Abstract
HDL particles may enter atherosclerotic lesions having an acidic intimal fluid. Therefore, we investigated whether acidic pH would affect their structural and functional properties. For this purpose, HDL2 and HDL3 subfractions were incubated for various periods of time at different pH values ranging from 5.5 to 7.5, after which their protein and lipid compositions, size, structure, and cholesterol efflux capacity were analyzed. Incubation of either subfraction at acidic pH induced unfolding of apolipoproteins, which was followed by release of lipid-poor apoA-I and ensuing fusion of the HDL particles. The acidic pH-modified HDL particles exhibited an enhanced ability to promote cholesterol efflux from cholesterol-laden primary human macrophages. Importantly, treatment of the acidic pH-modified HDL with the mast cell-derived protease chymase completely depleted the newly generated lipid-poor apoA-I, and prevented the acidic pH-dependent increase in cholesterol efflux. The above-found pH-dependent structural and functional changes were stronger in HDL3 than in HDL2. Spontaneous acidic pH-induced remodeling of mature spherical HDL particles increases HDL-induced cholesterol efflux from macrophage foam cells, and therefore may have atheroprotective effects.
Keywords: fusion, atherosclerosis, chymase, high density lipoprotein
HDL are composed of varying subpopulations of particles that are highly heterogeneous in size, physicochemical properties, metabolism, and in their anti-atherogenic functions (1, 2). The particles undergo dynamic remodeling in circulation by several plasma factors, such as LCAT (3), cholesteryl ester transfer protein (CETP) (4), phospholipid transfer protein (PLTP) (5), hepatic lipase, and endothelial lipase (6). Such physiological HDL remodeling can lead to destabilization of HDL structure, and in some cases also to the release of lipid-free/lipid-poor apoA-I from the particles and to particle fusion (7). Similarly, serum opacity factor (8) has been found to destabilize HDL particles with ensuing fusogenic formation of larger particles and concomitant release of lipid-free/lipid-poor apoA-I. Thus, generation of larger HDL particles coupled with the release of apoA-I appears to be a seminal characteristic of circulating HDL particles when exposed to various factors involved in their physiological remodeling.
The ability of HDL to remove excess cholesterol from macrophages in the arterial intima, i.e., their ability to initiate the macrophage-specific reverse cholesterol transport pathway, is a key anti-atherogenic action of HDL. The various subclasses of HDL particles have distinct abilities to stimulate cellular cholesterol efflux, the mature spherical α-migrating HDL particles preferring ABCG1-mediated and the lipid-poor preβ-migrating apoA-I preferring ABCA1-mediated cholesterol efflux (9). Importantly, the ability of HDL to remove cholesterol from macrophage foam cells may be compromised when HDL particles are modified by enzymatic or nonenzymatic processes such as oxidation, glycosylation, nitrosylation/chlorination, or by proteolysis (10).
Acidic pH of the extracellular fluid is a common characteristic of inflammatory tissue sites (11, 12) and, importantly, it has also been observed in human atherosclerotic lesions (13). Accordingly, circulating HDL particles entering such lesions may be exposed to acidic pH. On the basis of the fact that HDL particles are sensitive to various physiological perturbations, we hypothesized that the structural and functional properties of the particles are also sensitive to changes in pH of the medium in which they are suspended. Our data show that at acidic pH, HDL particles undergo spontaneous remodeling, with formation of lipid-poor apoA-I displaying preβ mobility and fusion of the α-migrating HDL particles, and that the generated mixture of preβ-HDL and fused α-HDL possesses an enhanced ability to promote cholesterol efflux from cultured human macrophage foam cells. The enhanced efflux-inducing capacity was shown to be primarily due to the de novo generation of the preβ-HDL particles.
MATERIALS AND METHODS
Isolation of human lipoproteins
LDL (d = 1.019–1.050 g/ml), HDL2 (d = 1.063–1.125 g/ml), and HDL3 (d = 1.125–1.210 g/ml) were prepared from freshly isolated plasma of healthy volunteers obtained from the Finnish Red Cross by sequential flotation ultracentrifugation using KBr for density adjustment (14). Ultracentrifugation was carried out in a Beckman OptimaTM TLX system table top ultracentrifuge using a Beckman fixed-angle rotor (TLA-100.3) at 541,000 g. This method subfractionates HDL from plasma in only 6 h, and it yielded HDL preparations that were almost totally devoid (<0.7% of total HDL) of the minor preβ-migrating component composed of lipid-poor apoA-I found to be present in the HDL3 subfraction isolated by the traditional lengthy ultracentrifugation protocol (typically 5% to 8% of total HDL) (15). The HDL2 and HDL3 subfractions were washed by reflotation for 2 h at densities of 1.125 and 1.21 g/ml, respectively. Lipoprotein purity was assessed by size-exclusion chromatography (SEC), agarose gel electrophoresis, and nondenaturing gradient polyacrylamide gel electrophoresis (NDGGE). No detectable activities of LCAT, PLTP, or CETP were present in the HDL2 or HDL3 preparations, when determined using assays described previously in detail (16, 17). Lipoprotein stock solutions were dialyzed against PBS, pH 7.4, containing 1 mM EDTA, filtered, purged with nitrogen, and then stored at 4°C, and used within 2 weeks, during which no changes in protein or lipid composition or particle size were observed. The amounts of lipoproteins are expressed in terms of their total protein concentrations, which were determined by bicinchoninic acid protein assay kit (Pierce; Rockford, IL) using BSA as the standard. LDL was acetylated by repeated additions of acetic anhydride (18), and acetyl-LDL was radiolabeled by treatment with [3H]cholesteryl linoleate ([1,2-3H]cholesteryl linoleate, Amersham Pharmacia) (19). The specific activities of the [3H]CE acetyl-LDL preparations ranged from 50 to 100 dpm/ng protein. Lipid-free apoA-I was purified according to a previously published procedure (20). The results are presented for lipoprotein preparations derived from a single donor, and similar data were also obtained for lipoprotein preparations derived from three other donors.
SEC of acidic pH-treated HDL particles
HDL2 or HDL3 (0.1–2 mg protein/ml) was incubated in either 20 mM MES (pH 5.5, 5.75, 6.0, 6.25, or 6.5), 20 mM PIPES (pH 6.5 or 7.0), or 20 mM Tris (pH 7.0 or 7.5) buffer containing 150 mM NaCl, 2 mM MgCl2, and 2 mM CaCl2 for the indicated times. All incubations were carried out at 37°C. After incubation, the samples were centrifuged at 10,000 g for 10 min, and the particle sizes of the HDL-containing supernatants were analyzed using a high-resolution SEC Superose HR6 column connected to the Amersham-Pharmacia (GE Healthcare) AKTA chromatography system. Typically, a 50 µl aliquot was injected into the column and eluted with PBS buffer at a flow rate of 0.5 ml/min, and fractions (0.5 ml) were collected for protein and lipid analyses. Kinetics of the elution profile changes in SEC were analyzed using the Unicorn 5.2 software. The particle size was assessed by a calibration curve (R2 = 0.98) using a gel filtration calibration kit (GE Healthcare).
Analysis of HDL composition
Proteins were resolved in 12.5% SDS-PAGE under reducing conditions, transferred to nitrocellulose, and immunoblotted either with anti-human apoA-I polyclonal antibody (21) or with anti-human apoA-II monoclonal antibody (22). The amounts of total cholesterol were measured using the Amplex Red cholesterol assay kit (Invitrogen) according to the manufacturer's instructions. Phospholipids were measured by a fluorometric assay (23) using Amplex Red reagent to enhance its sensitivity. Essentially, in this method, choline-containing phospholipids (PCs) are hydrolyzed by phospholipase D (Sigma P0065) into free choline, which is further oxidized with choline oxidase to yield H2O2. The formed H2O2 then reacts stoichiometrically with the Amplex Red reagent (10-acetyl-3,7-dihydroxyphenoxazine) in the presence of HRP, and ultimately forms the fluorescent compound resorufin. Finally, the fluorescence was measured with a VICTOR3 multilabel plate reader (Perkin Elmer; Finland) using an excitation wavelength of 544 nm and an emission wavelength of 595 nm.
Analysis of HDL particle size and quantitation of preβ-HDL
HDL samples (8 µg as total protein) were loaded onto self-prepared 4–30% polyacrylamide gradient gels (8.0 cm × 8.0 cm) and run at 125 V under nondenaturing conditions overnight at 4°C to reach equilibrium and then stained with Coomassie blue. HDL particle size was determined based on the use of high-molecular-weight electrophoresis calibration standards as molecular size markers. Particle size was also assessed by negative staining electron microscopy (24). For this purpose, HDL samples (3 μl from HDL stock, 1 mg/ml) were dried on carbon-coated grids, after which 3 μl of 1% potassium phosphotungstate, pH 7.4, was added and also dried on the grids. The samples were viewed and photographed in a JEOL 1200EX electron microscope at the Institute for Biotechnology, Department of Electron Microscopy, Helsinki, Finland. For the determination of the size distribution of the lipoprotein particles, the diameters of 100 randomly selected particles were measured from the electron micrographs.
