the major coat protein p8, and requires use of sodium periodate, which can undesirably oxidize cysteines. Enzymatic ligations offer another genetic approach, as demonstrated with biotin ligase12 and sortase A.13,14 These techniques offer more specificity than prior chemical-labeling approaches, but also require genetic engineering of phage DNA, which may be undesired or unfeasible in certain contexts. Our goal was to develop a simple yet reliable chemical strategy that did not require prior genetic engineering.
To provide a facile, controlled method for modifying filamentous phage with hundreds or even thousands of new functional groups, we have applied a 2-step transamination/oxime formation technique.15–18 This reaction sequence has been shown to be highly selective for N-terminal groups and does not lead to the transamination of lysine ε-amines. Using high-throughput solid phase screening methods, we have previously determined that this reaction proceeds most readily when N-terminal alanine residues are present, and that it can be accelerated by proximal lysine side chains.19 The phage p8 monomers possess a solvent-exposed N-terminal alanine and a lysine at the i+7 position, making this an especially promising substrate for this reaction.
This modification strategy was developed using filamentous fd phage that display single-chain antibody fragments (scFvs) on their p3 minor coat proteins. These scFvs recognize either epidermal growth factor receptor (EGFR) or human epidermal growth factor receptor 2 (HER2), and were identified using phage display.20–22 The overexpression of these receptors is associated with many different breast cancer serotypes, thus providing a motivation for the installation of imageable groups23,24 on these phage for use in diagnostic applications.25 In parallel we also used fd phage bearing an scFv targeting botulinum toxin serotype A (anti-BoNT) as a negative control.22
Results and Discussion
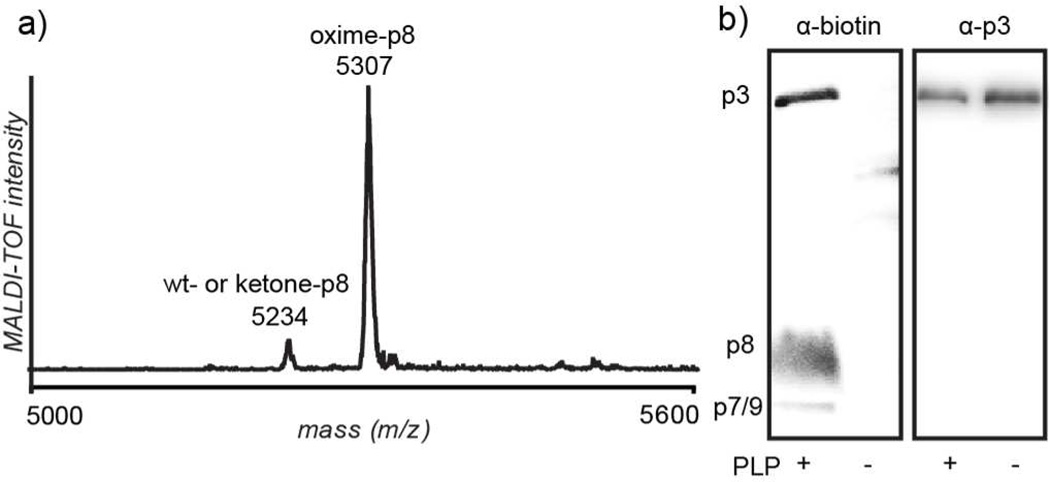
To introduce ketones into the coat proteins, phage were transaminated using a 100 mM solution of pyridoxal 5’-phosphate (PLP) at pH 6.5 for 13 h. The excess PLP was then removed by precipitating the phage, after which they were exposed to various alkoxyamine compounds in pH 6.5 buffer for up to 24 h. Aniline catalysis was used to accelerate oxime formation, as has been previously reported by Dawson and coworkers.26 The specific reaction times and alkoxyamine concentrations were selected based on the levels of modification sought. To estimate the overall extent of p8 modification, a sample of ketone-labeled fd phage was reacted with 2-(aminooxy)acetic acid. Analysis of the coat proteins was achieved using MALDI-TOF mass spectrometry, revealing that the vast majority of the p8 proteins formed the oxime product (Figure 2a, Supporting Information Figure S1), indicating that each fd phage can be loaded with thousands of molecules. Only one addition per p8 was observed, indicating N-terminal specificity even in the presence of five p8 lysines. The overall protein recovery for the transamination and oxime formation steps ranged from 55–95%, with 80% being a typical value. Unfortunately, despite many attempts, protein digest experiments failed to give any cleaved species for the p8 protein, presumably due to its very low solubility and propensity for aggregation once removed from the assembled structure.
