Abstract
Bile acids (BAs) are a group of chemically related steroids recognized as regulatory molecules whose profiles can change in different physio-pathological situations. We have developed a sensitive, fast, and reproducible ultraperformance liquid chromatography/multiple reaction monitoring/mass spectrometry method to determine the tissue and sera BA profiles in different species (human, rat, and mouse) by quantifying 31 major and minor BA species in a single 21-min run. The method has been validated according to FDA guidelines, and it generally provides good results in terms of intra- and interday precision (less than 8.6% and 16.0%, respectively), accuracy (relative error measurement between –11.9% and 8.6%), and linearity (R2 > 0.996 and dynamic ranges between two and four orders of magnitude), with limits of quantification between 2.5 and 20 nM. The new analytical approach was applied to determine BA concentrations in human, rat, and mouse serum and in liver tissue. Our comparative study confirmed and extended previous reports, showing marked interspecies differences in circulating and hepatic BA composition. The targeted analysis revealed the presence of unexpected minoritary BAs, such as tauro-alpha-Muricholic acid in human serum, thus allowing us to obtain a thorough profiling of human samples. Its great sensitivity, low sample requirements (25 µl of serum, 5 mg of tissue), and comprehensive capacity to profile a considerable number of BAs make the present method a good choice to study BA metabolism in physiological and pathological situations, particularly in toxicological studies.
Keywords: lipidomics, metabolomics, targeted analysis
Bile acids (BAs) are major components of bile formed from cholesterol through various enzymatic reactions in hepatocytes. Before being excreted into bile canaliculi, primary BAs synthesized in the liver (i.e., cholic acid [CA] and chenodeoxycholic acid [CDCA] in humans and alpha-muricholic acid [αMCA] and beta-muricholic acid [βMCA] in rodents) (1, 2) are mainly conjugated with taurine or glycine amino acids through the terminal side-chain carboxylic group present in the BA structure. Once in the intestine, primary BAs are deconjugated and converted into secondary BAs by microbiota. Then, most BAs are reabsorbed back to the liver, conjugated by hepatocytes, and re-excreted into bile to complete enterohepatic circulation (3).
In the past, BAs were considered to be mere detergents required for the solubilization and absorption of dietary fats. However, BAs are now recognized as regulatory molecules capable of activating specific receptors. BAs are physiological ligands for the farnesoid X receptor, an intracellular BA sensor that controls the expression of the genes involved in BA synthesis, metabolism, and transport in order to minimize the deleterious effects of their accumulation (4, 5). BAs are also able to bind other nuclear receptors (e.g.,. PXR or VDR) and the G-protein coupled receptor TGR5 and can activate several cell signaling pathways (i.e., JNK, ERK, or AKT) (5–8). By activating these receptors and signaling cascades, BAs regulate not only their own homeostasis but also fatty acid, lipoprotein, glucose, and energy metabolism (5, 7, 9). The primary signaling function of BAs seems to be the regulation of metabolic flux in the liver and the gastrointestinal tract during the feed/fast cycle; however, they are also involved in the control of cell proliferation and inflammatory processes (5, 9).
The BA pool constitutes a large mixture of chemically related steroids, including isomeric forms, with diverse physicochemical properties and biological functions. Physiological concentrations of circulating BAs in healthy subjects are low; a number of conditions, including hepatobiliary and intestinal diseases or drug-induced liver injury, can alter BA homeostasis, leading to increased BA levels (10–12). Accumulation of BAs is considered a hallmark of chronic cholestatic liver disease; more specifically, it has been shown that its deleterious toxic effects depend on the profile of accumulated BAs (13). In particular, hydrophobic species (i.e., CDCA, lithocholic acid [LCA], or DCA) and their conjugates have been reported to be highly cytotoxic, and their accumulation can damage hepatic cells from inducing mitochondrial dysfunction, oxidative stress, or apoptosis (14). Furthermore, BAs have been associated with damage in other organs, such as the colon, where BAs may promote colon carcinogenesis (15).
The relevance of the physiological functions associated with BAs, their involvement in pathological processes, and their potential pharmaceutical applications have led to a growing interest in elucidating BA patterns in different biological matrices (16). Changes in BA profiles can be used as biomarkers of disease (10, 17, 18), and, therefore, accurate and sensitive techniques are required for the comprehensive profiling BAs, including those found at relatively low concentrations in physiological conditions. A number of LC/MS methods have been developed for this purpose, but their extensive sample processing and their limited capacity to detect minor BAs (11, 19–25) have restrained their use. In fact, detection of low-concentrated or unusual BAs in human samples, is an important target of BA profiling given their important role in some pathological situations (10, 13, 26).
Here we present a fast, sensitive, and simple ultraperformance liquid chromatography (UPLC) mass spectrometry (MS) method to comprehensively perform BA profiling of human, mouse, and rat serum and in liver tissue. Our targeted method allows the simultaneous quantification of 31 BAs in a single analysis, including major, minor, and species-specific forms present in human, mouse, and rat, thus allowing the detection of a priori unexpected BAs considered (erroneously) exclusive of certain species. Such features represent key advantages when compared with other MS methods previously published, thus constituting great value in exploring biomarkers and biochemical mechanisms involving changes in BAs profiles.
MATERIALS AND METHODS
Chemicals
Methanol, water, and acetonitrile were of LC/MS grade and were purchased from Fisher Scientific (Loughborough, UK). Formic acid, activated charcoal, cholic acid (5β-cholanic acid-3α,7α,12α-triol, CA), glycocholic acid [5β-cholanic acid-3α,7α,12α-triol -N-(carboxymethyl)-amide, GCA], taurocholic acid[5β-cholanic acid-3α,7α,12α-triol-N-(2-sulphoethyl)-amide, TCA], chenodeoxycholic acid (5β-cholanic acid-3α,7α-diol, CDCA), glycochenodeoxycholic acid [5β-cholanic acid-3α,7α-diol-N-(carboxymethyl)-amide, GCDCA), taurochenodeoxyxholic acid (5β-cholanic acid-3α,7α-diol-N-(2-sulphoethyl)-amide, TCDCA), deoxyxholic acid (5β-cholanic acid-3α,12α-diol, DCA), glycodeoxycholic acid [5β-cholanic acid-3α,12α-diol-N-(carboxymethyl)-amide, GDCA], taurodeoxyxholic acid [5β-cholanic acid-3α,12α-diol-N-(2-sulphoethyl)-amide, TDCA], ursodeoxycholic acid (5β-cholanic acid-3α, 7β-diol, UDCA), lithocholic acid (5β-cholanic acid-3α-ol, LCA), and taurolithocholic acid [5β-cholanic acid-3α-ol-N-(2-sulphoethyl)-amide, TLCA] were purchased from Sigma-Aldrich Química SA (Madrid, Spain). Tauroursodeoxycholic acid [5β-cholanic acid-3α,7β-diol-N-(2-sulphoethyl)-amide, TUDCA] was purchased from Calbiochem/Merck (Mollet del Vallès, Spain). α-muricholic acid (5β-cholanic acid-3α,6β,7α-triol, αMCA), tauro-α-muricholic acid [5β-cholanic acid-3α,6β,7α-triol-N-(2-sulphoethyl)-amide, TαMCA], β-muricholic acid (5β-cholanic acid-3α,6β,7β-triol, βMCA), tauro-β-muricholic acid [5β-cholanic acid-3α,6β,7β-triol-N-(2-sulphoethyl)-amide, TβMCA], ω-muricholic acid (5β-cholanic acid-3α,6α,7β-triol, ωMCA), tauro-ω-muricholic acid [5β-cholanic acid-3α,6α,7β-triol-N-(2-sulphoethyl)-amide, TωMCA], hyocholic acid (5β-cholanic acid-3α,6α,7α-triol, HCA), glycohyocholic acid [5β-cholanic acid-3α,6α,7α-triol-N-(carboxymethyl)-amide, GHCA], taurohyocholic acid [5β-cholanic acid-3α,6α,7α-triol-N-(2-sulphoethyl)-amide, THCA], glycoursodeoxycholic acid [5β-cholanic acid-3α,7β-diol-N-(carboxymethyl)-amide, GUDCA], glycolithocholic acid [5β-cholanic acid-3α-ol-N-(carboxymethyl)-amide, GLCA], hyodeoxycholic acid (5β-cholanic acid-3α,6α-diol, HDCA), glycohyodeoxycholic acid [5β-cholanic acid-3α,6α-diol-N-(carboxymethyl)-amide, GHDCA], taurohyodeoxycholi acid [5β-cholanic acid-3α,6α-diol-N-(2-sulphoethyl)-amide, THDCA], murocholic acid(5β-cholanic acid-3α, 6β-diol, MuroCA), dehydrocholic acid (5β-colanic acid-3,7,12-trione, DHCA), glycodehydrocholic acid [5β-colanic acid-3,7,12-trione-N-(carboxymethyl)-amide, GDHCA], taurodehydrocholic acid [5β-colanic acid-3,7,12-trione-N-(2-sulphoethyl)-amide, TDHCA], and deuterated internal standards (IS) lithocholic acid-2,2,4,4-D4 (LCA-D4), deoxycholic acid-2,2,4,4-D4 (DCA-D4), cholic acid-2,2,4,4-D4 (CA-D4), glycochenodeoxyxholic acid-2,2,4,4-D4 (GCDCA-D4), and glycocholic acid-2,2,4,4-D4 (GCA-d4) were purchased from Steraloid inc (Newport, RI).
