Abstract
Upon apoptotic stimuli, lysosomal proteases, including cathepsins and chymotrypsin, are released into cytosol due to lysosomal membrane permeabilization (LMP), where they trigger apoptosis via the lysosomal-mitochondrial pathway of apoptosis. Herein, the mechanism of LMP was investigated. We found that caspase 8-cleaved Bid (tBid) could result in LMP directly. Although Bax or Bak might modestly enhance tBid-triggered LMP, they are not necessary for LMP. To study this further, large unilamellar vesicles (LUVs), model membranes mimicking the lipid constitution of lysosomes, were used to reconstitute the membrane permeabilization process in vitro. We found that phosphatidic acid (PA), one of the major acidic phospholipids found in lysosome membrane, is essential for tBid-induced LMP. PA facilitates the insertion of tBid deeply into lipid bilayers, where it undergoes homo-oligomerization and triggers the formation of highly curved nonbilayer lipid phases. These events induce LMP via pore formation mechanisms because encapsulated fluorescein-conjugated dextran (FD)-20 was released more significantly than FD-70 or FD-250 from LUVs due to its smaller molecular size. On the basis of these data, we proposed tBid-PA interactions in the lysosomal membranes form lipidic pores and result in LMP. We further noted that chymotrypsin-cleaved Bid is more potent than tBid at binding to PA, inserting into the lipid bilayer, and promoting LMP. This amplification mechanism likely contributes to the culmination of apoptotic signaling.
Keywords: Bid, chymotrypsin, lysosome, enzymology
Physiological and pathological cell death have been classified according to morphological criteria into at least three categories: type I cell death or apoptosis; type II cell death or autophagic cell death; and type III cell death or necrosis (1, 2). Lysosomes were known to be involved in autophagy (3, 4) and, after the massive rupture of lysosomes, in necrosis (5). In response to lethal stimuli, partial and selective lysosomal membrane permeabilization (LMP) occurs in certain lysosomes (6), and the redistributed lysosomal proteases induce apoptosis (7). Several lysosomal cathepsins (e.g., cathepsin B, D, L, etc.) have been implicated in apoptosis (8, 9). Recently, we found that, besides being a digestive enzyme, chymotrypsin is a novel member of lysosomal proteases (10). Chymotrypsin release as a consequence of LMP in a small proportion of lysosomes appears to play an important role in cell apoptosis, irrespective of whether the apoptosis is triggered by extrinsic or intrinsic mediators (11). Further investigation indicated that, during the initiation of apoptosis, both the activation of caspase 8 and the existence of Bid are necessary for the induction of LMP, but the precise mechanism of LMP remains to be elucidated.
The activation of caspase 8 leads to the proteolysis of Bid and the formation of caspase 8-cleaved Bid (tBid), which induces mitochondrial outer membrane permeabilization (MOMP) and results in the amplification of apoptotic signaling (12, 13). The mechanism of MOMP has been studied extensively (14). Mitochondrial membranes are enriched with cardiolipin, an acidic phospholipid that provides specificity for the targeting of tBid to mitochondria, leading to MOMP and ultimately to apoptosis (15). Whether there are similar mechanisms underlying tBid-dependent LMP during apoptosis remains unclear.
To reveal the precise mechanisms of tBid-dependent LMP, we began the present study by analyzing the phospholipid composition of lysosomal membrane. Besides sphingolipids and cholesterol, these membranes are rich in acidic phospholipids, especially phosphatidic acid (PA), phosphatidylserine (PS), and phosphatidylinositol (PI), but without cardiolipin. We demonstrated that tBid alone is sufficient to induce LMP in intact cells or in isolated lysosomes. Bax might cause an enhancement of tBid-induced LMP, but neither Bax nor Bak is crucial for this LMP. By using a cell-free model that involved large unilamellar vesicles (LUVs) mimicking the phospholipid composition of lysosomes, we found that tBid interacts with PA, which facilitates its insertion into model membrane and causes its conformation change. Size-exclusion chromatography and cross-linking assay indicated that tBid forms homo-oligomers in PA-containing membranes. 31P-NMR analysis showed that tBid promotes the formation of highly curved nonbilayer lipid phases via its interaction with PA. On the basis of these data and our previous reports (16, 17), we proposed that tBid-PA interactions in the lysosomal membranes formed “lipidic pores,” which lead to the permeabilization of lysosomal or lysosomal-like membranes. As a result of LMP, lysosomal proteases are redistributed into the cytosol. We also found that chymBid, the proteolytic product of Bid by chymotrypsin released from lysosomes, exhibits more potent activity than that of tBid in binding with PA, inserting into the lipid bilayer, and promoting efficient leakage from lysosomal membranes. These events may play an important signal amplification role during the culmination of the mitochondrial-lysosomal apoptotic pathway.
MATERIALS AND METHODS
Chemicals and antibodies
1,2-Dioleoyl-sn-glycero-3-phosphate (PA), 1,2-dioleoyl-snglycero-3-[phospho-rac-(1-glycerol)] (PG), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (PE), 1,2-dioleoyl-sn-glycero-3-phosphocholine (PC), 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (PS), and 1,2-dioleoyl-sn-glycero-3-phospho-(1’-myo-inositol) (PI) were purchased from Avanti Polar Lipids (Alabaster, AL). N-(1-pyrenyl)maleimide (PM) and acridine orange (AO) were from Invitrogen (Eugene, OR). Fluorescein-conjugated dextran (FD)-20, FD-70, or FD-250 and Thesit were from Sigma-Aldrich (St. Louis, MO). Bis(2-(3-sulfo-N-succinimidyloxycarbonyloxy)ethyl)sulfone (Sulfo-BSOCOES) was purchased from Pierce (Rockford, IL). BioPORTER protein delivery reagent was purchased from Genlantis (San Diego, CA). 3-[(3-cholamidopropyl)-dimethyl-ammonio]1-propane sulfonate (CHAPS) was purchased from J. T. Baker Inc. (Princeton, NJ). Antibodies against Bid, cathepsin B, COX IV, and GAPDH were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibody against cytochrome c was purchased from BD Bioscience (Sparks, MD). Antibody against LAMP-1 was purchased from Cell Signaling Technology (Danvers, MA). Other reagents were manufactured in China and were of analytical grade.
Expression and labeling of recombinant proteins
Recombinant full-length murine Bid with an N-terminal His6 tag and recombinant human activated caspase 8 were obtained as we described previously (16). Recombinant human chymotrypsin was obtained and activated as we described previously (11).Truncated Bid processed by caspase 8 (tBid; includes N-terminal fragment 1–59 and C-terminal fragment 60–195) was obtained by adding caspase 8 to the full-length Bid and then incubating overnight at 4°C (10). chymBid, which includes C-terminal fragment 68–195 only (the N-terminal fragment is degraded during chymotrypsin treatment), was obtained by adding chymotrypsin to the full-length Bid and then incubating at 30°C for 2 h (11). In some experiments, tBid was labeled with fluorescent probe N-(1-pyremyl)maleimide (PM) (18). Before labeling, tBid was dialyzed exhaustively against Tris buffer (20 mM Tris-HCl [pH 7.5] and 50 mM NaCl) and then labeled with PM dye (3 mol of PM to 1 mol of tBid) in the same buffer with rotation at 30°C for 2 h in the dark. After the reaction, the protein sample was dialyzed against Tris buffer (20 mM Tris-HCl [pH 7.5] and 50 mM NaCl).
Recombinant full-length human Bax with no additional amino acid residues was expressed in Escherichia coli and purified as an intein/chitin-binding domain fusion as described previously (19). The plasmid pTYB1-Bax was kindly provided by Prof. Yigong Shi (Tsinghua University, China). Lysis of E. coli was achieved by ultrasonication, and at no point was the protein exposed to detergents that alter the native Bax conformation. After affinity chromatography with a chitin column (New England Biolabs, Ipswich, MA), intein self-cleavage and release of Bax from its fusion partner was initiated by incubation with buffer containing 100 mM 2-mercaptoethanol for 36 h. Then full-length human Bax was further purified by an ion-exchange chromatography on a mono-Q column (Amersham Pharmacia Biotech, Piscataway, NJ).
Analysis of phospholipids of lysosomal membranes
Lysosomes were purified from the livers of Sprague-Dawley rats according to a method described previously (20) with minor modifications. All steps were carried out at 4°C unless otherwise noted. Briefly, several livers were minced, rinsed with sucrose/PIPES buffer (250 mM sucrose, 20 mM PIPES [pH 7.2]), resuspended in 10 volumes of sucrose/PIPES buffer, and homogenized by two brief pulses from a Brinkman Polytron homogenizer. The homogenate was centrifuged for 10 min at 540 g to remove nuclei and particulates. CaCl2 (final concentration, 1 mM) was added to the supernatant, followed by incubation for 5 min at 37°C to disrupt the mitochondria. The supernatant was centrifuged for 10 min at 18,000 g, and the heavy membrane pellet was retained and resuspended in sucrose/PIPES buffer, centrifuged again for 10 min at 18,000 g, and resuspended in Percoll (40% w/v) in sucrose/PIPES. The Percoll solution was centrifuged for 30 min at 44,000 g to form a gradient, and 1-ml fractions were collected from the bottom of the tube and assayed for mitochondrial contamination by using the lysosomal and mitochondrial enzyme markers as described below. The lysosomal fractions were pooled, diluted in sucrose/PIPES (1:10 v/v) to decrease the Percoll, and pelleted by centrifugation at 17,000 g for 10 min. The lysosomes were washed, resuspended in an equal volume of sucrose/PIPES, and used immediately. Because the purity of lysosomes was crucial in this study, we analyzed the isolated lysosomes by immunoblotting with antibodies against LAMP1 (lysosomal marker), COX IV (mitochondrial marker), and GAPDH (cytosolic marker) to determine the mitochondrial or cytosolic contaminations. Only lysosomes without any detectable mitochondrial or cytosolic markers were used in the following experiments.