Figure 2.
Analysis of filamentous phage modified with small molecules. a) Matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) spectrum showing p8 oxime formation following reaction with 2-(aminooxy)acetic acid (expected mass increase: 73 m/z, observed: 73 m/z). Non-transaminated fd proteins exposed to the same alkoxyamine resulted in no oxime product formation (see Supporting Information Figure S1). b) Western blot of M13KE coat protein labeling with biotin followed by blotting with neutravidin-HRP or α-p3 antibodies. Coat protein molecular weights are as follows: p3, 46.5 kD; p6, 12.4 kD; p7, 3.6 kD; p8, 5.2 kD; p9, 3.7 kD. Labeling of p7 and p9 cannot be distinguished due to their similar molecular weights (3.6 and 3.7 kD, respectively). p6 is not observed, congruent with an N-terminus inaccessible for modification.
The only by-product was a small amount of a covalent adduct of the protein with the PLP, which presumably formed through an aldol addition of the N-terminal pyruvamide to the pyridoxal aldehyde group. This PLP adduct was not visible following reaction of phage with aminooxy-derivatized molecules via MALDI-TOF mass spectrometry analysis, possibly due to its poor ionization or insufficient quantity. It was, however, identified by mass spectrometry after disassembling the phage using RP-HPLC to isolate the PLP adduct-p8 from wt- and ketone-p8 species (Supporting Information Figures S2 and S3). The negative charge of the phosphate group resulted in the earlier elution via RP-HPLC. This species has been observed in transamination reactions previously, and since it possesses a ketone group, it can still participate in oxime formation.16 This, in addition to its very low abundance, renders it insignificant for most applications.
The small p3-to-p8 ratio for fd phage prevented p3 detection by mass spectrometry and western blotting. Instead, we turned to the use of M13KE phage, which are fd analogs with smaller genomes. They require a smaller number of p8 proteins to tile the length of the phage, and therefore have a higher ratio of p3 to p8 proteins. The M13KE and wt-fd coat proteins are identical, except for a single D12N point mutation in p8.27,28 To detect the modifications with improved sensitivity, transaminated M13KE was exposed to biotin-ONH2 and analyzed via western blotting with neutravidin-HRP (Figure 2b). All of the coat proteins with accessible N-termini, including p3, showed labeling. The α-p3 blot in Figure 2b shows that both lanes contain approximately the same concentration of phage, while the neutravidin-HRP (α-biotin) blot shows that only PLP-reacted phage are biotin labeled.
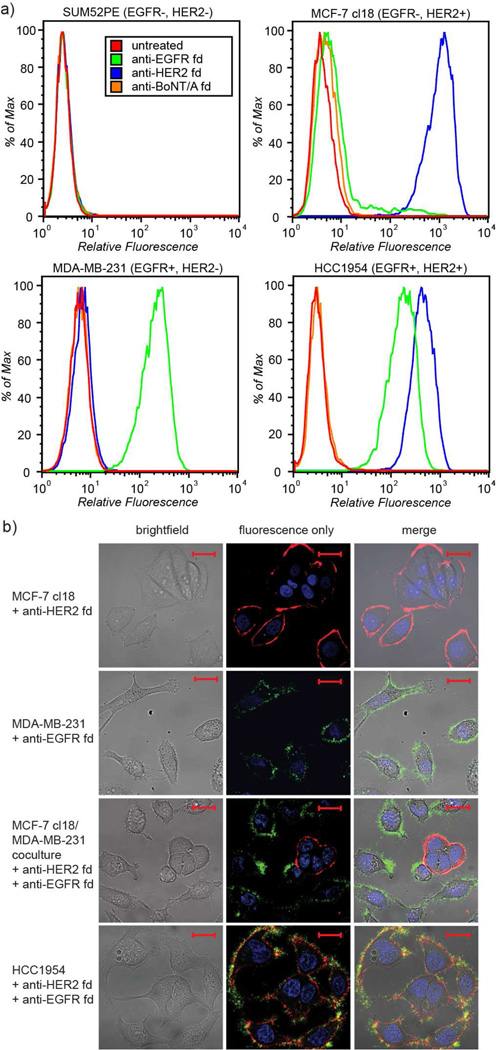
To verify the ability of the modified phage to bind their targets, samples of transaminated anti-EGFR, anti-HER2, and anti-BoNT fd phage were reacted with Alexa Fluor® 488 or 647 C5-aminooxyacetamide (AF488/647-ONH2) dyes. For the cell microscopy experiments described below, approximately 6–8% of the p8 proteins (~300 copies/phage, as determined using UV/vis) were labeled with the fluorophores. We have increased the levels of fluorophore modification with AF488 to levels of up to 80% by using