Abstract
ABCG1 is an ABC half-transporter that exports cholesterol from cells to HDL. This study set out to investigate differences in posttranslational processing of two human ABCG1 protein isoforms, termed ABCG1(+12) and ABCG1(− 12), that differ by the presence or absence of a 12 amino acid peptide. ABCG1(+12) is expressed in human cells and tissues, but not in mice. We identified two protein kinase A (PKA) consensus sites in ABCG1(+12), absent from ABCG1(− 12). Inhibition of PKA with either of two structurally unrelated inhibitors resulted in a dose-dependent increase in cholesterol export from cells expressing ABCG1(+12), whereas ABCG1(− 12)-expressing cells were unaffected. This was associated with stabilization of the ABCG1(+12) protein, and ABCG1(+12)-S389 was necessary to mediate these effects. Mutation of this serine to aspartic acid, simulating a constitutively phosphorylated state, resulted in accelerated degradation of ABCG1(+12) and reduced cholesterol export. Engineering an equivalent PKA site into ABCG1(− 12) rendered this isoform responsive to PKA inhibition, confirming the relevance of this sequence. Together, these results demonstrate an additional level of complexity to the posttranslational control of this human ABCG1 isoform that is absent from ABCG1(− 12) and the murine ABCG1 homolog.
Keywords: ATP binding cassette transporters, cholesterol efflux, high-density lipoprotein, protein stability, phosphorylation
ABCG1 is a member of the ATP binding cassette (ABC) transporter family of proteins that are involved in translocating substrates across cellular membranes. This half-transporter functions as a homodimer to export cholesterol from cells to HDL, which is the first step of the reverse cholesterol transport pathway (1, 2). Although no human phenotype has been described as being associated with ABCG1 ablation, studies in animals and cell culture models suggest that its main role lies in the protection of macrophages and other vascular cells from cholesterol accumulation, a process that is thought to be an important step in the development of atherosclerosis (3, 4).
We have previously shown that ABCG1 can potentially synergise with ABCA1, another ABC transporter, whereby ABCA1 donates cholesterol and phospholipids to lipid-free apolipoprotein A-I, which in turn can be further lipidated by ABCG1 (5). This cooperative function of the two transporters may be important in the developing atherosclerotic plaque, where there might be restricted access to larger HDL particles. Understanding how these ABC transporters function in macrophages and how they can be stimulated will provide opportunities to increase reverse cholesterol transport in the vessel wall. The transcriptional regulation of ABCG1 as a well-established target gene of the liver X receptor (LXR) is reasonably well understood, with cholesterol accumulation increasing and cholesterol removal reducing gene expression (6). However, the posttranslational control of the transporter is relatively unexplored at present.
One aspect that makes ABCG1 a complex and intriguing transporter is the fact that more than one protein isoform is expressed in human cells. Several N-terminal variants have been proposed based on different translational start sites, but these have not been characterized to date (7, 8). Another interesting isoform variation arises from alternative splicing of the ABCG1 gene in a section of the cytoplasmic domain between the ATP cassette and the transmembrane region (7). Alternative splicing in this region leads to either the presence or the absence of a 12 amino acid (AA) internal segment in the ABCG1 protein, which we have previously termed ABCG1(+12) and ABCG1(− 12), respectively (9). An extensive characterization of the expression of these two isoforms in cells and whole tissues showed that mRNA of both isoforms is expressed in human macrophages, vascular smooth-muscle cells, and endothelial cells, as well as in whole human liver, lung, spleen and brain. The ABCG1(+12):ABCG1(− 12) ratio varied from approximately 0.4 to 1.0, depending upon the type of cells or tissue. Using two-dimensional gel electrophoresis, we showed that both protein isoforms are expressed in human macrophages at comparable levels (9). Interestingly, the exon that encodes for the 36 base pair region in the ABCG1 gene seems to be absent in a number of other mammalian species, including rodents (9). Moreover, our previous work indicated potential differences in posttranslational processing of ABCG1(+12) and ABCG1(− 12). Protein turnover studies showed that the basal half-life of the isoforms individually expressed in CHO-K1 cells was different, with ABCG1(+12) displaying a much shorter half-life than ABCG1(− 12) (9). The cholesterol export activity of cells expressing ABCG1(− 12) was always higher under basal conditions than those expressing ABCG1(+12) (9). The mechanisms underlying these observations are unknown and deserve attention, particularly considering that most studies currently undertaken to elucidate the function and regulation of ABCG1 with respect to diseases such as atherosclerosis, diabetes, and Alzheimer's disease are performed in murine models, which do not express ABCG1(+12). Hence, any potential regulatory aspects that are exclusive to ABCG1(+12) are overlooked when studying ABCG1 in a murine model.
Here, we set out to investigate the underlying mechanism for the difference in activity of the two protein isoforms due entirely to this relatively small sequence variation. We focused on the importance of potential phosphorylation sites in and around the 12 AA section in ABCG1(+12) and report that a serine residue adjacent to the 12 AA section is a likely target for protein kinase A (PKA) in ABCG1(+12), but not in ABCG1(− 12), suggesting differential posttranslational regulation of the two protein isoforms.
MATERIALS AND METHODS
Reagents
Primers, H89, KT5720, calphostin C, protein kinase C (PKC) fragment 19-36 (PKC19-36), PD98059, KT-93, DT-2, protease inhibitor cocktail (P8340) and phosphatase inhibitor cocktail (P5726), BSA (essentially FA-free), NADH, and sodium pyruvate were purchased from Sigma. QuikChange site-directed mutagenesis kits were purchased from Stratagene. Zeocin and all cell culture media and reagents were purchased from Invitrogen. BCA protein reagents were purchased from Pierce. The LXR agonist T0901317 was from Cayman Chemicals. Anti-ABCG1 polyclonal antibody was from Novus. Anti-tubulin monoclonal antibody from Sigma. Secondary anti-mouse and anti-rabbit antibodies were from Jackson Laboratories and Sigma. Plasmid purification kits and lipofectamine transfection reagents were from Invitrogen. [1α ,2α (n)-3H]cholesterol was from Perkin Elmer. ECL reagents were from Millipore and Amersham. Reagents for pouring SDS-PAGE gels, including acrylamide, Tris-HCL, glycine, SDS, and TEMED were purchased from Amresco.
Cell culture
CHO-K1 cells were cultured in Ham's F12 medium containing 10% (v/v) heat-inactivated fetal calf serum (FCS), l-glutamine (2 mM), penicillin (100 U/ml), and streptomycin (100 μg/ml). ABCG1-overexpressing cells were cultured as CHO-K1 cells with the addition of 200 µg/ml Zeocin. RAW264.7 murine macrophages were cultured in DMEM containing 10% (v/v) FCS and glutamine, penicillin, and streptomycin at concentrations as described above. For upregulation of LXR target genes, RAW264.7 macrophages were incubated in the above media containing 10% FCS plus T0901317 (1 µM) for 24 h. Control cells were treated with vehicle 0.01% (v/v) DMSO.
