European Phragmites australis is one of four main cp-DNA haplotype clusters present worldwide. The European gene pool extends from North America to Far East Asia and South Africa. Extensive gene flow occurs only within the temperate region of Europe.
Abstract
Background and aims
Two Phragmites australis taxa are recognized in Europe: P. australis ssp. altissimus, also known as Phragmites isiaca, in the Mediterranean region and P. australis in the temperate region. Another taxonomic group in the Mediterranean is Phragmites frutescens. European genotypes are diverse genetically, cytologically and morphologically, and are related to African, Asiatic and American genotypes. We investigated chloroplast DNA (cpDNA) diversity in Europe and defined the current borders of the European gene pool.
Methodology
We analysed chloroplast variation with parsimony and genetic distance methods, and compared it with that of nuclear amplified fragment length polymorphism and microsatellites. We also investigated the phenological pattern of 188 genotypes collected worldwide in a common garden in Denmark. We assumed that non-flowering genotypes could indicate climatic, geographic and/or reproductive barriers to dispersal and would have been recorded in the genetic pattern as groups genetically isolated from, or within, the European pool.
Principal results
The European P. australis gene pool extends from North America to the Far East and South Africa. However, African and North American genotypes are differentiating from the European genotypes. Mediterranean P. australis is genetically different from temperate P. australis and shares several similarities with Phragmites mauritianus in Africa and Phragmites karka in Asia. Phragmites frutescens shares the cpDNA sequences with both these tropical species. Two DNA bands can distinguish Mediterranean P. australis from P. frutescens and P. mauritianus and from temperate P. australis, and reveal possible hybrids among these species in the Mediterranean region. Phenological data confirmed possible gene flow within the temperate region of Europe, whereas the Mediterranean genotypes did not set inflorescences in Denmark, suggesting reproductive barriers between temperate and Mediterranean P. australis.
Conclusions
European P. australis appears as one of four main Phragmites groups known in the world. Further research is needed to understand the implications of long-distance dispersal at the population level.
Introduction
The genus Phragmites Adans. (Poaceae) is cosmopolitan with a distribution ranging from the tropics to cold temperate areas of both hemispheres (Clevering and Lissner 1999). The Phragmites species are tall (up to 6 m), perennial grasses predominantly found in littoral zones of lakes, along rivers and in shallow freshwater and brackish wetlands (Brix 1999). At present, five species are recognized, of which Phragmites australis (Cav.) Trin ex Steudel is the most widely distributed (Clevering and Lissner 1999). There is, however, morphological and cytological variation within Phragmites species, and in particular within the cosmopolitan P. australis, that has resulted in the description of a number of subspecies and varieties, whose taxonomic rank has been discussed and frequently revised. In Europe, two P. australis subspecies were proposed by Clayton (1968): P. australis ssp. australis, distributed in the temperate regions of both hemispheres in the Old and the New World, and P. australis ssp. altissimus (Benth) Clayton, distributed along the shores of the Mediterranean, extending to Iran, Arabia, Kenya, Ethiopia and the southern edge of the Sahara (Clayton 1967, 1968). The name P. australis ssp. altissimus has been accepted in the world checklist of selected plant families in the Kew database, where the holotype is conserved. The epithet ‘isiaca’, referring to the borders of the ancient Egyptian Empire, is also frequently used for the Mediterranean Phragmites both at the subspecific and the varietal level, to define the limit of the distribution range of European Phragmites (Hultén and Fries 1986). The Flora Europeae (Tutin et al. 1980) cites Phragmites isiaca Kunth. as a large variant of P. australis, with which it intergrades, but dismisses the specific or subspecific rank of P. isiaca. The recent recognition of Phragmites frutescens H. Scholz. in the Mediterranean region (Greuter and Scholz 1996; Scholz and Böhling 2000; Mutlu 2002) has increased the uncertainties about the identity of Mediterranean Phragmites. The morphological similarities of P. frutescens to P. mauritianus Kunth., rather than to European P. australis, suggested a tropical origin of this species from P. mauritianus (Greuter and Scholz 1996). Lambertini et al. (2006) confirmed genetic similarities of P. frutescens with P. mauritianus.
The subspecific rank for P. australis ssp. australis as proposed by Clayton (1968) has not been formally accepted. Phragmites australis is the former Phragmites communis Trin. and has progressively incorporated Phragmites taxa collected worldwide. Conert (1961) merged 64 taxa into P. communis and recognized 11 varieties within P. communis based on leaf anatomy and cytology. All varieties were present in Europe, and most of them in a limited geographic area, such as Germany. Even though Conert's classification system has been abandoned, it reflects very well the often gradual morphological variation in size, colour and leaf structure present in Europe, sometimes observable even among adjacent stands (Rolletschek et al. 1999). In addition to P. communis var. communis, defined by the purple colour of the inflorescences, Conert described the European diversity in plant size (var. humilis and var. pumila, as opposed to var. arundinaceaea and pseudodonax), colour (var. flavescens, with light-brown inflorescences and var. striato-picta, with yellow–green-striped leaves), presence of hairs (var. hirsuta), leaf stiffness (var. stenophylla) and rhizome size (var. stolonifera). Several genetic studies at the population level conducted throughout Europe have consistently shown that genetic variance is distributed within, rather than among, populations, and that populations can be genetically very different from each other even at a local scale (Lambertini et al. 2008; Fér and Hroudová 2009; Paul et al. 2011). However, isolation-by-distance patterns have failed to be detected (Lambertini et al. 2008; Fér and Hroudová 2009; Lukacs 2009; Krzakowa and Michalak 2010; Paul et al. 2011) and, in general, lack of genetic structure seems to characterize the European continent (Lambertini et al. 2006). Phylogeographically, European Phragmites belongs to a group that extends throughout Eurasia and North America, as well as in the Southern Hemisphere in Africa and Australia (Saltonstall 2002; Lambertini et al. 2006). The lack of structure within the European group, as opposed to other geographically distinct P. australis groups, suggests largely reticulate interrelationships within a metapopulation with indeterminable borders.
When relationships among genotypes are analysed at the continental scale, the idea of a single population of randomly mating individuals seems to work well for European Phragmites (Lambertini et al. 2006, 2008). However, genetic distances among adjacent stands may be high and comparable to those observed among stands hundreds, if not thousands, of kilometres apart (Lambertini et al. 2008). This implies dispersal of pollen and/or seeds across different climatic areas, as well as establishment and fitness under very different environmental conditions. The cryptic invasion of a Euroasiatic cp-lineage in North America, described as haplotype M (Saltonstall 2002), shows that a single maternal lineage can establish in a variety of habitats and populate an entire continent. In contrast to many invasive clonal species (Geng et al. 2007; Lambertini et al. 2010), the invasive spreading of P. australis in North America seems to be driven by seed dispersal (McCormick et al. 2010), and seed viability has been shown to be positively related to population-level genetic diversity (Kettenring et al. 2011). Genetic diversity within the Euroasiatic lineage appears therefore to be a key factor explaining its broad ecological amplitude. In the European range, P. australis shows a reticulate pattern of relationships, whereas in North America the Euroasiatic P. australis haplotype M is reported to be genetically distinct and reproductively isolated from the native cp-lineages of P. australis ssp. americanus Saltonstall, P.M. Petersoon & Soreng (Saltonstall 2011). In the Gulf Coast of the USA, however, the nuclear alleles of the Euroasiatic lineage are introgressed in another distinctive cp-lineage, known as haplotype I (Saltonstall 2002), which has recently been revealed to be an interspecific hybrid between P. mauritianus and a population of P. australis in the Mediterranean region (Lambertini et al. 2012).
The first aim of this study was to analyse chloroplast DNA (cpDNA) diversity in Europe and the phylogeographic relationships of European haplotypes at the global scale. The second aim was to identify the current borders of the European gene pool in order to determine the range within which gene flow is likely to be occurring, and thus understand the intercontinental relationships of European Phragmites. Given the broad distribution of European-related P. australis (Saltonstall 2002; Lambertini et al. 2006), we used a global set of samples representing all continents and all species of the genus. Differently from previous studies we compared the global pattern of phylogeographic relationships inferred by cpDNA with that of nuclear DNA. We also examined phenology in a common garden in Denmark of 188 Phragmites genotypes from all over the world, in order to have a model system in which to observe the implications of long-distance interrelationships and their congruence with molecular markers. The hypothesis behind the common garden was that genotypes releasing pollen and seeds in Denmark can contribute alleles to the European gene pool. On the contrary, non-flowering genotypes may reveal climatic, geographic and/or reproductive barriers to dispersal and would be recorded in the genetic pattern as groups genetically isolated from, or within, the European gene pool.
Materials and methods
Study area
A living collection of Phragmites genotypes was established in the experimental garden of Aarhus University beginning in 2001. The collection represents all continents and all species of the genus, but most samples are from Europe. The clones established in the garden are listed in [Additional Information—Table 1] of the present study. This collection was sampled for DNA and used for a phenological investigation.