UPLC-MS analysis
UPLC separation was performed in an Acquity UPLC system (Waters, UK) equipped with an Acquity UPLC BEH C18 (1.7 μm, 2.1 × 100 mm; Waters) column. The temperatures of the column and the autosampler were set at 65°C and 4°C, respectively. The sample injection volume was 4 µl. Eluents consisted in 0.1% formic acid in water (eluent A) and 0.1% formic acid in acetonitrile (eluent B). The flow rate was set at 0.5 ml/min. A 21-min elution gradient was performed as follows: during the first 0.5 min, eluent composition was set at 95% A and 5% B, which was linearly changed to 75% A and 25% B in 5 min; then the proportion of B was increased to 40% in the next 10.5 min, followed by a further increase to 95% B reached at min 17.5 and kept for 1.5 min. Finally, the initial conditions were recovered and maintained for 2 min for column conditioning.
The MS analysis was performed using a Waters Xevo TQ-S mass spectrometer (Waters) equipped with an ESI source in the negative-ion mode working in the multiple reaction monitoring (MRM) mode. A capillary voltage of 2 kV, a source temperature of 120°C and a desolvation temperature of 380°C were used. Desolvation and the cone gas flow were set as 800 liters/h and 150 liters/h, respectively, and the collision gas was 0.25 ml/min. Transitions, cone voltages, and collision energies were automatically tuned for each BA using the Quanoptimizer software (Waters. The data station operating software used was MassLynx 4.1 (Waters).
Preparation of standard solutions and calibration curves
The standard stock solution of each individual BA and deuterated IS were prepared at a concentration of 2 mg/ml in methanol.
A mixed stock solution of all the BAs was prepared in methanol:water (50:50, v/v) at a final concentration of 100 μM for each compound. A mixed stock solution of all the IS was also prepared in methanol:water (50:50, v/v) at a final concentration of 100 μM, except LCA-D4, whose concentration was 200 μM. Quality controls (QCs) were prepared by an appropriate methanol:water (50:50, v/v) dilution of the different standard stock solutions.
The working solutions of the individual BA standards and IS were obtained by diluting stock solutions in methanol:water (50:50, v/v). Standards calibration curves, with concentrations in the 0.62–10,000 nM range, were prepared by serial half dilutions of the BA mix stock solution. IS concentrations were kept constant at all the calibration points at 1 µM for LCA-D4 and at 0.5 µM for CA-D4, DCA-D4, GCA-D4, and GCDCA-D4.
Sample collection
Rat and mouse studies.
Six-week-old male OFA rats (200–240 g) and 6-week-old male C57BL/6 mice (20–22 g) were purchased from Charles River (Barcelona, Spain) and acclimatized to laboratory conditions for at least 7 days. Animals were housed (12-h light-dark cycle, 21–25°C, 30–70% humidity, woodchip bedding) and fed ad libitum with a standard chow diet (Scientific Animal Food and Engineering, Augy, France).
Rats were anesthetized with sodium thiobarbital (0.1 g/kg), and mice were anesthetized with a mixture of ketamine/diazepam/atropine (50/5/1 mg/kg). In rats, blood was collected by cardiac puncture, whereas it was obtained from the abdominal aorta in mice. After coagulation and centrifugation (1,000 g for 10 min at 4°C), serum samples were aliquoted and stored at −80°C until the analysis. Livers were removed, rinsed in PBS, divided into small portions, flash-frozen in liquid N2, and stored at −80°C until the analysis. Serum and liver samples were obtained from the same animals. All the experimental protocols were approved by the Institutional Animal Ethics Committee.
Human studies.
Blood samples were collected from healthy human volunteers by standard venipuncture. After sample centrifugation, sera were aliquoted and stored at −80°C until analysis. Human liver samples were obtained from cadaveric liver grafts. These samples were obtained at the end of the bench surgery during the cold ischemia time and were immediately frozen in liquid N2 and stored at −80°C until analysis. Informed consent was obtained in all cases, and the experimental procedures were conducted in accordance with the ethical standards of the Declaration of Helsinki. The study was approved by the Institutional Ethics Committee.
Sample preparation
Serum samples.
First, 50 µl of serum samples were allowed to thaw on ice and were subsequently spiked with 25 µl of a 1/100 dilution of the deuterated IS stock solution. Afterward, 225 µl of cold methanol were added for protein precipitation, and samples were then vortexed 3 × 10 s and maintained at −20°C for 20 min. After centrifugation at 10,000 g for 10 min at 4°C, supernatants were transferred to clean tubes and dried in a Savant speedvac concentrator (Thermo Electron Corporation). The residue was then reconstituted by adding 50 µl of methanol:water (50:50, v/v), and was centrifuged at 10,000 g for 1 min at 4°C. The supernatant was transferred into 350 μl 96-well plates for its analysis.
Liver samples.
Frozen tissue samples (5–100 mg) were placed in 2 ml tubes containing CK14 ceramic beads (Precellys, Saint Quentin en Yvelines, France). For each 100 mg of tissue, 600 µl of cold methanol and 200 µl of a 1/100 dilution of the IS stock solution were added. Then, liver tissues were homogenized twice for 25 s at 6,000 rpm at 4°C in a Precellys 24 Dual system equipped with a Criolys cooler (Precellys). Tubes were centrifuged at 3,000 g for 5 min at 4°C, and supernatants were transferred to clean tubes. A second BA extraction was performed by 400 µl of cold methanol. Finally, the two extraction supernatants were pooled, aliquoted, and stored at −80°C until the analysis.