Preparation of constructs and stable cell lines
Constructs overexpressing either ABCG1(− 12) or ABCG1(+12) were prepared and stably overexpressed in CHO-K1 cells as previously described (5, 9). Single base pair mutations were introduced in the construct encoding for ABCG1(+12) using a QuikChange site-directed mutagenesis kit, to introduce serine-to-alanine changes in T378, S388, S389, or S390 respectively, or a serine-to-aspartic acid change at S389. An insertion mutant was created in the ABCG1(− 12) construct by inserting six base pairs that encoded for RK in position 375 using the polymerase incomplete primer extension method (10). Primers were designed according to the manufacturer's instructions and sequences are available upon request. Mutations were confirmed by sequence verification.
Stably overexpressing cell lines using individual constructs with point mutations or insertions were produced in CHO-K1 cells as previously described (5, 9). Briefly, CHO-K1 cells were seeded at 4 × 105 in 60 mm dishes and transfected using lipofectamine reagent (5 µg DNA and 20 µl lipofectamine). After 24 h, media were refreshed and 1 mg/ml zeocin was added. After several days of selection, surviving cells were subjected to single-cell dilution in 96-well plates. Surviving colonies originating from single cells were expanded and screened for ABCG1 expression. Positive clones were maintained routinely in 200 µg/ml zeocin in Ham's F12 medium containing 10% FCS and additions as specified under “cell culture.” The presence of mutated ABCG1 was confirmed after isolation of RNA from individual cell lines and sequence verification as described (5). Cell lines were selected on the basis of having comparable ABCG1 protein expression, measured by SDS-PAGE, between parental and mutated ABCG1. Cell lines were maintained and handled individually to avoid cross-contamination.
ABCG1 mRNA and protein expression
ABCG1 mRNA expression in overexpressing cell lines was measured via real-time PCR as previously described in detail in Gelissen et al. (5). For measurement of ABCG1 protein levels as well as housekeeping genes, cells were washed and lysed in 1% IgePal dissolved in 50 mM Tris-HCL, 150 mM NaCl (pH 7.8) with the addition of 5 µl/ml each of a protease inhibitor cocktail and phosphatase inhibitor cocktail (Sigma). Equal amounts of cell protein per lane were separated using 8% SDS-PAGE gels. After transfer, membranes were blotted using anti-ABCG1 (1:2,500) or tubulin (1:3,000) antibodies. HRP-conjugated anti-mouse or anti-rabbit secondary antibodies were used at 1:10,000, and blots were visualized using ECL. Protein expression levels were quantified using Image J software (version 1.45).
Cholesterol efflux and cell viability assay
Cholesterol efflux assays were performed as described previously (5) with minor modifications. Briefly, cells were radiolabeled overnight with [1α ,2α (n)-3H]cholesterol (1–2 µCi/ml) in serum-containing medium. The labeling medium was removed, and the cells were washed twice with PBS, then incubated for up to 90 min in equilibration medium (serum-free F12 containing 0.1% essentially FA-free BSA). The cells were washed again and preincubated for 30 min in serum-free F12 containing BSA (as above) with vehicle only or kinase inhibitors at the indicated concentrations. Concentrations were chosen depending on data available from published studies that showed no cytotoxicity, but effects on phosphorylation targets, in CHO-K1 cells, as well as published IC50 values. In the case of calphostin C, the compound was photo-activated for 30 min by placing the cells with inhibitor or vehicle under visible light. After the preincubation, the cells were incubated for a further 4 h with media containing the specified inhibitor (or vehicle control) plus the indicated concentrations of HDL2 for 4 h. Radioactivity was measured separately in cells and media, and efflux was calculated as the proportion in the media relative to the total media plus cellular pool. HDL2 was prepared by ultracentrifugation as described (5). Cell viability was measured after incubation with inhibitors for the same times as cholesterol efflux assays using a spectrophotometric lactate dehydrogenase viability assay (11).
Cholesterol export assays in RAW264.7 macrophages were performed as above with one modification. Cells were incubated with or without the addition of the LXR agonist T0901317 (1 µM) to the labeling and equilibration media to upregulate ABCA1 and ABCG1 expression. After labeling, cells were equilibrated for 1 h, followed by preincubation with H89 and efflux performed as described above.
RESULTS
Identification of potential phosphorylation sites
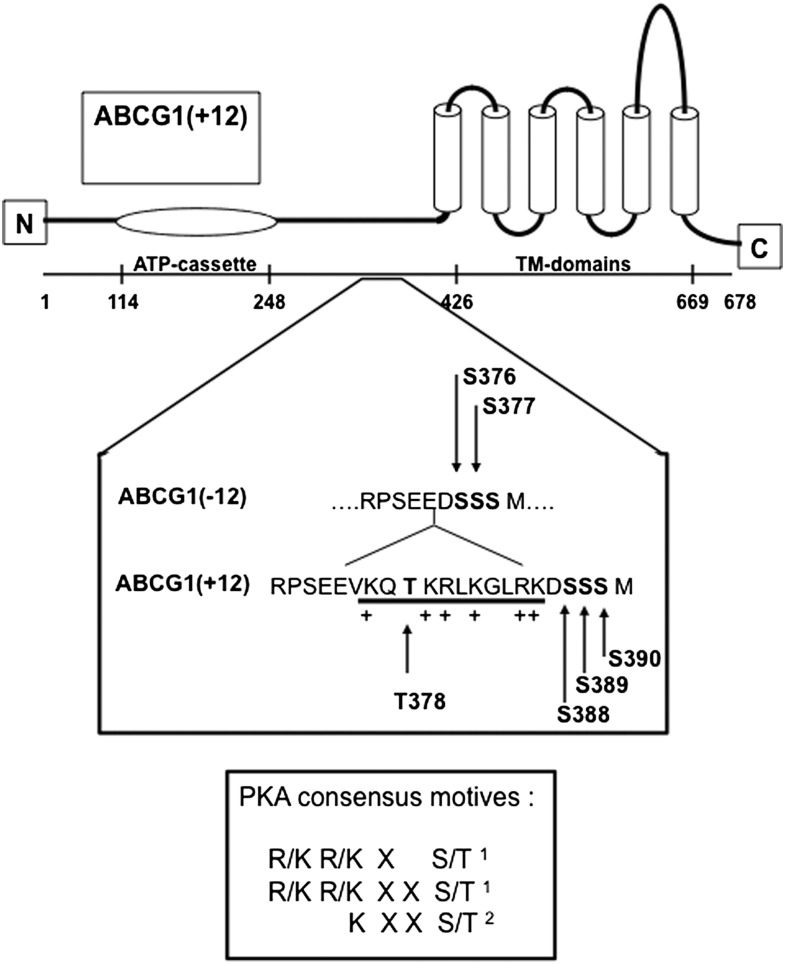
Inspection of the protein sequence in and around the 12 AA region of ABCG1(+12) (Fig. 1) revealed a threonine within the 12 AA section (T378) and three serines in close proximity (S388, S389, and S390). Analysis of the sequence of ABCG1(− 12) and ABCG1(+12) using the phosphorylation prediction programs NetPhosK 1.0 and NetPhos 2.0 [www.cbs.dtu.dk/services/NetPhos (12)] suggested that T378 in ABCG1(+12) may be a potential target for PKC-mediated phosphorylation. Furthermore, S388 and S389 are both part of PKA consensus motifs[see Fig. 1, (13, 14)]. The absence of the 12 AA section in ABCG1(− 12) removes the positively charged arginine and lysine prior to S388 and S389, hence the identical serines in ABCG1(− 12), i.e., S376 and S377, are not part of the PKA consensus motifs. This implies that the presence of the 12 AA section in the ABCG1 protein may result in differences in the phosphorylation state and hence posttranslational processing of the two ABCG1 isoforms. Therefore T378, S388, and S389 in ABCG1(+12) were chosen for further investigation, with S390 included as a control.