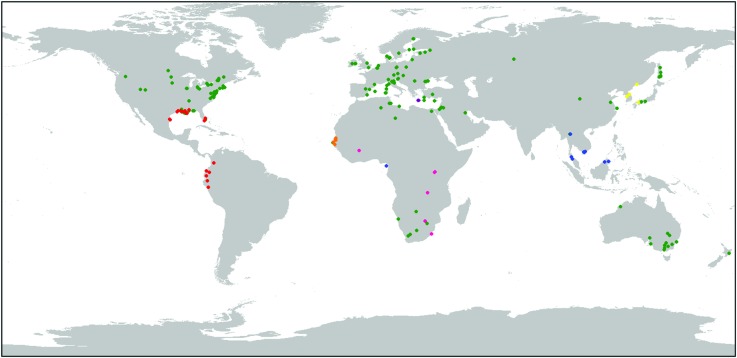
For the molecular work we included additional reference samples (not cultivated in the garden) from the Mediterranean area and the Middle East (from Sardinia-Italy, Libya and Jordan) (Lambertini et al. 2012), from Louisiana in the US Gulf Coast (Lambertini et al. 2012) and from Asia (China and Vietnam) (Lambertini et al. in preparation). The distribution of the samples used in the study is shown in Fig. 1. Previous studies (Lambertini et al. 2006, 2008) have shown that interrelationships in European Phragmites are sensitive to the inclusion or exclusion of outgroups. Outgroups are necessary in any phylogenetic analysis to provide a point of comparison and polarize ancestral and derived characters. The inclusion of a single outgroup or a few distantly related ougroups may, however, not be helpful, as homologies and alignment might become uncertain. Therefore, we included a considerable number of outgroups which should increase the likelihood that the European basal node is correctly placed and that characters are correctly polarized.
Fig. 1.
Distribution of the Phragmites samples used in the study. The sample set includes the species P. australis (green), P. mauritianus (pink), P. frutescens (purple), P. karka (blue), P. japonicus (yellow), hybrids of P. mauritianus × P. australis (red) and hybrids of P. australis × P. mauritianus (orange).
DNA extraction
DNA was isolated from apical leaves dried in silica gel with the Qiagen DNeasy Plant Mini Kit as described in Lambertini et al. (2006). The additional samples from the Mediterranean area, Gulf Coast and Asia were extracted with the EZNA SP Plant DNA Kit (Omega Bio-Tek), after grinding the dry leaves in a mortar with liquid nitrogen.
cpDNA sequences
Seventy-three clones from the Aarhus and Mediterranean collections, selected to represent Europe from the Atlantic Coast to the Danube delta in longitude, and from northern Sweden to Jordan in latitude, were sequenced. The Danube delta, represented by several genotypes in our garden, was more densely sampled (20 samples) because of the high genetic diversity detected in this region in previous studies (Lambertini et al. 2008, 2012).
trnT-trnL chloroplast intergenic spacer (Taberlet et al. 1991; Saltonstall 2001) and rbcL-psaI chloroplast intergenic spacer (Saltonstall 2001, 2002) were amplified. Ten picomoles of forward and reverse primers were added to 10 µL of 2×Mastermix (VWR Amplicon) and 2 µL of template DNA. Sterile water was added to reach the final volume of 20 µL. Amplification was run in a Peltier Thermal Cycler PTC-200-MJ Research programmed with 94 °C for 3 min, 40 cycles of 94 °C for 30 s, 50 °C for 40 s, 72 °C for 1 min, followed by 72 °C for 7 min. The amplified product was sequenced with forward and reverse primers in an ABI sequencer. As most of the variation in these regions is represented by tandem repeats, we amplified and sequenced those samples differing from GenBank accessions multiple times, to make sure that repetitive sequences were not due to amplification or sequencing artefacts.
Sequences were aligned with the program BioEdit ver. 7.0 Sequence Alignment Editor (Hall 1999). trnT-trnL and rbcL-psaI sequences in NCBI (Saltonstall 2002; Hauber et al. 2011; Vachon and Freeland 2011; Freeland and Vachon 2012; Lambertini et al. 2012) were included in the alignment to increase the number of possible outgroups.
New cpDNA haplotypes (hereafter referred as haplotypes) were classified following Saltonstall (2002) (K. Saltonstall, pers. commun.) and deposited in GenBank. Variation in the number of mononucleotide repeats in the trnT-trnL sequence has previously been attributed to intra-haplotypic variation (Saltonstall 2002) and, as such, has not been used to define haplotypes. However, given the frequency of these variants in the European data set, and the possible phylogeographic value of this variation (Chu et al. 2011, 2012; Hauber et al. 2011; Vachon and Freeland 2011; Freeland and Vachon 2012; Lambertini et al. 2012; Saltonstall and Lambertini 2012), we treated all haplotypic variants as independent haplotypes. The capital letter symbolizing the haplotypes (e.g. haplotype M, L, O, etc.) stands for haplotypes whose trnT-trnL and rbcL-psaI sequences have been deposited in GenBank, according to Saltonstall (2002). The variation among these haplotypes is given by indels, substitutions and number of tandem repeat units of 31- and 33-bp motifs (minisatellites). Haplotypes defined by a capital letter followed by a number (e.g. M1, M2, M3, L1, O2, etc.) (following Hauber et al. 2011) differ in the number of repeats at one (or more) microsatellite loci in the trnT-trnL and/or the rbcL-psaI region from the haplotypes deposited in GenBank (Saltonstall 2002), and are defined as cp-microsatellite variants in the present study. These cp-microsatellite variants have not been deposited in GenBank because the classification system used in the present study is preliminary. The analysis of a larger cp-variation in microstatellites worldwide might produce a different classification system (K. Saltonstall and C. Lambertini, in preparation).
In total, 69 different sequences were obtained combining our data set with the sequences downloaded from GenBank. To avoid overestimating the number of mutations associated with repeated sequences, we coded repeat motifs (microsatellites and minisatellites) as multistate characters (Doyle 1992). In this way, differences due to the number of repeats are counted as one character, independently of their number. Indels were coded with two states (present/ absent) and indels consisting of minisatellites were coded with two characters (present + no. of repeats or absent + missing data). The final matrix counted 1713 characters, of which 27 were multistate characters.
n-DNA markers
Amplified fragment length polymorphisms (AFLPs) and microsatellites (or simple sequence repeat — SSRs) data were obtained from previous global studies (Lambertini et al. 2006, 2012). The AFLP data set, obtained from the amplification of three primer combinations (E-ACT + M-CTT, E-CAG + M-ATG, E-CGT + M-CAG), produced a matrix of 265 taxa and 105 polymorphic fragments.
The microsatellite data, representing loci PaGT4, PaGT8, PaGT9, PaGT13, PaGT14 and PaGT22 (Saltonstall 2003), were used as binary data and entered in a matrix of 150 taxa and 79 characters.
Waxy bands
The primers used to amplify the grass-waxy gene (F-for: TGCGAGCTCGACAACATCATGCG and M-bac: GGCGAGCGGCGCGATCCCTCGCC; Mason-Gamer et al. 1998) amplify additional products. Two bands of ∼200 and 100 bp have previously been found to be unique to Phragmites in the Mediterranean region (200 bp) and to P. frutescens and P. mauritianus (100 bp), and to have phylogeographic resolution (Lambertini et al. 2012). We therefore screened the presence/absence pattern of these bands in the European sample set. Template DNA (1 µL) was added to 10 µL 2× Mastermix (VWR Amplicon), 10 pmol of forward and reverse primers, and sterile water to reach the final volume of 20 µL. PCR conditions were 94 °C for 3 min, 40 cycles of 94 °C for 30 s, 62 °C for 40 s, 72 °C for 40 s, followed by 72 °C for 7 min. The amplified product was run on a 1.5 % agarose gel for 1 h 30 min at 125 V, 84 mA, and stained with ethidium bromide. Bands of 200 and 100 bp were isolated in a subset of the Mediterranean samples with the EZNA Gel Extraction kit (Q-spin column) (Omega Bio-Tek) and sequenced with forward and reverse primers.
DNA data analysis
Chloroplast DNA sequences, AFLPs and SSRs were analysed separately. The three data sets were subject to parsimony analysis with PAUP ver. 4.0b10 (Swofford 2002). A heuristic search was performed with 37 % jack knife deletion, 1000 replicates, jac resampling and fast-stepwise addition (Farris et al. 1996). Individual pairwise genetic distances were also calculated, choosing the option ‘total character difference’. The matrix of genetic distances was imported in GenAlex ver. 6.4 (Peakall and Smouse 2006) and analysed in a principal coordinate analysis (PCoA). To increase the resolution of the European part of the PCoA, we plotted individual coordinate values in Excel graphs. Most of the Australian genotypes were excluded from the AFLP matrix of genetic distances because the PCoA failed to complete the analysis, as 250 is the maximum number of samples that can be run by the program. However, as the Australian genotypes are closely related among each other, the placement of the main clusters on the PCoA is not affected by the exclusion of part of the genotypes of this group.
A spectrum of genetic distances (Rozenfeld et al. 2007) was constructed with AFLP data to compare genetic diversity structure in North America, Europe and a restricted population in the Po Plain, Italy (Lambertini et al. 2008). Genetic distances (‘total character difference’, calculated with PAUP) within the three groups were plotted against their frequencies within each group. Samples from the native American cp-lineages and from the US Gulf Coast were excluded from the North American population, and samples from the Mediterranean region, Africa and Asia were excluded from the European group. In total, the North American population was represented by 30 samples, the European by 103 and the Po Plain by 46.
Phenology
Denmark is in the temperate area of the Northern Hemisphere. The climate is oceanic with an average temperature of 0 °C (32 °F) in the coldest month (February) and 17 °C (63 °F) in the warmest month (July). Annual average precipitation is ∼700 mm without dry periods. There are up to 18 daily hours of sunlight during the summer.