a ten-fold excess of aniline26 (100 mM for AF488), albeit with some levels of decreased solubility. The modified phage bound to their appropriate cell surface receptors with excellent specificity, as revealed using flow cytometry (Figure 3a and Supporting Information Figures S4–S7). The negative control anti-BoNT phage showed no binding. In terms of cell viability, these data also indicated that only 0.25% to 3.0% of the cells had died during the exposure to the phage-based imaging agents, which was in line with untreated cell samples.
Figure 3.
Fluorophore modified fd phage cell binding results. a) Flow cytometry with AF488 labeled phage (applied at 0.8 nM) indicated selective recognition of EGFR and HER2 epitopes. The legend for all histograms is shown in SUM52PE inset. Gating data are shown in Supporting Information Figures S4–S7. b) Live cell confocal microscopy images of fluorescently labeled anti-HER2 and anti-EGFR fd showed marker-specific binding to breast cancer cell lines. Fluorescence is as follows: DAPI (blue), anti-HER2 fd (red), anti-EGFR fd (green). Scale bars represent 20 µm. Control and larger images with all fluorescence channels are shown in Supporting Information Figures S8–S12.
The selective binding capabilities of the EGFR and HER2 targeted phage were also confirmed in microscopy experiments. A panel of breast cancer cells was treated with the phage, and visualized using live cell confocal microscopy. These images (Figure 3b and Supporting Information Figures S8–S12) demonstrated the retention of excellent specificities and binding capabilities of fd for their targeted receptors following chemical modification. Upon increased incubation times (>2h), phage targeting overexpressed markers were observed to be internalized by the respective cells. Preliminary results indicate that this occurs via receptor-mediated endocytosis; however, further experiments to clarify this behavior are in progress.
The ability of these fd to image receptor overexpression in vitro, even when different cell types are mixed, portends well for their use in vivo. In anticipation of future in vivo applications, we investigated the attachment of poly(etheylene glycol) (PEG) polymers to the phage capsids. PEG has been shown to reduce non-specific binding, decrease immunogenicity, and increase the solubility of attached molecules.29 Ketone-labeled fd were reacted with 2 kDa O-(methoxypoly(ethylene glycol))-hydroxylamine (PEG2k-ONH2),30 and the percentage of p8 proteins that were modified was quantified using RP-HPLC (Supporting Information Figure S13). By varying the reaction times, samples with differing levels of PEG2k-labeled p8s were prepared. Presumably higher concentrations of the PEG2k-ONH2 could achieve shorter modification times, but we avoided using them to prevent precipitation of the phage (as was purposely done in the protein purification steps). For phage previously labeled with fluorophores, there were no observed changes in the absorption or emission properties of the dyes upon addition of the chains. The added PEG chains also caused no morphological changes that could be observed by TEM (Supporting Information Figure S14).