Fig. 1.
Diagram of ABCG1 protein with detailed sequence of ABCG1(− 12) and ABCG1(+12) around the 12 AA section. Arrows indicate threonine and serine residues identified as potential phosphorylation sites in ABCG1(+12) and the equivalent sites in ABCG1(− 12). PKA consensus sites are indicated in the box (1 and 2 refer to references 14 and 13, respectively).
PKC inhibition reduces cholesterol export from cells expressing either ABCG1(− 12) or ABCG1(+12)
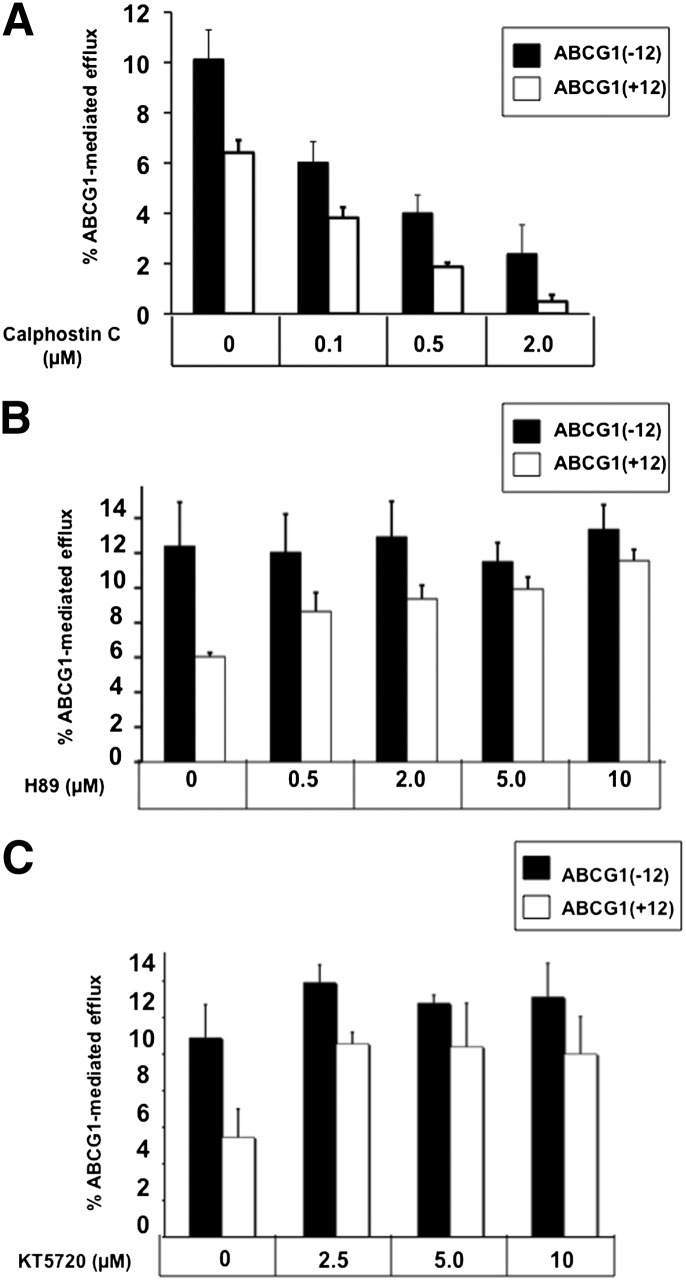
We previously found that the cholesterol export activity and ABCG1 protein levels of cells expressing ABCG1(+12) were significantly lower than those expressing ABCG1(− 12), despite identical ABCG1 mRNA expression (9). To assess whether this difference in activity was related to posttranslational processing involving phosphorylation of a potential PKC site at T378 in ABCG1(+12), cells expressing the individual isoforms of ABCG1(− 12) or ABCG1(+12) were incubated with increasing concentrations of the PKC inhibitor calphostin C. As previously shown (9), cholesterol export from cells expressing ABCG1(+12) was lower than that of ABCG1(− 12)-expressing cells (Fig. 2A, see 0 µM calphostin C). Inhibition of PKC reduced cholesterol efflux from both ABCG1(− 12) and ABCG1(+12), and these effects were already seen at low inhibitor concentrations (0.1 µM calphostin C). A second, structurally unrelated PKC inhibitor, PKC19-36, also reduced cholesterol export from both cell lines (data not shown). Together, these results showed that activity of both ABCG1 isoforms was affected equally by PKC inhibition, suggesting that PKC phosphorylation of sites other than T378 in ABCG1(+12) might be involved in the regulation of ABCG1 function. Considering that the aim of this study was to investigate the differential activity of the two isoforms, these results (although interesting) were not further investigated here.
Fig. 2.
Effect of PKC and PKA inhibitors on cholesterol efflux from cells expressing ABCG1(− 12) or ABCG1(+12). A: Cells were preincubated with the indicated concentrations of the PKC inhibitor calphostin C in media containing BSA (0.1%) for 30 min, then for a further 4 h with calphostin C at the indicated concentrations, in the presence of HDL (10 µg/ml). Cholesterol efflux was measured as described in Materials and Methods. Data are mean ± SEM from three independent experiments, each performed with triplicate cultures. Significant reductions in cholesterol efflux from both ABCG1(− 12) and ABCG1(+12) cells with calphostin C at 0.1, 0.5, and 2 µM (P < 0.05). B: Cells were preincubated with the indicated concentrations of H89 in media containing BSA (0.1%) for 30 min, then for a further 4 h with the addition of HDL (10 µg/ml). Cholesterol efflux was measured as described in Materials and Methods. Data are mean ± SEM of three independent experiments performed in triplicate cultures. Two-way ANOVA showed a significant isoform effect (P < 0.05); significant increases of cholesterol efflux from ABCG1(+12) cells only with H89 at 2, 5, and 10 µM (P < 0.02). C: Cells were preincubated with the indicated concentrations of KT5720 in media containing BSA (0.1%) for 30 min, then for a further 4 h with the addition of HDL (10 µg/ml). Cholesterol efflux was measured as described in Materials and Methods. Data are mean ± SEM of three independent experiments performed in triplicate cultures. Significant increase in cholesterol efflux from ABCG1(+12) cells only at 2.5 µM (P < 0.05).