The Phragmites garden at Aarhus University (56°13′43″N, 10°07′37″E) included 188 different genotypes at the time of the experiment. Each genotype was planted in a separate pot of about 1 m diameter, and the pots were ordered in a square matrix at about 2 m distance from each other. The plot of about 250 × 350 cm2 was outdoors in an open area without trees. The plants were grown in commercial soil mixed with quartz sand (about 50 % of each), watered with phreatic water every second day during the growing season to keep the water level just below the soil level, and fertilized with a commercial NPK fertilizer with microelements once a week. All above-ground biomass was harvested and burnt at the end of the vegetative season to eliminate seed dispersal from the garden.
All clones had been in the garden for at least 5 years before phenology was investigated. Given the weight of the pots, it was not possible to move them within the plot to control margin effect. However, given the large distance between pots, all genotypes had chances to receive pollen both from within the garden and from the wild. We recorded the reproductive condition of each genotype in 2005 and 2006 on a weekly basis from June to September. We were interested in knowing which genotypes of the garden, including European and non-European genotypes, could release pollen and seeds in Europe, and how dispersed or non-dispersed alleles contributed to the pattern of phylogeographic relationships within Europe and at the global scale. For each genotype we registered the presence or absence of: (i) inflorescences, (ii) mature inflorescences (open flowers with exposed stigmas and/or anthers) and (iii) seeds. In November 2006, we recorded whether or not genotypes were releasing seeds. Seed release was defined as the presence of depleted inflorescences (visible bare inflorescence axes) or easily detachable seeds upon touch. Cross- versus self-pollination was not considered relevant to the aim of the present study as alleles would have been present in the gene pool in both cases.
Results
Phylogeographic relationships inferred by cpDNA sequences
We found four previously recognized haplotypes (haplotypes K, L, M and O) and four novel ones (haplotypes AH, AJ, AK and AL) in Romania, Northern Europe and the Mediterranean area (Table 1). The majority of rbcL-psaI sequences were identical to the rbcL-psaI haplotype 4 (Saltonstall 2002; NCBI accession no. AY0116335), whereas the trnT-trnL sequences showed considerable variation. The limited variation in the rbcL-psaI region was restricted to two base substitutions which distinguished haplotype AJ and haplotype AK. Conversely, the trnT-trnL sequences were constant within P. frutescens and assignable to haplotype 5 (Saltonstall 2002; NCBI accession no. AY016328), while rbcL-psaI sequences detected two different haplotypes within the two P. frutescens accessions analysed in this study, haplotype I and haplotype U (Table 1).
Table 1.
European and Mediterranean haplotypes identified by cpDNA. The haplotypes are in alphabetical order. cp-microsatellite variants are indicated with a letter following the trnL-trnT and rbcL-psaI sequences deposited in GenBank by Saltonstall (2002). For example, trnT-trnL sequences 4a, 4c, 4d, etc., differ from sequence 4 in GenBank in the number of repeats at the microsatellite loci. This variation in cp- microsatellites is also indicated in the number following the haplotype letter (e.g. M1, M3, M4, etc.).
| Sample ref. | Species | Country | Locality | trnT-trnL sequence | rbcL-psaI sequence | trnT-trnL+ rbcL-psaI haplotype | (#) NCBI accession no. |
|---|---|---|---|---|---|---|---|
| Pa68AL | P. australis | Algeria | Guebbour, south of Hassi Messaoud | 4d | 4 | M4 | |
| Pa67BE | P. australis | Belgium | Schelde-estuarie, Burcht, Antwerp | 4 | 4 | M | |
| Pa146BE | P. australis | Belgium | Schelde, Konkelschoor, Berlare | 5c | 4 | L1 | |
| Pa671CY | P. australis | Cyprus | Coral Beach, Pafos | 4 | 4 | M | |
| PaH2EG | P. australis | Egypt | Abu Raqman, 37 Km S of Cairo | 4a | 4 | M1 | |
| Pa83EE | P. australis | Estonia | Lake Vortsjarv | 4b | 21b# | AJ | HQ664451 |
| Pa159EE | P. australis | Estonia | Lake Peipsi | 7a | 4 | O2 | |
| Pa54FI | P. australis | Finland | Husöviken, Åland | 7c | 4 | O1 | |
| Pa160FI | P. australis | Finland | Mariehamn, Åland | 5c | 4 | L1 | |
| Pa217FI | P. australis | Finland | Raisionlahti, Turku | 7a | 4 | O2 | |
| Pa70FR | P. australis | France | Campignol, Narbonne | 4c | 4 | M3 | |
| Pa64DE | P. australis | Germany | Chiemsee | 19 | 4 | AD | |
| Pa640DE | P. australis | Germany | Lusatia lignite mining area, Plessa Lake | 4 | 4 | M | |
| Pa670GB | P. australis | Great Britain | Blacktoft Sands | 4 | 4 | M | |
| Pa57GR | P. australis | Greece | Crete, Georgioupolis | 4 | 4 | M | |
| Pa853GR | P. australis | Greece | Crete, Kalives | 4e | 4 | M5 | |
| Pa748GR | P. australis | Greece | Kos, Kos City | 4 | 4 | M | |
| Pf87GR | P. frutescens | Greece | Crete, Dramia | 5i | 3 | I2 | |
| Pf196GR | P. frutescens | Greece | Crete, Georgioupolis | 5i | 9 | U3 | |
| Pa163NL | P. australis | Holland | Verdronken Land van Saeftinghe | 4 | 4 | M | |
| Pa602NL | P. australis | Holland | Slotermeer | 7c | 4 | O1 | |
| Pa77HU | P. australis | Hungary | Lake Ferto | 4 | 4 | M | |
| Pa58IE | P. australis | Ireland | Kilcock | 4 | 4 | M | |
| Pa164IE | P. australis | Ireland | Lake Ree | 4 | 4 | M | |
| Pa91IL | P. australis | Israel | Dead Sea, south-west coast | 4a | 4 | M1 | |
| Pa75IT | P. australis | Italy | Gorgona | 7a | 4 | O2 | |
| Pa770IT | P. australis | Italy | Po Delta, Polesine Camerini | 4 | 4 | M | |
| Pa771IT | P. australis | Italy | Po Delta, Barricata | 4 | 4 | M | |
| Pa755IT | P. australis | Italy | Riccione | 4 | 4 | M | |
| Pa757IT | P. australis | Italy | Sardinia, S. Antioco, Canisoni Beach | 4d | 4 | M4 | |
| Pa762IT | P. australis | Italy | Sardinia, S. Antioco, Cala Lunga | 4 | 4 | M | |
| Pa763IT | P. australis | Italy | Sardinia, S. Antioco, Cala Lunga | 4d | 4 | M4 | |
| Pa10IT | P. australis | Italy | Sardinia, Valledoria | 4 | 4 | M | |
| Pa11IT | P. australis | Italy | Sardinia, Valledoria | 4d | 4 | M4 | |
| Pa12IT | P. australis | Italy | Sardinia, Isola Rossa | 4 | 4 | M | |
| Pa732JO | P. australis | Jordan | Wadi Shuaeb | 4a | 4 | M1 | |
| Pa733JO | P. australis | Jordan | Dead Sea, near Wadi Mujib | 4d | 4 | M4 | |
| Pa734JO | P. australis | Jordan | Azraq Wetlands | 4d | 4 | M4 | |
| Pa735JO | P. australis | Jordan | Wadi Shuaeb/ Fahais near Amman | 4d | 4 | M4 | |
| Pa701LB | P. australis | Libya | Gabron Lake, Erg Ubari dune, Vadi El Agial V | 4d | 4 | M4 | |
| Pa710LB | P. australis | Libya | Mafu Lake, Erg Ubari dune, Vadi El Agial Val | 4a | 4 | M1 | |
| Pa85LI | P. australis | Lithuania | Silute | 4 | 4 | M | |
| Pa81RO | P. australis | Romania | Lake Obretinu-Mare | 4a | 4 | M1 | |