Total lipids were obtained from lysosomal membranes by a two-step lipid extraction protocol (21), dried by evaporation under nitrogen, and stored dry at –20°C. Before analysis, the samples were diluted with hexane-isopropanol (3: 1, V/V). Sample aliquots (20 µl) were injected on an Astec Diol 5 mm diol-bonded silica normal-phase spherical HPLC column (250 × 4.6 mm), separated with a mobile phase of acetonitrile-methanol-85% phosphoric acid (100: 10: 0.8, V/V) flowing at 1.0 ml/min, and measured with a DAD detector at 205 nm.
Analysis of tBid-induced lysosomal membrane permeabilization
Purified rat liver lysosomes were suspended in buffer containing 250 mM sucrose and 20 mM HEPES (pH 7.2) and then incubated with the indicated concentrations of tBid for 30 min, followed by centrifugation at 18,000 g for 10 min. The pellets (lysosomes) were subjected to immunoblot analysis to detect the binding of tBid to lysosomal membranes. The supernatant was subjected to immunoblot analysis to assess the release of lysosomal contents. The percentage of cathepsin B released into the supernatant was used as an index of lysosomal membrane permeabilization (LMP). In some experiments, lysosomes were preincubated with 1 μg/μl PA or PC for 15 min before measuring the binding of tBid to lysosomal membranes and the tBid-induced LMP.
Analysis of tBid-induced mitochondrial outer membrane permeabilization
Mitochondria were purified from the livers of Sprague-Dawley rats according to a method described previously (12) with minor modifications. Mitochondria were suspended in buffer containing 400 mM mannitol, 5 mM succinate, 10 mM KH2PO4, and 50 mM Tris-HCl (pH 7.2) and then incubated with the indicated concentrations of tBid for 30 min, followed by centrifugation at 12,000 g for 10 min. The supernatant and the pellets (mitochondria) were subjected to immunoblotting analysis to detect the release of mitochondrial cytochrome c (10).
Assay for liposomal membrane permeabilization
Lipids were mixed in the appropriate ratio from stocks dissolved in chloroform. The organic solvent was removed by evaporation under a stream of nitrogen gas, followed by incubation for 2 h in a vacuum to ensure complete solvent removal. Lipid films were resuspended in HEPES buffer (10 mM HEPES [pH 7.4], 50 mM NaCl, and 0.2 mM EDTA) and subjected to 10 freeze-thaw cycles. Large unilamellar vesicles (LUVs) were then formed by extrusion through 100-nm Nucleopore polycarbonate membranes. To prepare FITC-dextran (FD)-containing liposomes, 5 mg/ml of FITC-labeled FD-20, FD-70, or FD-250 (numbers = mol. wt. of dextran /1,000) was added to the HEPES buffer, and nonentrapped fluorophores were removed by centrifugation at 100,000 g for 30 min with a Beckman TL-100.3 ultracentrifuge.
The leakage assay was performed as described previously (16). After incubation of increasing amounts of tBid with LUVs (phospholipids concentration, 100 μM) in 100 μl of HEPES buffer at 30°C for 30 min, the vesicles were sedimented by centrifugation (25 min, 35,000 rpm, 4°C). Half of the supernatant (S) was sampled out, and 350 μl HEPES buffer containing 0.1% Thesit was added to the remainder (R) in the centrifugation tube to dilute and dissolve the sample pellets. The control followed the same procedure except for the addition of buffer instead of protein. The FITC-dextran contained in each of these solutions was quantified using a Hitachi F-4500 spectrofluorometer with 5 nm bandwidths centered at 497 and 620 nm for excitation and emission, respectively. Fluorescent intensity was corrected for self-quenching according to a standard curve of fluorescence versus FITC-dextran concentration. The percentage of leakage was determined by
where B is the extent of leakage by the control.
Assay for protein-lipid interactions
The interaction between tBid and phospholipids was studied by surface plasmon resonance (22). The analysis was performed at 25°C using a BIAcore 2000 instrument equipped with a CM5 research-grade sensor chip. tBid proteins were immobilized on the sensor chip surface of one flow cell, and that of the second flow cell was left unmodified and served as a control. The lipids were suspended in buffer (10 mM HEPES [pH 7.4], 50 mM NaCl, 0.2 mM EDTA), and the kinetics of binding responses in this buffer were recorded as the lipids were injected and flowed across the two cells.
Insertion of tBid into monolayers and bilayers
Detection of the insertion of tBid into monolayers containing phosphatidic acid was performed by methods we described previously (17). Briefly, 3 ml HEPES buffer was added into the mini-trough as a subphase followed by dropping the phospholipids on the buffer surface to form a monomolecular lipid layer, and the surface pressure was measured with a film balance. After the initial surface pressure stabilized at its plateau value, the appropriate amount of protein was injected to the mixing chamber with a magnetic stir bar through a 0.7 cm2 hole in the edge from where it rapidly diffused into the monolayer-spreading disk to cause an increase in surface pressure. All measurements were performed at room temperature.
Fluorescence measurements were used to analyze the binding of tBid to LUV bilayer membranes (17, 18). tBid was labeled with PM to form PM-labeled tBid and then incubated with PC/PE LUVs (PC, 80%; PE, 20%) or PA-containing LUVs (PC, 70%; PE, 20%; PA, 10%) in HEPES buffer (10 mM HEPES [pH 7.4], 50 mM NaCl, and 0.2 mM EDTA) at 30°C for 30 min. Fluorescence measurements were performed with a Hitachi F-4500 fluorescence spectrometer set to 340 nm excitation and 376 nm emission with 2.5 nm slit widths.
Assay of tBid oligomerizationminus
The oligomerization of tBid was measured by size-exclusion chromatography. Experiments were performed in a Superdex-200 (1.5 × 45 cm) column equilibrated with 100 mM KCl, 10 mM HEPES, and 0.2 mM EDTA (pH 7.0) with or without 2% (w/v) CHAPS (J. T. Baker Inc.) at a 1 ml/min flow rate. The column was calibrated using protein gel filtration standards (Bio-Rad). Samples of 300 μl were loaded onto the column followed by collection of 2-ml elution fractions. Aliquots of individual fractions were subjected to SDS-PAGE in 15% Tris-glycine gels, followed by visualization of tBid using anti-Bid antibody.
The oligomerization of tBid was further confirmed by a cross-linking assay. tBid was incubated with LUVs in buffer containing 20 mM HEPES (pH 7.0), 0.5 M NaCl, and 1 mM EDTA at 37°C for 30 min. At the end of the incubation, the samples were centrifuged at 35,000 g under cold, and the pellets and supernants were collected. Sulfo-BSOCOES (in dimethyl sulfoxide) was added to the pellets and the supernatants separately from a 10-fold stock solution to a final concentration of 10 mM. After incubation for 30 min at room temperature, the cross-linker was quenched by the addition of 1 M Tris-HCl (pH 7.5) to a final concentration of 20 mM. Then the samples were lysed and subjected to SDS-PAGE. Western blotting was performed to analyze the oligomerization of tBid.
31P-NMR analysis
31P-NMR was used to detect the formation of nonbilayer microdomains of phospholipids (23). Liposomes containing PC and PA were incubated with different concentrations of tBid or chymBid for 30 min. NMR spectra were recorded at 25°C on a Varian Inova 600 MHz NMR spectrometer equipped with an indirect detection probe. The proton resonance frequency was 599.82 MHz, and that for 31P was 242.80 MHz. A standard single pulse sequence was used with composite pulse decoupling operating on protons during the acquisition period. Typical parameters used were: spectral width, 4.8 kHz; time domain data points, 16,000; recycle delay, 1.2 s; number of scans, 10,000. All free induction decays were multiplied by an exponential function with a 100 (200) Hz line broadening factor before Fourier transformation.
Delivery of tBid into cells and analysis of lysosomal membrane permeabilization
Wild-type and Bax−/−/Bak−/− mouse embryonic fibroblasts (MEFs) were generous gifts from Prof. Quan Chen (Institute of Zoology, CAS) and were maintained in DMEM supplemented with 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% heat-inactivated FBS (v/v). Recombinant tBid was delivered into MEFs by using the BioPORTER protein delivery kit (Sigma) according to the manufacturer's instructions. Briefly, 50 μl of tBid solution (1 μM) or a PBS control was used to hydrate the dried BioPORTER. The solution was pipetted up and down and incubated at room temperature for 5 min. Finally, the volume of the complexes was brought to 0.5 ml with serum-free medium and then transferred directly into the MEFs. After incubation for 5 h, 2 ml of serum-containing medium was added. The cells were incubated overnight and then subjected to the LMP assays.
Lysosomal permeabilization was determined in intact healthy or apoptotic cells by staining with 5 μg/ml AO for 15 min. AO-emitted red (lysosomal) and green (nuclear and cytosolic) fluorescence was analyzed with an inverted laser scanning confocal microscope (model FV500; Olympus, Tokyo, Japan) or a flow cytometer (FACSCalibur; BD, Mountain View, CA).
Data analysis
All data are expressed as the mean ± SD unless otherwise indicated. Differences between groups were compared by ANOVA followed by post hoc Bonferroni tests to correct for multiple comparisons. Differences were considered to be statistically significant at p < 0.05.