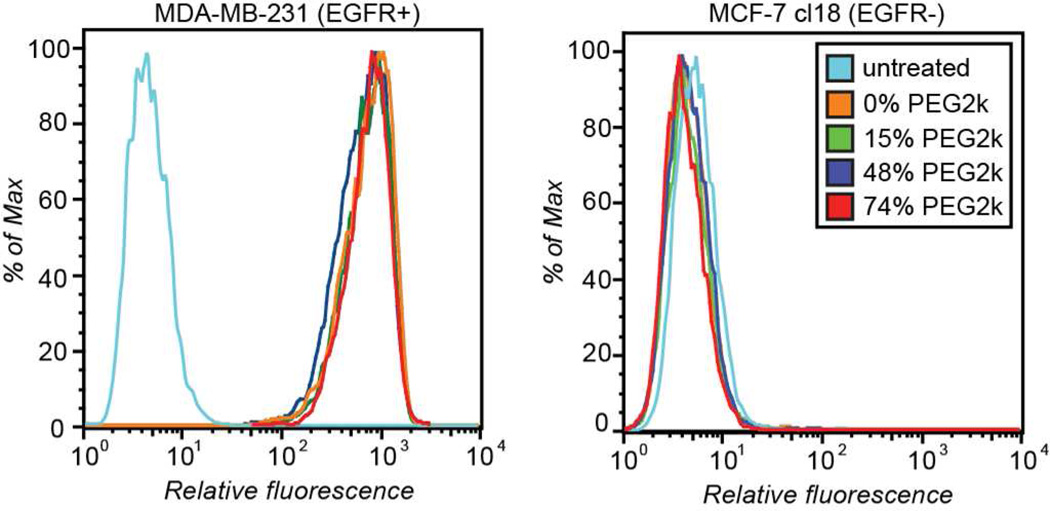
Zeta potential measurements were obtained in order to determine the ability of the PEG polymers to shield fd charge (Supporting Information Figure S15). An increase in negative charge was noted following PLP modification, presumably due to the loss of the cationic N-terminal amino groups on the p8 monomers. As anticipated, the negative charge decreased with increasing levels of PEG modification. At 67% p8 labeling the zeta potential was −5.5±7.3 mV, nearly an order of magnitude less than that of ketone-labeled fd. The binding abilities of PEG-labeled fd were also evaluated by flow cytometry (Figure 4 and Supporting Information Figures S16–S18). The PEG-labeled anti-EGFR fd continued to bind MDA-MB-231 (EGFR positive) cells, while none bound the MCF-7 cl18 cells (EGFR negative).
Figure 4.
Flow cytometry analysis of AF488 labeled anti-EGFR fd possessing various levels of PEG modifications. The target cells were MDA-MB-231 (EGFR+, left) and the control cells were MCF-7 cl18 (EGFR−, right). Phage concentrations were 0.8 nM. Gating data are shown in Supporting Information Figure S16. As a negative control, anti-BoNT fd labeled with nearly identical levels of PEG were incubated with both cell lines, and showed no binding (see Supporting Information Figures S17–S18).
Summary
The chemical modifications to fd phage described herein were used to produce highly selective fluorescent imaging agents that can be readily adapted for use with MRI, PET, and other detection modalities. Furthermore, the binding molecule, phage length, and labeling molecule type can be adjusted for a variety of in vitro and in vivo applications.31 By combining the ability of phage display to obtain genetically encodable binding molecules with the N-terminal transamination/oximation method for appending chemical functionality, a much wider variety of well-defined multifunctional materials can now be accessed.
Methods
Unless otherwise noted, all chemical reagents were purchased from Aldrich. Alexa Fluor® 488 and 647 C5-aminooxyacetamide, bis(triethylammonium) salt, N-(aminooxyacetyl)-N’-(D-biotinoyl) hydrazine, trifluoroacetic acid salt, and neutravidin-HRP were purchased from Invitrogen. Anti-M13 p3 was purchased from New England Biolabs. O-(Methoxypoly(ethylene glycol))-hydroxylamine (PEG2k-ONH2) was prepared as previously described.30 Cells were maintained according to ATCC recommended guidelines. For specific instrumentation and detailed experimental information see the Supporting Information.
Fd production
Fd displaying anti-EGFR, -HER2, and -BoNT scFvs used in these experiments have been reported previously,20–22 and were generated using standard techniques.32
Transamination
Fd (75–128 nM) were transaminated using 100 mM PLP in 25 mM phosphate buffer at pH 6.5 for 13 h at rt. Excess PLP was removed via a series of precipitations with 20% PEG8k/2.5M NaCl, supernatant removal, and resuspension in PBS.