Aliquots of 150 μl of each homogenate were evaporated to dryness in a Savant speedvac concentrator and later reconstituted in 50 µl of methanol:water (50:50, v/v), centrifuged at 10,000 g for 1 min at 4°C, and transferred into 350 μl volume 96-well plates for further analysis.
Method validation
The bioanalytical method used in this study was developed in terms of linearity, accuracy, and precision following the compliance criteria described by the FDA Guidance for industry: bioanalytical method validation (27).
The lower limit of quantification (LLOQ) was defined as the lowest concentration at which the analyte could be quantified with a coefficient of variation (CV) below 20% and below ±20% deviation from the nominal value. The limit of detection (LOD) was determined as the lowest concentration at which the analyte response was at least three times the blank response. Calibration curves were generated by plotting the peak area ratio of the respective compound to the corresponding internal standard versus the nominal concentration (Table 1). The line of best fit was determined by linear-weighted (1/×) least-squares regression. The linearity acceptance criterion for the correlation coefficient was 0.99 or better. Each back calculated standard concentration should be within ±15% deviation from the nominal value, except for the LLOQ, for which the maximum acceptable deviation was ±20%.
TABLE 1.
Mass spectrometer setup for the quantification of selected bile acids
| Bile Acid | Transition | Cone | Collision Energy | Retention Time | Internal Standard |
| m/z | V | eV | min | ||
| CA | 407.3 > 407.3 | 120 | 10 | 13.90 | CA-D4 |
| αMCA | 407. 3 > 407.3 | 120 | 10 | 10.92 | CA-D4 |
| βMCA | 407. 3 > 407.3 | 120 | 10 | 11.30 | CA-D4 |
| ωMCA | 407. 3 > 407.3 | 120 | 10 | 10.57 | CA-D4 |
| HCA | 407. 3 > 407.3 | 120 | 10 | 12.71 | CA-D4 |
| CDCA | 391.3 > 391.3 | 120 | 10 | 17.13 | DCA-D4 |
| DCA | 391.3 > 391.3 | 120 | 10 | 17.22 | DCA-D4 |
| UDCA | 391.3 > 391.3 | 120 | 10 | 14.00 | DCA-D4 |
| HDCA | 391.3 > 391.3 | 120 | 10 | 14.41 | DCA-D4 |
| MuroCA | 391.3 > 391.3 | 120 | 10 | 12.69 | DCA-D4 |
| LCA | 375.3 > 375.3 | 90 | 10 | 17.74 | LCA-D4 |
| DHCA | 401.2 > 401.2 | 90 | 10 | 9.14 | CA-D4 |
| GCA | 464.3 > 74 | 80 | 40 | 11.08 | GCA-D4 |
| GHCA | 464.3 > 74 | 80 | 40 | 9.71 | GCA-D4 |
| GCDCA | 448.3 > 74 | 80 | 40 | 14.61 | GCDCA-D4 |
| GDCA | 448.3 > 74 | 80 | 40 | 15.32 | GCDCA-D4 |
| GUDCA | 448.3 > 74 | 80 | 40 | 10.71 | GCDCA-D4 |
| GHDCA | 448.3 > 74 | 80 | 40 | 10.97 | GCDCA-D4 |
| GLCA | 432.3 > 74 | 80 | 40 | 17.31 | LCA-D4 |
| GDHCA | 458.3 > 74 | 80 | 40 | 6.98 | GCA-D4 |
| TCA | 514.3 > 80 | 130 | 60 | 9.10 | GCA-D4 |
| TαMCA | 514.3 > 80 | 130 | 60 | 6.73 | GCA-D4 |
| TβMCA | 514.3 > 80 | 130 | 60 | 6.80 | GCA-D4 |
| TωMCA | 514.3 > 80 | 130 | 60 | 6.59 | GCA-D4 |
| THCA | 514.3 > 80 | 130 | 60 | 7.80 | GCA-D4 |
| TCDCA | 498.3 > 80 | 130 | 60 | 11.85 | GCDCA-D4 |
| TDCA | 498.3 > 80 | 130 | 60 | 12.55 | GCDCA-D4 |
| TUDCA | 498.3 > 80 | 130 | 60 | 8.61 | GCDCA-D4 |
| THDCA | 498.3 > 80 | 130 | 60 | 8.71 | GCDCA-D4 |
| TLCA | 482.3 > 80 | 130 | 60 | 16.17 | LCA-D4 |
| TDHCA | 508.2 > 80 | 130 | 60 | 5.87 | GCA-D4 |
| LCA-D4 | 379.3 > 379.3 | 90 | 10 | 17.74 | |
| DCA-D4 | 395.3 > 395.3 | 120 | 10 | 17.22 | |
| CA-D4 | 411.3 > 411.3 | 120 | 10 | 13.90 | |
| GCDCA-D4 | 452.4 > 74 | 80 | 40 | 14.61 | |
| GCA-D4 | 468.4 > 74 | 80 | 40 | 11.08 |
Intraday and interday precision and accuracy were determined by analyzing individual BAs in three different QCs corresponding to low (40 nM), medium (312 nM), and high (2500 nM) representative concentrations. The IS concentration was maintained constant in all the QCs (i.e., 1 μM for LCA-D4 and 0.5 μM for CA-D4, DCA-D4, GCA-D4, and GCDCA-D4). Each QC was analyzed five times in three different experimental samples batches. The acceptance criteria for precision within and between batches, expressed as CV, were ±20% for the LLOQ and ±15% or better for the other concentrations. Accuracy is expressed as the relative measurement error (RME) and was calculated using the following formula: RME (%) = 100 × (calculated concentration − nominal concentration)/nominal concentration. The acceptance criteria were ±20% for within and between batches for the LLOQ and ±15% or better for the other concentrations.
To account for possible matrix effects in BA recoveries and method accuracy, pooled samples were obtained for each specimen type (i.e., serum and tissue) and animal species (i.e., rat, mouse, and human). Then endogenous BAs were stripped off by activated charchoal incubation, as previously described (21). Next, known amounts of individual BAs were added to the prepared BA free matrices and to a blank matrix consisting in methanol:water (50:50, v/v). Three different concentrations were used for each BA: low (40 nM), medium (312 nM), and high (2500 nM). ISs were added to each sample, and their concentrations were kept constant in all the samples (1/200 dilution of the stock IS solution). Finally, spiked samples were evaporated to dryness, resuspended in methanol:water (50:50, v/v), centrifuged at 10,000 g for 1 min at 4°C, and transferred to 350 μl 96-well plates for their analysis. Each spiked sample was injected five times, and RME values were calculated.