Agarose gel electrophoresis (0.6% gel) was carried out using the Paragon electrophoresis system according to the instructions of the manufacturer (Beckman Coulter, Inc.). Proteins were transferred from the agarose gel to the polyvinylidene difluoride (PVDF) membrane by pressure blotting. ApoA-I was identified using a monoclonal anti-human apoA-I antibody (Abcam; UK), followed by an HRP-conjugated anti-rabbit IgG as the secondary antibody (Dako; Denmark). To further examine the effect of pH on the formation of preβ-HDL, HDL2 or HDL3 (0.1–1 mg/ml) was incubated in either 20 mM MES (pH 5.5–6.5), 20 mM PIPES (pH 6.5 or 7.0), or 20 mM Tris (pH 7.5) buffer containing 150 mM NaCl, 2 mM MgCl2, and 2 mM CaCl2 at 37°C for different periods of time (0–48 h). The preβ-HDL and α-HDL contents were quantified by two-dimensional crossed immunoelectrophoresis (25), and the amount of preβ-HDL was expressed as a percentage of the sum of the preβ- and α-mobile areas.
Circular dichroism spectroscopy
Samples of HDL3 or LDL (50 μg/ml) were analyzed by circular dichroism (CD) before and after their incubation at pH 7.5 or 5.5. The treated samples were placed in 0.1 cm quartz cuvettes, and CD spectra were recorded on a JASCO J-715 spectropolarimeter (Japan Spectroscopic Co.; Tokyo, Japan) in the region of 190–250 nm with a step size of 0.5 nm, scan speed of 50 nm per min, band width of 1 nm, and 1 s response. The cell-holder compartment was thermostatically maintained at 37 ± 0.1°C. For each sample, five spectra were averaged, and blank measurements were subtracted. The kinetics of apolipoprotein unfolding were monitored by recording CD signals at 222 nm. Molar ellipticity ([Θ]) was calculated from the equation: [Θ] = (MRW)* Θ/10lc, where Θ is a measured ellipticity in degrees, l is the cuvette path length (0.1 cm), c is the protein concentration (g/ml), and the mean residue weight (MRW) is obtained from the molecular weight and the number of amino acids. The α-helix contents were calculated from the equation using [Θ] at 222 nm: percent α-helix = [(-[Θ] 222+ 3,000)/(36,000 + 3,000)] * 100 (26, 27).
Proteolysis of acidic pH-modified HDL by chymase
HDL2 or HDL3 (1 mg/ml) was incubated in either 20 mM MES (pH 5.5) or 20 mM Tris (pH 7.5) buffer containing 150 mM NaCl, 2 mM MgCl2, and 2 mM CaCl2 at 37°C. After 24 h incubation, samples were dialyzed extensively against 5 mM Tris (pH 7.4) containing 150 mM NaCl, 1 mM EDTA. Aliquots of the dialyzed HDL fractions (1 mg/ml) were then incubated in the absence or presence of 200 BTEE units/ml of recombinant human chymase (kindly provided by Teijin Ltd. Hino, Tokyo, Japan). The enzymatic activity of chymase was measured and 1 U of chymase activity was defined, as described by Woodbury et al., (28). After incubation, the samples were kept on ice, and 50 µl aliquots were analyzed by SEC. Aliquots (200 µl) were withdrawn, and soybean trypsin inhibitor (SBTI; final concentration: 100 µg/ml) was added to the samples to fully inhibit chymase activity (15). Aliquots of the various chymase-treated preparations and their corresponding nontreated HDL preparations were then added to macrophage foam cell cultures, and their abilities to promote cholesterol efflux were examined.
Primary monocyte-macrophage cultures
Human monocytes were isolated from buffy coats (Finnish Red Cross Blood Transfusion Center; Helsinki, Finland) by centrifugation in Ficoll-Paque gradient as described (29). Washed cells were suspended in DMEM supplemented with 100 U/ml penicillin and 100 µg/ml streptomycin, counted, and seeded on 24 well-plates (1.5 million cells per well). After 1 h, nonadherent cells were removed and the medium was replaced with macrophage-serum-free medium (Gibco) supplemented with 1% penicillin-streptomycin and 10 ng/ml of granulocyte macrophage colony-stimulating factor (Biosite; San Diego, CA). The medium was then changed every 2 to 3 days throughout the culture period.
Cholesterol efflux from macrophage foam cells
The monocyte-derived macrophages were incubated in DMEM (pH 7.4) containing 25 μg/ml of [3H]CE-acetyl-LDL for 24 h to induce the formation of foam cells. To measure cholesterol efflux, macrophages were washed with PBS, and fresh media containing the various cholesterol acceptors (25 µg protein/ml) were added. After incubation for 16 h, the media were collected centrifuged at 300 g for 10 min to remove cellular debris, and the radioactivity in the supernatants was determined by liquid scintillation counting. Cells were solubilized with 0.2 M NaOH, and analyzed for radioactivity. Cholesterol efflux was expressed as the percentage radioactivity in the medium relative to the sum of total radioactivity present in the medium and the cells. Cholesterol efflux to the incubation medium in the absence of any added cholesterol acceptors was considered as basal efflux, and was subtracted from the efflux values obtained in the presence of acceptors.
Statistical analysis
Results are reported as mean ± SD. Statistical significance (P < 0.05) was determined by two-tailed Student's t-test.
RESULTS
Effect of pH on HDL particle size
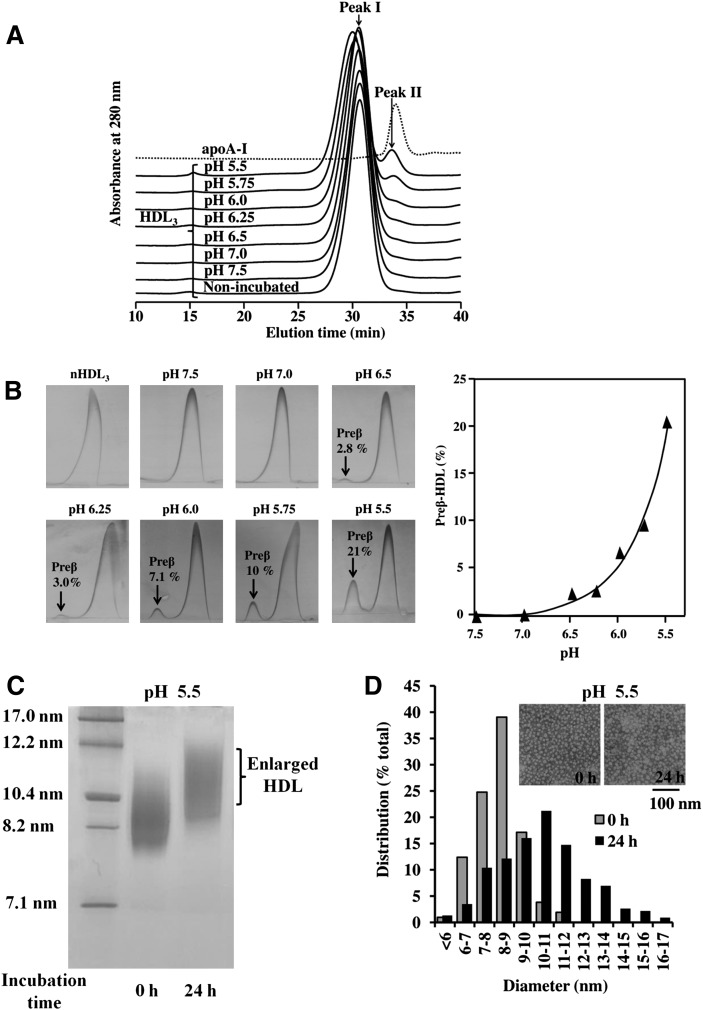
HDL3 and HDL2 were incubated at different pH values (5.5-7.5) for 24 h, after which aliquots of the incubation media were analyzed by SEC. Figure 1A shows that by using this methodology, non-incubated HDL3 (nHDL3) eluted as a single peak, and that the same applied to HDL3 preparations incubated for 24 h at pH 7.5, 7.0, or 6.5. In contrast, incubation of HDL3 at pH 6.0 and below led to substantial changes in the elution profile of HDL3, the major peak (I) having a shorter elution time and a minor peak (II) appearing only slightly earlier than did the isolated and purified lipid-free apoA-I, which served as a standard. Thus, the SEC analysis revealed that when compared with the original Peak I (nHDL3), the particles in the major peak had an increased size, whereas the particles in the newly appeared Peak II were much smaller. Analysis of pH-treated HDL3 using the highly sensitive two-dimensional crossed-immunoelectrophoresis revealed that incubation at pH 6.5 and below resulted in the formation of preβ-migrating particles and that the lower the pH, the higher was the amount of preβ-HDL formed (Fig. 1B). To estimate the sizes of the HDL3 particles during size conversion, the fractions of the major peak were pooled and analyzed by 4–30% NDGGE. This analysis confirmed the presence of enlarged HDL particles after incubation at pH 5.5 (Fig. 1C). Compared with HDL3, similar but less-remarkable effects were observed when HDL2 was incubated at pH 5.5 (see supplementary Fig. I). In contrast, LDL, which lacks apoA-I but contains a nonexchangeable and nonreleasable apolipoprotein, apoB-100, was fully resistant to the above-shown acidic pH-induced effects on HDL (see supplementary Fig. IIA).