Reaction with biotin and western blotting
The final reaction concentrations were: 296 nM M13KE, 10 mM phosphate buffer (pH 6.2), 10 mM aniline, and 16 mM biotin-ONH2. After 15 h at rt the reaction was quenched by adding DL-glyceraldehyde to a final concentration of 33.3 QM, followed by SDS-PAGE. A 1:10,000 dilution of anti-p3 and a 1:2,000 dilution of Neutravidin-HRP were used for blotting experiments. A Genscript 1-hour western kit was used for detection.
Reaction with 2-(aminooxy)acetic acid
24 nM fd was reacted with 5 mM 2-(aminooxy)acetic acid in 100 mM anilinium acetate, pH 4.7, for 21 h at rt. Reaction conditions were different from those used for other labeling reactions because the objective of this experiment was to estimate the percentage of transaminated p8s. For this reason, low pH and high aniline concentrations were used to maximize p8 labeling. Other experiments used higher pHs and lower aniline concentrations to better control the percent of p8s modified.
Reaction with fluorophores
The final reaction concentrations were: 185 nM fd, 20 mM phosphate buffer (pH 6.2), 10 mM (for AF488) or 100 mM aniline (for AF647), and 1 mM AF488/647-ONH2. Exposure to these conditions for 45 min at rt resulted in 2% p8 modification (for phage to be reacted with PEG subsequently). Otherwise reactions continued for 16–18 h at rt, resulting in 6–8% levels of dye modification. After the reaction, the excess fluorophore was removed using the purification described above.
Flow cytometry
500,000 cells in 100 µL 1% FBS/DPBS were mixed with 100 µL 0.8 nM fd and incubated at 4 °C. After 1 h, each sample was diluted to 1 mL with 1% FBS/DPBS, and the cells were washed before resuspending in 200 µL 1% FBS/DPBS.
Cell microscopy
2 mL of 25,000 cells/mL in 35 mm glass bottom dishes (Mattek) were grown at 37 °C with 5% CO2 for 72–96 h. The cells were washed with PBS, 150 QL of 0.8 nM fd in 1% FBS/DPBS was added before incubating at 37 °C with 5% CO2 for 1 h. The cells were washed three times with PBS followed by addition of 1 mL phenol red-free media with 10% FBS. DAPI was added to 1 µM prior to imaging.
Reaction with PEG2k
37 nM fd, 20 mM PEG2k-ONH2, 20 mM phosphate buffer (pH 6.2) and 10 mM aniline. Following the appropriate reaction times, samples were washed over an Illustra Nap-5 gel filtration column, eluting with PBS.
Supplementary Material
Acknowledgements
These studies were generously supported by the DOD Breast Cancer Research Program (BC061995) and the NCI SPORE in Breast Cancer (P50-CA58207). ZMC was supported by the Berkeley Chemical Biology Graduate Program (NRSA Training Grant 1 T32 GMO66698). MEF is supported by a DOD BCRP postdoctoral fellowship (BC100159). HC was supported by the Dow Foundation Sustainable Products and Solutions Program.