RESULTS
tBid binds to lysosomal membranes and induces LMP directly
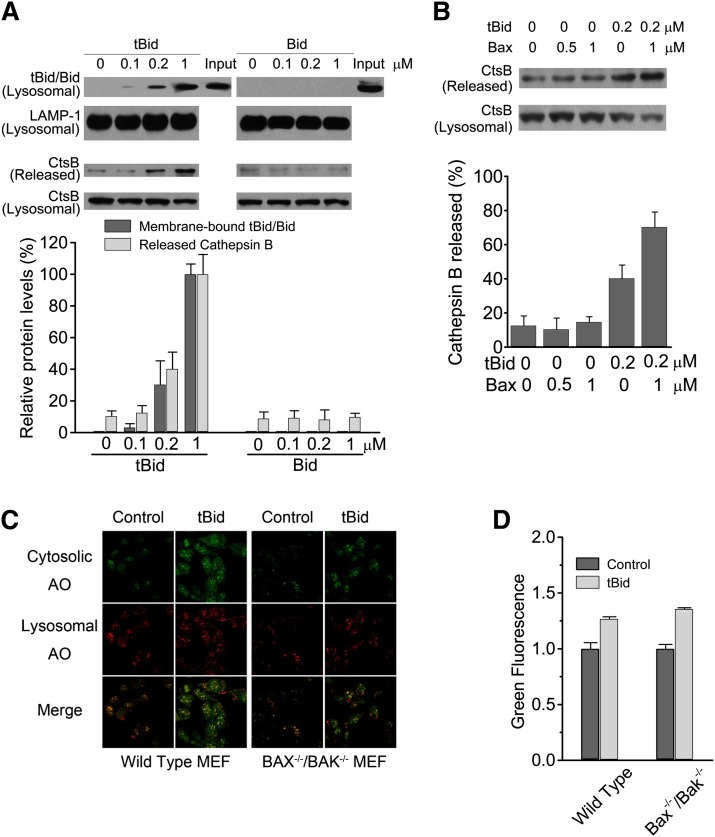
We first measured the ability of tBid to induce LMP in a cell-free system. Lysosomes were isolated from rat livers, and the purity was assessed by Western blotting (Supplementary Fig. I). Lysosomes without detectable mitochondrial or cytosolic contaminations were incubated with tBid and the LMP was quantified by analyzing the percentage of the lysosomal cathepsin B (a well-accepted marker enzyme of lysosomes) that was released into the supernatant. Incubation of tBid with lysosomes caused its significant translocation to the lysosomal membranes and resulted in the loss of lysosomal membrane integrity in a dose-dependent manner (Fig. 1A). In contrast, uncleaved Bid could not bind to lysosomes and showed no LMP-inducing effects. The addition of Bax could potentiate the LMP induced by tBid, but Bax by itself could not induce significant LMP (Fig. 1B).
Fig. 1.
tBid binds to lysosomal membranes and induces LMP directly. A: tBid, but not full-length Bid, binds to lysosomal membranes and induces LMP directly. Indicated concentrations of tBid (Bid cleaved with caspase 8) or full-length Bid was incubated with rat liver lysosomes in buffer containing 250 mM sucrose and 20 mM HEPES (pH 7.2) at 37°C for 30 min. The binding of Bid to lysosomal membranes and the release of lysosomal cathepsin B (a marker of LMP) was detected by immunoblotting. B: Bax potentiates LMP induced by tBid. Indicated concentrations of tBid and Bax were incubated with rat liver lysosomes, and the release of lysosomal cathepsin B was detected by immunoblotting. C: tBid induces LMP in intact cells. Recombinant tBid protein was introduced into wild-type and Bax−/−/Bak−/− mouse embryonic fibroblasts with BioPORTER reagent, and the redistribution of lysosomotropic fluorophore AO was observed with laser confocal scanning microscopy. D: tBid induces LMP in intact cells. Recombinant tBid protein was introduced into wild-type and Bax−/−/Bak−/− mouse embryonic fibroblasts with BioPORTER reagent, and alterations in AO fluorescence were analyzed with flow cytometry.
We further investigated whether tBid is capable of inducing LMP in intact cells. We introduced the recombinant tBid protein into MEFs with BioPORTER reagent by methods we described previously (10). With the help of BioPORTER reagent, recombinant tBid could be uptaken into cells via endocytosis and then partially released from the endosomes into the cytosol, where they interact with intracellular structures and molecules. The lysosomotropic fluorophore AO was used to determine the degree of LMP. AO is a cell-permeable dye that gives rise to red fluorescence at high concentrations and green fluorescence at low concentrations. AO accumulates by proton trapping in intact lysosomes because it becomes positively charged in the acidic lysosomal milieu. After LMP, AO is released from lysosomes into the cytosol, where it emits enhanced green fluorescence that can be monitored by fluorescence microscopy. After delivery of tBid into wild-type MEFs, a clear reduction of red fluorescence with a concomitant increase in green fluorescence could be observed, suggesting that tBid induced LMP at the cellular level (Fig. 1C, left panel). To further explore whether tBid induced LMP independent of Bax or Bak, two proapoptotic Bcl-2 proteins that are essential for tBid-induced MOMP, the effect of tBid on LMP in Bax−/−/Bak−/− MEF cells was measured. Albeit slightly attenuated, a similar decrease in red fluorescence and increase in green fluorescence were also observed in Bax−/−/Bak−/− MEF cells (Fig. 1C, right panel). These data indicated that tBid is sufficient to trigger LMP at the cellular level. Similar results were obtained by flow cytometry (Fig. 1D). Although Bax and Bak might enhance the role of tBid to some extent, they are not necessary for the induction of LMP.
tBid induces the permeabilization of model membranes, mimicking the phospholipid composition of lysosomal membranes
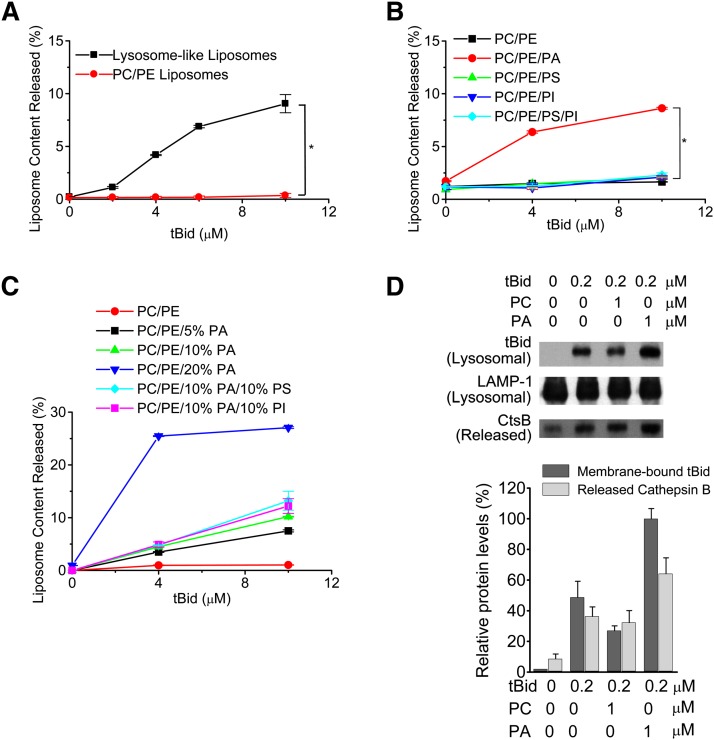
Because the above results demonstrated that tBid could interact with lysosomes and induce the permeabilization of lysosomal membrane without Bax or Bak, we used a model membrane system (large unilamellar vesicles [LUVs]) mimicking the phospholipid composition of lysosomal membranes (Table 1) to examine the interaction between tBid and lysosomal membrane lipids. tBid induced the leakage of FITC-dextrans from LUVs, mimicking the phospholipid composition of lysosomal membranes in a dose-dependent manner (Fig. 2A). Because the lysosomal membranes were rich in acidic phospholipids, including phosphatidic acid (PA, 6.3%), phosphatidylserine (PS, 5.4%), and phosphatidylinositol (PI, 6.1%) (Table 1) but without cardiolipin, we further investigated the influence of lysosomal membrane-enriched acidic phospholipids (PA, PS, and PI) on the permeabilization of model membranes. We found that only PA increased the tBid-induced membrane permeabilization significantly. PS, PI, or their combinations showed no effect on the tBid-induced membrane permeabilization (Fig. 2B), whereas PA increased tBid-induced membrane permeabilization in a concentration-dependent manner. Combinations of PA with PS or PI could not further potentiate the tBid-induced membrane permeabilization (Fig. 2C), suggesting that PA is the key molecule that bridges tBid and membrane permeabilization in LUVs mimicking the phospholipid composition of lysosomal membranes.
TABLE 1.
Analysis of phospholipids of rat liver lysosomal membranes
| Phospholipid | PC | PE | PA | PS | PI |
| % | 57.5 | 24.7 | 6.3 | 5.4 | 6.1 |
Fig. 2.