Fig. 1.
Effect of pH on HDL3 size and preβ-HDL formation. A: SEC Profile of HDL3 at different pHs. HDL3 (1 mg/ml) was incubated in either 20 mM MES (pH 5.5–6.5), 20 mM PIPES (pH 6.5 or 7.0), or 20 mM Tris (pH 7.0 or 7.5) for 24 h, after which HDL samples (1 mg/ml) or purified apoA-I (0.5 mg/ml) were centrifuged at 10,000 g for 10 min. Aliquots of the supernatants (50 µl for HDL3 and 80 µl for apoA-I) were injected into a Superose HR6 column and eluted with PBS buffer (pH 7.4). Main fractions are labeled as Peaks I and II. nHDL3 = nonincubated HDL3. Each profile is representative of at least two independent experiments. B: Detection of preβ-HDL by two-dimensional crossed immunoelectrophoresis. HDL3 (1 mg/ml) was incubated at different pH values (5.5–7.5) for 24 h, after which α-HDL and preβ-HDL contents were measured using two-dimensional crossed immunoelectrophoresis. The amounts of preβ-HDL are expressed as a percentage of the sum of the preβ- and α-mobile areas. The left panel shows two-dimensional crossed immunoelectrophoresis, and the right panel shows preβ-HDL formation plotted as a function of pH. C: NDGGE analysis. Fractions in Peak I of HDL3 at 0 h (nHDL3) and 24 h at pH 5.5 were collected, and aliquots (8 µg) were analyzed by 4–30% NDGGE. D: Size distribution and electron microscopy of HDL3 at acidic pH. HDL3 (1 mg/ml) was incubated at pH 5.5 for 24 h, after which samples were negatively stained and photographed in the electron microscope as described in Materials and Methods. Data are representative of two independent experiments.
We next used negative staining electron microscopy to visualize the particle morphology both before and after incubation of HDL3 at acidic pH. Before the incubation, HDL3 appeared as spherical particles with a mean diameter of 8.3 ± 1.2 nm (range 6–11 nm) (Fig. 1D). Incubation of HDL3 at pH 5.5 caused a significant change in the particle size distribution (mean diameter 10.8 ± 2.3 nm; range 7–16 nm), and revealed a remarkable increase in the population of larger particles. Indeed, we found that 60% of the acidic pH-treated particles were within the size range of 9–13 nm. We also observed that large particles with diameters of 12–16 nm, not present in the nHDL3, appeared. Taken together, these results show that upon exposure to pH 5.5, of the ultracentrifugally isolated HDL3 and HDL2, the HDL3 particles in particular undergo remodeling, which consists of two stages: release of smaller apoA-I-containing particles with preβ-mobility and formation of enlarged α-migrating HDL particles.
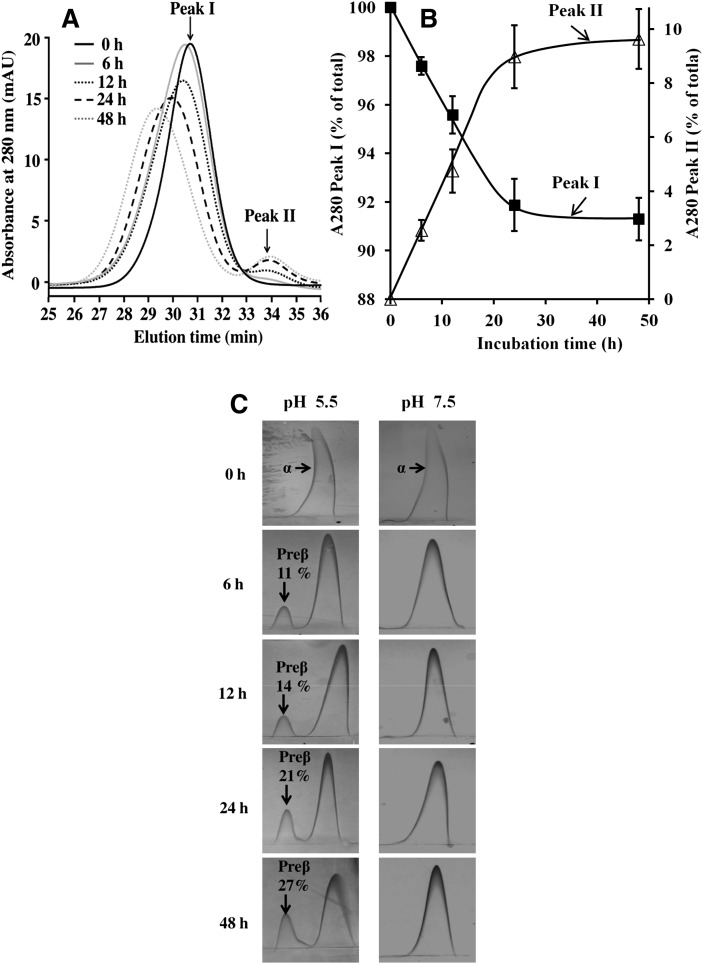
Acidic pH-induced HDL remodeling and preβ-HDL generation as a function of time
Next, we investigated the rate of the formation of enlarged HDL particles and the detachment of apoA-I. For this purpose, HDL3 was incubated at pH 5.5 and aliquots of supernatants at different time points up to 48 h were analyzed by SEC. The formation of enlarged HDL particles (Peak I) and the release of apoA-I with preβ-mobility (Peak II), were both time-dependent (Fig. 2A, B). To obtain more-precise information about the time-dependent effect of acidic pH on HDL size, the average sizes of particles in Peaks I and II were calculated using gel filtration calibration of proteins with known Stokes diameters. Native HDL3 particles were 8.8 ± 0.1 nm in diameter, and incubation at pH 5.5 progressively increased their size (Table 1). The size of the particles in Peak II remained unchanged (7.5 ± 0.2 nm), a finding consistent with previous reports on the size of preβ1-HDL (30, 31). To quantitatively determine the generation of preβ-HDL, the HDL particles were analyzed by two-dimensional crossed-immunoelectrophoresis. Incubation of HDL3 at pH 5.5 resulted in the formation of preβ-HDL in a time-dependent manner (Fig. 2C, left). After 6 h incubation at pH 5.5, the fraction of the newly generated preβ-HDL fraction amounted to 11% of the total HDL, and it increased to 27% after 48 h incubation. In contrast, at neutral pH 7.5, even after 48 h incubation, the majority of HDL3 migrated in the α-position and only a negligible amount (always <1%) of preβ-HDL was detected (Fig. 2C; right). Similar, but less-profound, data were also found for HDL2 (see supplementary Fig. IIIA, B), again revealing that HDL3 is more susceptible to acidic pH remodeling than is HDL2.
Fig. 2.
Time-dependent effect of acidic pH on HDL3. A: SEC profile of HDL3 at different incubation times. HDL3 (1 mg/ml) was incubated in 20 mM MES (pH 5.5) for the indicated times. Aliquots of the supernatants (50 µl) were analyzed by SEC after centrifugation, as described in Fig. 1A. Each profile is representative of three to six independent experiments. B: Kinetics of acidic pH-induced remodeling according to Peak I disappearance and Peak II appearance in panel A. Peak areas were calculated by integration of the 280 nm absorbance using the Unicorn 5.2 software. The data (mean ± SD) are expressed as a percentage of the sum of the 280 nm absorbance of Peak I and Peak II. C: Time-dependent effect of acidic pH on preβ-HDL level. HDL3 (1 mg/ml) was incubated for various periods of time at pH 7.5 or pH 5.5, after which α-HDL and preβ-HDL contents were measured using two-dimensional crossed immunoelectrophoresis. The amounts of preβ-HDL are expressed as a percentage of the sum of the preβ- and α-mobile areas.
TABLE 1.
Particle size of HDL3 at pH 5.5 as a function of time
HDL3 (1 mg/ml) was incubated in 20 mM MES (pH 5.5) for the indicated times. The samples were analyzed by SEC, and particle sizes of Peaks I and II were calculated using a gel filtration calibration kit.
P < 0.05 versus 0 h (nHDL3).