PKA inhibition increases cholesterol export from ABCG1(+12)- but not ABCG1(− 12)-overexpressing CHO-K1 cells
As mentioned above, S388 and S389 in ABCG1(+12) were identified as potential PKA targets in ABCG1(+12), but these are absent from ABCG1(− 12). To assess whether the difference in activity between cells expressing ABCG1(− 12) and ABCG1(+12) involved phosphorylation of these sites by PKA, cells were incubated with increasing concentrations of the PKA inhibitor H89 (Fig. 2B). Incubation with H89 resulted in a dose-dependent increase in cholesterol export from cells expressing ABCG1(+12) but not from cells expressing ABCG1(-12). This effect was already evident at low H89 concentrations (0.5 μ M) and brought cholesterol export levels from ABCG1(+12) cells to levels almost equivalent to ABCG1(− 12) cells by 5 µM. Background cholesterol efflux from CHO-K1 cells was unaffected by H89, whereas treatment with the inhibitor did not affect ABCG1 mRNA levels (data not shown). Treatment of cells with a structurally unrelated PKA inhibitor, KT5720, also increased cholesterol export from cells expressing ABCG1(+12) but not from cells expressing ABCG1(− 12) (Fig. 2C), supporting the results observed for H89 (Fig. 2B). Cell viability was not affected by incubation with either inhibitor (data not shown).
Macrophages of human origin express both ABCG1 isoforms; hence, measuring an effect of a PKA inhibitor on cholesterol export from individual human isoforms is technically challenging. However, mice do not express ABCG1(+12), and murine ABCG1 has high sequence homology to ABCG1(− 12) (9). To assess whether the activity of this single isoform is affected by PKA inhibition, experiments were performed using RAW264.7 murine macrophages, incubated with T0901317 to induce ABCG1 protein expression. HDL-mediated cholesterol efflux from these cells was unaffected by treatment with H89 for 4 h at 10 μ M (data not shown), suggesting that the murine ABCG1(− 12) isoform behaves similarly to its human counterpart.
Both H89 and KT5720 have documented off-target effects at higher concentrations (15), with both inhibitors potentially affecting the mitogen-activated protein kinase (MAPK) signaling pathway. KT5720 has been shown to affect the MAPK pathway, whereas H89 inhibits extracellular signal-regulated kinase (ERK)1/2 signaling, which is part of the MAPK pathway (15). To exclude that the effects seen by H89 and KT5720 on ABCG1(+12) activity were due to off-target effects on ERK1/2, experiments were performed with the specific ERK1/2 inhibitor PD98059. Incubation of both ABCG1(− 12)- and ABCG1(+12)-expressing cells with PD98059 did not affect cholesterol export (see supplementary Fig. IIIA), suggesting that the effects of H89 and KT5720 on ABCG1(+12) are not due to collateral effects on ERK1/2. We also excluded two other kinases, specifically PKG and calcium/calmodulin-dependent protein kinase (CaMK)II. PKG has the same consensus sequence as PKA, and H89 at high concentrations has been suggested to affect PKG activity (15). Although there is no documented crossover of H89 on CaMKII, the consensus sequence for this kinase is similar to that of PKA (14). Using specific inhibitors for both PKG and CaMKII (DT-2 and KN-93, respectively), the involvement of these two kinases was excluded, inasmuch as neither of the two inhibitors had any effect on cholesterol export from cells expressing ABCG1(+12) or ABCG1(− 12) (see supplementary Fig. II).
Altogether, these results showed that the activity of ABCG1(+12)-expressing cells could be increased by incubation with two independent PKA inhibitors, whereas ABCG1(− 12) was unaffected.
PKA inhibition increases ABCG1(+12) protein stability, but not that of ABCG1(− 12)
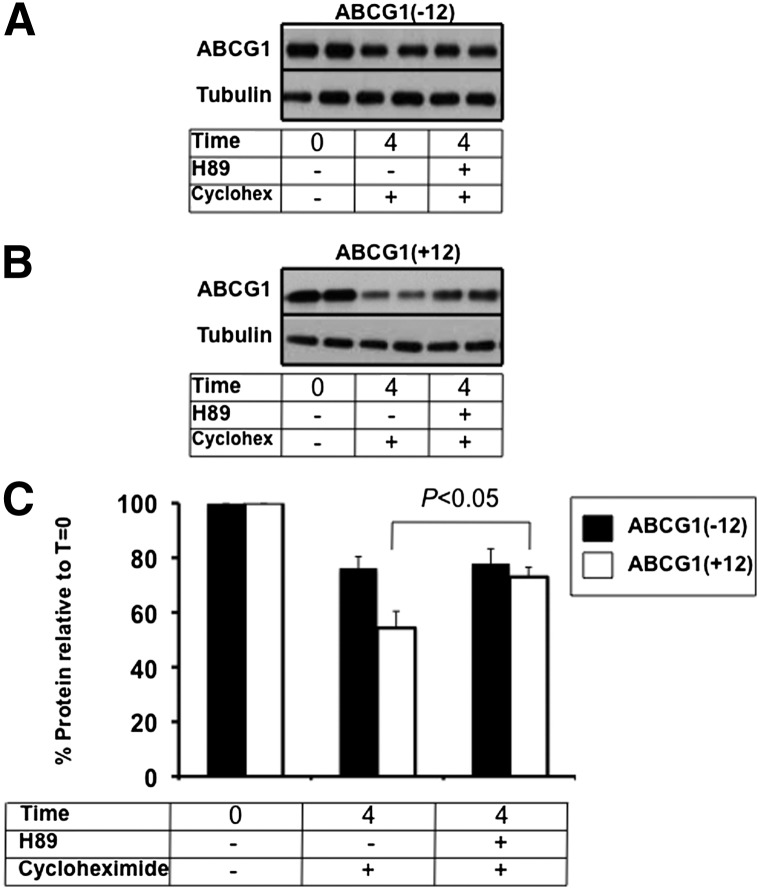
Previously, we reported that ABCG1 protein levels in cells expressing ABCG1(+12) were significantly lower than those in cells expressing ABCG1(− 12), despite identical ABCG1 mRNA expression (9). We furthermore showed that the half-life of ABCG1(+12) was shorter than that of ABCG1(− 12). To determine whether the increase in cholesterol export induced by H89 in ABCG1(+12)-expressing cells was due to differences in ABCG1 protein turnover, cells expressing each individual isoform were incubated with H89 in the presence of cycloheximide to inhibit protein synthesis. Initial time course experiments implied that differences in protein expression between the two isoforms were apparent at T = 4 h without any sign of cytotoxicity (data not shown); hence, this time point was employed for further experiments. Degradation of ABCG1(+12) after 4 h was more extensive than that of ABCG1(− 12) (Fig. 3A, B; compare T = 4 with cycloheximide and Fig. 3C) which is in agreement with our previous observations using 35S-protein degradation studies performed under similar conditions (9). Addition of H89 resulted in a significant protein stabilization for ABCG1(+12) but not for ABCG1(− 12), suggesting that PKA is probably involved in the degradation of ABCG1(+12) but not of ABCG1(− 12). Considering that stabilization of this pool coincides with an increase in cholesterol export (Fig. 2B), this pool must be significant in terms of its contribution to overall cholesterol export function.