PA plays an essential role in tBid-induced permeabilization of model membranes. A: tBid induces the permeabilization of model membranes, mimicking the phospholipid composition of lysosomal membranes. FD-20 containing LUVs mimicking the phospholipid composition of lysosomal membranes (PC, 66%; PE, 16.2%; PA, 6.3%; PS, 5.4%; PI, 6.1%) or FD-20 containing PC/PE LUVs (PC, 80%; PE, 20%) were incubated with indicated concentrations of tBid in HEPES buffer (10 mM HEPES [pH 7.4], 50 mM NaCl, and 0.2 mM EDTA) at 30°C for 30 min. The release of encapsulated FD-20 (an index of membrane permeabilization) was determined. * P < 0.01. B: PA is necessary for the tBid-induced permeabilization of model membranes. FD-20 containing LUVs were prepared with PC (74%), PE (20%) and PA (6%), PS (6%), PI (6%), or a combination of PS (3%) and PI (3%), respectively, and the tBid-induced membrane permeabilization was determined. * P < 0.01. C: PA potentiates tBid-induced permeabilization of model membranes in a dose-dependent manner. FD-20 containing LUVs were prepared with PC (75, 70, or 60%, respectively), PE (20%), and the indicated concentrations of PA, PI, or PS, and the tBid-induced membrane permeabilization was determined. D: PA potentiates tBid-induced LMP. Rat liver lysosomes were incubated with PC or PA at 37°C for 15 min and then incubated with tBid at 37°C for 30 min. The binding of Bid to lysosomal membranes and the release of lysosomal cathepsin B were detected by immunoblotting.
We next investigated the influence of PA on tBid-induced LMP in isolated lysosomes. Lysosomes from rat liver were incubated with different phospholipids, such as PC or PA, to a final concentration of 1 μg/μl, and spontaneous diffusion of the phospholipids into the lysosomes then occurred. This spontaneous lipid transfer from aqueous solution to lysosomal membranes usually reached near equilibrium within 15 min (24). The binding of tBid to lysosomes was analyzed, and the occurrence of LMP was measured. The addition of PA could enhance the translocation of tBid onto the lysosomes and potentiate the occurrence of LMP (Fig. 2D). In contrast, the uncharged (neutral) phospholipid PC slightly inhibits the binding of tBid to lysosomal membranes and decreased the LMP, probably by reducing the PA ratio in lysosomal membrane.
tBid interacts with PA and inserts into model membranes
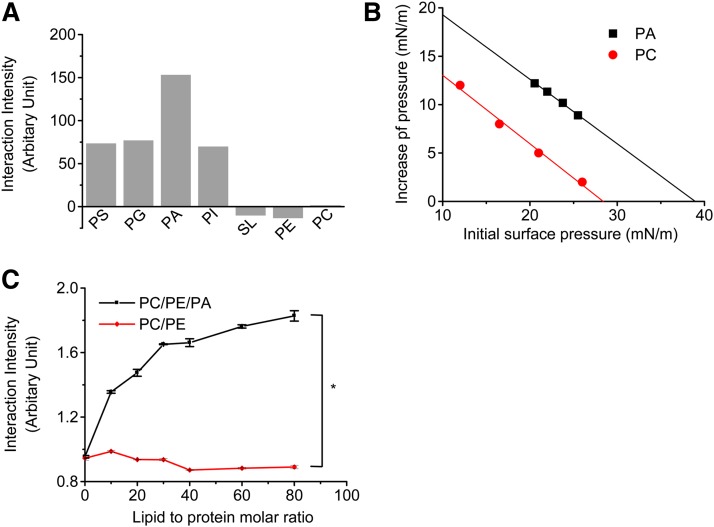
Although the above data show that PA plays an important role in tBid-induced LMP, the underlying mechanism remained to be elucidated. It has been well accepted that cardiolipin, which is present exclusively in mitochondrial membranes, mediates the targeting of tBid to mitochondria by providing a unique structure of the membrane (15). We therefore asked if tBid binds to lysosomal membranes via interacting with certain lipid molecules, such as PA. We used a sensitive BIAcore assay to investigate the direct interaction between tBid and the different phospholipids that are present in lysosomal membranes. Results showed that all three kinds of acidic phospholipids enriched in lysosomal membranes can directly interact with tBid; among them, PA shows the strongest interaction capacity (Fig. 3A). In contrast, no significant interaction between tBid and uncharged PC and PE was observed. Sphingolipids, another type of lipids that are enriched in lysosomal membranes, also did not interact with tBid.
Fig. 3.
tBid interacts with PA and inserts into PA-containing lipid membranes. A: Quantitative determination of interaction between tBid and phospholipids. tBid was immobilized on a CM5 chip, and the lipids were suspended in buffer containing 10 mM HEPES (pH 7.4), 50 mM NaCl, and 0.2 mM EDTA. The kinetics of binding responses were recorded at 25°C using a BIAcore 2000 instrument. B: tBid inserts into PA monolayer model membrane systems. Detection of the insertion of tBid into PA or PC monolayers was performed with a film balance. C: Fluorescence measurements of tBid-PA interactions. tBid was labeled with PM to form PM-labeled tBid and then incubated with PC/PE LUVs (PC, 80%; PE, 20%) or PA-containing LUVs (PC, 70%; PE, 20%; PA, 10%) in HEPES buffer (10 mM HEPES [pH 7.4], 50 mM NaCl, and 0.2 mM EDTA) at 30°C for 30 min. Fluorescence measurements was performed with a Hitachi F-4500 fluorescence spectrometer set to 340 nm excitation and 376 nm emission with 2.5 nm slit widths. * P < 0.01.
Next, we investigated whether tBid can insert into model membrane systems. Usually, an increase in surface pressure is observed when a protein inserts into a phospholipid monolayer. The πc of tBid in PA monolayers was 39 mN/m, which was much greater than the equivalence pressure, indicating that tBid could insert into PA monolayers (Fig. 3B). In contrast, tBid could not insert into PC monolayers.
The insertion of tBid to bilayer membranes of PA-containing LUVs was measured by fluorescence measurements. Fig. 3C indicates that tBid had more potential to insert into PA-containing phospholipid bilayers but not neutral PC bilayers, and this result is consistent with the results obtained in studies of the phospholipid monolayer.
tBid forms homo-oligomers, triggers the formation of nonbilayer lipid phases, and induces membrane permeabilization via pore formation
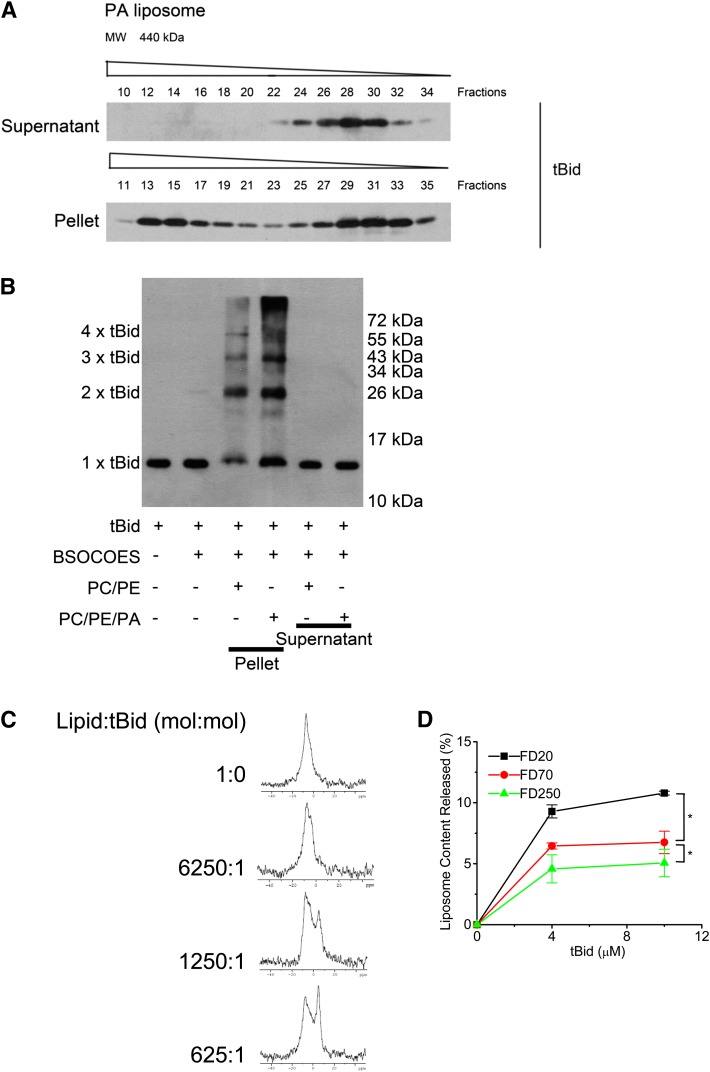
The above data provided direct evidence that tBid can insert into membranes containing PA. We next studied the oligomeric status of membrane-bound tBid by size-exclusion chromatography assays on Superdex 200. After incubation of tBid with liposomes containing PA, lipid-containing and lipid-free fractions were separated by centrifugation, and the amounts and pattern of liposome-bound tBid (pellet) and free tBid (supernatant) were analyzed by SDS-PAGE and immunoblotting. All samples were treated with 2% (w/v) CHAPS to allow for membrane solubilization. Free tBid (supernatant) eluted close to its calculated monomeric mass (Fig. 4A). However, liposome-bound tBid (pellet) shifted the elution profile of the majority of the protein to high-molecular-weight fractions, indicating that tBid forms oligomeric complexes with molecular weight as high as 400 kDa in the presence of PA.