Distribution of proteins and lipids in acidic pH-modified HDL
We next examined whether incubation of HDL at pH 5.5 would exert differential effects on HDL protein and lipid distribution. Before incubation, nHDL3 elution profiles for protein, total cholesterol, and phospholipids fully coincided, and incubation at acidic pH induced similar shifts in all the elution profiles of the main peak (data not shown). However, the small HDL particles (Peak II) appeared to contain only minor amounts of phospholipids and cholesterol. Detailed analysis indicated that of the total protein, 88 ± 4% and 12 ± 0.6% were in Peaks I and II, respectively, and that the corresponding values for total phospholipids were 94 ± 3% and 6.4 ± 0.3%, and for total cholesterol, 95 ± 3% and 5.4 ± 0.3%. Compared with nHDL3, the enlarged HDL3 particles (Peak I) contained less protein and slightly more lipids, whereas the preβ-HDL particles (Peak II) were rich in protein and had lower amounts of lipids (Table 2). Immunoblot analysis (data not shown) revealed the presence of both apoA-I and apoA-II in the enlarged HDL particles (Peak I), whereas only apoA-I was detected in the preβ-HDL particles (Peak II). Assuming that the average molecular mass of PC is 750 Da and that the arithmetical mean of the summated molecular mass of cholesteryl ester and of cholesterol is 550 Da, we could calculate that each apoA-I in the small HDL particles had bound, on average, 12 molecules of PC and 8 molecules of cholesterol. Thus, under acidic conditions, apoA-I was released as lipid-poor apoA-I- PC-cholesterol complexes.
TABLE 2.
Composition of nHDL3, larger HDL3 (Peak I), and small HDL3 (Peak II)
| Sample | Protein | Phospholipid | Total Cholesterol | Protein/Lipid Ratio |
| % | % | % | ||
| nHDL3 | 50.4 ± 0.8 | 31.5 ± 1.0 | 18.1 ± 0.8 | 1.0 |
| Peak I | 48.3 ± 1.2a | 33.1 ± 0.9a | 19.2 ± 0.5a | 0.9 |
| Peak II | 67.4 ± 2.0b | 22.1 ± 1.9b | 10.5 ± 0.7b | 2.1 |
HDL3 was incubated at pH 5.5 for 24 h, and the incubation mixtures were applied to a high-resolution SEC Superose HR6 column to obtain Peak I and Peak II. Then their protein, total phospholipids, and total cholesterol were determined as described in Materials and Methods.
P < 0.05 versus nHDL3.
P < 0.001 versus nHDL3.
HDL remodeling at acidic pH depends on particle concentration and is irreversible
Next, we examined the effect of HDL concentration on the size conversion of the particles observed at pH 5.5. As the concentration of HDL3 increased from 0.1 to 2 mg protein/ml, Peak I showed a gradual shift toward shorter elution times, with progressively increased release of lipid-poor apoA-I in peak II (see supplementary Fig. IVA), suggesting greater interactions between enlarged HDL particles at higher concentrations. Although Peak II was not detectable by SEC at concentrations below 0.25 mg/ml, two-dimensional crossed-immunoelectrophoresis analysis revealed substantial amounts of preβ-HDL formed (see supplementary Fig. IVB). Thus, our data indicated that acidic pH-induced HDL remodeling may occur even at the low concentration of HDL typically found in interstitial fluids (32).
To examine whether the process of acidic pH-induced HDL remodeling is reversible, HDL subfractions were incubated for 24 h at pH 5.5 to trigger particle remodeling, and then the incubation mixtures were neutralized by extensive dialysis against neutral buffer of pH 7.5. After an additional 24 h incubation at pH 7.5, the acidic pH-induced changes in SEC profiles of HDL3 and of HDL2 were still present, demonstrating the irreversible nature of HDL interconversion at acidic pH (data not shown).
Unfolding of HDL proteins is the initiating step, followed by apoA-I release with ensuing enlargement of the apoA-I-deficient HDL particles
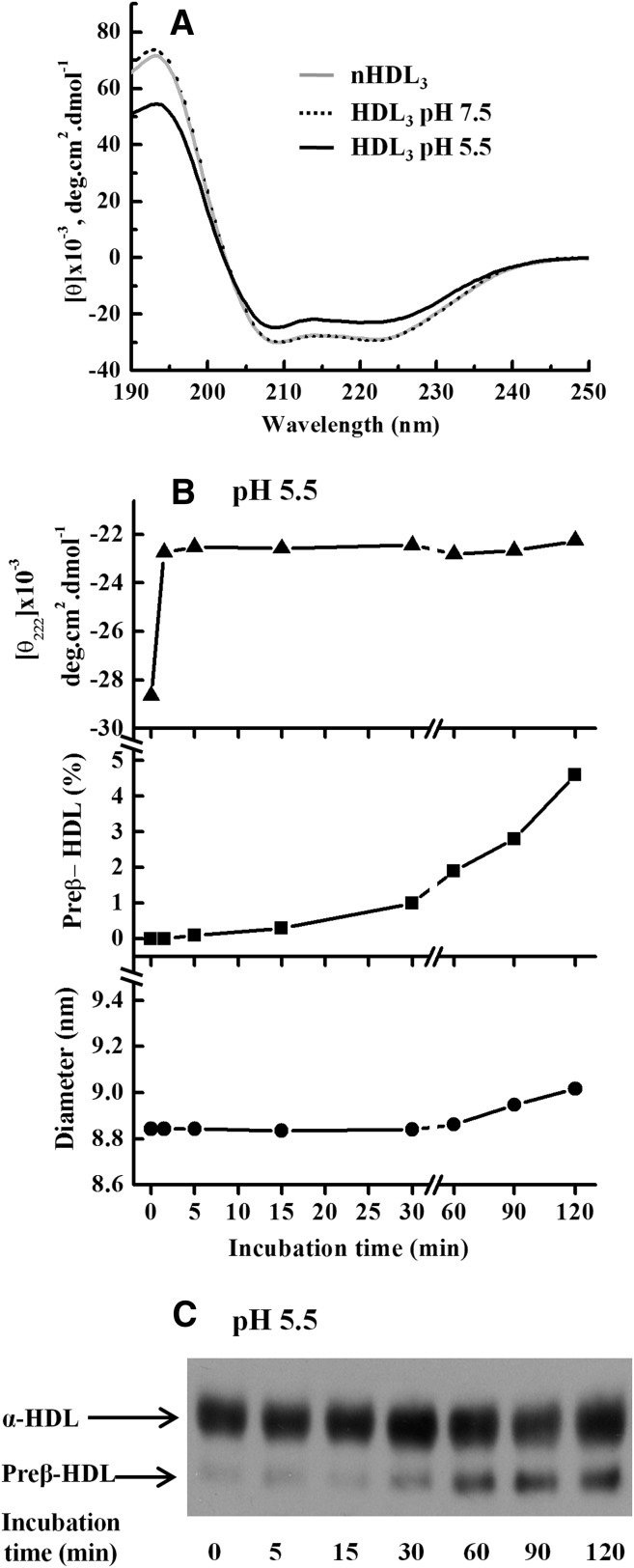
To gain insight into the mechanism by which acidic pH induces remodeling of HDL, we investigated the effect of pH on the conformational characteristics of HDL apolipoproteins. First, far-UV CD spectra were recorded for nHDL and for HDL after incubation at pH 7.5 or pH 5.5 for 24 h. No significant changes in CD spectra within this time period were observed at pH 7.5, when compared with spectra obtained for nHDL3. In contrast, CD spectra of HDL3 at pH 5.5 showed reduced mean residue ellipticity signal intensity (Fig. 3A), indicating unfolding of HDL apolipoproteins. To better discern the process of acidic pH-induced HDL remodeling, we monitored the time courses of HDL unfolding, preβ-HDL release, and particle size at early time points. We found that at pH 5.5, the unfolding of apolipoproteins on HDL3 occurred immediately after acidification of the medium, and that unfolding was already complete when the first measurement was performed, i.e., not later than 1 min after the acidification (Fig. 3B, top). This finding well accords with previous reports on the chemical and thermal denaturation of HDL (33–35). In contrast, no changes in CD spectra were found when LDL was incubated at pH 5.5 (see supplementary Fig. IIB), indicating that the acidic pH-induced unfolding does not apply to apoB-100. When aliquots of the same HDL3 samples were analyzed for the presence of preβ-HDL and size changes, we found that preβ-HDL particles appeared in the HDL3 fraction after incubation at pH 5.5 for 30 min (Fig. 3B, middle; Fig. 3C), i.e., before any significant changes in HDL3 size occurred, which, again, were observed only after 60 min of incubation (Fig. 3B, bottom). Thus, the data indicated that after acidification, unfolding of HDL apolipoproteins is the first step, followed by apoA-I release and subsequent enlargement of the HDL particles.
Fig. 3.