Fig. 3.
Effect of H89 on ABCG1(− 12) and ABCG1(+12) protein levels. ABCG1(− 12) (A) or ABCG1(+12) cells (B) were incubated with or without H89 in the presence of cycloheximide (10 μg/ml) for 4 h in 0.1% BSA-containing media and were harvested. Equal amounts of cell protein were loaded per lane, and ABCG1 and housekeeping (tubulin) levels were measured by SDS-PAGE as described in Materials and Methods. Data are representative of at least three independent experiments. C: Bands were quantified using Image J software, and results were expressed relative to the starting levels at T = 0 for each experiment. Results are mean ± SEM of duplicate cultures from three independent experiments, each performed in duplicate cultures.
Serine 389 of ABCG1(+12) is essential for the effects of PKA inhibition on cholesterol export from CHO-K1 cells expressing ABCG1(+12)
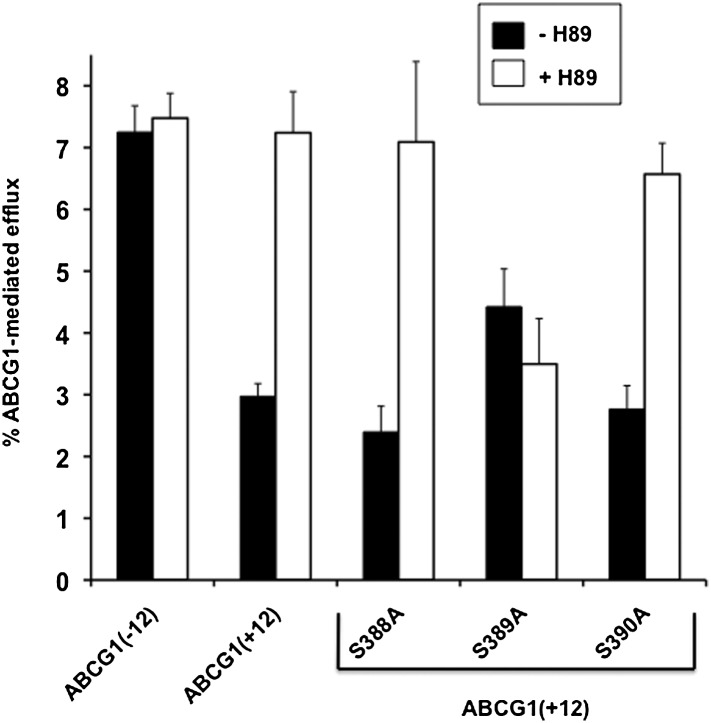
Next, we determined whether any of the potential phosphorylation sites described earlier were actively involved in the increased ABCG1 activity upon incubation with PKA inhibitors. Individual cell lines stably overexpressing ABCG1(+12) with single point mutations at S388, S389, or S390 (changed to alanine) were produced, and cholesterol export to HDL was tested. Comparison of ABCG1 mRNA expression among the lines showed that expression was similar among the lines, with the exception of the S389A mutant ,which had approximately half the mRNA expression compared with the other cell lines (see supplementary Fig. IIIA).
Basal cholesterol export from ABCG1 parent or mutant-expressing cells was similar and unaffected by H89 treatment (data not shown). HDL-induced cholesterol export from cells expressing ABCG1(+12)-S388A and ABCG1(+12)-S390A was similar to that of cholesterol export from wild-type ABCG1(+12) cells (Fig. 4). Furthermore, upon incubation with H89, cholesterol export from cells expressing the S388A or S390A mutants was increased to a level similar to that of the parental ABCG1(+12). However, HDL-mediated cholesterol efflux from the S389A mutant was on average slightly higher than the parental line, despite having lower mRNA expression, whereas cholesterol export from this mutant was completely unaffected by H89 treatment (Fig. 4). Altogether, these data suggest that S389 is necessary to the mediation of the increased cholesterol export seen from cells expressing ABCG1(+12) upon inhibition of PKA.
Fig. 4.
Effect of single point mutations on cholesterol efflux from cells expressing ABCG1(+12), with or without PKA inhibition. CHO-K1 cells stably expressing wild-type ABCG1(+12) with or without the single point mutations at S388, S389, or S390 were generated as described in Materials and Methods. ABCG1 mRNA levels are presented in supplementary Fig. IIIA. ABCG1(− 12), ABCG1(+12), and individual point mutant cell lines were preincubated with or without 10 µM H89 in media containing BSA (0.1%) for 30 min, then for a further 4 h with or without addition of HDL (10 µg/ml). Cholesterol efflux was measured as described in Materials and Methods. Data are mean ± SEM of three independent experiments, each performed in triplicate cultures. H89 significantly increased cholesterol export from cells expressing ABCG1(+12), S388A, and S390A (P < 0.05), but not ABCG1(− 12) or S389A.