| Pa84RO | P. australis | Romania | Lake Oborny | 4 | 4 | M | |
| Pa624RO | P. australis | Romania | Lake Obretinu-Mare | 4 | 21b | AJ | |
| Pa625RO | P. australis | Romania | Lake Obretinu-Mare | 4 | 4 | M | |
| Pa643RO | P. australis | Romania | Lake Razim | 3 | 21b | AI | |
| Pa647RO | P. australis | Romania | Lake Razim | 4 | 4 | M | |
| Pa650RO | P. australis | Romania | Lake Razim | 4 | 4 | M | |
| Pa651RO | P. australis | Romania | Lake Razim | 4 | 4 | M | |
| Pa652RO | P. australis | Romania | Lake Razim | 4 | 4 | M | |
| Pa654RO | P. australis | Romania | Lake Razim | 4 | 22# | AK | JQ265822 |
| Pa655RO | P. australis | Romania | Lake Razim | 4 | 4 | M | |
| Pa656RO | P. australis | Romania | Lake Razim | 3 | 21b | AI | |
| Pa657RO | P. australis | Romania | Lake Razim | 3 | 21b | AI | |
| Pa658RO | P. australis | Romania | Lake Razim | 4 | 22 | AK | |
| Pa659RO | P. australis | Romania | Lake Razim | 4 | 22 | AK | |
| Pa660RO | P. australis | Romania | Lake Razim | 4 | 22 | AK | |
| Pa661RO | P. australis | Romania | Lake Obretinu-Mare | 5c | 4 | L1 | |
| Pa662RO | P. australis | Romania | Lake Obretinu-Mare | 5c | 4 | L1 | |
| Pa169RU | P. australis | Russia | St Petersbourg | 4 | 4 | M | |
| Pa52ES | P. australis | Spain | El Garxal (Ebro) | 3 | 4 | K | |
| Pa72ES | P. australis | Spain | Gallocanta N | 4 | 4 | M | |
| Pa74ES | P. australis | Spain | L'Encanyissada (Ebro) | 4 | 4 | M | |
| Pa95ES | P. australis | Spain | Gallocanta NW | 4 | 4 | M | |
| Pa300ES | P. australis | Spain | Mallorca, Alcudia | 4c | 4 | M3 | |
| Pa681ES | P. australis | Spain | Laguna de la Mata, Torrevieja | 4 | 4 | M | |
| Pa141SE | P. australis | Sweden | Luleå | 21# | 4 | AH | JQ265821 |
| Pa615SE | P. australis | Sweden | Tåkern | 5f | 4 | L2 | |
| Pa637SE | P. australis | Sweden | Gammelstaden | 21 | 4 | AH | |
| Pa53TN | P. australis | Tunisia | Ras Taguermes, Djerba | 4a | 4 | M1 | |
| Pa97TN | P. australis | Tunisia | Ras Taguermes, Djerba | 22# | 4 | AL | HQ449553 |
| Pa89TK | P. australis | Turkey | Aksehir | 4 | 21a | AJ1 |
Based on the database of sequences accumulated from our previous and ongoing studies, many of the European haplotypes appeared to have intercontinental distribution, ranging from Africa to Europe, Far East Asia and North America. The most striking example is haplotype M, which is present on all four continents. Haplotype M presents several cp-microsatellite variants that are mainly distributed in the Mediterranean region. One such variant, haplotype M1 or Delta-type (Hauber et al. 2011; Lambertini et al. 2012), is also present in Romania and the Mississippi River delta. Haplotype K is present in Spain and in a population extending throughout South Africa, Namibia and Botswana. Haplotype AI, previously described as Greeny3-type (Lambertini et al. 2012), is closely related to haplotype K (differing in one single substitution in the rbcL-psaI region) and is present in Romania and in the US Gulf Coast. Haplotype AD or Greeny2-type (Lambertini et al. 2012), present in Germany, is also shared with the population in the Gulf Coast. Haplotype O presents several cp-microsatellite variants in Far East Asia. Two of these variants occur in Northern Europe and the Mediterranean region. Two cp-microsatellite variants of haplotype L were found in Europe. Haplotype L1 appears widely spread across the continent (Belgium, Finland and Romania), whereas haplotype L2 was found only in Sweden. Among the novel haplotypes identified in this study, haplotype AJ (NCBI accession no. HQ664451) was found in Estonia and Romania, and one of its cp-microsatellite variants was found in Turkey. Haplotype AH (NCBI accession no. JQ265821) was found exclusively in Northern Sweden (>65°N) and haplotype AL (NCBI accession no. HQ449553) only on Djerba island in Tunisia. Haplotype AI (Greeny3-type), previously described in the US Gulf Coast, appeared to be frequent in the Danube delta in Romania.
Phragmites frutescens haplotypes are shared with the species P. mauritianus in tropical Africa (cp-microsatellite variant I2) and with P. karka in tropical Asia (cp-microsatellite variant U3). Both haplotypes I and U are present in Asia, where they show cp-microsatellite variation. Interestingly, only one of the variants, here indicated as I2, is present in Greece (P. frutescens), Uganda and Burkina Faso (P. mauritianus), and in the US Gulf Coast and South America (P. mauritianus× P. australis, described as the Land-type by Lambertini et al. 2012).
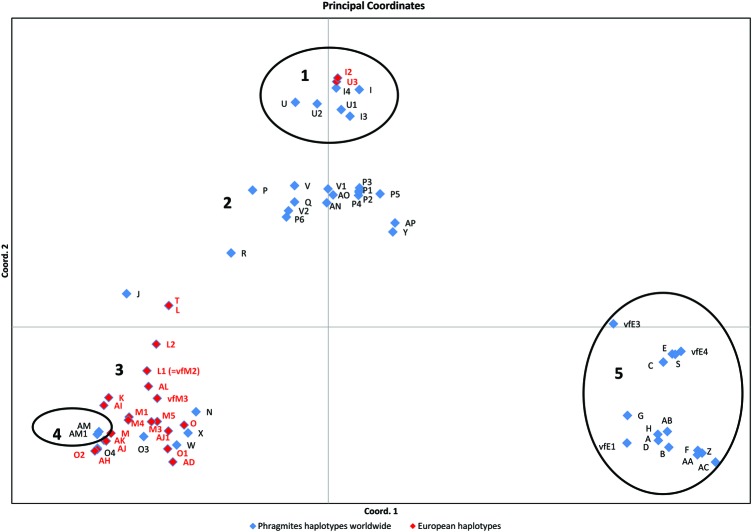
Despite frequent long-distance dispersal and lack of support as a monophyletic clade (Fig. 2), European P. australis haplotypes appear very closely related among each other. The PCoA clearly separates four main clusters of haplotypes within the genus, which largely overlap in geographic distribution. European P. australis and P. frutescens appear in two distantly related clusters and they co-exist in the Mediterranean region.
Fig. 2.
Chloroplast DNA diversity in Phragmites worldwide. Coord. 1 accounts for 45 % and Coord. 2 for 24 % of the variation. Cluster 1 is supported by the parsimony analysis (jack knife = 70) and includes specimens assigned to P. mauritianus, P. frutescens and P. karka, and hybrids P. mauritianaus× P. australis. The geographic distribution of the cluster ranges from tropical Africa, to the Mediterranean, tropical Asia, tropical America and the US Gulf Coast. Cluster 2 includes specimens assigned to P. mauritianus, P. karka and P. australis. The distribution is South Africa, tropical Asia, Far East, Australia and South America. Cluster 3 includes specimens assigned to P. australis and hybrids P. australis× P. mauritianus. The distribution is Europe, Africa, temperate Asia and North America. Cluster 4 is supported by the parsimony analysis (jack knife = 76) and includes specimens assigned to P. japonicus. Distribution: Far East Asia. Cluster 5 is supported by the parsimony analysis (jack knife = 89) and includes specimens assigned to P. australis ssp. americanus. The distribution is limited to North America. The classification of haplotypes vfM2, vfM3, vfE1, vfE3 and vfE4 (Vachon and Freeland 2011; Freeland and Vachon 2012) does not follow the same classification system as Saltonstall (2002) and followed in the present study. According to the present study, vfM2 corresponds to haplotype L1, vfM3 should have a new letter, vfE1 (deposited in GenBank both as E1 and E2) corresponds to a cp-variant of haplotype AB (Meadows and Saltonstall 2007), haplotype vfE3 (Vachon and Freeland 2011; Freeland and Vachon 2012) corresponds to a cp-microsatellite variant of haplotype E (Saltonstall 2002), whereas haplotype E4 (Vachon and Freeland 2011; Freeland and Vachon 2012) should have a new letter. The distribution ranges are defined by combining our collection sites with Fig. 1 in Saltonstall (2002).