Kinetics of acidic pH-induced HDL3 remodeling. A: Far-UV CD analysis of HDL3. HDL3 (1 mg/ml) was incubated in 20 mM MES (pH 5.5) or 20 mM Tris (pH 7.5) for 24 h. Aliquots of treated HDL3 and nHDL3 (non-incubated HDL3) were diluted to 50 µg/ml for CD measurement as described in Materials and Methods. B: Sequence of HDL3 remodeling at acidic pH. HDL3 (1 mg/ml) was incubated in 20 mM MES (pH 5.5) for the indicated times. Aliquots of samples were used for CD measurement at 222 nm (top), for preβ-quantitation by two-dimensional crossed immuneoelectrophoresis (middle), and for size by SEC (bottom). C: Detection of preβ-HDL by agarose gel electrophoresis. Aliquots of samples (5 µg) in panel B were also analyzed by agarose gel electrophoresis as described in Materials and Methods. Data are representative of at least two independent experiments.
HDL remodeling at acidic pH enhances their ability to facilitate cholesterol efflux from cultured macrophage foam cells
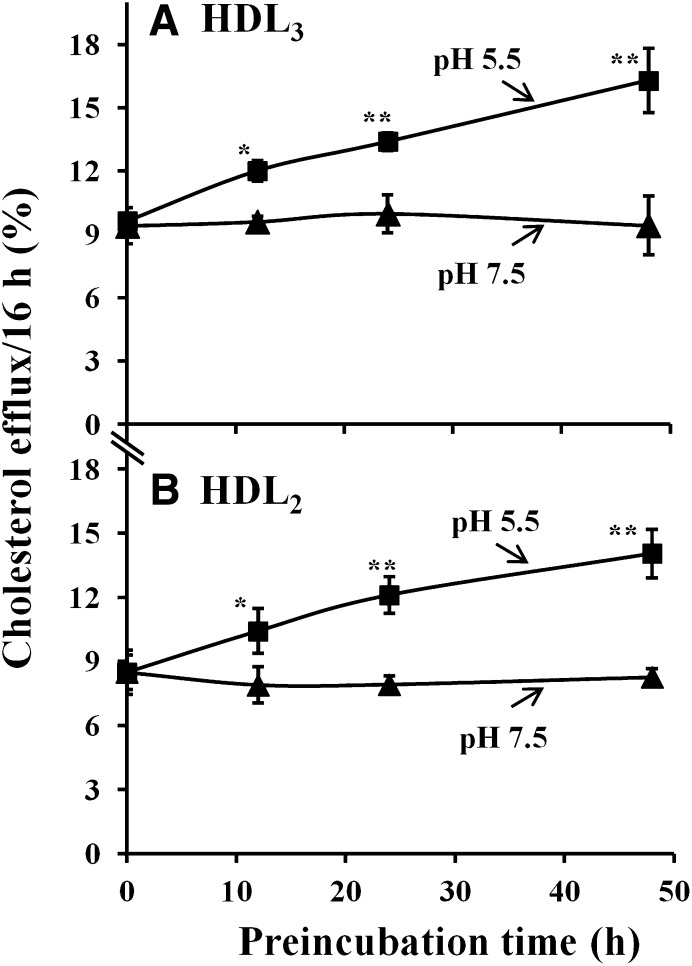
As indicated in the above experiments, incubation of HDL at pH 5.5 resulted in changes of their structural properties. This prompted us to study the ability of acidic pH-modified HDL to induce cholesterol efflux from cultured macrophage foam cells. HDL3 and HDL2 were preincubated at pH 5.5 or pH 7.5 for different periods of time (0–48 h) and were then used as acceptors for cholesterol efflux by incubating them for 16 h at pH 7.4 with the cholesterol-loaded cells. Interestingly, preincubation of HDL3 or HDL2 (Fig. 4A, B) at pH 5.5 increased their capacity to induce cholesterol efflux: the longer the preincubation time of HDL particles at acidic pH, the higher was the rate of cholesterol efflux induced by the acidic pH-treated HDL. In sharp contrast, preincubation of HDL3 or HDL2 at pH 7.5 did not change their ability to induce cholesterol efflux, when compared with nHDL particles (0 h preincubation). Thus, our data demonstrated that acidic pH induces conversion of HDL to particle subpopulations with higher capacity to induce macrophage cholesterol efflux.
Fig. 4.
Time-dependent effect of acidic pH on HDL cholesterol efflux from cultured macrophage foam cells. A: HDL3 or B: HDL2 (1 mg/ml) was preincubated in 20 mM MES (pH 5.5) or 20 mM Tris (pH 7.5) for various times (0–48 h). Aliquots (final concentration: 25 µg/ml) were added to [3H]cholesterol-loaded macrophage foam cells. After incubation for 16 h with the different cholesterol acceptors, the media were collected and centrifuged to remove cellular debris, and the radioactivity of each supernatant was determined by liquid scintillation counting. Cells were solubilized by 0.2 M NaOH, and radioactivity was determined in the cell lysates. Efflux of cholesterol was calculated as dpmmedium/(dpmcells+ dpmmedium)×100. Results are representative as mean ± SD of triplicate wells from a donor. *P < 0.05 versus HDL at 0 h; **P < 0.01 versus HDL at 0 h. Similar data were observed with macrophages derived from three different donors.
Chymase treatment abolishes enhanced cholesterol efflux induced by acidic pH-modified HDL
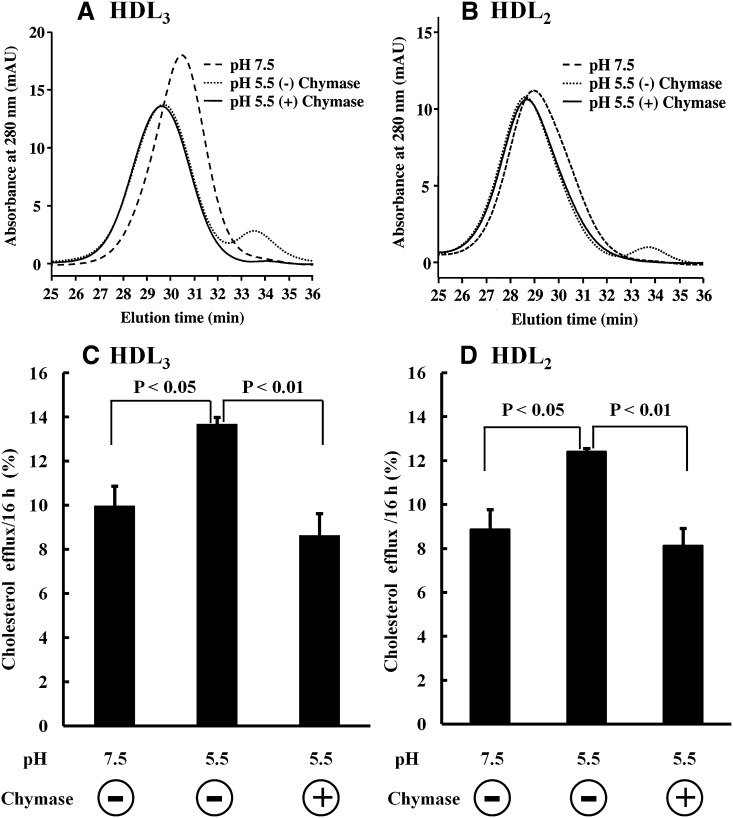
Previously, we have demonstrated that particles with preβ-mobility are responsible for the ABCA1-dependent cholesterol efflux from macrophage foam cells induced by human plasma and HDL3, and, moreover, that selective proteolytic depletion of the preβ-particles by mast cell chymase results in loss of the ABCA1-dependent cholesterol efflux induced by plasma or by HDL3 (15, 36, 37). These observations allowed us to test the relative contribution of the preβ-particles to the observed acidic pH-dependent enhancement of cholesterol efflux by HDL3 and HDL2. Acidic pH-modified HDL3 and HDL2 were neutralized by extensive dialysis at pH 7.4, and incubated in the absence or presence of chymase. SEC data showed that chymase treatment completely depleted the lipid-poor apoA-I particles (Peak II) in acidic pH-treated HDL3 (Fig. 5A) and HDL2 (Fig. 5B), and also abolished their ability to enhance cholesterol efflux (Fig. 5C, D). Taken together, at acidic pH, HDL particles undergo remodeling into larger particles with formation of lipid-poor apoA-I particles with preβ-mobility, the latter of which mainly covered the enhanced ability of the acid pH-modified HDL to induce cholesterol efflux.
Fig. 5.
Chymase effect on acidic pH-modified HDL. A, C: HDL3 or B, D: HDL2 (1 mg/ml) was incubated in either 20 mM MES (pH 5.5) or 20 mM Tris (pH 7.5). After 24 h incubation, samples were dialyzed extensively against 5 mM Tris (pH 7.4) containing 150 mM NaCl and 1 mM EDTA. Dialyzed HDL3 or HDL2, which were treated at pH 5.5, were divided into two identical aliquots and then incubated with and without chymase (200 BTEE U/ml). As a control, dialyzed HDL3 or HDL2, which were incubated at pH 7.5, was also incubated without chymase under the same conditions. After 6 h incubation, aliquots were taken and placed in ice, and aliquots (50 µl) were injected into the Superose HR6 gel filtration column for SEC analysis. Separately, aliquots (200 µl) were withdrawn, and SBTI (final concentration: 100 µg/ml) was added to all samples to fully inhibit chymase. Aliquots (final concentration: 25 µg/ml) were added to [3H]cholesterol-loaded macrophage foam cells cultured in medium containing SBTI (100 µg/ml). Cholesterol efflux was measured after 16 h incubation, as described in Fig. 4. Results are representative as mean ± SD of triplicate wells from a donor. Similar data were observed with macrophages derived from three different donors.