RESULTS
The UPLC-MRM-MS method
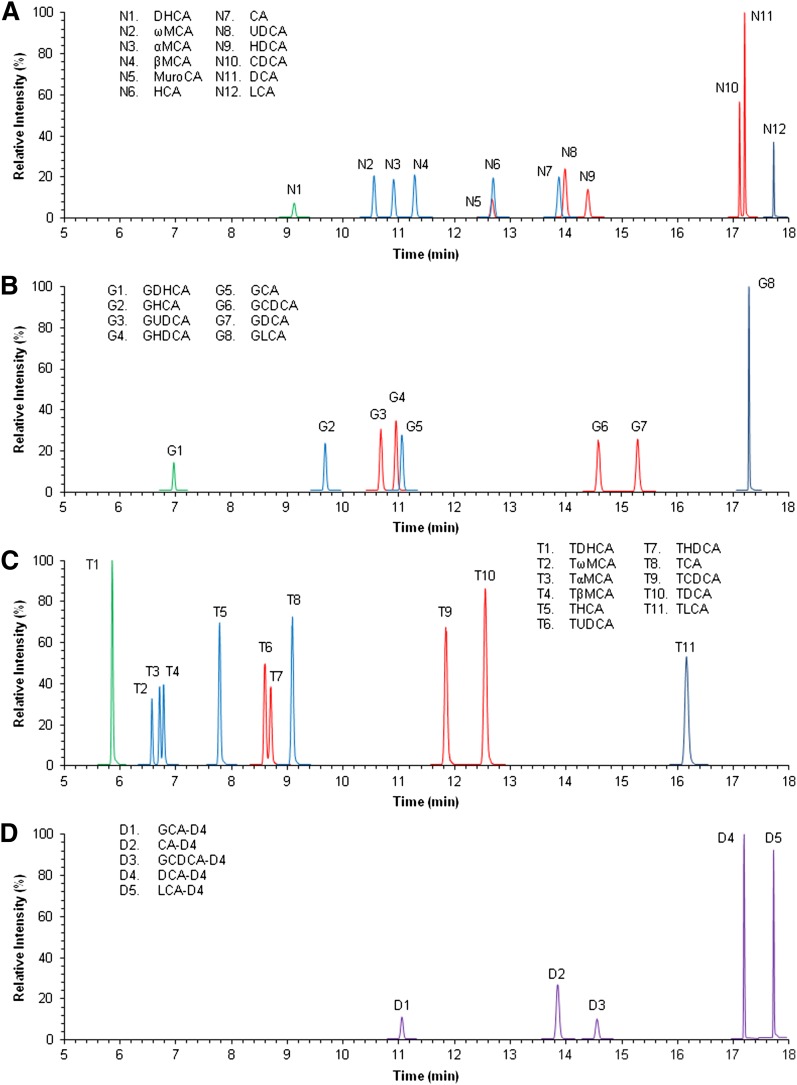
An UPLC-MRM-MS method for targeted BA profiling in different biological matrices has been developed in our laboratory. The method includes the simultaneous quantification of twelve nonconjugated, eight glycine-conjugated, and eleven taurine-conjugated BAs, and five additional deuterated BAs (IS). Given the existence of isobaric structures in the BA pool, appropriate chromatographic separation before MS detection is a particular critical issue for analyzing BAs. Here the successful separation of the 31 BAs was accomplished in 21 min by using a C18 reversed-phase column and acidified water and acetonitrile as eluents (Fig. 1). Representative chromatograms of the BA profiles for the rat, mouse, and human liver and serum specimens are depicted in supplementary Figures I–VI.
Fig. 1.
Chromatographic separation of the BAs standard mix solution. A: Nonconjugated BAs. Coeluting BAs (i.e., N5/N6) get separated and individually quantified by BA-specific MRM transition (Table 1). B: Glycine-conjugated BAs. C: Taurine-conjugated BAs. D: Deuterium-labeled internal standards. All the BAs were separated and detected in a single analytical run. Green: DHCA; blue: tri-hydroxylated BAs; red: di-hydroxylated BAs; dark blue: mono-hydroxylated BAs.
MS conditions, including the MRM transitions for each compound, were automatically set up by the direct infusion of each individual standard. Given the stability of the steroid nucleus, the identification of any characteristic product ions for the nonconjugated BAs was not possible. Hence, m/z values of 375.3, 391.3, 407.3, and 401.2 were respectively selected as both precursor and product ions for mono-, di-, and tri-hydroxylated BAs and DHCA. The use of the MRM operating mode instead of a single ion recording mode enabled the selection of a second mass filter to help eliminate bias due to possible analytical interferences, thus improving the signal-to-noise ratio (28). Regarding the conjugated BAs, taurine or glycine residues are easily fragmented; thus, it was possible to select different m/z values for the product ions and the precursor ions. In the glycine-conjugated species, an m/z value of 74 (loss of glycine) was selected as the common product ion for all the glycine conjugates, and m/z values of 432.3, 448.3, 464.3, and 458.3 were selected as the precursor ions for mono-, di-, and tri-hydroxylated and GDHCA bile acids, respectively. Regarding the taurine-conjugated BAs, m/z values of 482.3, 498.3, 514.3, and 508.2 were respectively selected as the precursor ions for mono-, di-, and tri-hydroxylated and TDHCA bile acids, and an m/z value of 80 (the SO3− anion from the taurine moiety) was selected as the product ion. The deuterated BAs used in our study as IS showed similar fragmentation profiles to their corresponding nondeuterated counterparts (Table 1).
Serum and tissue liver sample preparation
Most methods used to target BAs in serum involve the removal of proteins by precipitation using organic solvents, followed by a solid-phase extraction (SPE) (12, 22, 24, 28–33). On the other hand, preparation of liver tissue involves a homogenization step, usually followed by an SPE step (17, 34). SPE is used to concentrate the compounds of interest and to eliminate possible analytical interferences. After testing the behavior of different SPE matrices, low recovery efficiencies were obtained for a number of BAs (data not shown). Thus, to minimize sample handling and any possible bias, serum samples were processed by simple protein precipitation with methanol, followed by solvent evaporation and a final reconstitution of the pellet immediately before its analysis. Regarding liver sample preparation, a previous homogenization step before protein precipitation was carried out. In both procedures, five deuterated ISs (CA-D4, DCA-D4, LCA-D4, GCA-D4, and GCDCA-D4) were added at the beginning of the sample treatment procedure to account for any possible bias during the sample processing procedure.
The reproducibility of the serum processing method was assayed by analyzing six different preparations of one commercially available pooled human serum (Sigma-Aldrich) four times. The CV (%) in the quantification of individual BAs ranged from 1.1 to 11.6 in the four injection replicates and from 0.8 to 8.1 for the six serum preparations (see supplementary Table I). These results demonstrate the reproducibility and reliability of the serum sample preparation procedure used in our study.
One important question about the tissue analysis, particularly in the case of the liver where different structural and functional domains are present, is assessing whether a small, singular portion of tissue (i.e., a liver biopsy) is representative of the whole organ. To assess the representativeness of a liver biopsy in quantitative BA profiling, seven liver biopsies were randomly taken from different lobes of a rat liver, and each BA was further analyzed. In brief, the accuracy and precision quantification results show appropriate accuracies for all the BAs, with only a slight deviation in the determination of LCA and GLCA, two of the minor species present in the BA pool (supplementary Table II).
Method validation
The UPLC-MS method was validated in terms of accuracy, precision, and linearity. The possible matrix effects and bias during the analysis were also studied. Overall accuracy and precision were appropriate for all the determinations. Intraday and interday accuracies, measured as RME, ranged from −11.9% to 8.6%. Precision, measured as CV (%), was below 8.6 and 16.0 for intraday and interday, respectively (Table 2). In terms of linearity, the regression coefficients for all the calibration curves of the BAs were higher than 0.996. Calibration curve concentrations were increased to 10 μM, although linearity was maintained at higher concentrations for some BAs.
TABLE 2.