Fig. 4.
tBid forms homooligomers, triggers the formation of nonbilayer lipid phase, and induces membrane permeabilization via pore formation. A: Size-exclusion chromatography of tBid homo-oligomers in PA-containing LUVs. LUVs (PC, 70%; PE, 20%; PA, 10%) were incubated with tBid in buffer containing 20 mM HEPES (pH 7.0), 0.5 M NaCl, and 1 mM EDTA at 37°C for 30 min and subjected to centrifugation at 35,000 g under cold. The supernatant and pellet were analyzed for tBid homooligomers with a Superdex-200 (1.5 × 45 cm) column equilibrated with 100 mM KCl, 10 mM HEPES, and 0.2 mM EDTA (pH 7.0) at a 1 ml/min flow rate. B: Immunoblotting analysis of tBid homo-oligomers in PA-containing LUVs. tBid was incubated with PA-containing LUVs (PC, 70%; PE, 20%; PA, 10%) or control PC/PE LUVs (PC, 80%; PE, 20%) in buffer containing 20 mM HEPES (pH 7.0), 0.5 M NaCl, and 1 mM EDTA at 37°C for 30 min and subjected to centrifugation at 35,000 g under cold. The supernatant and pellet was cross-linked with Sulfo-BSOCOES, and the oligomerization of tBid was analyzed by immunoblotting. C: tBid induces the formation of nonbilayer lipid phase. LUVs (PC, 80%; PA, 20%) were incubated with different concentrations of tBid for 30 min. 31P NMR spectra were recorded at 25°C on a Varian Inova 600 MHz NMR spectrometer equipped with an indirect detection probe. D: Pore formation induced by tBid in PA-containing membranes. FD-20, FD-70, or FD-250 (numbers = mol. wt. of dextran /1,000) was encapsulated in PA-containing LUVs (PC, 70%; PE, 18%; PA, 10%), and the tBid-induced release of LUV contents was measured. * P < 0.05.
The homo-oligomerization of tBid on PA-containing liposomes was further confirmed by a cross-linking assay. tBid was incubated with liposomes composed of PC or PC/PA, followed by centrifugation to separate the membranous fraction from the soluble fraction. Both fractions were treated with the sulfo-BSOCOES cross-linker and analyzed by SDS-PAGE and immunoblotting. After adding tBid to PA-containing liposomes, a substantial amount of tBid was incorporated into the liposomal membrane (Fig. 4B, lane 4). Once tBid appeared in the membrane, the 14 kDa (monomer), the 28 kDa (dimer), the 42 kDa (trimer), the 56 kDa (tetramer), and the higher-molecular-weight band (homo-oligomers) cross-linked complexes also appeared. Significantly less tBid was incorporated into the liposomal membrane containing PC only, a result that is consistent with those obtained in the film balance and BIAcore assays. Meanwhile, as more tBid incorporated into the membrane, the intensity of the homooligomer band increased. The cross-linked bands that appear in the membranous fractions were not detected in soluble fractions (Fig. 4B, lanes 5 and 6).
It has been well accepted that PA forms nonbilayer structures in certain conditions (25), so we next questioned whether there is any change in lipid polymorphism during LMP as a consequence of tBid-PA interaction. The shape of the 31P NMR spectrum is known to be different for lipids in the bilayer phase and in nonbilayer phases, such as the inverted hexagonal phase (23). A typical polymorphic transition of phospholipids (PC-PA, 4:1) could be induced after the addition of tBid (Fig. 4C). In the absence of protein, the 31P NMR spectrum had a peak upfield of PA, which was set to 0 ppm. By adding tBid into the lipidic mixture, the major peak shifted downfield below the chemical shift of PA. Moreover, at lower tBid concentration, there was still significant intensity in the power pattern of the pure lipid mixture above 0 ppm that disappeared at higher tBid concentrations. The spectra indicated that there is a gradual shifting in the lipid layer from bilayer to hexagonal phase as the tBid concentration increased. In other words, in conjunction with the formation of tBid homo-oligomers in liposomes as a consequence of tBid-PA interaction, a change of the lipid polymorphism also occurred in the lipid bilayer of this model system.
To clarify whether the oligomerization of tBid and the formation of nonbilayer lipid phases result in the formation of “pores,” we studied the leakage induced by tBid from the LUVs encapsulated with FITC-dextrans of different molecular weights. The tBid-induced leakage of FD-20, FD-70, and FD-250 separately encapsulated in LUVs composed of PA and PC was assayed. The results revealed that the leakage extent of FD-20 was higher than that of FD-70 or FD-250 (Fig. 4D). These results suggest that leakage occurred due to the formation of discrete pores rather than a channel (fissure) mechanism because the leakage efficiency of markers with different molecular weights would be the same in the latter case.
chymBid is more potent than tBid in triggering the permeabilization of membranes
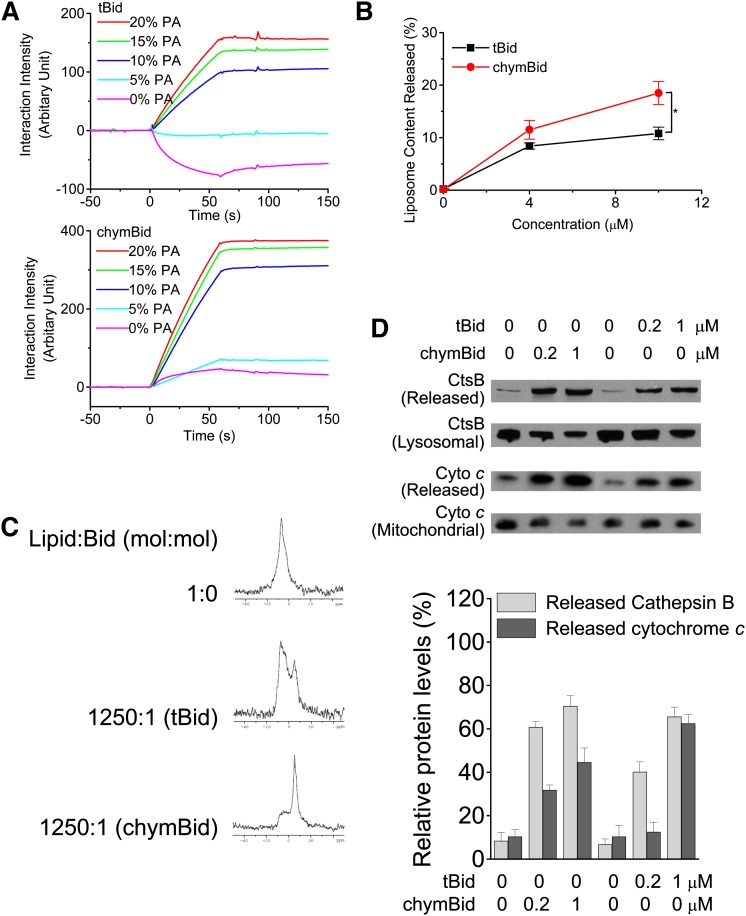
We previously reported that chymotrypsin is not just a digestive enzyme but is also stored in lysosomes and involved in apoptosis (10). Chymotrypsin cleaves Bid rapidly to form chymBid, which is capable of inducing MOMP and causing apoptosis. Further study indicated that Bid activation initiated by caspase-8 might be reinforced by chymotrypsin as a consequence of LMP, thereby causing a positive feedback loop leading to an accumulation of cleaved Bid that resulted in MOMP (11). The structural difference between tBid and chymBid is clear in that the former still contains the N-terminal fragment (23, 26), whereas this fragment has been cleaved in the latter case. We compared the ability of tBid and chymBid to trigger the permeablization of various membranes. chymBid bound to PA-containing liposomes much more strongly than tBid (Fig. 5A) and was more efficient at promoting the leakage of FITC-dextrans from LUVs (Fig. 5B). 31P-NMR indicated that chymBid shifted the lipid layer from a bilayer to hexagonal phase much more readily than tBid (Fig. 5C). The effects of chymBid or tBid on the induction of LMP and MOMP were also investigated, and chymBid was found to be more effective in inducing the release of cathepsin B from lysosomes and cytochrome c from mitochondria in comparison with tBid (Fig. 5D). Taken together, these results indicated that chymBid, after its binding with PA, is more potent in triggering the permeabilization of lysosomal membranes than tBid.
Fig. 5.
chymBid is more potent than tBid in triggering the permeabilization of membranes. A: chymBid shows stronger interaction with PA. chymBid or tBid was immobilized on a CM5 chip, and binding responses were recorded at 25°C using a BIAcore 2000 instrument. B: chymBid and tBid induce the permeabilization of model membranes containing PA. LUVs containing FD-20 (PC, 70%; PE, 20%; PA, 10%) were incubated with the indicated concentrations of chymBid or tBid at 30°C for 30 min, and the membrane permeabilization was determined. * P < 0.05. C: chymBid and tBid induces the formation of nonbilayer lipid phase. LUVs (PC, 80%; PA, 20%) were incubated with chymBid or tBid for 30 min, and 31P NMR spectra were recorded at 25°C. D: chymBid and tBid induce the permeabilization of lysosomal and mitochondrial membranes. Rat liver lysosomes or mitochondria were incubated with the indicated concentrations of chymBid and tBid at 37°C for 30 min, and the release of lysosomal cathepsin B or mitochondrial cytochrome c was detected by immunoblotting.
DISCUSSION
The initiation of LMP is one of the crucial steps leading to cell death. Complete disruption of lysosomes leads to uncontrolled necrotic cell death; in contrast, partial and selective LMP induces apoptotic cell death. The multitude of parallel pathways make the lysosome-dependent apoptosis a highly complex process, and thus the precise mechanisms of LMP in apoptosis are poorly understood. Based on our previous findings that caspase 8 activation and Bid expression are necessary for the induction of LMP (11), and inspired by the similarities between tBid-dependent LMP and MOMP, we studied the process of LMP in a relatively simplified experimental system with purified lysosomes and artificial model membrane systems mimicking the lysosomal membranes. The results of the present investigation indicated that PA, one of the major acidic phospholipids enriched in lysosomal membrane, is essential for tBid-induced permeabilization of lysosomal and lysosomal-like membranes. PA facilitates the targeting and insertion of tBid into lipid bilayers, where it changes conformation, forms homooligomers, and triggers the formation of nonbilayer lipid phases (hexagonal lipid phases). These events lead to the formation of lipidic pores and the induction of LMP.