DISCUSSION
In the present study, we have characterized the effect of acidic pH on structural and functional properties of ultracentrifugally isolated human HDL3 and HDL2 particles. The data indicate that at acidic pH, HDL particles belonging to either subclass undergo remodeling into larger particles with formation of lipid-poor apoA-I with preβ-mobility. Importantly, when compared with native HDL, the ability of acidic pH-modified HDL to promote cholesterol efflux from cultured macrophage foam cells was enhanced. To the best of our knowledge, the present study is the first report revealing the effect of acidic pH on both the structural and functional properties of human HDL.
The critical importance of pH on HDL stability became evident as we observed that in contrast to the effect of acidic pH, at neutral pH, the ultracentrifugally isolated HDL subfractions were stable and totally resistant to changes in particle size. The size conversion taking place at acidic pH was confirmed by four different methods, i.e., SEC, NDGGE, agarose gel electrophoresis, and electron microscopy (Figs. 1 and 2). Considering the changes in the size of the acidic pH-modified HDL particles, we could calculate that the formation of larger particles (11–13 nm) had resulted from the fusion of two to three native HDL particles (7–9 nm), consistent with fusion of HDL particles induced by chemical and thermal denaturation (38, 39). Further analysis indicated that about 50% of the particles analyzed individually by negative staining electron microscopy had an increased diameter; i.e., they were fused particles containing the core lipid of two or more native HDL particles. The enlarged particles, like the native HDL particles, contained both apoA-I and apoA-II as their major proteins and they migrated in α-position, while the smaller newly formed preβ-migrating particles contained only apoA-I, revealing selective release of this apolipoprotein from particle surfaces. This finding is consistent with the known lower lipophilicity of apoA-I, when compared with that of apoA-II (40, 41).
The acidic pH-mediated HDL remodeling was a time- and concentration-dependent process (Fig. 2 and supplementary Figs. IIIA, IVA). The gradual increases in the quantities of both the lipid-poor apoA-I particles and the fused particles (Fig. 2A, supplementary Figs. IIIA, IVA) strongly suggest that these two processes are mechanistically connected. Previous reports have shown that exposure of HDL to chemical or thermal modification induces HDL apolipoprotein unfolding and subsequent particle fusion with concomitant apoA-I dissociation from the particles (35, 38, 42, 43). Also, chaotropic detergent perturbation has been shown to transfer apoA-I from HDL particles to the aqueous phase with concomitant formation of larger HDL-like particles (38). However, no further experimental attempts have been made to define whether apoA-I dissociation precedes fusion under chemically induced and thermally induced HDL remodeling. Importantly, our kinetic studies have provided the first experimental evidence demonstrating that during acidic pH-induced HDL remodeling, the initiating step is the unfolding of HDL apolipoproteins, followed by apoA-I release and subsequent fusion of the apoA-I-deficient HDL particles (Fig. 3). Given the dominance of apoA-I in mature HDL particles, the changes in the CD spectra at acidic pH are likely to be attributable mainly to conformational changes of apoA-I rather than of other apolipoproteins present in HDL particles (44). The indispensability of the presence of apoA-I for HDL particle stability well agrees with the concept that HDL fusion can be triggered by an imbalance between the hydrophobic core and the particle surface, particularly when the latter becomes protein-deficient (39). Actually, heat-induced and chemically induced HDL fusion (38, 42, 43) have been ascribed to dissociation of apoA-I. ApoA-I release has also been proposed to be responsible for PLTP-mediated HDL fusion (5, 45). In contrast, LDL particles, which solely contain a nonreleasable apoB-100 on the particle surface, were totally resistant to acidic pH-dependent particle modification (see supplementary Fig. II). However, like the HDL particles, fusion of LDL particles has been shown to require reduction in the apolipoprotein content of the particle surface; i.e., in the case of LDL, release of protease-generated apoB-100 peptide fragments from the surface of proteolyzed LDL particles is the fusion-triggering mechanism (46). Taken together, release of apoA-I with ensuing generation of larger fused HDL particles appears to be a common feature of HDL in response to its physicochemical perturbations and also during its physiological remodeling.
As indicated by SEC and two-dimensional crossed-immunoelectrophoresis (Fig. 2 and supplementary Fig. III), the remodeling of HDL3 is faster than that of HDL2 at acidic pH, an observation similar to that seen in the remodeling of HDL2 and HDL3 by guanidine denaturation (38) or by the serum opacity factor (8). The greater susceptibility of HDL3 to acidic pH remodeling suggests that these particles are less stable than HDL2 particles. The faster remodeling of HDL3 particles may simply be due to their smaller sizes, which make them less able to accommodate perturbations (38). Another susceptibility-increasing factor could be the phospholipid:protein ratio of HDL particles, which is related to particle size and is also known to affect particle stability (47, 48). Indeed, compositional analysis of the HDL subfractions used in this study indicated that the phospholipid:protein ratio in the HDL3 preparations was only about 67% of that in HDL2 preparations, i.e., 0.6 and 0.9, respectively. Also, PLA2 treatment of HDL, which results in decreased levels of particle phospholipids, does enhance the CETP-mediated dissociation of apoA-I (48), whereas enrichment of HDL with phospholipids using the detergent dilution method (49) prevents serum opacity factor-induced apoA-I release (8). Finally, several human plasma factors (LCAT, PLTP, and CETP) consume or transfer lipids and induce remodeling of HDL (3–5), which can lead to destabilization of the HDL structure and, in some cases, also to the release of lipid-free/lipid-poor apoA-I and to particle fusion (7). We were unable to detect any measurable activities of LCAT, PLTP, or CETP in the HDL preparations either before or after incubation at acidic pH, thus excluding any significant involvement of these physiological factors in the observed remodeling of HDLs at acidic pH.
Regarding the functionality of the remodeled HDL, we found that acidic pH produces a progressive increase in the ability of HDL to induce cholesterol efflux from macrophage foam cells (Fig. 4A, B). Similarly, the serum opacity factor-induced remodeling of HDL has been shown to exhibit an enhanced ability to promote cholesterol efflux (50). Using mast cell chymase, which selectively and completely depleted preβ-HDL in acidic pH-modified HDL (Fig. 5A, B), we found that the gained capacity of the acidic pH-modified HDL to induce cholesterol efflux was fully abrogated (Fig. 5C, D). Thus, the data demonstrate that the preβ-HDL particles generated upon incubation of HDL at acidic pH mainly accounted for the increased cholesterol efflux capacity.
As suggested by the present findings, in an acidic environment, HDL remodeling may lead to enhanced cholesterol efflux. Atherosclerotic lesions have been shown to exhibit heterogeneity of extracellular pH, and pH values below 6.0 have been detected in some lipid-rich areas by fluorescent microscopic imaging (13). In advanced atherosclerotic lesions, acidic environments develop in the deeper hypoxic areas as a result of local production and secretion of protons and other extracellular fluid-acidifying agents (51). Microelectrode measurements of pH in microenvironments of activated cultured macrophages have demonstrated pH values as low as 3.6, and a rapid decrease in pH values, ranging from 0.5 to 2.0 pH units per h after activation of the cells has been observed (52). Importantly, when activated by oxidized LDL, macrophages can strongly acidify their local environment (53). Similarly, upon contact with aggregated LDL, macrophages can generate acidic extracellular surface-connected domains in which pH values below 6.5, and even down to pH 5.0, are present (54).
Based on the above information, we can envision that in the hypoxic areas of atherosclerotic lesions, acidic pH-triggered release of apoA-I from HDL at pH values of less than 6.5 may occur in the immediate vicinity of activated macrophages (55). Actually, generation of lipid-poor apoA-I in the immediate vicinity of macrophages appears to be optimal because, for the cholesterol efflux-inducing effect of apoA-I, interaction between apoA-I and macrophage surface lipid transporters (notably ABCA1) is required. However, we need to be cautious when extrapolating the present in vitro findings to in vivo conditions. Atherosclerotic arterial intima possesses a variety of powerful proteolytic enzymes potentially capable of degrading preβ-HDL particles in the extracellular milieu of the tissue, at either neutral or acidic pH (10). Notably, activated macrophages secrete many powerful proteases with an acidic pH optimum, and it is known that lipid-poor apoA-I particles with preβ-mobility are extremely sensitive to proteolytic degradation (37, 56). Thus, the benefit of generating lipid-poor apoA-I particles in an acidic extracellular microenvironment generated by activated macrophages may easily get lost. Furthermore, the enhanced cholesterol efflux induced by acidic pH-modified HDL might be partially counteracted by reduction in the ability of human macrophages to release cholesterol via the ABCA1 pathway in acidic culture conditions (27). Accordingly, we do not know the ultimate balance, either increase or decrease of ABCA1-dependent cholesterol efflux in acidic regions present in advanced atherosclerotic lesions. Treating advanced acidic lesions with infusions containing mimics of apoA-I resistant to proteolysis (57) would be one strategy to support the cholesterol efflux-inducing potential of spontaneously generated apoA-I in such acidic environments.