Phylogeographic relationships inferred by nuclear markers
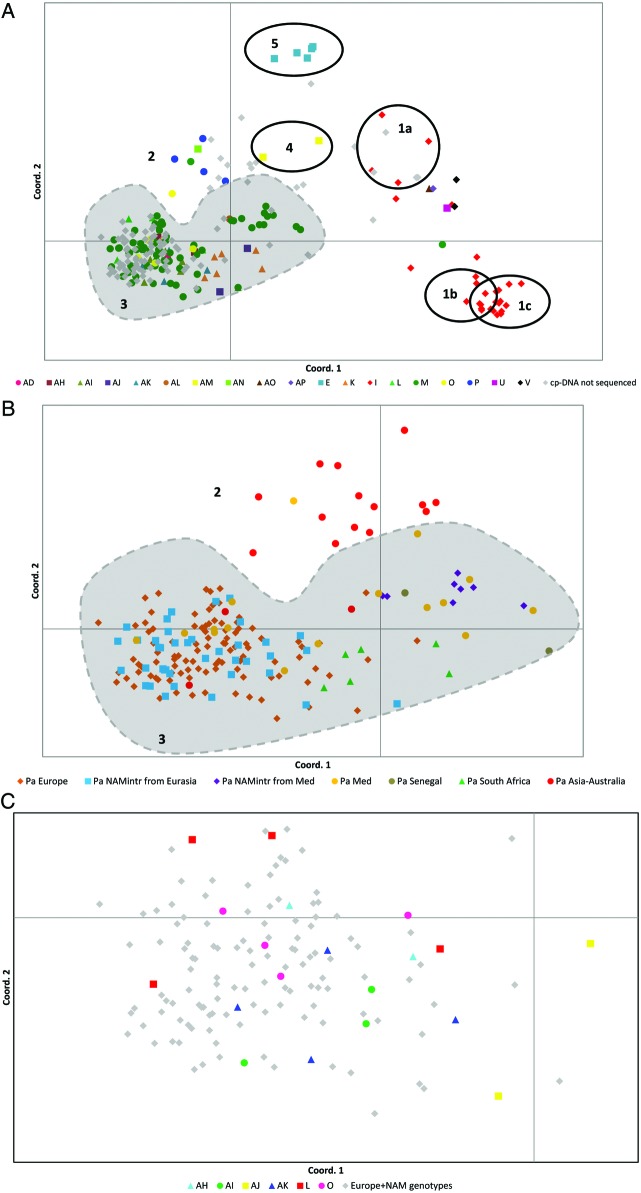
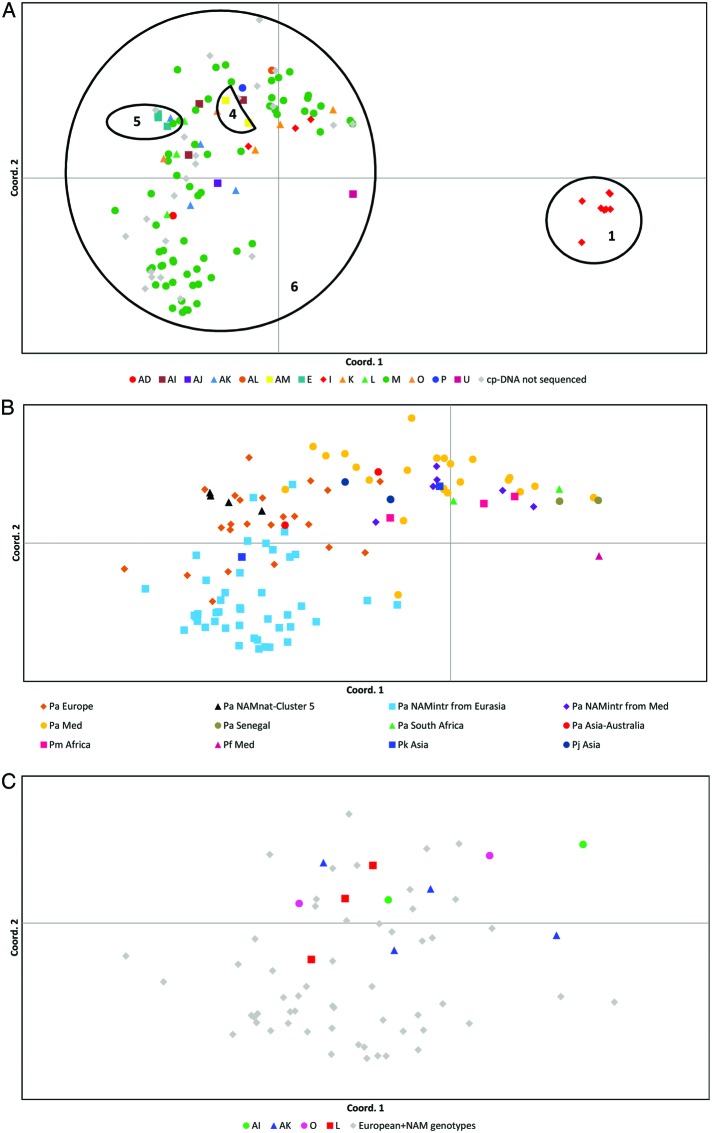
The pattern of interrelationships revealed by nuclear markers mirrors the haplotypic structure detected by cpDNA sequences at the global scale, as the same groups are supported in parsimony analysis (Figs 3A and 4A). However, parsimony analysis of the AFLP data split the cluster containing haplotypes I and U (cluster 1, Fig. 2) into three geographic groups based in Asia (cluster 1a), South America (cluster 1b) and the US Gulf Coast (cluster 1c), and left P. frutescens in the Mediterranean, P. mauritianus in Africa and a part of the P. karka genotypes in Asia without support (Fig. 3A). The parsimony analysis of SSR data (Fig. 4A) did not support the P. karka group (cluster 1a in Fig. 3A) in Asia. The geographic differentiation within this cluster of haplotypes (I and U) and the lack of support in some regions reveal that different pools of nuclear alleles evolved in different parts of the range. On the contrary, the cluster of European P. australis haplotypes (cluster 3 in Figs 2 and 3A) appears intact despite its intercontinental distribution (Europe, North America, Africa and Asia) and its large variation in closely related haplotypes (Fig. 3C). SSR alleles did not separate European P. australis based on haplotypes (Fig. 4A and C).
Fig. 3.
Amplified fragment length polymorphism diversity in Phragmites worldwide. Coord. 1 accounts for 43 % and Coord. 2 for 17 % of the variation. The clusters of genotypes are enumerated according to Fig. 2. (B) and (C) focus on European P. australis. (A) The different colours represent the different haplotypes in Fig. 1 and include their cp-microsatellite variants. Chloroplast DNA cluster 1 (Fig. 2) is split into clusters 1a, 1b and 1c in (A). Cluster 1a is supported by the parsimony analysis (jack knife = 72) and includes genotypes assigned to P. karka distributed in tropical Africa. Cluster 1b is supported by the parsimony analysis (jack knife = 70) and includes hybrid genotypes of P. mauritianus× P. australis in tropical America. Cluster 1c is supported by the parsimony analysis (jack knife = 74) and includes hybrid genotypes P. mauritianus× P. australis in the US Gulf Coast. The genotypes between cluster 1a and 1b/1c are genotypes of P. mauritianus, P. frutescens and P. karka in tropical and South Africa, Mediterranean and tropical Asia belonging to cpDNA cluster 1 (Fig. 2). Cluster 2 includes P. australis genotypes in the Far East. Cluster 3 includes P. australis genotypes in Europe, Africa, North America and Asia and P. australis× P. mauritianus genotypes in tropical Africa. Cluster 4 is supported by the parsimony analysis (jack knife = 64) and includes genotypes of P. japonicus in the Far East. Cluster 5 is supported by the parsimony analysis (jack knife = 98) and includes genotypes of P. australis ssp. americanus in North America. (B) The different colours indicate different continents. North America is represented by two colours in order to distinguish between two different introductions from Eurasia (haplotype M) and from the Mediterranean region (haplotype M1). Panel C focuses on the temperate region of Europe. The different colours indicate different haplotypes.
Fig. 4.
SSR diversity in Phragmites worldwide. Coord. 1 accounts for 43 % and Coord. 2 for 20 % of the variation. The clusters of genotypes are enumerated according to Figs 2 and 3A. (B) and (C) focus on European P. australis. (A) The parsimony analysis splits the genotypes into two groups (jack knife = 98). Cluster 1 includes hybrid genotypes of P. mauritianus× P. australis in tropical America and the US Gulf Coast. A new cluster 6 includes all remaining genotypes and joins AFLP clusters 1b, 2 and 3 (A). Cluster 4 (P. japonicus in the Far East, jack knife = 83) and cluster 5 (P. australis ssp. americanus in North America, jack knife = 80) are supported by the parsimony analysis within cluster 6. (B) focuses on cluster 6. The different colours indicate different continents and different species. (C) focuses on the temperate region of Europe. The different colours indicate different haplotypes.
Differently from the cpDNA sequences, nuclear alleles detected geographic structure within the European group. Both AFLP and SSR PCoAs separated the Mediterranean and African samples from the European gene pool (Figs 3B and 4B). Mediterranean P. australis appeared more closely related to Senegal and South African P. australis samples than to European P. australis. SSRs indicated relationships of Mediterranean P. australis also with African P. mauritianus and Asiatic P. karka samples, no longer supported as a monophyletic group by the parsimony analysis (Fig. 4B). The AFLPs split the Mediterranean group into two groups, one related to Europe and one to Africa (Fig. 3B), whereas the SSRs showed one single Mediterranean group (Fig. 4B).
A nuclear DNA band of about 200 bp amplified by waxy primers appeared exclusive to the Mediterranean genotypes (Fig. 5) and could distinguish P. australis in the Mediterranean region from European P. australis in the temperate region (absent band), as well as from P. frutescens, sympatric in the Mediterranean area. Phragmites frutescens, like P. mauritianus, has a shorter band of about 100 bp (Fig. 5). Samples carrying both bands of 200 and 100 bp had previously been interpreted as hybrids between P. mauritianus and P. australis (Lambertini et al. 2012). The distribution of the double band, previously detected in Senegal, South America and the US Gulf Coast, appeared also to include the Mediterranean area (Fig. 5, lane 9). However, the sequence of the 100-bp band isolated from this sample from Sardinia (Italy) did not match any of the five sequences of P. mauritianus previously isolated (Lambertini et al. 2012).
Fig. 5.
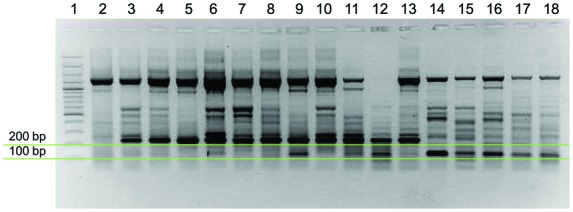
Diversity in grass-waxy sequences in the Mediterranean region and in Africa. The 200-bp band distinguishes Mediterranean (lanes 3–10) from European temperate P. australis (lane 2). The genotype in lane 2 is from Mallorca, Spain, and appears as one undifferentiated (yellow) genotype clustering together with temperate European and North American P. australis in Fig. 3B. It also released seeds in Denmark. The 100-bp band marks P. mauritianus (lanes 15–17) and P. frutescens (lanes 18–19). Samples with 200- and 100-bp bands (lanes 9, 11 and 12) have been interpreted as hybrids between P. australis and P. mauritianus (Lambertini et al. 2012). Senegal (lanes 11–13) appears as a hybrid zone with hybrids and non-hybrids. Sardinia (Italy) (lanes 9–10) also appears as a hybrid zone. Lanes: (1) gene ruler, (2) Pa300ES Spain, (3) Pa68AL Algeria, (4) Pa97TN Tunisia, (5) Pa710LB Libya, (6) Pa732JO Jordan, (7) Pa671CY Cyprus, (8) Pa57GR Greece, (9) Pa759IT Sardinia, Italy, (10) Pa764IT Sardinia, Italy, (11) PaH5SN Senegal, (12) PaH6SN Senegal, (13) Pa102SN Senegal, (14) Pm106ZA South Africa, (15) Pm73UG Uganda, (16) Pm93UG Uganda, (17) Pf87GR Greece and (18) Pf196GR Greece.