The precision, accuracy, and linear regression parameters obtained for each BA
| Intraday Validation |
Interday Validation |
Linear Regression Parametersb |
||||||||||||||
| Precision |
Accuracy |
Precision |
Accuracy |
|||||||||||||
| Bile Acid | La | M | H | L | M | H | L | M | H | L | M | H | ULOQ | LLOQ | LOD | R2 |
| nM | nM | nM | ||||||||||||||
| CA | 4.8 | 4.3 | 1.6 | 1.5 | 3.8 | 5.4 | 11.1 | 12.7 | 10.0 | 1.3 | 3.1 | 6.3 | 5,000 | 5 | 2.5 | 0.9992 |
| αMCA | 7.2 | 4.3 | 2.6 | 2.8 | 3.0 | 0.5 | 14.7 | 13.3 | 12.4 | 3.4 | 2.6 | 2.3 | 5,000 | 5 | 1.2 | 0.998 |
| βMCA | 6.9 | 4.6 | 3.2 | 2.8 | 1.8 | 3.0 | 13.3 | 13.3 | 13.4 | 3.3 | 0.7 | 4.6 | 5,000 | 5 | 1.2 | 0.998 |
| ωMCA | 5.5 | 3.4 | 1.4 | 2.6 | 2.4 | 1.8 | 12.3 | 13.1 | 13.0 | 3.5 | 1.7 | 3.2 | 5,000 | 5 | 1.2 | 0.998 |
| HCA | 4.8 | 4.2 | 3.4 | 3.4 | 2.5 | 4.5 | 9.0 | 9.9 | 9.9 | 3.8 | 1.5 | 5.2 | 5,000 | 2.5 | 1.2 | 0.998 |
| CDCA | 6.7 | 5.0 | 1.9 | −0.9 | −7.1 | 2.6 | 11.4 | 11.8 | 6.8 | −0.3 | −10.1 | 4.1 | 10,000 | 10 | 2.5 | 0.998 |
| DCA | 3.7 | 3.7 | 2.4 | 1.0 | −1.0 | −1.2 | 12.1 | 13.2 | 13.4 | 1.3 | −2.1 | −1.5 | 5,000 | 5 | 2.5 | 0.998 |
| UDCA | 4.4 | 3.7 | 3.3 | 3.1 | −10.2 | 1.7 | 14.8 | 12.3 | 10.3 | 3.4 | −11.9 | 1.1 | 5,000 | 5 | 1.2 | 0.996 |
| HDCA | 3.3 | 4.0 | 3.1 | 4.1 | −10.0 | 4.4 | 14.8 | 13.1 | 12.3 | 4.4 | −11.8 | 4.6 | 5,000 | 20 | 2.5 | 0.995 |
| MuroCA | 2.8 | 4.5 | 4.4 | 2.2 | −10.7 | 5.9 | 16.0 | 14.0 | 13.4 | 4.0 | −11.4 | 5.3 | 5,000 | 20 | 2.5 | 0.996 |
| LCA | 4.1 | 4.0 | 1.5 | 1.3 | 1.0 | −2.3 | 5.7 | 7.2 | 3.7 | 0.5 | 0.0 | −2.3 | 10,000 | 10 | 5 | 0.998 |
| DHCA | 5.6 | 3.9 | 2.0 | 3.5 | 2.0 | −2.0 | 9.6 | 11.5 | 9.6 | 3.9 | 1.6 | −1.6 | 10,000 | 20 | 10 | 0.998 |
| GCA | 4.0 | 1.2 | 1.1 | −1.1 | 1.7 | 3.1 | 10.5 | 8.1 | 9.5 | −1.2 | 1.1 | 4.3 | 10,000 | 5 | 2.5 | 0.9997 |
| GHCA | 4.0 | 1.3 | 1.6 | −2.8 | 6.7 | −0.6 | 8.8 | 7.1 | 6.2 | −2.8 | 6.4 | −1.4 | 10,000 | 20 | 5 | 0.9992 |
| GCDCA | 3.2 | 0.9 | 0.8 | −0.6 | 0.7 | 0.4 | 7.8 | 6.7 | 8.3 | −0.5 | 0.0 | 0.4 | 10,000 | 10 | 5 | 0.9998 |
| GDCA | 4.7 | 2.7 | 1.9 | −2.0 | 5.4 | 3.6 | 8.1 | 6.5 | 5.6 | −2.8 | 3.9 | 3.4 | 10,000 | 10 | 1.2 | 0.999 |
| GUDCA | 2.8 | 4.3 | 1.0 | −0.7 | −0.4 | 2.3 | 13.5 | 12.5 | 10.3 | −0.2 | −1.1 | 2.4 | 10,000 | 10 | 5 | 0.9997 |
| GHDCA | 3.4 | 4.5 | 1.1 | 0.7 | 1.2 | 2.1 | 12.1 | 12.7 | 10.0 | 1.0 | 0.2 | 0.7 | 10000 | 10 | 5 | 0.9997 |
| GLCA | 8.6 | 5.1 | 6.6 | −4.7 | 5.0 | −5.8 | 15.4 | 15.8 | 11.3 | −5.2 | 3.7 | −7.3 | 1,250 | 20 | 10 | 0.996 |
| GDHCA | 4.8 | 2.3 | 1.5 | 0.3 | 2.5 | −1.8 | 6.5 | 8.5 | 6.1 | 0.0 | 3.7 | −1.5 | 5,000 | 20 | 5 | 0.999 |
| TCA | 7.5 | 4.5 | 3.8 | 0.3 | 2.1 | 2.2 | 11.8 | 10.0 | 11.4 | −0.2 | 1.1 | 2.3 | 10,000 | 2.5 | 1.2 | 0.9991 |
| TαMCA | 6.5 | 3.8 | 3.9 | −1.5 | 4.1 | 2.4 | 10.8 | 12.4 | 11.1 | −1.7 | 4.0 | 3.1 | 10,000 | 10 | 1.2 | 0.9992 |
| TβMCA | 6.6 | 5.3 | 2.8 | −2.2 | 2.4 | 2.6 | 11.9 | 13.6 | 10.2 | −3.3 | 1.8 | 4.3 | 10,000 | 10 | 1.2 | 0.998 |
| TωMCA | 6.0 | 3.4 | 3.8 | −2.1 | 3.4 | 1.9 | 11.4 | 10.0 | 9.5 | −3.3 | 4.1 | 3.1 | 10,000 | 20 | 1.2 | 0.9993 |
| THCA | 6.4 | 3.9 | 4.3 | −2.9 | 1.0 | 6.0 | 11.2 | 10.6 | 10.3 | −4.3 | 0.9 | 8.6 | 10,000 | 10 | 1.2 | 0.999 |
| TCDCA | 5.5 | 4.3 | 3.5 | −2.8 | 0.5 | 7.8 | 6.1 | 6.1 | 5.8 | −3.4 | −1.8 | 8.5 | 10,000 | 10 | 2.5 | 0.998 |
| TDCA | 5.1 | 4.3 | 4.9 | 0.5 | −0.2 | 1.0 | 7.5 | 7.9 | 8.4 | 0.5 | −1.4 | 0.2 | 10,000 | 2.5 | 0.6 | 0.998 |
| TUDCA | 4.8 | 3.9 | 3.4 | −2.1 | −0.2 | 4.5 | 5.7 | 5.9 | 5.9 | −3.0 | −1.4 | 3.4 | 10,000 | 10 | 2.5 | 0.9991 |
| THDCA | 5.4 | 5.8 | 5.1 | −5.0 | −4.9 | 6.2 | 5.7 | 8.6 | 7.6 | −6.4 | −4.4 | 6.9 | 10,000 | 5 | 1.2 | 0.997 |
| TLCA | 6.2 | 5.3 | 5.7 | −1.7 | 0.3 | 1.1 | 15.7 | 10.2 | 14.5 | −3.7 | −0.9 | −0.1 | 5,000 | 20 | 5 | 0.997 |
| TDHCA | 5.5 | 3.7 | 3.4 | −0.1 | 3.2 | −0.6 | 8.4 | 6.1 | 6.9 | −0.6 | 5.1 | 1.1 | 10,000 | 20 | 2.5 | 0.9989 |
Precision is expressed as a coefficient of variation (%). Accuracy is expressed as the relative measurement error and is calculated using the following formula: RME (%) = 100 × (calculated concentration − nominal concentration)/nominal concentration.