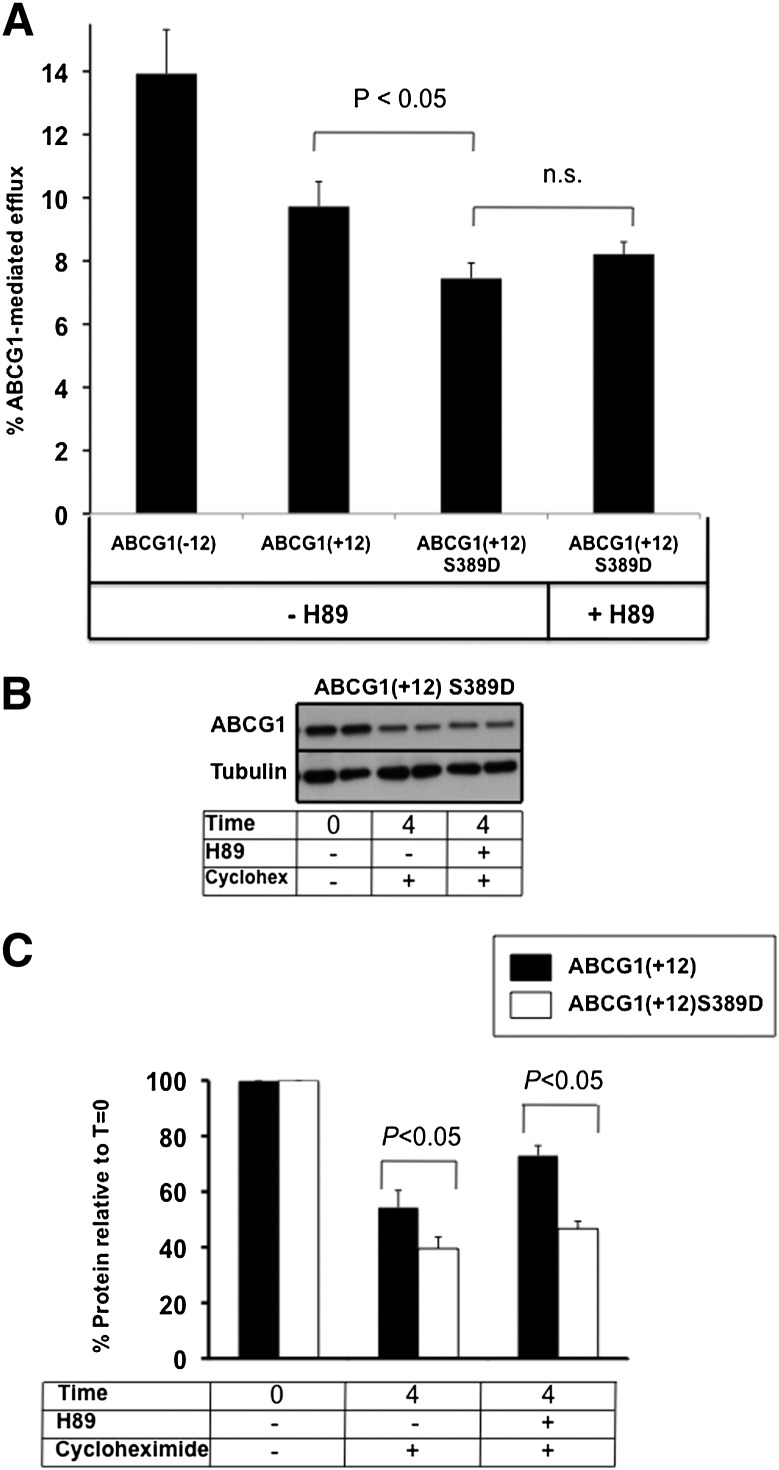
To confirm that phosphorylation of S389 was indeed involved in these findings, this AA was mutated to an aspartic acid in order to produce a phosphomimetic mutant, imitating a permanently phosphorylated state. ABCG1 mRNA expression was identical between the parental and mutant cell lines (see supplementary Fig. IIIB).Figure 5A shows that basal cholesterol export from ABCG1(+12)-S389D-expressing cells was reduced compared with that of the wild-type ABCG1(+12), whereas incubation with H89 did not affect the cholesterol export capacity to HDL. Protein degradation of this mutant was more extensive compared with that of the parent ABCG1(+12), and stabilization with H89 did not occur (Fig. 5B, with quantification of three independent experiments in Fig. 5C). This finding provides further evidence of the involvement of this site in the increased protein degradation of ABCG1(+12).
Fig. 5.
Effect of H89 on ABCG1(+12)-S389D cholesterol export and protein levels. A: Cholesterol export was measured as described in the legend to Fig. 2 and Materials and Methods from ABCG1(+12)-S389D-overexpressing cells and compared with the parental ABCG1(+12) cell line, with and without the addition of H89 (10 μ M). B: ABCG1(+12)-S389D-overexpressing cells were incubated with or without H89 in the presence of cycloheximide (10 μg/ml), as described in the legend to Fig. 4. Data are representative of at least three independent experiments. C: Bands were quantified using Image J software, and results were expressed relative to the starting levels at T = 0 for each experiment. Results are mean ± SEM of duplicate cultures from three independent experiments, all performed in duplicate cultures. Data for ABCG1(+12) wild-type are same as those presented in Fig. 4C.
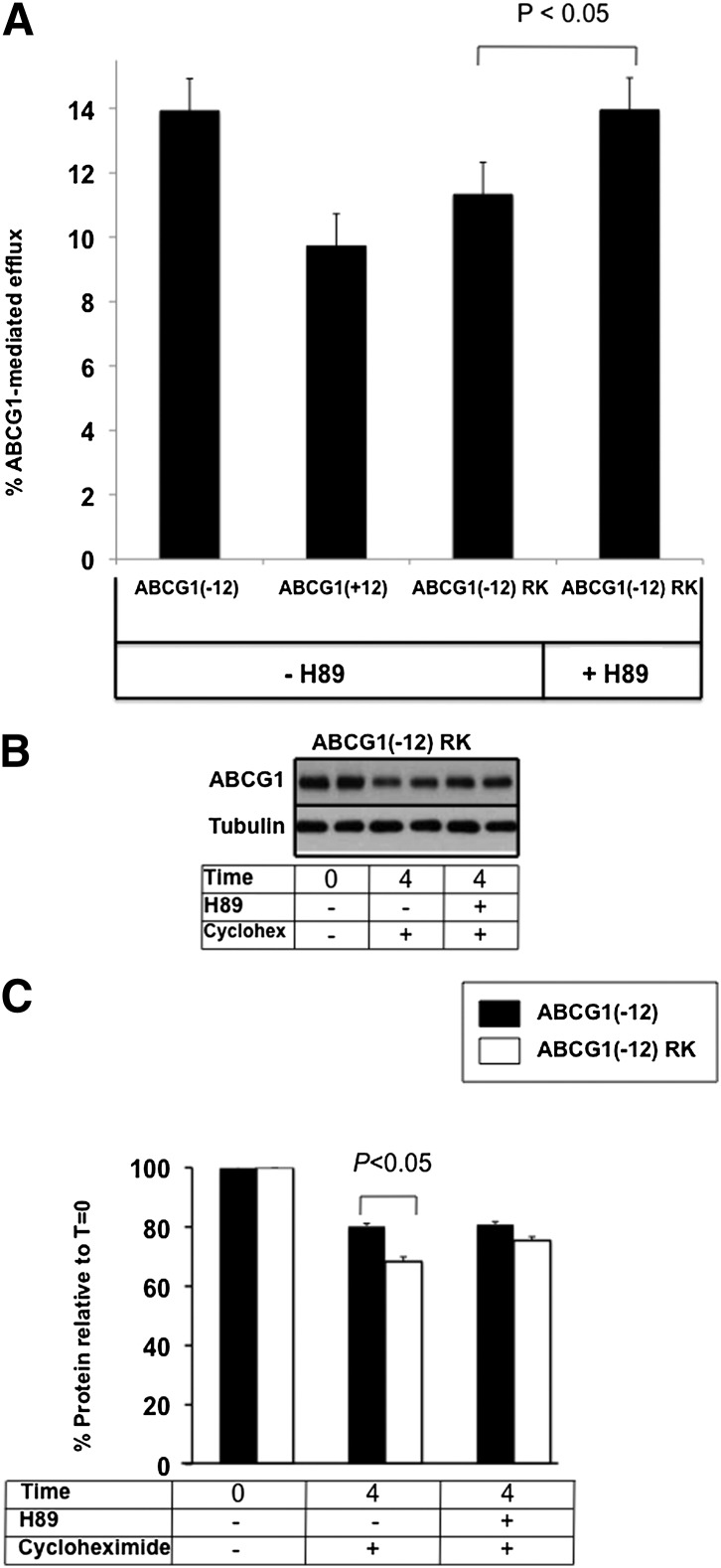
ABCG1(− 12) also contains the equivalent serine in position 377 (see Fig. 1). However, due to the absence of the two nearby positive charges, this residue is an unlikely PKA target. We constructed an additional mutant, in which we inserted the positively charged AA RK at positions − 3 and − 4 relative to S377 in ABCG1(− 12) to determine whether this produced an ABCG1(− 12) protein that resembled ABCG1(+12) in terms of protein degradation and cholesterol export capacity.Figure 6A shows that the basal cholesterol export capacity of this mutant compared with that of the wild-type ABCG1(− 12) was somewhat lower, possibly due to slightly lower ABCG1 mRNA expression in the mutant cell line (see supplementary Fig. IIIB). Most importantly, treatment with H89 increased the mutants’ cholesterol export capacity, similar to the effect seen in ABCG1(+12) in Fig. 2B, where wild-type ABCG1(− 12) was unaffected. Protein degradation of this ABCG1(− 12) RK insertion mutant was modestly but significantly accelerated compared with that of wild-type ABCG1(− 12) (Fig. 6B, with quantification of three independent experiments in Fig. 6C). Altogether, these data provide further evidence that these two positive charges are essential to make this serine a target for phosphorylation and subsequent degradation.