Supplementary Material
Acknowledgments
The authors thank Maija Atuegwu, Hanna Lähteenmäki, Mari Jokinen, and Sari Nuutinen for their excellent technical assistance.
Footnotes
Abbreviations:
- CD
- circular dichroism
- CETP
- cholesteryl ester transfer protein
- NDGGE
- nondenaturing gradient polyacrylamide gel electrophoresis
- nHDL
- non-incubated HDL
- PC
- choline-containing phospholipids
- PLTP
- phospholipid transfer protein
- PVDF
- polyvinylidene difluoride
- SEC
- size-exclusion chromatography
Wihuri Research Institute is maintained by the Jenny and Antti Wihuri Foundation. This study was also supported by the Academy of Finland (M.J.) and the Sigrid Juselius Foundation (M.L-R).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of four figures.
REFERENCES
- 1.Navab M., Reddy S. T., Van Lenten B. J., Fogelman A. M. 2011. HDL and cardiovascular disease: atherogenic and atheroprotective mechanisms. Nat. Rev. Cardiol. 8: 222–232 [DOI] [PubMed] [Google Scholar]
- 2.Meurs I., Van Eck M., Van Berkel T. J. 2010. High-density lipoprotein: key molecule in cholesterol efflux and the prevention of atherosclerosis. Curr. Pharm. Des. 16: 1445–1467 [DOI] [PubMed] [Google Scholar]
- 3.Liang H. Q., Rye K. A., Barter P. J. 1996. Remodelling of reconstituted high density lipoproteins by lecithin:cholesterol acyltransferase. J. Lipid Res. 37: 1962–1970 [PubMed] [Google Scholar]
- 4.Rye K. A., Hime N. J., Barter P. J. 1997. Evidence that cholesteryl ester transfer protein-mediated reductions in reconstituted high density lipoprotein size involve particle fusion. J. Biol. Chem. 272: 3953–3960 [DOI] [PubMed] [Google Scholar]
- 5.Lusa S., Jauhiainen M., Metso J., Somerharju P., Ehnholm C. 1996. The mechanism of human plasma phospholipid transfer protein-induced enlargement of high-density lipoprotein particles: evidence for particle fusion. Biochem. J. 313: 275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lund-Katz S., Phillips M. C. 2010. High density lipoprotein structure-function and role in reverse cholesterol transport. Subcell. Biochem. 51: 183–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lie J., de Crom R., Jauhiainen M., van Gent T., van Haperen R., Scheek L., Jansen H., Ehnholm C., van Tol A. 2001. Evaluation of phospholipid transfer protein and cholesteryl ester transfer protein as contributors to the generation of pre beta-high-density lipoproteins. Biochem. J. 360: 379–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillard B. K., Courtney H. S., Massey J. B., Pownall H. J. 2007. Serum opacity factor unmasks human plasma high-density lipoprotein instability via selective delipidation and apolipoprotein A-I desorption. Biochemistry. 46: 12968–12978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asztalos B. F., de la Llera-Moya M., Dallal G. E., Horvath K. V., Schaefer E. J., Rothblat G. H. 2005. Differential effects of HDL subpopulations on cellular ABCA1- and SR-BI-mediated cholesterol efflux. J. Lipid Res. 46: 2246–2253 [DOI] [PubMed] [Google Scholar]
- 10.Lee-Rueckert M., Kovanen P. T. 2011. Extracellular modifications of HDL in vivo and the emerging concept of proteolytic inactivation of prebeta-HDL. Curr. Opin. Lipidol. 22: 394–402 [DOI] [PubMed] [Google Scholar]
- 11.Lardner A. 2001. The effects of extracellular pH on immune function. J. Leukoc. Biol. 69: 522–530 [PubMed] [Google Scholar]
- 12.Bellocq A., Suberville S., Philippe C., Bertrand F., Perez J., Fouqueray B., Cherqui G., Baud L. 1998. Low environmental pH is responsible for the induction of nitric-oxide synthase in macrophages. Evidence for involvement of nuclear factor-kappaB activation. J. Biol. Chem. 273: 5086–5092 [DOI] [PubMed] [Google Scholar]
- 13.Naghavi M., John R., Naguib S., Siadaty M. S., Grasu R., Kurian K. C., van Winkle W. B., Soller B., Litovsky S., Madjid M., et al. 2002. pH Heterogeneity of human and rabbit atherosclerotic plaques; a new insight into detection of vulnerable plaque. Atherosclerosis. 164: 27–35 [DOI] [PubMed] [Google Scholar]
- 14.McPherson P. A., Young I. S., McKibben B., McEneny J. 2007. High density lipoprotein subfractions: isolation, composition, and their duplicitous role in oxidation. J. Lipid Res. 48: 86–95 [DOI] [PubMed] [Google Scholar]
- 15.Lee M., Metso J., Jauhiainen M., Kovanen P. T. 2003. Degradation of phospholipid transfer protein (PLTP) and PLTP-generated pre-beta-high density lipoprotein by mast cell chymase impairs high affinity efflux of cholesterol from macrophage foam cells. J. Biol. Chem. 278: 13539–13545 [DOI] [PubMed] [Google Scholar]
- 16.Jauhiainen M., Dolphin P. J. 1986. Human plasma lecithin-cholesterol acyltransferase. An elucidation of the catalytic mechanism. J. Biol. Chem. 261: 7032–7043 [PubMed] [Google Scholar]
- 17.Jauhiainen M., Metso J., Pahlman R., Blomqvist S., van Tol A., Ehnholm C. 1993. Human plasma phospholipid transfer protein causes high density lipoprotein conversion. J. Biol. Chem. 268: 4032–4036 [PubMed] [Google Scholar]
- 18.Basu S. K., Goldstein J. L., Anderson G. W., Brown M. S. 1976. Degradation of cationized low density lipoprotein and regulation of cholesterol metabolism in homozygous familial hypercholesterolemia fibroblasts. Proc. Natl. Acad. Sci. USA. 73: 3178–3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown M. S., Dana S. E., Goldstein J. L. 1975. Receptor-dependent hydrolysis of cholesteryl esters contained in plasma low density lipoprotein. Proc. Natl. Acad. Sci. USA. 72: 2925–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pussinen P. J., Jauhiainen M., Metso J., Pyle L. E., Marcel Y. L., Fidge N. H., Ehnholm C. 1998. Binding of phospholipid transfer protein (PLTP) to apolipoproteins A-I and A-II: location of a PLTP binding domain in the amino terminal region of apoA-I. J. Lipid Res. 39: 152–161 [PubMed] [Google Scholar]
- 21.Siggins S., Jauhiainen M., Olkkonen V. M., Tenhunen J., Ehnholm C. 2003. PLTP secreted by HepG2 cells resembles the high-activity PLTP form in human plasma. J. Lipid Res. 44: 1698–1704 [DOI] [PubMed] [Google Scholar]
- 22.Pussinen P. J., Jauhiainen M., Ehnholm C. 1997. ApoA-II/apoA-I molar ratio in the HDL particle influences phospholipid transfer protein-mediated HDL interconversion. J. Lipid Res. 38: 12–21 [PubMed] [Google Scholar]
- 23.Nanjee M. N., Gebre A. K., Miller N. E. 1991. Enzymatic fluorometric procedure for phospholipid quantification with an automated microtiter plate fluorometer. Clin. Chem. 37: 868–874 [PubMed] [Google Scholar]
- 24.Oorni K., Hakala J. K., Annila A., Ala-Korpela M., Kovanen P. T. 1998. Sphingomyelinase induces aggregation and fusion, but phospholipase A2 only aggregation, of low density lipoprotein (LDL) particles. Two distinct mechanisms leading to increased binding strength of LDL to human aortic proteoglycans. J. Biol. Chem. 273: 29127–29134 [DOI] [PubMed] [Google Scholar]
- 25.van Haperen R., van Tol A., Vermeulen P., Jauhiainen M., van Gent T., van den Berg P., Ehnholm S., Grosveld F., van der Kamp A., de Crom R. 2000. Human plasma phospholipid transfer protein increases the antiatherogenic potential of high density lipoproteins in transgenic mice. Arterioscler. Thromb. Vasc. Biol. 20: 1082–1088 [DOI] [PubMed] [Google Scholar]
- 26.Tanaka M., Dhanasekaran P., Nguyen D., Ohta S., Lund-Katz S., Phillips M. C., Saito H. 2006. Contributions of the N- and C-terminal helical segments to the lipid-free structure and lipid interaction of apolipoprotein A-I. Biochemistry. 45: 10351–10358 [DOI] [PubMed] [Google Scholar]