PA is an anionic lipid consisting of a negatively charged phosphomonoester headgroup attached to a hydrophobic diacylglycerol backbone. It is present in all organisms and serves as a key intermediate in the synthesis of neutral lipids and glycerophospholipids, including PS, PE, PC, and PI. In addition, PA is emerging as an important signaling lipid (27, 28). By binding its effector proteins such as RGS4 (29), Son of sevenless (Sos) (30), Raf-1 (31), and mammalian target of rapamycin (mTOR) (32, 33), PA recruits them to membranes and regulates their activity in different cellular pathways (34). It has been proposed that the binding of proteins to PA is generally specified through nonspecific electrostatic interactions between clusters of positively charged amino acids in the protein and the negatively charged headgroup of PA. However, rarely do PA effectors also bind other abundant anionic lipids. Our results indicated that PA accounts for the targeting of tBid to lysosomal membranes. This specificity for PA might be due to its unique structure. PA contains a phosphomonoester headgroup rather than the phosphodiesters in other phospholipids. With evidence provided by 31P-NMR, Kooijman et al. (35) proposed a model in which hydrogen bonding of the effector protein to the phosphomonoester of PA enables it to carry a greater negative charge at physiological range pH. Thus, PA effectors favor binding to PA over other anionic lipids because of the higher negative charge of PA and the stronger electrostatic interactions.
During MOMP, cardiolipin, which is present in mitochondrial membranes, mediates the targeting of tBid to mitochondria (15, 36), where tBid can promote the oligomerization of Bak (37–39) or Bax (26, 39), which is a key step in membrane permeabilization. tBid also elicits the conformational alteration of membrane-bound Bcl-2 (40, 41) and Bcl-xl (42) and thus regulates MOMP. tBid binds to mitochondrial outer membranes through electrostatic interactions. The α-helices 4, 5, and 6 (amino acids 103–162) of tBid were initially proposed to be its membrane-binding domain (15). However, it has been more recently proved that α-helix 6 (α6) of tBid is necessary, albeit not sufficient, for membrane binding (43, 44). This helix, together with helix α7, forms an antiparallel hairpin structure that is surrounded by the other six amphipathic α-helices. A 33 amino acid domain, encompassing α6 and α7, behaved as the minimum domain in tBid that was sufficient for its membrane binding (45). Moreover, α8 is complementary with helix α6–α7 because it provides an increasing positive charge (46, 47). Taken together, the α6–α8 fragment of tBid is a good candidate for its binding to mitochondrial outer membranes through electrostatic interactions. In the case of LMP, PA mediates the targeting of tBid to lysosomes and induces Bax- and Bak-independent membrane permeabilization. However, the precise mechanisms remain elusive.
By using solid-state 1H/15N NMR, Gong et al. (48) reported that all α-helices of tBid appear to interact with the membrane in a parallel fashion. By Monte Carlo simulations, Veresov et al. (49) reported that upon association with the membrane, significant conformational changes occurs in tBid primarily due to rotations within the loops between helices. Garcia-Saez et al. (46) reported that tBid evolves into a membrane-inserted species upon apoptotic signal triggering and that the hairpin-forming domains of tBid (H6-H7) showed pore activity. We herein provide evidence that membrane permeabilization occurred as a result of the insertion of tBid into PA-containing model membranes, probably via a “pore-formation” mechanisms because the smaller encapsulated contents were more prone to tBid-induced leakage (17) when the leakage efficiency of encapsulated contents with different molecular weights would be the same if the release were due to the channel mechanism.
We then examined the nature of the “pore” formed by tBid. A membrane pore is traditionally presumed to be composed by protein complexes. Pore-forming proteins have been classified into two distinct classes. One class, whose secondary structures are predominantly β-sheets, forms crystallizable porin-like transmembrane β-barrel pores. Another class, whose secondary structures are predominantly α-helical segments, forms pores without a crystallizable protein assembly. Proapoptotic Bcl-2 family proteins Bax and Bak belong to the α-pore-forming proteins. Their α-helical segments form a transmembrane barrel-stave assembly stabilized by hydrophobic interactions. We have tried to detect the “pore structure” framed by tBid homooligomers in PA-containing membranes by cryoelectron microscopy but have been unsuccessful. tBid forms homooligomers in model membranes in the presence of PA (Fig. 4A, B) or cardiolipin (50) and causes the permeabilization of lysosomal and lysosome-like membranes, but it is not an α-pore-forming protein because no transmembrane helix insertion occurred during its binding to membranes (48, 49). We therefore proposed that, instead of framing “proteinaceous pores,” tBid homo-oligomers might induce the formation of “lipidic pores” to cause permeabilization of PA-containing membranes. Lipidic pores are pores whose walls were partially lined by lipid headgroups. Lipid membranes act as barriers that separate compartments in aqueous solvents. Their structure corresponds to a lamellar bilayer, which self-organizes due to the hydrophobic effect. However, certain proteins or peptides can disrupt the bilayer structure of the membrane by forming nonbilayer (nonlamellar) lipid structures, leading to the membrane permeabilization via “lipidic pores” (51). By 31P NMR measurement, we found that tBid-PA interaction promotes the formation of nonbilayer lipid phases (hexagonal lipid phases). PA is a typical nonbilayer lipid; the small cross-sectional area of its headgroup gives it a cone-shaped structure (52, 53), which prevents tight packing of PA's headgroup with the headgroups of neighboring lipids and exposes the hydrophobic acyl layer of the bilayer surrounding PA. This loose packing provides an excellent site for insertion of helices of tBid. The interaction between tBid and PA disrupted the lipid bilayer order by causing the formation of highly curved nonbilayer phases, and, as a result of this membrane remodeling, “lipidic pores” formed. Similar to the “lipidic pore” formed by tBid/Bax during MOMP (14, 23, 54–56), this tBid- and PA-dependent “lipidic pore” accounts for the tBid-induced permeabilization of lysosomal or lysosome-like membranes.
After the induction of LMP by tBid, several lysosomal proteases, including cathepsins and chymotrypsin, were relocated into the cytosol, where they process Bid rapidly. In our previous report, we demonstrated that chymotrypsin, but not cathepsins, account for the rapid processing of Bid in neutral pH conditions (11). We found in the present investigation that chymBid was more potent than tBid at binding to PA, inserting into the lipid bilayer, and promoting the membrane permeabilization of lysosomal or mitochondrial membranes. This amplification mechanism likely contributes to the induction of LMP and the culmination of apoptotic signaling. To investigate the difference between tBid and chymBid, we analyzed and compared the processing of full-length Bid by caspase 8 or chymotrypsin. The truncated carboxyl-terminal fragment of tBid (amino acids 60–195) harboring the full apoptogenicity of the molecule remains associated with the N-terminal inert fragment (amino acids 1–59) after cleavage by caspase 8. Dissociation of fragment 60–195 from fragment 1–59 occurs during targeting of tBid to mitochondria. The dissociation of fragment 60–195 from fragments 1–59 is essential for tBid-induced mitochondrial damage because fragment 1–59 inhibits the membrane destabilizing effect of tBid (57, 58). This dissociation is likely to occur during targeting of the tBid 60–195 fragment to mitochondria and may be driven by the electrostatic interactions between fragment 60–195 and the acidic phospholipids (especially cardiolipin) in the mitochondrial membranes (43). However, when Bid was proteolytically cleaved by chymotrypsin to form chymBid, only a carboxyl-terminal fragment (fragment 68–195) was generated. The amino terminal fragment that might inhibit the carboxyl-terminal fragments of Bid was degraded during the chymotrypsin-mediated proteolytic process. The greater efficacy of chymBid in interacting with PA, binding to membranes, and permeabilizing the membranes might be due to the specific Bid processing caused by chymotrypsin.
One of the primary questions remaining to be addressed is how to achieve a partial, selective permeabilization of lysosomes that allows for the translocation of only a moderate amount of lysosomal enzyme to the cytosol to culminate apoptosis rather than the complete breakdown of lysosomes, as seen with induction of a necrotic process. Due to the heterogeneous nature of lysosomes, only a small proportion of lysosomes underwent LMP after proapoptotic stimulation (10, 11). Larger lysosomes were reported to be more prone to LMP than smaller lysosomes (6), but the basis for this observation is unknown. Do phospholipase D signaling and PA dynamics determine the fate of lysosomes? Related studies are in progress.