Footnotes
Supporting Information Available: Full characterization of conjugates, chromatograms, and additional flow cytometry data are provided. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Smith GP, Petrenko VA. Phage Display. Chem. Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 2.Kehoe JW, Kay BK. Filamentous Phage Display in the New Millennium. Chem. Rev. 2005;105:4056–4072. doi: 10.1021/cr000261r. [DOI] [PubMed] [Google Scholar]
- 3.Mao CB, Solis DJ, Reiss BD, Kottmann ST, Sweeney RY, Hayhurst A, Georgiou G, Iverson B, Belcher AM. Virus-Based Toolkit for the Directed Synthesis of Magnetic and Semiconducting Nanowires. Science. 2004;303:213–217. doi: 10.1126/science.1092740. [DOI] [PubMed] [Google Scholar]
- 4.Lee YJ, Yi H, Kim WJ, Kang K, Yun DS, Strano MS, Ceder G, Belcher AM. Fabricating Genetically Engineered High-Power Lithium-Ion Batteries Using Multiple Virus Genes. Science. 2009;324:1051–1055. doi: 10.1126/science.1171541. [DOI] [PubMed] [Google Scholar]
- 5.Nam YS, Shin T, Park H, Magyar AP, Choi K, Fantner G, Nelson KA, Belcher AM. Virus-Templated Assembly of Porphyrins into Light-Harvesting Nanoantennae. J. Am. Chem. Soc. 2010;132:1462–1463. doi: 10.1021/ja908812b. [DOI] [PubMed] [Google Scholar]
- 6.Merzlyak A, Indrakanti S, Lee S-W. Genetically Engineered Nanofiber-Like Viruses for Tissue Regenerating Materials. Nano Lett. 2009;9:846–852. doi: 10.1021/nl8036728. [DOI] [PubMed] [Google Scholar]
- 7.Arap W, Pasqualini R, Ruoslahti E. Cancer Treatment by Targeted Drug Delivery to Tumor Vasculature in a Mouse Model. Science. 1998;279:377–380. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- 8.Yacoby I, Benhar I. Targeted Filamentous Bacteriophages as Therapeutic Agents. Expert Opin. Drug Delivery. 2008;5:321–329. doi: 10.1517/17425247.5.3.321. [DOI] [PubMed] [Google Scholar]
- 9.Hilderbrand SA, Kelly KA, Niedre M, Weissleder R. Near Infrared Fluorescence-Based Bacteriophage Particles for Ratiometric pH Imaging. Bioconjugate Chem. 2008;19:1635–1639. doi: 10.1021/bc800188p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li K, Chen Y, Li S, Nguyen HG, Niu Z, You S, Mello CM, Lu X, Wang Q. Chemical Modification of M13 Bacteriophage and Its Application in Cancer Cell Imaging. Bioconjugate Chem. 2010;21:1369–1377. doi: 10.1021/bc900405q. [DOI] [PubMed] [Google Scholar]
- 11.Ng S, Jafari MR, Matochko WL, Derda R. Quantitative Synthesis of Genetically Encoded Glycopeptide Libraries Displayed on M13 Phage. ACS Chemical Biology. 2012 doi: 10.1021/cb300187t. asap article. [DOI] [PubMed] [Google Scholar]
- 12.Chen I, Choi Y-A, Ting AY. Phage Display Evolution of a Peptide Substrate for Yeast Biotin Ligase and Application to Two-Color Quantum Dot Labeling of Cell Surface Proteins. J. Am. Chem. Soc. 2007;129:6619–6625. doi: 10.1021/ja071013g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hess GT, Cragnolini JJ, Popp MW, Allen MA, Dougan SK, Spooner E, Ploegh HL, Belcher AM, Guimaraes CP. M13 Bacteriophage Display Framework That Allows Sortase-Mediated Modification of Surface-Accessible Phage Proteins. Bioconjugate Chemistry. 2012 doi: 10.1021/bc300130z. asap article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: a Versatile Method for Protein Labeling. Nat. Chem. Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- 15.Gilmore JM, Scheck RA, Esser-Kahn AP, Joshi NS, Francis MB. N-Terminal Protein Modification Through a Biomimetic Transamination Reaction. Angew. Chem. Int. Ed. 2006;45:5307–5311. doi: 10.1002/anie.200600368. [DOI] [PubMed] [Google Scholar]
- 16.Scheck RA, Dedeo MT, Iavarone AT, Francis MB. Optimization of a Biomimetic Transamination Reaction. J. Am. Chem. Soc. 2008;130:11762–11770. doi: 10.1021/ja802495w. [DOI] [PubMed] [Google Scholar]