L, low concentration (40 nM, each BA); M, medium concentration (312 nM, each BA); H, high concentration (2,500 nM, each BA).
ULOQ, upper limit of quantification; LLOQ, lower limit of quantification; LOD, limit of detection.
Wide dynamic linear ranges of quantification, from two to four orders of magnitude depending on the BA, and high sensitivity (LLOQ ranging from 2.5 to 20 nM and LOD from 0.6 to 10 nM) were achieved by the combined used of UPLC tandem modern MS instruments operating in the MRM mode.
Any possible matrix effects in BA recoveries during sample preparation and analysis were also studied. To this end, endogenous BAs were stripped off from the different biological matrices by incubation with activated charcoal. Afterward, known amounts of BAs were added to each stripped matrix and to blanks. The accuracy of BA quantification was calculated for each matrix at three different concentrations (i.e., low, medium, and high). Accuracy values ranged from −19.1% to 29.0% (expressed as RME), with only 7 of the 651 determinations beyond the ±20% range (see supplementary Table III).
Bile acids profiling in human, rat, and mouse serum and in liver specimens
The UPLC-MRM-MS method was satisfactorily applied to quantify the major and minor BAs present in human, rat, and mouse serum and in liver samples. The quantitative results for the serum and liver BAs profiling are respectively summarized in Table 3 and Table 4. In terms of total amount of BAs, the human species was seen to have the lowest BA content in liver and serum, rat had the highest concentration in serum, and mouse showed the highest BA content in the liver. As expected, the proportion of nonconjugated BAs was higher in serum than in the liver samples in all three species (supplementary Figure VII). Glycine-conjugated BAs were the most abundant in the human liver, whereas tauro-BAs were the predominant conjugated forms in rodents, with glycine-conjugates being practically absent in mouse.
TABLE 3.
Bile acid concentrations in the human, rat, and mouse serum samples
| Bile Acid | Human Serum | Rat Serum | Mouse Serum |
| CA | 440 ± 651 | 12,000 ± 5,600 | 1,240 ± 450 |
| αMCA | <LODa | 2,800 ± 1,300 | 142 ± 95 |
| βMCA | <LOD | 2,600 ± 2,700 | 1,080 ± 550 |
| ωMCA | <LOD | 1,360 ± 700 | 1,450 ± 970 |
| HCA | 24 ± 19 | 78 ± 31 | 15 ± 12 |
| CDCA | 380 ± 410 | 1,700 ± 600 | 42 ± 23 |
| DCA | 320 ± 120 | 420 ± 250 | 390 ± 220 |
| UDCA | 43 ± 27 | 200 ± 120 | 74 ± 45 |
| HDCA | <LLOQb | 230 ± 130 | 58 ± 31 |
| MuroCA | <LOD | 45 ± 20 | 45 ± 21 |
| LCA | <LOD | 54 ± 33 | 22 ± 11 |
| DHCA | <LOD | <LOD | <LOD |
| GCA | 85 ± 55 | 730 ± 500 | 3.5 ± 0.5 |
| GHCA | 6.8 ± 3.3 | 0.13 ± 0.35 | <LOD |
| GCDCA | 450 ± 210 | 74 ± 33 | 0.08 ± 0.20 |
| GDCA | 104 ± 44 | 48 ± 27 | <LOD |
| GUDCA | 76 ± 40 | 4.1 ± 1.9 | <LOD |
| GHDCA | 2.1 ± 3.4 | 8.1 ± 4.0 | <LOD |
| GLCA | 17 ± 20 | 2.3 ± 1.5 | 0.42 ± 0.49 |
| GDHCA | <LOD | <LOD | <LOD |
| TCA | 14 ± 12 | 660 ± 390 | 260 ± 110 |
| TαMCA | 2.9 ± 2.7 | 150 ± 210 | 102 ± 46 |
| TβMCA | <LOD | 160 ± 170 | 230 ± 140 |
| TωMCA | <LLOQ | 18 ± 11 | 290 ± 170 |
| THCA | 1.3 ± 1.1 | 1.44 ± 0.62 | 2.8 ± 2.7 |
| TCDCA | 69 ± 56 | 118 ± 73 | 12.3 ± 7.0 |
| TDCA | 21 ± 18 | 40 ± 21 | 55 ± 23 |
| TUDCA | 2.7 ± 2.7 | 6.9 ± 5.3 | 17 ± 12 |
| THDCA | <LOD | 16.1 ± 6.3 | 18.8 ± 7.7 |
| TLCA | 0.33 ± 0.52 | 2.5 ± 1.4 | 0.33 ± 0.61 |
| TDHCA | <LOD | <LOD | <LOD |
| Nonconjugated | 1,200 ± 1,100 | 22,000 ± 11,000 | 4,600 ± 2,300 |
| Glycine-conjugated | 740 ± 310 | 870 ± 550 | 4.08 ± 0.80 |
| Taurine-conjugated | 110 ± 88 | 1,480 ± 760 | 990 ± 500 |
| Total | 2,100 ± 1,100 | 24,000 ± 11,000 | 5,500 ± 2,800 |
The results are expressed in nM as mean ± SD (n = 8).
Under the limit of detection.
Under the limit of quantification.
TABLE 4.