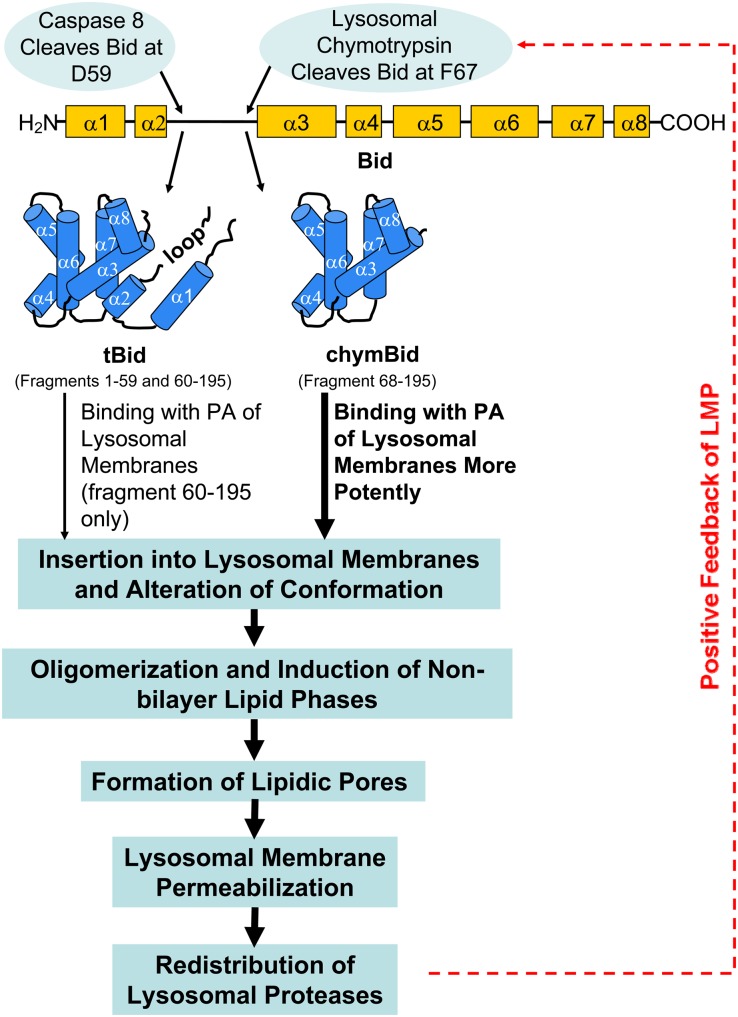
In summary, we herein hypothesized a novel mechanism underlying the occurrence of LMP during apoptosis. Our data suggested that PA, an acidic phospholipid enriched in lysosomal membranes, is essential for LMP. During the initiation phase of LMP, tBid formed by activated caspase 8 is targeted and inserted to the lysosomal membranes by interaction with PA, where tBid changes conformation, forms homooligomers, and triggers the formation of nonbilayer lipid phases, which account for the formation of lipidic pores and the consequent initiation of LMP. After the occurrence of LMP, different lysosomal proteases are redistributed from a small proportion of lysosomes into the cytosol, among them chymotrypsin, which rapidly cleaves Bid to form chymBid. chymBid shows stronger interaction with PA, triggers the formation of more nonbilayer lipid phases, and causes more significant LMP (Fig. 6). This amplification of LMP culminates in the lysosome-dependent apoptotic signaling.
Fig. 6.
Schematic diagram of tBid- and chymBid-induced LMP. During the initiation phase of LMP, tBid formed by activated caspase 8 is targeted to the lysosomal membranes by interaction with PA, where tBid changes conformation, forms homo-oligomers, and triggers the formation of nonbilayer lipid phases, which account for the formation of “lipidic pores” and the consequent initiation of LMP. As a result of LMP, different lysosomal proteases are redistributed into the cytosol, among them chymotrypsin, which rapidly cleaves Bid to form chymBid. chymBid shows stronger interaction with PA, triggers the formation of more nonbilayer lipid phases, and causes more significant LMP. This amplification of LMP culminates in the lysosome-dependent apoptotic signaling.
Supplementary Material
Acknowledgments
The authors thank Prof. Quan Chen (Institute of Zoology, CAS) for providing wild-type and Bax−/−/Bak−/− MEF cells, Prof. Yigong Shi (Tsinghua University) for providing pTYB1-Bax plasmid, Profs. Xuejun C. Zhang and Pingsheng Liu (Institute of Biophysics, CAS) and Huiru Tang (Wuhan Institute of Physics and Mathematics, CAS) for valuable discussions and suggestions, and Wenmin Zhong, Teng Zhou, and Qianqian Chen (Institute of Biophysics, CAS) and Wei Li (Institute of High Energy Physics, CAS) for technical assistance.
Footnotes
Abbreviations:
- AO
- acridine orange
- chymBid
- chymotrypsin-cleaved Bid
- CHAPS
- 3-[(3-cholamidopropyl)-dimethyl-ammonio]-1-propane sulfonate
- FD
- fluorescein-conjugated dextran
- LMP
- lysosomal membrane permeabilization
- LUV
- large unilamellar vesicle
- MEF
- mouse embryonic fibroblast
- MOMP
- mitochondrial outer membrane permeabilization
- PA
- phosphatidic acid
- PC
- phosphotidylcholine
- PE
- phosphotidylethanolamine
- PG
- phosphotidylglycerol
- PI
- phosphotidylinositol
- PIPES
- piperazine-1,4-bis(2-ethanesulfonic acid)
- PM
- N-(1-pyrenyl)maleimide
- PS
- phosphatidylserine
- Sulfo-BSOCOES
- bis(2-(3-sulfo-N-succinimidyloxycarbonyloxy)ethyl)sulfone
- tBid
- caspase 8-cleaved Bid
This work is supported by National Basic Research Program of China grants 2010CB833701 and 2012CB934003 and by National Natural Science Foundation of China grants 31070736 and 31100595.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure.
REFERENCES
- 1.Clarke P. G. 1990. Developmental cell death: morphological diversity and multiple mechanisms. Anat. Embryol. (Berl.). 181: 195–213 [DOI] [PubMed] [Google Scholar]
- 2.Galluzzi L., Maiuri M. C., Vitale I., Zischka H., Castedo M., Zitvogel L., Kroemer G. 2007. Cell death modalities: classification and pathophysiological implications. Cell Death Differ. 14: 1237–1243 [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21: 2861–2873 [DOI] [PubMed] [Google Scholar]
- 4.Denton D., Nicolson S., Kumar S. 2012. Cell death by autophagy: facts and apparent artefacts. Cell Death Differ. 19: 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guicciardi M. E., Leist M., Gores G. J. 2004. Lysosomes in cell death. Oncogene. 23: 2881–2890 [DOI] [PubMed] [Google Scholar]
- 6.Ono K., Kim S. O., Han J. 2003. Susceptibility of lysosomes to rupture is a determinant for plasma membrane disruption in tumor necrosis factor alpha-induced cell death. Mol. Cell. Biol. 23: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boya P., Kroemer G. 2008. Lysosomal membrane permeabilization in cell death. Oncogene. 27: 6434–6451 [DOI] [PubMed] [Google Scholar]
- 8.Repnik U., Stoka V., Turk V., Turk B. 2012. Lysosomes and lysosomal cathepsins in cell death. Biochim. Biophys. Acta. 1824: 22–33 [DOI] [PubMed] [Google Scholar]
- 9.Masson O., Bach A. S., Derocq D., Prebois C., Laurent-Matha V., Pattingre S., Liaudet-Coopman E. 2010. Pathophysiological functions of cathepsin D: targeting its catalytic activity versus its protein binding activity? Biochimie. 92: 1635–1643 [DOI] [PubMed] [Google Scholar]
- 10.Miao Q., Sun Y., Wei T., Zhao X., Zhao K., Yan L., Zhang X., Shu H., Yang F. 2008. Chymotrypsin B cached in rat liver lysosomes and involved in apoptotic regulation through a mitochondrial pathway. J. Biol. Chem. 283: 8218–8228 [DOI] [PubMed] [Google Scholar]
- 11.Zhao K., Zhao X., Tu Y., Miao Q., Cao D., Duan W., Sun Y., Wang J., Wei T., Yang F. 2010. Lysosomal chymotrypsin B potentiates apoptosis via cleavage of Bid. Cell. Mol. Life Sci. 67: 2665–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo X., Budihardjo I., Zou H., Slaughter C., Wang X. 1998. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 94: 481–490 [DOI] [PubMed] [Google Scholar]
- 13.Li H., Zhu H., Xu C. J., Yuan J. 1998. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 94: 491–501 [DOI] [PubMed] [Google Scholar]
- 14.Tait S. W., Green D. R. 2010. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11: 621–632 [DOI] [PubMed] [Google Scholar]
- 15.Lutter M., Fang M., Luo X., Nishijima M., Xie X., Wang X. 2000. Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat. Cell Biol. 2: 754–761 [DOI] [PubMed] [Google Scholar]
- 16.Zhai D., Miao Q., Xin X., Yang F. 2001. Leakage and aggregation of phospholipid vesicles induced by the BH3-only Bcl-2 family member, BID. Eur. J. Biochem. 268: 48–55 [DOI] [PubMed] [Google Scholar]
- 17.Yan L., Miao Q., Sun Y., Yang F. 2003. tBid forms a pore in the liposome membrane. FEBS Lett. 555: 545–550 [DOI] [PubMed] [Google Scholar]
- 18.Lovell J. F., Billen L. P., Bindner S., Shamas-Din A., Fradin C., Leber B., Andrews D. W. 2008. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 135: 1074–1084 [DOI] [PubMed] [Google Scholar]
- 19.Suzuki M., Youle R. J., Tjandra N. 2000. Structure of Bax: coregulation of dimer formation and intracellular localization. Cell. 103: 645–654 [DOI] [PubMed] [Google Scholar]
- 20.Stoka V., Turk B., Schendel S. L., Kim T. H., Cirman T., Snipas S. J., Ellerby L. M., Bredesen D., Freeze H., Abrahamson M., et al. 2001. Lysosomal protease pathways to apoptosis. Cleavage of bid, not pro-caspases, is the most likely route. J. Biol. Chem. 276: 3149–3157 [DOI] [PubMed] [Google Scholar]
- 21.Fraldi A., Annunziata F., Lombardi A., Kaiser H. J., Medina D. L., Spampanato C., Fedele A. O., Polishchuk R., Sorrentino N. C., Simons K., et al. 2010. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J. 29: 3607–3620 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Liu J., Durrant D., Yang H. S., He Y., Whitby F. G., Myszka D. G., Lee R. M. 2005. The interaction between tBid and cardiolipin or monolysocardiolipin. Biochem. Biophys. Res. Commun. 330: 865–870 [DOI] [PubMed] [Google Scholar]
- 23.Epand R. F., Martinou J. C., Fornallaz-Mulhauser M., Hughes D. W., Epand R. M. 2002. The apoptotic protein tBid promotes leakage by altering membrane curvature. J. Biol. Chem. 277: 32632–32639 [DOI] [PubMed] [Google Scholar]