- 17.Dixon HBF. N-Terminal Modification of Proteins. J. Protein Chem. 1984;3:99–108. [Google Scholar]
- 18.Snell EE. The Vitamin B6 Group. V. The Reversible Interconversion of Pyridoxal and Pyridoxamine by Transamination Reactions. J. Am. Chem. Soc. 1945;67:194–197. [Google Scholar]
- 19.Witus LS, Moore T, Thuronyi BW, Esser-Kahn AP, Scheck RA, Iavarone AT, Francis MB. Identification of Highly Reactive Sequences for PLP-Mediated Bioconjugation Using a Combinatorial Peptide Library. J. Am. Chem. Soc. 2010;132:16812–16817. doi: 10.1021/ja105429n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y, Drummond DC, Zou H, Hayes ME, Adams GP, Kirpotin DB, Marks JD. Impact of Single-Chain Fv Antibody Fragment Affinity on Nanoparticle Targeting of Epidermal Growth Factor Receptor-Expressing Tumor Cells. J. Mol. Biol. 2007;371:934–947. doi: 10.1016/j.jmb.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connell D, Becerril B, Roy-Burman A, Daws M, Marks JD. Phage Versus Phagemid Libraries for Generation of Human Monoclonal Antibodies. J. Mol. Biol. 2002;321:49–56. doi: 10.1016/s0022-2836(02)00561-2. [DOI] [PubMed] [Google Scholar]
- 22.Amersdorfer P, Wong C, Smith T, Chen S, Deshpande S, Sheridan R, Marks JD. Genetic and Immunological Comparison of Anti-Botulinum Type A Antibodies from Immune and Non-Immune Human Phage Libraries. Vaccine. 2002;20:1640–1648. doi: 10.1016/s0264-410x(01)00482-0. [DOI] [PubMed] [Google Scholar]
- 23.Hooker JM, O’Neil JP, Romanini DW, Taylor SE, Francis MB. Genome-Free Viral Capsids as Carriers for Positron Emission Tomography Radiolabels. Mol. Imaging Biol. 2008;10:182–191. doi: 10.1007/s11307-008-0136-5. [DOI] [PubMed] [Google Scholar]
- 24.Datta A, Hooker JM, Botta M, Francis MB, Aime S, Raymond KN. High Relaxivity Gadolinium Hydroxypyridonate-Viral Capsid Conjugates: Nanosized MRI Contrast Agents. J. Am. Chem. Soc. 2008;130:2546–2552. doi: 10.1021/ja0765363. [DOI] [PubMed] [Google Scholar]
- 25.Milanezi F, Carvalho S, Schmitt FC. EGFR/HER2 in Breast Cancer: a Biological Approach for Molecular Diagnosis and Therapy. Expert Rev. Mol. Diagn. 2008;8:417–434. doi: 10.1586/14737159.8.4.417. [DOI] [PubMed] [Google Scholar]
- 26.Dirksen A, Hackeng TM, Dawson PE. Nucleophilic Catalysis of Oxime Ligation. Angew. Chem., Int. Ed. 2006;45:7581–7584. doi: 10.1002/anie.200602877. [DOI] [PubMed] [Google Scholar]
- 27.van Wezenbeek PM, Hulsebos TJ, Schoenmakers JG. Nucleotide Sequence of the Filamentous Bacteriophage M13 DNA Genome: Comparison with Phage Fd. Gene. 1980;11:129–148. doi: 10.1016/0378-1119(80)90093-1. [DOI] [PubMed] [Google Scholar]
- 28.Noren KA, Noren CJ. Construction of High-Complexity Combinatorial Phage Display Peptide Libraries. Methods. 2001;23:169–178. doi: 10.1006/meth.2000.1118. [DOI] [PubMed] [Google Scholar]
- 29.Pasut G, Veronese FM. PEG Conjugates in Clinical Development or Use as Anticancer Agents: an Overview. Adv. Drug Delivery Rev. 2009;61:1177–1188. doi: 10.1016/j.addr.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 30.Schlick TL, Ding Z, Kovacs EW, Francis MB. Dual-Surface Modification of the Tobacco Mosaic Virus. J. Am. Chem. Soc. 2005;127:3718–3723. doi: 10.1021/ja046239n. [DOI] [PubMed] [Google Scholar]
- 31.Specthrie L, Bullitt E, Horiuchi K, Model P, Russel M, Makowski L. Construction of a Microphage Variant of Filamentous Bacteriophage. J. Mol. Biol. 1992;228:720–724. doi: 10.1016/0022-2836(92)90858-h. [DOI] [PubMed] [Google Scholar]
- 32.Barbas CF, Burton DR, Scott JK, Silverman GJ. Phage Display: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.