Bile acid concentrations in the human, rat and mouse liver samples
| Bile Acid | Human liver | Rat liver | Mouse liver |
| CA | 6.5 ± 9.2 | 3,200 ± 3,900 | 3,000 ± 3,000 |
| αMCA | <LODa | 1,300 ± 1,300 | 530 ± 450 |
| βMCA | <LOD | 4,400 ± 5,600 | 3,400 ± 1,800 |
| ωMCA | <LOD | 1,080 ± 970 | 1,900 ± 1,500 |
| HCA | <LOD | 17 ± 14 | 15 ± 17 |
| CDCA | <LLOQb | 170 ± 200 | 25 ± 12 |
| DCA | 11.7 ± 8.1 | 66 ± 62 | 50 ± 42 |
| UDCA | <LOD | 91 ± 86 | 36 ± 15 |
| HDCA | <LOD | 140 ± 100 | 49 ± 13 |
| Murocholic acid | <LOD | 66 ± 40 | 170 ± 320 |
| LCA | <LLOQ | 16 ± 14 | <LLOQ |
| DHCA | <LOD | <LOD | <LOD |
| GCA | 2,700 ± 2,000 | 14,000 ± 6,700 | 330 ± 260 |
| GHCA | 2.7 ± 6.8 | 32 ± 25 | <LLOQ |
| GCDCA | 5,300 ± 3,700 | 16,70 ± 770 | 1.7 ± 3.3 |
| GDCA | 2,800 ± 3,000 | 920 ± 530 | 9.0 ± 3.6 |
| GUDCA | 250 ± 270 | 330 ± 120 | 7.3 ± 4.6 |
| GHDCA | <LOD | 130 ± 78 | <LLOQ |
| GLCA | 240 ± 370 | 67 ± 24 | 2.2 ± 4.7 |
| GDHCA | <LOD | <LOD | <LOD |
| TCA | 1,100 ± 770 | 48,000 ± 17,000 | 78,000 ± 50,000 |
| TαMCA | <LOD | 18,200 ± 4,600 | 20,000 ± 11,000 |
| TβMCA | <LOD | 21,000 ± 16,000 | 93,000 ± 49,000 |
| TωMCA | <LOD | 3,600 ± 1,800 | 67,000 ± 39,000 |
| THCA | <LOD | 141 ± 47 | 740 ± 550 |
| TCDCA | 2,800 ± 2,300 | 8,900 ± 3,900 | 5,300 ± 3,100 |
| TDCA | 1,100 ± 1,400 | 2,700 ± 1,500 | 7,000 ± 3,000 |
| TUDCA | 64 ± 74 | 2,300 ± 1,500 | 5,900 ± 3,700 |
| THDCA | <LOD | 920 ± 280 | 2,900 ± 1,800 |
| TLCA | 140 ± 180 | 370 ± 150 | 300 ± 100 |
| TDHCA | <LOD | <LOD | <LOD |
| Nonconjugated | 43 ± 48 | 11,000 ± 12,000 | 9,200 ± 6,600 |
| Glycine-conjugated | 11,300 ± 7,600 | 17,000 ± 7,900 | 350 ± 270 |
| Taurine-conjugated | 5,100 ± 4,100 | 106,000 ± 42,000 | 280,000 ± 16,000 |
| Total | 16,000 ± 11,000 | 133,000 ± 48,000 | 290,000 ± 160,000 |
The results are expressed in fmol/mg of tissue as mean ± SD (n = 8).
Under the limit of detection.
Under the limit of quantification.
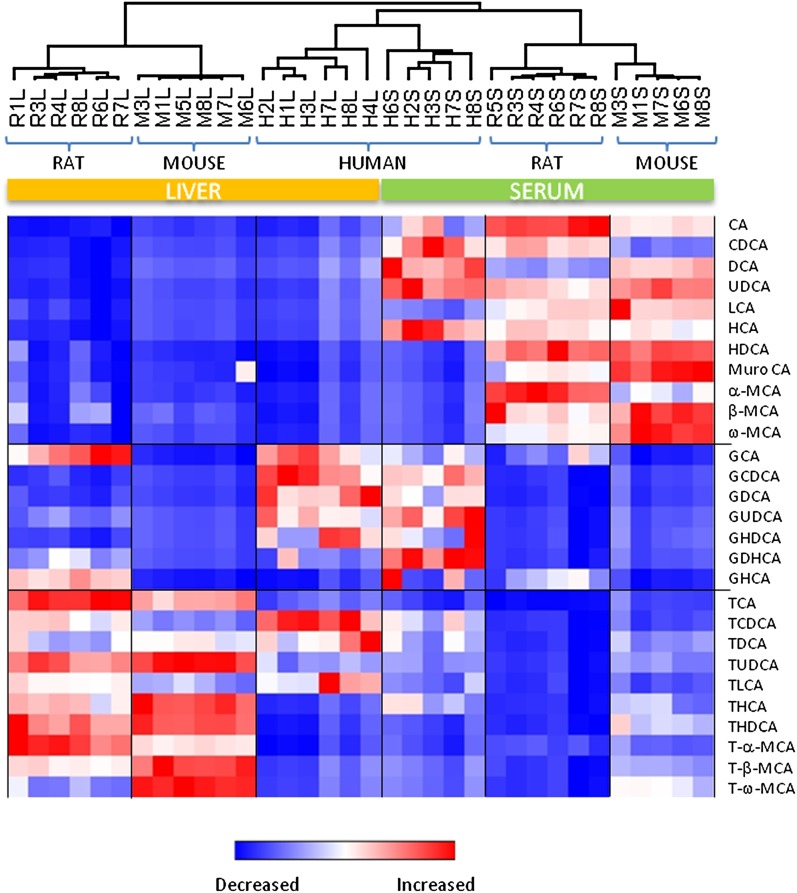
To gain a more in-depth insight into intra- and interspecies BA patterns, an unsupervised cluster analysis of the BA quantitative results was performed (Fig. 2). Despite the fact that a different profile of individual BAs was found between rat and mouse serum, nonconjugated BAs represented more than 80% in both species. CA was the most abundant form in rat serum (50% of the total), followed by αMCA (12%), βMCA (10%), and CDCA (7%), whereas CA (24%), ωMCA (24%), βMCA (19%), and DCA (7%) were the major BAs in mouse serum. Unlike what was observed in rodents, not only nonconjugated BAs (53% of the total) but also glycine-conjugated BAs (42%) were highly abundant in human serum, with GCDCA being the most abundant form (24% of the total), followed by DCA (19%), CA (16%), and CDCA (15%) nonconjugated BAs.
Fig. 2.
An unsupervised hierarchical clustering analysis based on the quantitative analysis of the 31 BAs. Each row shows the data for a specific BA, and each column shows the BAs profiles for the different species. The clustering analysis efficiently distinguishes between serum and liver samples and, more importantly, between the studied species.
A comparison made of the BA profiles in liver tissue between human and rodents revealed marked differences in not only the relative abundance of the taurine and glycine conjugates but also in the levels of specific BAs. Trihydroxylated taurine-conjugated BAs (TωMCA, TαMCA, TβMCA, and TCA) were the predominant forms in rodents (representing 67% and 89% of the total pool in rat and mouse livers, respectively), with higher proportions of TωMCA and TβMCA in mouse and of TαMCA and TCA in rat. This pattern was similar to that observed in rat and mouse serum with the homologous nonconjugated BAs. The taurine and glycine conjugates of CDCA, CA, and DCA were the predominant species in the human liver (accounting for 95% of the total BA hepatic pool). GCDCA and GDCA, two major BAs in the human liver, were minor forms in rodents, whereas the taurine conjugates of MCAs (not present in the human liver) were the most abundant hepatic BAs in rodents.
DISCUSSION
The main aim of the study was to develop a single UPLC-MRM-MS analytical method for the reliable quantification of the major and minor BAs present in different biological matrices (i.e., serum and liver tissue) and in different species (i.e., human, mouse, and rat). The BA pool in the biological samples presents isobaric structures that share identical fragmentation patterns. Therefore, such isomers can only be properly detected by the mass spectrometer if they have been previously well resolved during the liquid chromatography (LC) step. The combined use of UPLC speed, compared with conventional LC, and the increased peak capacity of small packed columns (<2 μm) enabled the achievement of suitable BA chromatographic separation (Fig. 1 and supplementary Figures I–VI). The use of MS operating in the MRM mode enabled the accomplishment of high sensitivity, reliable quantification, and a wide dynamic range. The proposed instrumental configuration offers a key advantage over other MS methods (13, 21, 35), allowing the profiling of a larger number of BAs. Moreover, the high sensitivity attained, if compared with other previously reported methods (20–24, 28, 30, 36), considerably reduces sample requirements because now 25 μl of serum and 5 mg of liver tissue suffice to perform proper BA profiling. This fact is particularly important when the amount of sample is limited (i.e., mouse serum or human liver tissue). Furthermore, the representativeness of such a small liver biopsy in relation to the whole organ is demonstrated by the good precision and accuracy values obtained during the BA quantification of seven liver biopsies from different parts of a specific rat liver (see supplementary Table II).