Fig. 6.
Effect of H89 on ABCG1(− 12) RK cholesterol export and protein levels. A: Cholesterol export was measured as described in the legend to Fig. 2 and Materials and Methods from ABCG1(− 12) RK-overexpressing cells and compared with the parental ABCG1(− 12) cell line, with and without the addition of H89 (10 μ M). B: ABCG1(− 12) RK-overexpressing cells were incubated with or without H89 in the presence of cycloheximide (10 μg/ml), as described in the legend to Fig. 4. Significant increase with H89 treatment in ABCG1(-12)RK cells (p < 0.03) at T = 4. Data are representative of at least three independent experiments. C: Bands were quantified using Image J software, and results were expressed relative to the starting levels at T = 0 for each experiment. Results are mean ± SEM of duplicate cultures for three independent experiments, each performed in duplicate cultures. Data for ABCG1(− 12) wild-type are the same as those presented in Fig. 4C.
DISCUSSION
In this study, we set out to investigate the underlying mechanisms for a difference in activity of the two major ABCG1 protein isoforms that occur in humans. These proteins differ only by 12 AAs; however, this segment has the potential to impact the primary and secondary protein characteristics, owing to the presence of six charged residues. We reported previously that the predicted isoelectric point of the proteins in human macrophages is significantly different [6.5 for ABCG1(− 12) vs. 8.0 for ABCG1(+12)], making it possible to separate them using two-dimensional gel electrophoresis (9). Furthermore, prediction of the secondary protein structure [http://bioinf.cs.ucl.au.uk/psipred; (16)] suggests that the area between AA 350 and AA 400 may form an additionalα -helix in ABCG1(+12) that is absent from ABCG1(− 12). Hence, a small change in protein sequence has the potential to have a major impact on protein structure and function.
This paper presents the first evidence for a difference in posttranslational processing of the two ABCG1 isoforms involving phosphorylation sites that have direct implications on ABCG1 activity. Only a few studies to date have addressed the involvement of phosphorylation in the regulation of ABCG1 function. Nagelin et al. (17, 18) concluded that serine phosphorylation is involved in the regulation of ABCG1 protein degradation. The authors showed that ABCG1 degradation induced by 12/15 lipoxygenase, a lipid oxidation enzyme implicated in atherogenesis, is mediated via serine phosphorylation through the p38- and JNK2-dependent pathways. These studies were performed primarily in murine macrophages (expressing the ABCG1(− 12) homolog), but the authors also confirmed their findings using overexpressed human ABCG1(+12) (18). Any involvement of PKA or PKC in the 12/15 lipoxygenase-mediated degradation of ABCG1 was excluded in these studies (18). Here, we showed that pharmacological inhibition of PKA, using two structurally unrelated inhibitors, substantially increased cholesterol export mediated by ABCG1(+12) but not by ABCG1(− 12). Measurement of protein degradation by using cycloheximide to block protein synthesis showed that PKA inhibition stabilized ABCG1(+12) protein but left ABCG1(− 12) unaffected. A number of potential other kinases were excluded, and we furthermore identified the likely phosphorylation site responsible for these results as the middle serine, namely S389, by site-directed mutagenesis studies.
The current data make extensive use of pharmacological inhibitors, which are often questioned for their specificity. In this case, the aims were to look for distinct differences in activity resulting from the presence or absence of only a small section of the ABCG1 protein. Two structurally unrelated inhibitors were used for both PKC and PKA, which is generally recommended for such experiments (19). Also, cholesterol efflux from parent CHO-K1 cells was unaffected, which is another suggested criteria for specificity (19). A number of alternate kinases with identical recognition motifs or which were documented alternate kinase targets of the inhibitors used, i.e., PKC, ERK1/2, PKG, and CaMKII, were excluded. We further identified ABCG1(+12)-S389 as the site responsible, which is synonymous with a classic PKA phosphorylation site (13, 14). Due to these numerous lines of evidence, we are confident of the involvement of PKA in the regulation of ABCG1(+12) activity through S389.
The mechanism of the PKA-dependent degradation that we identified for ABCG1(+12) is currently under investigation. However, considering that there are a large number of lysine residues in the 12 AA section of this isoform, the involvement of ubiquitination is currently being investigated. Ubiquitination and proteasomal degradation are known to affect the protein stability of murine ABCG1 (20), indicating that sites other than those in the 12 AA section of ABCG1(+12) can be ubiquitinated. However, incubation of our cell systems with MG132, a proteasomal inhibitor, does not seem to affect isoform-specific ABCG1-mediated cholesterol export (data not shown). Another possibility under investigation is the localization of the pool of ABCG1 that is rescued by inhibition of PKA; we suggest that this small pool must be essential for cholesterol export. ABC transporters, such as the cystic fibrosis conductance regulatory transporter (CFTR), which transports chloride ions at the cell surface, and ABCA1 have been shown to use multi-faceted trafficking pathways. There is strong evidence that CFTR undergoes regulated trafficking between intracellular organelles and the cell surface, including recycling between endosomes and the plasma membrane (21). Interestingly, PKA is involved in the regulation of CFTR's itinerary, whereby phosphorylation via PKA results in decreased internalization of plasma membrane pools and increased insertion into the plasma membrane from intracellular stores, leading to overall increased cell surface expression (reviewed in Ref. 21). The exact cellular localization of ABCG1, as well as its site of action, is currently under debate. A recent study by Tarling and Edwards (22) showed that ABCG1 is localized in endosomes, and the authors suggested that it functions entirely intracellularly. However, a study by Wang et al. (23) found ABCG1 in endosomes as well as in the golgi and plasma membrane in macrophages, and furthermore showed increased ABCG1 plasma membrane expression upon LXR activation (23). We are currently investigating whether stabilization with H89 alters ABCG1 localization in our models.
In conclusion, we have shown that the posttranslational processing of the ABCG1 isoforms ABCG1(+12) and ABCG1(− 12) is distinct. Considering that the majority of current studies on the function of ABCG1 are performed in murine models that lack ABCG1(+12), important conclusions regarding the regulation of this transporter and hence, extrapolation to the human situation may be overlooked. We recommend that future studies, when performed in murine models, be extended to include human models wherever possible to ensure that extrapolation of results is valid.