In addition to the Mediterranean group, the PCoA of AFLPs (Fig. 3B) separated the Asiatic genotypes from the European ones. The red group of Asiatic samples (Fig. 3B) appears differentiated from Europe both by cpDNA sequences (cluster 2, Fig. 2) and nuclear AFLPs (cluster 2, Fig. 3A and B). A few Asiatic samples, however, had European AFLP alleles (Fig. 3B). The SSRs also differentiated a group of North American genotypes within the European group (Fig. 4B). This group in North America includes genotypes from the Atlantic Coast. As this group is not recognizable on the PCoA of AFLP data, we compared the spectrum of AFLP genetic distances between the whole North American introduced population and the European population. Pairwise genetic distances among North American genotypes appeared shorter (mode = 14 AFLP fragments) than those among European samples (mode = 19 AFLP fragments). This also held when the North American's spectrum was compared with that of a geographically restricted population within the temperate region of Europe, like the Po Plain in Northern Italy (mode = 16 AFLP fragments).
Phenology
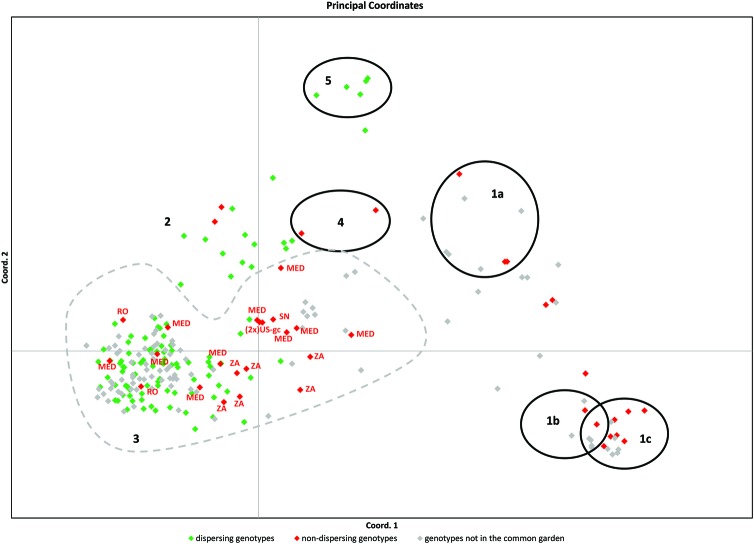
Of the 188 genotypes present in the experimental area, only 83 reproduced in 2005 and 2006 and released seeds in 2006 [see Additional Information—Table 1]. Phylogeographically, the genotypes that released seeds and/or pollen in Denmark belonged to cluster 3 (genotypes from Europe, Asia and the Euroasiatic introduced genotypes to North America), cluster 2 (genotypes from Far East Asia, mostly from Sakhalin Island in Russia, and one single genotype from Australia) and from cluster 5 (genotypes of native North American P. australis ssp. americanus). Genotypes of P. mauritianus (including its hybrids in the US Gulf Coast), P. frutescens, P. karka (cluster 1), P. japonicus Steud (cluster 4) did not set inflorescences in Denmark. Within the European cluster (cluster 3), P. australis genotypes from South Africa, Senegal, the Mediterranean region and American genotypes introduced from the Mediterranean region also did not flower. Two genotypes from Romania and three genotypes from the Mediterranean region more closely related to the temperate group of European P. australis also did not set inflorescences in Denmark (Fig. 6).
Fig. 6.
Amplified fragment length polymorphism relationships among dispersing and non-dispersing genotypes in the common garden at Aarhus University. Dispersing genotypes are the genotypes that released seeds and/or pollen in 2005 and/or 2006. The red dots in the European gene pool belong to genotypes from South Africa, Senegal, the Mediterranean region, North America (introduced from the Mediterranean region) and Romania.
Compared with 2005, the pattern of reproduction observed differed in November 2006 [see Additional Information—Table 1]. Genotypes from Northern Europe, South Africa and Australia that did not produce inflorescences in 2005, flowered in 2006, and the Northern European and Australian genotypes released seeds in 2006. On the contrary, genotypes that produced seeds in 2005 produced only inflorescences in November 2006 (genotypes from Northern Europe, Romania, Central Russia and Sakhalin Island, and one native North American genotype from Utah).The duration of the flowering period ranged from the end of August to mid-September for clones from Sakhalin Island, to flowering at the beginning of August and lasting until the end of September for most European genotypes. The longer flowering period of the European plants was the result of continuous inflorescence production and maturation among the ramets. In general, there was a shift in flowering times from north to south for the genotypes of the Northern Hemisphere. The few genotypes of the Southern Hemisphere that set seeds in Denmark started flowering at the end of August, and the first spikelets opened in early September (Australia). Many late-flowering genotypes did not produce seeds in Denmark but they flowered and released pollen in 2005 and 2006.
Discussion
Chloroplast DNA sequences have identified four main clusters of haplotypes with continental or intercontinental distribution. This structure is also identifiable by nuclear markers. Haplotypes from two of these clusters are present in Europe and are sympatric in the Mediterranean region. In Europe, the two groups are recognized taxonomically, one being P. australis and the other P. frutescens. In total, 12 P. australis haplotypes (K, L, M, O, T, AD, AH, AI, AJ, AK, AL and vfM3) and two P. frutescens haplotypes (I and U) have been reported to date in Europe, together with several cp-microsatellite variants. Haplotypes L, M, O and T had previously been documented in Europe by Saltonstall (2002), and haplotypes vfM2 and vfM3 by Vachon and Freeland (2011). The two haplotypes indicated in the present study as vfM2 and vfM3, found by Vachon and Freeland (2011) in Great Britain and described in GenBank as M2 and M3 do not follow the alphabetical system introduced by Saltonstall (2002). According to the present study, vfM2 corresponds to haplotype L1, whereas vfM3 should have a new letter. Differently from Saltonstall (2002), Vachon and Freeland (2011) and Saltonstall and Lambertini (2012), we also considered cp-microsatellite variation in the analyses, because cp-microsatellite variation appeared restricted within limited regions in the range of some haplotypes, and may thus be phylogeographically informative. cp-microsatellite variants of haplotype M are frequent in the Mediterranean region, but only one such variant, haplotype M, is spread across temperate Europe, Asia and North America. Another variant M1 (Delta-type) has recently been found in the US Gulf Coast (Hauber et al. 2011; Lambertini et al. 2012) and nuclear markers have confirmed its origin in the Mediterranean region (Lambertini et al. 2012). The presence of the same variant M1 also in Romania, as shown in the present study, is not surprising as the population in the US Gulf Coast shares several genetic similarities with the population in the Danube delta (Lambertini et al. 2012). We would expect a random pattern of distribution and relationships if cp-microsatellite variation in haploytpe M were merely due to homoplasy. This, however, cannot be generalized to all haplotypes. The inclusion of cp-microsatellite variants in the analysis and the different codification system that we used for repeated sequences can explain differences with the global haplotypic structure previously presented by Saltonstall (2002), Vachon and Freeland (2011), Chu et al. (2011), and Saltonstall and Lambertini (2012). This is the first study that compares cpDNA and nuclear variation at the global scale, and the fact that the major groups can be identified by both DNA sources indicates reliability of the molecular markers used and strongly supports the phylogeographic model presented in this study.
European P. australis (in sensu lato) appears as a group that extends across Europe, Africa, Asia and North America. Africa is differentiated from Europe. The Mediterranean region appears more similar to African Phragmites in nuclear DNA rather than to temperate European Phragmites. Differentiation is also ongoing in North America. The gap in our sampling from East Europe to the Far East does not allow us to set the phylogeographic limit of the European gene pool in Asia. However, Far East Phragmites appears different from European Phragmites both in haplotypic composition and nuclear markers. A recent study of Phragmites cpDNA variation in China (An et al. 2012) identified haplotypes M, O and P together with several of their variants, and reported that haplotype M is distributed in West China, whereas haplotype O is widely distributed across all the country and haplotype P is restricted to East China. The study of Chu et al. (2011) also identified several P-related haplotypes in South Korea and some new P. japonicus haplotypes related to haplotypes X and W. In our data, haplotype P is represented in a separate cluster of haplotypes, which also has distinctive nuclear alleles (cluster 2, Figs 2, 3A and B). Haplotypes AM and AM1 (Fig. 2) belong to our P. japonicus samples and appear as a monophyletic group evolved within European P. australis.
More difficult to understand is the extension of the group to which P. frutescens belongs. Phragmites frutescens shares the cpDNA sequences with both P. mauritianus, taxonomically recognized as a distinctive species with distribution in tropical Africa, and with P. karka, whose distribution ranges from West Africa to tropical Asia (Clevering and Lissner 1999). This finding may have revealed an important role of P. frutescens in the evolutionary split between the two closely related tropical species. However, many more genotypes within the cluster of haplotypes I and U (cluster 1) need to be included in the analyses to understand the structure of this group.