- 24.Mantena S. K., King A. L., Andringa K. K., Eccleston H. B., Bailey S. M. 2008. Mitochondrial dysfunction and oxidative stress in the pathogenesis of alcohol- and obesity-induced fatty liver diseases. Free Radic. Biol. Med. 44: 1259–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kooijman E. E., Chupin V., Fuller N. L., Kozlov M. M., de Kruijff B., Burger K. N., Rand P. R. 2005. Spontaneous curvature of phosphatidic acid and lysophosphatidic acid. Biochemistry. 44: 2097–2102 [DOI] [PubMed] [Google Scholar]
- 26.Eskes R., Desagher S., Antonsson B., Martinou J. C. 2000. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol. Cell. Biol. 20: 929–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kooijman E. E., Burger K. N. 2009. Biophysics and function of phosphatidic acid: a molecular perspective. Biochim. Biophys. Acta. 1791: 881–888 [DOI] [PubMed] [Google Scholar]
- 28.Shin J. J., Loewen C. J. 2011. Putting the pH into phosphatidic acid signaling. BMC Biol. 9: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouyang Y. S., Tu Y., Barker S. A., Yang F. 2003. Regulators of G-protein signaling (RGS) 4, insertion into model membranes and inhibition of activity by phosphatidic acid. J. Biol. Chem. 278: 11115–11122 [DOI] [PubMed] [Google Scholar]
- 30.Zhao C., Du G., Skowronek K., Frohman M. A., Bar-Sagi D. 2007. Phospholipase D2-generated phosphatidic acid couples EGFR stimulation to Ras activation by Sos. Nat. Cell Biol. 9: 706–712 [DOI] [PubMed] [Google Scholar]
- 31.Rizzo M. A., Shome K., Watkins S. C., Romero G. 2000. The recruitment of Raf-1 to membranes is mediated by direct interaction with phosphatidic acid and is independent of association with Ras. J. Biol. Chem. 275: 23911–23918 [DOI] [PubMed] [Google Scholar]
- 32.Fang Y., Vilella-Bach M., Bachmann R., Flanigan A., Chen J. 2001. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 294: 1942–1945 [DOI] [PubMed] [Google Scholar]
- 33.Foster D. A. 2009. Phosphatidic acid signaling to mTOR: signals for the survival of human cancer cells. Biochim. Biophys. Acta. 1791: 949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stace C. L., Ktistakis N. T. 2006. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim. Biophys. Acta. 1761: 913–926 [DOI] [PubMed] [Google Scholar]
- 35.Kooijman E. E., Carter K. M., van Laar E. G., Chupin V., Burger K. N., de Kruijff B. 2005. What makes the bioactive lipids phosphatidic acid and lysophosphatidic acid so special? Biochemistry. 44: 17007–17015 [DOI] [PubMed] [Google Scholar]
- 36.Gonzalvez F., Pariselli F., Dupaigne P., Budihardjo I., Lutter M., Antonsson B., Diolez P., Manon S., Martinou J. C., Goubern M., et al. 2005. tBid interaction with cardiolipin primarily orchestrates mitochondrial dysfunctions and subsequently activates Bax and Bak. Cell Death Differ. 12: 614–626 [DOI] [PubMed] [Google Scholar]
- 37.Korsmeyer S. J., Wei M. C., Saito M., Weiler S., Oh K. J., Schlesinger P. H. 2000. Pro-apoptotic cascade activates BID, which oligomerizes BAK or BAX into pores that result in the release of cytochrome c. Cell Death Differ. 7: 1166–1173 [DOI] [PubMed] [Google Scholar]
- 38.Wei M. C., Lindsten T., Mootha V. K., Weiler S., Gross A., Ashiya M., Thompson C. B., Korsmeyer S. J. 2000. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 14: 2060–2071 [PMC free article] [PubMed] [Google Scholar]
- 39.Leber B., Lin J., Andrews D. W. 2007. Embedded together: the life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis. 12: 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng J., Tan C., Roberts G. J., Nikolaeva O., Zhang Z., Lapolla S. M., Primorac S., Andrews D. W., Lin J. 2006. tBid elicits a conformational alteration in membrane-bound Bcl-2 such that it inhibits Bax pore formation. J. Biol. Chem. 281: 35802–35811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng J., Ding J., Tan C., Baggenstoss B., Zhang Z., Lapolla S. M., Lin J. 2009. Oligomerization of membrane-bound Bcl-2 is involved in its pore formation induced by tBid. Apoptosis. 14: 1145–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Saez A. J., Ries J., Orzaez M., Perez-Paya E., Schwille P. 2009. Membrane promotes tBID interaction with BCL(XL). Nat. Struct. Mol. Biol. 16: 1178–1185 [DOI] [PubMed] [Google Scholar]
- 43.Gonzalvez F., Pariselli F., Jalmar O., Dupaigne P., Sureau F., Dellinger M., Hendrickson E. A., Bernard S., Petit P. X. 2010. Mechanistic issues of the interaction of the hairpin-forming domain of tBid with mitochondrial cardiolipin. PLoS ONE. 5: e9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petit P. X., Dupaigne P., Pariselli F., Gonzalvez F., Etienne F., Rameau C., Bernard S. 2009. Interaction of the alpha-helical H6 peptide from the pro-apoptotic protein tBid with cardiolipin. FEBS J. 276: 6338–6354 [DOI] [PubMed] [Google Scholar]
- 45.Hu X., Han Z., Wyche J. H., Hendrickson E. A. 2003. Helix 6 of tBid is necessary but not sufficient for mitochondrial binding activity. Apoptosis. 8: 277–289 [DOI] [PubMed] [Google Scholar]
- 46.Garcia-Saez A. J., Mingarro I., Perez-Paya E., Salgado J. 2004. Membrane-insertion fragments of Bcl-xL, Bax, and Bid. Biochemistry. 43: 10930–10943 [DOI] [PubMed] [Google Scholar]
- 47.Detmer S. A., Chan D. C. 2007. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 8: 870–879 [DOI] [PubMed] [Google Scholar]
- 48.Gong X. M., Choi J., Franzin C. M., Zhai D., Reed J. C., Marassi F. M. 2004. Conformation of membrane-associated proapoptotic tBid. J. Biol. Chem. 279: 28954–28960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veresov V. G., Davidovskii A. I. 2007. Monte Carlo simulations of tBid association with the mitochondrial outer membrane. Eur. Biophys. J. 37: 19–33 [DOI] [PubMed] [Google Scholar]
- 50.Grinberg M., Sarig R., Zaltsman Y., Frumkin D., Grammatikakis N., Reuveny E., Gross A. 2002. tBID Homooligomerizes in the mitochondrial membrane to induce apoptosis. J. Biol. Chem. 277: 12237–12245 [DOI] [PubMed] [Google Scholar]
- 51.Fuertes G., Gimenez D., Esteban-Martin S., Sanchez-Munoz O. L., Salgado J. 2011. A lipocentric view of peptide-induced pores. Eur. Biophys. J. 40: 399–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kooijman E. E., Chupin V., de Kruijff B., Burger K. N. 2003. Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic. 4: 162–174 [DOI] [PubMed] [Google Scholar]
- 53.van den Brink-van der Laan E., Killian J. A., de Kruijff B. 2004. Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim. Biophys. Acta. 1666: 275–288 [DOI] [PubMed] [Google Scholar]
- 54.Terrones O., Antonsson B., Yamaguchi H., Wang H. G., Liu J., Lee R. M., Herrmann A., Basanez G. 2004. Lipidic pore formation by the concerted action of proapoptotic BAX and tBID. J. Biol. Chem. 279: 30081–30091 [DOI] [PubMed] [Google Scholar]
- 55.Qian S., Wang W., Yang L., Huang H. W. 2008. Structure of transmembrane pore induced by Bax-derived peptide: evidence for lipidic pores. Proc. Natl. Acad. Sci. USA. 105: 17379–17383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schafer B., Quispe J., Choudhary V., Chipuk J. E., Ajero T. G., Du H., Schneiter R., Kuwana T. 2009. Mitochondrial outer membrane proteins assist Bid in Bax-mediated lipidic pore formation. Mol. Biol. Cell. 20: 2276–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kudla G., Montessuit S., Eskes R., Berrier C., Martinou J. C., Ghazi A., Antonsson B. 2000. The destabilization of lipid membranes induced by the C-terminal fragment of caspase 8-cleaved bid is inhibited by the N-terminal fragment. J. Biol. Chem. 275: 22713–22718 [DOI] [PubMed] [Google Scholar]
- 58.Chou J. J., Li H., Salvesen G. S., Yuan J., Wagner G. 1999. Solution structure of BID, an intracellular amplifier of apoptotic signaling. Cell. 96: 615–624 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.