- 27.Lee-Rueckert M., Lappalainen J., Leinonen H., Pihlajamaa T., Jauhiainen M., Kovanen P. T. 2010. Acidic extracellular environments strongly impair ABCA1-mediated cholesterol efflux from human macrophage foam cells. Arterioscler. Thromb. Vasc. Biol. 30: 1766–1772 [DOI] [PubMed] [Google Scholar]
- 28.Woodbury R. G., Everitt M. T., Neurath H. 1981. Mast cell proteases. Methods Enzymol. 80: 588–609 [DOI] [PubMed] [Google Scholar]
- 29.Saren P., Welgus H. G., Kovanen P. T. 1996. TNF-alpha and IL-1beta selectively induce expression of 92-kDa gelatinase by human macrophages. J. Immunol. 157: 4159–4165 [PubMed] [Google Scholar]
- 30.Duong P. T., Weibel G. L., Lund-Katz S., Rothblat G. H., Phillips M. C. 2008. Characterization and properties of pre beta-HDL particles formed by ABCA1-mediated cellular lipid efflux to apoA-I. J. Lipid Res. 49: 1006–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clay M. A., Barter P. J. 1996. Formation of new HDL particles from lipid-free apolipoprotein A-I. J. Lipid Res. 37: 1722–1732 [PubMed] [Google Scholar]
- 32.Sloop C. H., Dory L., Roheim P. S. 1987. Interstitial fluid lipoproteins. J. Lipid Res. 28: 225–237 [PubMed] [Google Scholar]
- 33.Reijngoud D. J., Phillips M. C. 1982. Mechanism of dissociation of human apolipoprotein A-I from complexes with dimyristoylphosphatidylcholine as studied by guanidine hydrochloride denaturation. Biochemistry. 21: 2969–2976 [DOI] [PubMed] [Google Scholar]
- 34.Reijngoud D. J., Phillips M. C. 1984. Mechanism of dissociation of human apolipoproteins A-I, A-11, and C from complexes with dimyristoylphosphatidylcholine as studied by thermal denaturation. Biochemistry. 23: 726–734 [DOI] [PubMed] [Google Scholar]
- 35.Jayaraman S., Gantz D. L., Gursky O. 2006. Effects of salt on the thermal stability of human plasma high-density lipoprotein. Biochemistry. 45: 4620–4628 [DOI] [PubMed] [Google Scholar]
- 36.Favari E., Lee M., Calabresi L., Franceschini G., Zimetti F., Bernini F., Kovanen P. T. 2004. Depletion of pre-beta-high density lipoprotein by human chymase impairs ATP-binding cassette transporter A1- but not scavenger receptor class B type I-mediated lipid efflux to high density lipoprotein. J. Biol. Chem. 279: 9930–9936 [DOI] [PubMed] [Google Scholar]
- 37.Lee M., von Eckardstein A., Lindstedt L., Assmann G., Kovanen P. T. 1999. Depletion of pre beta 1LpA1 and LpA4 particles by mast cell chymase reduces cholesterol efflux from macrophage foam cells induced by plasma. Arterioscler. Thromb. Vasc. Biol. 19: 1066–1074 [DOI] [PubMed] [Google Scholar]
- 38.Pownall H. J., Hosken B. D., Gillard B. K., Higgins C. L., Lin H. Y., Massey J. B. 2007. Speciation of human plasma high-density lipoprotein (HDL): HDL stability and apolipoprotein A-I partitioning. Biochemistry. 46: 7449–7459 [DOI] [PubMed] [Google Scholar]
- 39.Jayaraman S., Gantz D. L., Gursky O. 2004. Poly(ethylene glycol)-induced fusion and destabilization of human plasma high-density lipoproteins. Biochemistry. 43: 5520–5531 [DOI] [PubMed] [Google Scholar]
- 40.Lagocki P. A., Scanu A. M. 1980. In vitro modulation of the apolipoprotein composition of high density lipoprotein. Displacement of apolipoprotein A-I from high density lipoprotein by apolipoprotein A-II. J. Biol. Chem. 255: 3701–3706 [PubMed] [Google Scholar]
- 41.van Tornout P., Caster H., Lievens M. J., Rosseneu M., Assmann G. 1981. In vitro interaction of human HDL with human apolipoprotein A-II. Synthesis of apolipoprotein A-II-rich HDL. Biochim. Biophys. Acta. 663: 630–636 [DOI] [PubMed] [Google Scholar]
- 42.Mehta R., Gantz D. L., Gursky O. 2003. Human plasma high-density lipoproteins are stabilized by kinetic factors. J. Mol. Biol. 328: 183–192 [DOI] [PubMed] [Google Scholar]
- 43.Gao X., Jayaraman S., Gursky O. 2008. Mild oxidation promotes and advanced oxidation impairs remodeling of human high-density lipoprotein in vitro. J. Mol. Biol. 376: 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang R., Silva R. A., Jerome W. G., Kontush A., Chapman M. J., Curtiss L. K., Hodges T. J., Davidson W. S. 2011. Apolipoprotein A-I structural organization in high-density lipoproteins isolated from human plasma. Nat. Struct. Mol. Biol. 18: 416–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishida H. I., Klock D. G., Guo Z., Jakstys B. P., Nishida T. 1997. Phospholipid transfer protein can transform reconstituted discoidal HDL into vesicular structures. Biochim. Biophys. Acta. 1349: 222–232 [DOI] [PubMed] [Google Scholar]
- 46.Oorni K., Pentikainen M. O., Ala-Korpela M., Kovanen P. T. 2000. Aggregation, fusion, and vesicle formation of modified low density lipoprotein particles: molecular mechanisms and effects on matrix interactions. J. Lipid Res. 41: 1703–1714 [PubMed] [Google Scholar]
- 47.Tall A. R., Small D. M., Shipley G. G., Lees R. S. 1975. Apoprotein stability and lipid-protein interactions in human plasma high density lipoproteins. Proc. Natl. Acad. Sci. USA. 72: 4940–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rye K. A., Duong M. N. 2000. Influence of phospholipid depletion on the size, structure, and remodeling of reconstituted high density lipoproteins. J. Lipid Res. 41: 1640–1650 [PubMed] [Google Scholar]
- 49.Tchoua U., Gillard B. K., Pownall H. J. 2010. HDL superphospholipidation enhances key steps in reverse cholesterol transport. Atherosclerosis. 209: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tchoua U., Rosales C., Tang D., Gillard B. K., Vaughan A., Lin H. Y., Courtney H. S., Pownall H. J. 2010. Serum opacity factor enhances HDL-mediated cholesterol efflux, esterification and anti inflammatory effects. Lipids. 45: 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bjornheden T., Levin M., Evaldsson M., Wiklund O. 1999. Evidence of hypoxic areas within the arterial wall in vivo. Arterioscler. Thromb. Vasc. Biol. 19: 870–876 [DOI] [PubMed] [Google Scholar]
- 52.Silver I. A., Murrills R. J., Etherington D. J. 1988. Microelectrode studies on the acid microenvironment beneath adherent macrophages and osteoclasts. Exp. Cell Res. 175: 266–276 [DOI] [PubMed] [Google Scholar]
- 53.De Vries H. E., Ronken E., Reinders J. H., Buchner B., Van Berkel T. J., Kuiper J. 1998. Acute effects of oxidized low density lipoprotein on metabolic responses in macrophages. FASEB J. 12: 111–118 [DOI] [PubMed] [Google Scholar]
- 54.Haka A. S., Grosheva I., Chiang E., Buxbaum A. R., Baird B. A., Pierini L. M., Maxfield F. R. 2009. Macrophages create an acidic extracellular hydrolytic compartment to digest aggregated lipoproteins. Mol. Biol. Cell. 20: 4932–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Asplund A., Stillemark-Billton P., Larsson E., Rydberg E. K., Moses J., Hulten L. M., Fagerberg B., Camejo G., Bondjers G. 2010. Hypoxic regulation of secreted proteoglycans in macrophages. Glycobiology. 20: 33–40 [DOI] [PubMed] [Google Scholar]
- 56.Lindstedt L., Lee M., Oorni K., Bromme D., Kovanen P. T. 2003. Cathepsins F and S block HDL3-induced cholesterol efflux from macrophage foam cells. Biochem. Biophys. Res. Commun. 312: 1019–1024 [DOI] [PubMed] [Google Scholar]
- 57.Navab M., Anantharamaiah G. M., Hama S., Garber D. W., Chaddha M., Hough G., Lallone R., Fogelman A. M. 2002. Oral administration of an Apo A-I mimetic peptide synthesized from D-amino acids dramatically reduces atherosclerosis in mice independent of plasma cholesterol. Circulation. 105: 290–292 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.