Within the temperate European region (European P. australis in sensu strictu), there is no clear pattern of phylogeographic relationships. Several closely related haplotypes co-exist in the area, but haplotypic structure is not identifiable by nuclear markers. This suggests long-distance dispersal of genotypes and extensive gene flow within the temperate region. The phenological results appear in agreement with the European phylogeographic pattern. The relationships among genotypes releasing seeds and/or pollen and non-flowering genotypes in Denmark, shown in the PCoA of Fig. 6 as well as our previous studies (Lambertini et al. 2006, 2008), indicate reticulate relationships among the P. australis genotypes that released pollen and seeds, and differentiation of P. australis groups that either did not flower (Mediterranean clones) or are geographically distant and apparently isolated from Europe (American, Australian and South African clones). The common garden study in Europe shows that there are limits to sexual reproduction and seed dispersal across Europe, and these limits appear to select the European genetic variation. Genotypes from the Southern Hemisphere can establish in Denmark, but only a few can occasionally conclude their reproductive cycle in Denmark. The genotypes from Australia started to flower late in Denmark (September), but some of them released pollen and a few set seeds. Also one South African genotype started to flower in September but did not conclude its cycle. The fact that these geographically distant genotypes can occasionally reproduce in Denmark might explain some of the long-distance relationships seen in Europe, for example the phylogenetic relationship of haplotype K among South African, Spanish, Romanian and American genotypes. The Romanian and American genotypes have been assigned to haplotype AI (or Greeny3-type; Lambertini et al. 2012), but they share the trnT-trnL sequence which defines haplotype K. The European K-related genotypes have different nuclear alleles from the South African population, but not from the European gene pool. Another observation that suggests gene flow in Europe is given by the cytotypic variation within this cp-lineage. The South African individuals are octoploids whereas the Romanian genotypes are part of a mixed cytotype population in the Danube delta which includes octoploids (Pauca-Comanescu et al. 1999; Clevering et al. 2001). The same Romanian individuals analysed by flow cytometry by Clevering and Lissner (1999) were sequenced in the present study and cytotypic variation appeared distributed within every cp-lineage present in the region. Cytological incompatibilities might explain why the two Romanian genotypes do not reproduce in Denmark. Another possible explanation is that haplotype K was originally dispersed from Europe to South Africa where it eventually differentiated from the European pool. We cannot reconstruct the history of haplotype K based on this data set, but studies at the population level and experimental crosses can clarify gene flow possibilities and limitations. Given the north–south gradient in flowering times across Europe, reported also in other common garden studies (Clevering et al. 2001; Bastlová et al. 2004), non-flowering genotypes and strong asynchronies in flowering times within a geographically restricted population might indicate long-distance dispersal events.
With the exception of one genotype from Gorgona Island (Italy), which shared haplotype O with the North European genotypes and is likely to have been introduced from the mainland (F. Masi, pers. commun.), and one single genotype of M3 from Mallorca (Spain), none of the Mediterranean samples of either P. australis or P. frutescens flowered in Denmark, although inflorescences were observed on ramets by the authors in at least one P. australis population in Sardinia (Italy). This suggests either climatic barriers to dispersal of predominantly tropical and subtropical genotypes into the temperate regions of Europe, or breeding barriers that keep the Mediterranean genotypes isolated from temperate P. australis, as well as from P. frutescens. The nuclear waxy bands captured the diversity of the Mediterranean group (identified by the 200-bp waxy band), as well as its distinctiveness from European P. australis (lacking the 200-bp waxy band) and from P. frutescens (with the 100-bp waxy band). The 200-bp band presence/absence pattern appeared independent from cp-lineages, as genotypes of haplotype M with and without bands have been found in the Mediterranean region. The homologies and relationships based on the DNA sequences contained in the waxy bands have proven phylogenetically informative between the African and American genotypes (Lambertini et al. 2012) and need to be explored in the Mediterranean area. Several additional bands were amplified by grass-waxy primers in the Mediterranean samples, and these could provide further insights into the relationships of Mediterranean genotypes, as well as the DNA source of such bands. The occurrence of individuals with 200- and 100-bp bands, like the sample in lane no. 9 from Sardinia, and the hybrids from Senegal (Fig. 5), suggests interbreeding of Mediterranean P. australis with the tropical species P. mauritianus and/or with P. frutescens. SSR alleles detected similarities with both P. mauritianus and P. karka, and P. frutescens shares the cpDNA sequences with both P. mauritianus and P. karka. We could not find matching waxy sequences to conclusively demonstrate hybridization, but we also have a very small number of samples of these tropical species which span in distribution from South Africa, to Uganda, Burkina Faso, Thailand and Australia, and only two single individuals of P. frutescens from Crete in Greece. More variation is likely to be present in the Mediterranean region. We recently found a herbarium accession of P. frutescens in the herbarium at Bologna University (Italy). The specimen was collected in Misterbianco, close to Catania, in Sicily (Italy) in 1926 (accession no. BOLO0002314) and has seeds. A focused study in the Mediterranean region is necessary to fully understand the relationships of Mediterranean P. australis. The geographic distribution of the 200-bp band matches the distribution of P. australis var. isiaca (Hultén and Fries 1986). The plants growing in our living collection, showing the 200-bp band, also match the attribute ‘altissimus’ (very tall) given by Clayton (1967) to the Mediterranean subspecies. Both epithets appear therefore in agreement with the genetic identity of Mediterranean P. australis. However, this needs to be confirmed with phenotypes of var. isiaca and ssp. altissimus. The two P. frutescens plants in our collection are morphologically strikingly different from Mediterranean P. australis, as well as from any Phragmites species.
The introduced population of European Phragmites in North America also appears to be differentiated from Europe. The short genetic distances among the American genotypes, compared with European genotypes and with the genotypes of a restricted region in Europe, suggest founder effect and/or genetic drift. The individuals separated by SSR markers are located along the Atlantic Coast of the USA. This cluster of individuals, also detected by Lambertini et al. (2006), is located in the same region that was the gate of the Euroasiatic invasion (Saltonstall 2002).
Conclusions and forward look
This study reveals European P. australis as one distinct group out of four main cpDNA clusters known today in the world. The large scale of the study opens several windows onto the genetic dynamics within the European group and the relationships within the genus that stimulate further research.
The Mediterranean region appears an interesting spot of Phragmites diversity for evolutionary biologists, geneticists, taxonomists, ecologists and ecophysiologists. The Danube delta is another important region for its cytologic and haplotypic diversity, and could finally explain how chromosome numbers affect gene flow and diversification processes. Furthermore, given the genetic similarities of the Danube delta population to the population in the Mississippi delta (Lambertini et al. 2012), the resolution of the Romanian history will also provide further insights into the evolutionary processes taking place in the US Gulf Coast. The rest of Europe is no less interesting in the context of haplotypic diversity, morphology and long-distance relationships, and this should be taken into account in studies at the population scale. Phragmites australis has been farmed in Europe for thousands of years for roof thatching and more recently it is being extensively used in constructed wetlands. It would be particularly interesting to understand how human-assisted transport of genotypes has influenced the current pattern of phylogeographic relationships.
At the global scale, there is a large amount of diversity that is missing from this phylogeographic model. Phragmites diversity in Africa, Asia and South America is under-represented in our study and in the literature. The literature about China and the Far East is increasing, but there is still a big geographic gap to be covered with samples, which will complete the genetic picture and provide more outgroups (and hopefully statistical support) to understand the relationships among the main Phragmites phylogeographic groups.
Additional information
The following additional information is available in the on line version of this article:
Table 1. Phenological registrations of flowering times, seed setting and seed release in the common garden at Aarhus University, Denmark. Flowering times in 2005 and seed release in 2006 are reported. Every individual corresponds to a different genotype. Genotypes are divided in the main groups identified by the study and are listed from north to south within each group.
Accession numbers
Relevant NCBI entries:
For trnT-trnL sequences:
JQ265821: haplotype 21, sample Pa141SE, Luleå, Sweden
HQ449553: haplotype 22, sample Pa97TN, Djerba Tunisia
For rbcL-psaI sequences:
HQ664451: haplotype 21, sample Pa83EE, Lake Vortsjarv, Estonia
JQ265822: haplotype 22, sample Pa654RO, Lake Razim, Romania
Sources of funding
This study was supported by The Danish Council for Independent Research, Natural Sciences, via a grant to H.B.
Contributions by the authors
C.L. analysed the data and drafted the manuscript. H.B. participated in the design of the study and provided the funding. All authors commented on and approved the manuscript.
Conflict of interest statement
None declared.
Acknowledgements
We thank Kristin Saltonstall and an anonymous reviewer for their personal engagement in the review of the manuscript and for the constructive inputs. K. Saltonstall is also thanked for classifying the new haplotypes. Professor P.V. Arrigoni at the Department of Plant Biology, Florence University (Italy), is thanked for indicating the P. australis var. isiaca population in S. Antioco Island, Sardinia, Italy. Dennis Whigham is thanked for organizing this special issue of AoB PLANTS dedicated to P. australis in Europe and North America. Laura A. Meyerson and Melissa McCormick are thanked for inviting us as their ‘European partners’ in the genetic session. Karin M. Kettenring is thanked for commenting on an early draft of the manuscript. We also wish to thank Bologna Herbarium (BOLO) at the University of Bologna for providing samples for future studies. Special thanks to all colleagues and friends that sent Phragmites leaves, seeds or rhizomes from different parts of the world and contributed to the creation of this sample set.