Supplementary Material
Footnotes
Abbreviations:
- AA
- amino acid
- CaMK
- calcium/calmodulin-dependent protein kinase
- CFTR
- cystic fibrosis conductance regulatory transporter
- ERK
- extracellular signal-regulated kinase
- FCS
- fetal calf serum
- LXR
- liver X receptor
- MAPK
- mitogen-activated protein kinase
- PKA
- protein kinase A
- PKC
- protein kinase C
The authors thank the National Health and Medical Research Council (NHMRC) of Australia for funding this research.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three figures.
REFERENCES
- 1.Jessup W., Gelissen I., Gaus K., Kritharides L. 2006. Roles of ATP binding cassette transporters A1 and G1, scavenger receptor BI and membrane lipid domains in cholesterol export from macrophages. Curr. Opin. Lipidol. 17: 247–257 [DOI] [PubMed] [Google Scholar]
- 2.Ye D., Lammers B., Zhao Y., Meurs I., Van Berkel T. J., Van Eck M. 2011. ATP-binding cassette transporters A1 and G1, HDL metabolism, cholesterol efflux, and inflammation: important targets for the treatment of atherosclerosis. Curr. Drug Targets. 12: 647–660 [DOI] [PubMed] [Google Scholar]
- 3.Out R., Hoekstra M., Meurs I., deVos P., Kuiper J., Van Eck M., Van Berkel T. J. C. 2007. Total body ABCG1 expression protects against early atherosclerotic lesion development in mice. Arterioscler. Thromb. Vasc. Biol. 27: 594–599 [DOI] [PubMed] [Google Scholar]
- 4.Yvan-Charvet L., Pagler T. A., Seimon T. A., Thorp E., Welch C., Witzum J. L., Tabas I., Tall A. R. 2010. ABCA1 and ABCG1 protect against oxidative stress-induced macrophage apoptosis during efferocytosis. Circ. Res. 106: 1861–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gelissen I. C., Harris M., Rye K. A., Quinn C., Brown A. J., Kockx M., Cartland S., Packianathan M., Kritharides L., Jessup W. 2006. ABCA1 and ABCG1 synergize to mediate cholesterol export to apoA-I. Arterioscler. Thromb. Vasc. Biol. 26: 534–540 [DOI] [PubMed] [Google Scholar]
- 6.Tall A. R. 2008. Cholesterol efflux pathways and other potential mechanisms involved in the athero-protective effect of high density lipoproteins. J. Intern. Med. 263: 256–273 [DOI] [PubMed] [Google Scholar]
- 7.Croop J. M., Tiller G. E., Fletcher J. A., Lux M. L., Raab E., Goldenson D., Son D., Arciniegas S., Wu R. L. 1997. Isolation and characterization of a mammalian homolog of the Drosophila white gene. Gene. 185: 77–85 [DOI] [PubMed] [Google Scholar]
- 8.Schmitz G., Langmann T., Heimerl S. 2001. Role of ABCG1 and other ABCG family members in lipid metabolism. J. Lipid Res. 42: 1513–1520 [PubMed] [Google Scholar]
- 9.Gelissen I. C., Cartland S., Brown A. J., Sandoval C., Kim M., Dinnes D. L., Lee Y., Hsieh V., Gaus K., Kritharides L., et al. 2010. Expression and stability of two isoforms of ABCG1 in human vascular cells. Atherosclerosis. 208: 75–82 [DOI] [PubMed] [Google Scholar]
- 10.Klock H. E., Lesley S. A. 2009. The Polymerase Incomplete Primer Extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol. Biol. 498: 91–103 [DOI] [PubMed] [Google Scholar]
- 11.Kockx M., Guo D. L., Traini M., Gaus K., Kaj J., Wimmer-Kleikamp S., Rentero C., Burnett J. R., Le Goff W., Van Eck M., et al. 2009. Cyclosporin A decreases apolipoprotein E secretion from human macrophages via a protein phosphatase 2B-dependent and ATP-binding cassette transporter A1 (ABCA1)-independent pathway. J. Biol. Chem. 284: 24144–24154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blom N., Gammeltoft S., Brunak S. 1999. Sequence- and structure-based prediction of eukaryotic phosphorylation sites. J. Mol. Biol. 294: 1351–1362 [DOI] [PubMed] [Google Scholar]
- 13.Feramisco J. R., Glass D. B., Krebs E. G. 1980. Optimal spatial requirements for the location of basic residues in peptide substrates for the cyclic AMP-dependent protein kinase. J. Biol. Chem. 255: 4240–4245 [PubMed] [Google Scholar]
- 14.Schwartz D., Chou M. F., Church G. M. 2009. Predicting protein post-translational modifications using meta-analysis of proteome scale data sets. Mol. Cell. Proteomics. 8: 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray A. J. 2008. Pharmacological PKA inhibition: all may not be what it seems. Sci. Signal. 1: re4. [DOI] [PubMed] [Google Scholar]
- 16.Bryson K., McGiffin L. J., Marsden R. L., Ward J. J., Sodhi J. S., Jones D. T. 2005. Protein structure prediction servers at University College London. Nucl. Acids Res. 33: Web Server Issue W36–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagelin M. H., Srinivasan S. S., Lee J., Nadler J. L., Hedrick C. C. 2008. 12/15 Lipoxygenase activity increases the degradation of macrophage ATP-binding cassette transporter G1. Arterioscler. Thromb. Vasc. Biol. 28: 1811–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagelin M. H., Srinivasan S., Nadler J. L., Hedrick C. C. 2009. Murine 12/15 lipoxygenase regulates ATP-binding cassette transporter G1 protein degradation through p38- and JNK2-dependent pathways. J. Biol. Chem. 284: 31303–31314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen P. 2010. Guidelines for the effective use of chemical inhibitors of protein function to understand their roles in cell regulation. Biochem. J. 425: 53–54 [DOI] [PubMed] [Google Scholar]
- 20.Ogura M., Ayaori M., Terao Y., Hisada T., Iizuka M., Takiguchi S., Uto-Kondo H., Yakushiji E., Nakaya K., Sasaki M., et al. 2011. Proteasomal inhibition promotes ATP-binding cassette transporter A1 (ABCA1) and ABCG1 expression and cholesterol efflux from macrophages in vitro and vivo. Arterioscler. Thromb. Vasc. Biol. 31: 1980–1987 [DOI] [PubMed] [Google Scholar]
- 21.Ameen N., Silvis M., Bradbury N. A. 2007. Endocytic trafficking of CFTR in health and disease. J. Cyst. Fibros. 6: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarling E. J., Edwards P. A. 2011. ATP binding cassette transporter G1 (ABCG1) is an intracellular sterol transporter. Proc. Natl. Acad. Sci. USA. 108: 19719–19724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang N., Ranalletta M., Matsuura F., Peng F., Tall A. R. 2006. LXR-induced redistribution of ABCG1 to plasma membrane in macrophages enhances cholesterol mass efflux to HDL. Arterioscler. Thromb. Vasc. Biol. 26: 1310–1316 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.