References
- An J-X, Wang Q, Yang J, Liu J-Q. Phylogeographic analyses of Phragmites australis in China: native distribution and habitat preference of the haplotype that invaded North America. Journal of Systematics and Evolution. 2012;50:334–340. [Google Scholar]
- Bastlová D, Cižková H, Bastl M, Kvet J. Growth of Lythrum salicaria and Phragmites australis plants originating from a wide geographical area: response to nutrient and water supply. Global Ecology and Biogeography. 2004;13:259–271. [Google Scholar]
- Brix H. Genetic diversity, ecophysiology and growth dynamics of reed (Phragmites australis) Aquatic Botany. 1999;64:179–184. [Google Scholar]
- Chu H, Cho WK, Jo Y. Identification of natural hybrids in Korean Phragmites using haplotype and genotype analyses. Plant Systematics and Evolution. 2011;293:247–253. [Google Scholar]
- Clayton WD. Studies in Gramineae: XIV. Arundineae (Phragmites Adans.) Kew Bulletin. 1967;21:113–117. [Google Scholar]
- Clayton WD. The correct name of the common reed. Taxon. 1968;17:168–169. [Google Scholar]
- Clevering OA, Lissner J. Taxonomy, chromosome numbers, clonal diversity and population dynamics of Phragmites australis. Aquatic Botany. 1999;64:185–208. [Google Scholar]
- Clevering OA, Brix H, Lukavská J. Geographic variation in growth responses in Phragmites australis. Aquatic Botany. 2001;69:89–108. [Google Scholar]
- Conert HJ. Die Systematik und Anatomie der Arundineae. Frankfurt am Main: Cramer; 1961. [Google Scholar]
- Doyle JJ. Gene trees and species trees: molecular systematics as one-character taxonomy. Systematic Botany. 1992;17:144–163. [Google Scholar]
- Farris JS, Albert VA, Källersjö M, Lipscomb D, Kluge AG. Parsimony jackknifing outperforms neighbor-joining. Cladistics. 1996;12:99–124. doi: 10.1111/j.1096-0031.1996.tb00196.x. [DOI] [PubMed] [Google Scholar]
- Fér T, Hroudová Z. Genetic diversity and dispersal of Phragmites australis in a small river system. Aquatic Botany. 2009;90:165–171. [Google Scholar]
- Freeland J, Vachon N. Repetitive sequences in phylogeographic inference: a reply to Saltonstall and Lambertini (2012) Molecular Ecology Resources. 2012;12:586–589. doi: 10.1111/j.1755-0998.2012.03146.x. [DOI] [PubMed] [Google Scholar]
- Geng Y, Pan X, Xu C, Zhang W, Li B, Chen J, Lu B, Song Z. Phenotypic plasticity rather than locally adapted ecotypes allows the invasive alligator weed to colonize a wide range of habitats. Biological Invasions. 2007;9:245–256. [Google Scholar]
- Greuter W, Scholz H. Phragmites in Crete. Cenchrus frutescens and the nomenclature of the common reed (Gramineae) Taxon. 1996;45:521–523. [Google Scholar]
- Hall TA. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hauber DP, Saltonstall K, White DA, Hood CS. Genetic variation in the common reed. Phragmites australis Estuaries and Coasts. 2011;34:851–862. in the Mississippi River delta marshes: evidence for multiple introductions. [Google Scholar]
- Hultén E, Fries M. Atlas of North European vascular plants. North of the Tropic of Cancer. Koenigstein, Germany: Koeltz Scientific Books; 1986. [Google Scholar]
- Kettenring KM, McCormick MK, Baron HM, Whigham DF. Mechanisms of Phragmites australis invasion: feedbacks among genetic diversity, nutrients, and sexual reproduction. Journal of Applied Ecology. 2011;48:1305–1313. [Google Scholar]
- Krzakowa M, Michalak M. Genetic differentiation of common reed (Phragmites australis) populations from selected lakes of Pomerania (NW Poland), revealed by elecrophoretically detected peroxidase variability. Biodiversity: Research and Conservation. 2010;17:19–22. [Google Scholar]
- Lambertini C, Gustafsson MHG, Frydenberg J, Lissner J, Speranza M, Brix H. A phylogeographic study of the cosmopolitan genus Phragmites (Poaceae) based on AFLPs. Plant Systematics and Evolution. 2006;258:161–182. [Google Scholar]
- Lambertini C, Gustafsson MHG, Frydenberg J, Speranza M, Brix H. Genetic diversity patterns in Phragmites australis at the population, regional and continental scales. Aquatic Botany. 2008;88:160–170. [Google Scholar]
- Lambertini C, Riis T, Olesen B, Clayton JS, Sorrell BK, Brix H. Genetic diversity in three invasive clonal aquatic species in New Zealand. BMC Genetics. 2010;11:1–18. doi: 10.1186/1471-2156-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertini C, Mendelsshon IA, Gustafsson MGH, Olesen B, Riis T, Sorrell BK, Brix H. Tracing the origin of Gulf Coast Phragmites (Poaceae)—a story of long distance dispersal and hybridization. American Journal of Botany. 2012;99:538–551. doi: 10.3732/ajb.1100396. [DOI] [PubMed] [Google Scholar]
- Lukacs V. Hungary: 2009. Genetic diversity of the reed (Phragmites australis) studied by PCR-RAPD method. PhD Thesis. http://phd.lib.uni-corvinus.hu/408/2/lukacsviktoria_ten.pdf (6 January 2012) [Google Scholar]
- Mason-Gamer RJ, Weil CF, Kellogg EA. Granule-bound starch synthase: structure, function and phylogenetic utility. Molecular Biology and Evolution. 1998;15:1658–1673. doi: 10.1093/oxfordjournals.molbev.a025893. [DOI] [PubMed] [Google Scholar]
- McCormick MK, Kettenring KM, Baron HM, Whigham DF. Spread of invasive Phragmites australis in estuaries with differing degrees of development: genetic patterns, Allee effects and interpretation. Journal of Ecology. 2010;98:1369–1378. [Google Scholar]
- Meadows RE, Saltonstall K. Distribution of native and introduced Phragmites australis in freshwater and oligohaline tidal marshes of the Delmarva Peninsula and southern New Jersey. Journal of the Torrey Botanical Society. 2007;134:99–107. [Google Scholar]
- Mutlu B. Phragmites frutescens H. Scholz (Gramineae), a new record for the flora of Turkey. Hacettepe Journal of Biology and Chemistry. 2002;31:23–26. [Google Scholar]
- Pauca-Comanescu M, Clevering OA, Hanganu J, Gridin M. Phenotypic differences among ploidy levels of Phragmites australis growing in Romania. Aquatic Botany. 1999;64:223–234. [Google Scholar]
- Paul J, Kirk H, Freeland J. Genetic diversity and differentiation of fragmented reedbeds (Phragmites australis) in the United Kingdom. Hydrobiologia. 2011;665:107–115. [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolletschek H, Rolletschek A, Kuehl H, Kohl JG. Clone specific differences in a Phragmites australis stand. II. Seasonal development of morphological and physiological characteristics at the natural site and after transplantation. Aquatic Botany. 1999;64:247–260. [Google Scholar]
- Rozenfeld AF, Arnaud-Haond S, Hernandez-Garcia E, Eguiluz VM, Matias MA, Serrao E, Duarte CM. Spectrum of genetic diversity and networks of clonal organisms. Journal of the Royal Society Interface. 2007;4:1093–1102. doi: 10.1098/rsif.2007.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltonstall K. A set of primers for the amplification of noncoding regions of chloroplast DNA in the grasses. Molecular Ecology Notes. 2001;1:76–78. [Google Scholar]
- Saltonstall K. Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proceedings of the National Academy of Sciences of the USA. 2002;99:2445–2449. doi: 10.1073/pnas.032477999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltonstall K. Microsatellite variation within and among North American lineages of Phragmites australis. Molecular Ecology. 2003;12:1689–1702. doi: 10.1046/j.1365-294x.2003.01849.x. [DOI] [PubMed] [Google Scholar]
- Saltonstall K. Remnant native Phragmites australis maintains genetic diversity despite multiple threats. Conservation Genetics. 2011;12:1027–1033. [Google Scholar]
- Saltonstall K, Lambertini C. The value of repetitive sequences in chloroplast DNA for phylogeographic inference: a comment on Vachon & Freeland 2011. Molecular Ecology Resources. 2012;12:581–585. doi: 10.1111/j.1755-0998.2012.03146.x. [DOI] [PubMed] [Google Scholar]
- Scholz H, Böhling N. Phragmites frutescens (Gramineae) re-visited. The discovery of an overlooked, woody grass in Greece, especially Crete. Willdenowia. 2000;30:251–261. [Google Scholar]
- Swofford DL. PAUP. Phylogenetic analysis using parsimony (and other methods) Sunderland, MA: Sinauer Associates; 2002. Version 4. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Tutin TG, Heywood VH, Burges NA, Moore DM, Valentine DH, Walters SM, Webb DA. Flora Europeae. Cambridge University Press; 1980. [Google Scholar]
- Vachon N, Freeland JR. Phylogeographic inferences from chloroplast DNA: quantifying the effects of mutations in repetitive and non-repetitive sequences. Molecular Ecology Resources. 2011;11:279–285. doi: 10.1111/j.1755-0998.2010.02921.x. [DOI] [PubMed] [Google Scholar]