The method has been validated in terms of repeatability, precision, accuracy, and linearity by following the validation criteria established by the FDA (27). Overall, intraday and interday variations are under 16%, whereas accuracy, expressed as RME, is below 11% (Table 2). The validation results not only demonstrate the robustness of the method but also guarantee sufficient sensitivity and specificity for the reliable quantification of the 31 BAs present in the different biological samples. Profiling BAs is a potential tool to study liver-related pathological states (hepatobiliary diseases, hepatocarcinogenesis, steatosis, liver regeneration, and toxic damage). However, understanding the pathophysiological implications of altered BA homeostasis is limited by the fact that most studies are based on the quantitative analysis of a few predefined targets, with very little attention being paid to the less abundant forms (19). This fact is a major drawback, particularly for those studies that center on comparing the profiles of BAs (e.g., healthy and disease populations) where not only changes in the major BAs but also in the minor ones have been associated with several hepatobiliary diseases or liver damage conditions. For instance, significant alterations in the levels of some minor BAs (<1% of the total BA pool) have been reported in serum from patients with cholestasic liver diseases (13, 28), in plasma and urine from a genetic model of cholestasis in rats (31), in serum of rats treated with hepatotoxins (12), and in rat bile and liver samples during chemically induced hepatocarcinogenesis (37). Moreover, an in vitro study has suggested that LCA, a minor BA form, plays an important role in controlling the expression of several proteins involved in BA synthesis, metabolism, and transport (6). LCA is a promiscuous ligand capable of interacting with farnesoid X receptor, VDR, and PXR. It is considered a toxic BA with carcinogenic potential in the intestine and has shown cholestatic capacity in the liver of animals and humans. Overall, these findings evidence the interest of a method, like that presented herein, that is able to identify changes in a broad spectrum of BA species, including those present at relatively low concentrations.
BA composition in biological fluids (mainly serum, but also bile or urine) has been extensively investigated. The patterns of BAs that we obtained in serum samples from each species are in good agreement with those reported by other authors. However, our method provides a more specific and comprehensive profiling of BAs in rodents than previous reports in which α-, β-, and ω-MCA, and their taurine conjugates were not properly quantified (12, 23, 31). When using our method, not only primary and secondary BAs, the most commonly analyzed forms, but also free and conjugated 6αhydroxylated species can be accurately quantified. This allowed us to identify detectable levels of GHCA and THCA in human serum, thus extending previous studies that only showed the presence of the nonconjugated HCA homolog (10, 13). Furthermore, it was also possible to detect, for the first time, TαMCA in healthy human serum. These BAs, considered a priori unusual or nonhuman-specific BAs, have been detected by us in patients with severe liver diseases (unpublished results from our laboratory) and by others in different pathophysiological studies (26, 38, 39).
In contrast to circulating BAs, the profile of nonconjugated and conjugated BAs in liver tissue, and particularly in the human liver, has been poorly characterized. The liver plays a central role in the metabolic and transport processes that determine the BA body pool. By considering that BAs are signaling molecules involved in controlling the expression of different genes, any alteration of the intrahepatic BA content (concentration and/or composition) could affect the metabolic homeostasis of the liver and the whole organism. Defective canalicular export, due to impaired canalicular transport or a physical obstruction to bile flow, leads to intrahepatic BA accumulation, which may contribute to liver disease by inducing apoptosis and necrosis of hepatocytes (40). In addition, different studies in rodents have revealed significant alterations in the hepatic BA profile during hepatocarcinogenesis (37), liver regeneration (16), or high-cholesterol-diet feeding (20). We have recently reported altered levels of some BAs, including taurine and glycine conjugates, in the liver of patients suffering nonalcoholic fatty liver disease (17). However, to the best of our knowledge, comprehensive profiling of BAs in human liver tissue has not been performed. Our results show that GCDCA is the most abundant BA in the human liver (31% of the total pool), which represents a notable difference from rodents. Conjugated forms (glycine and taurine) of CDCA, a primary BA in humans, represent about 50% of the total BA pool in the human liver, whereas the total CA and MCA forms (α, β, or ω) are predominant in rodents (>80% of the total). These interspecies differences in the hepatic BA pool composition may be determinant in potential liver damage induced by specific BA accumulation in rodents and humans because CDCA is more toxic than CA or MCAs (9, 14).
In conclusion, we report the development of a new analytical method for the quantitative target profiling of circulating and hepatic BAs of human, mouse, and rat. The results clearly suggest that this method is a valuable tool for comparing BA profiles between different populations (e.g., healthy, disease or treated, nontreated), which is an especially important issue when searching for biomarkers and understanding liver damage mechanisms. As far as we are aware, it is one of the most comprehensive BA profiling tools described to date, and, more importantly, it allows an integrated comparative analysis of the major and minor BAs forms in human, rat, and mouse samples. Key advantages of the validated method include its high sensitivity, reproducibility, and low sample requirements. An important novelty of the current study is that it provides the first comparative analysis of circulating and hepatic BA profiles of human healthy donors and those of the two most commonly used experimental species in laboratory research with a single analytical method (9, 14). The observed interspecies differences in serum, and particularly in hepatic BA patterns, mean that special caution is needed with conclusions from animal studies on potential biomarkers or BA profiles characteristics of a particular disease when extrapolating them to the human liver.
Supplementary Material
Acknowledgments
The authors thank Dr. Laia Tolosa and Dr. Zacarías León for technical assistance and Dr. Roque Bort for valuable comments on the manuscript.
Footnotes
Abbreviations:
- αMCA
- alpha-muricholic acid
- BA
- bile acid
- βMCA
- beta-muricholic acid
- CA
- cholic acid
- CDCA
- chenodeoxycholic acid
- CV
- coefficient of variation
- DCA
- deoxycholic acid
- DHCA
- dehydrocholic acid
- GCA
- glycocholic acid
- GCDCA
- glycochenodeoxycholic acid
- GDCA
- glycodeoxycholic acid
- GDHCA
- glycodehydrocholic acid
- GHCA
- glycohyocholic acid
- GHDCA
- glycohyodeoxycholic acid
- GLCA
- glycolithocholic acid
- GUDCA
- glycoursodeoxycholic acid
- HCA
- hyocholic acid
- HDCA
- hyodeoxycholic acid
- IS
- internal standard
- LCA
- lithocholic acid
- LLOQ
- lower limit of quantification
- LOD
- limit of detection
- MRM
- multiple reaction monitoring
- MuroCA
- murocholic acid
- QC
- quality control
- RME
- relative measurement error
- SPE
- solid-phase extraction
- TαMCA
- tauro-alpha-Muricholic acid
- TβMCA
- tauro-beta-Muricholic acid
- TCA
- taurocholic acid
- TCDCA
- taurochenodeoxycholic acid
- TDCA
- taurodeoxycholic acid
- TDHCA
- taurodehydrocholic acid
- THCA
- taurohyocholic acid
- THDCA
- taurohyodeoxycholic acid
- TLCA
- taurolithocholic acid
- TUDCA
- tauroursodeoxycholic acid
- TωMCA
- tauro-omega-muricholic acid
- UDCA
- ursodeoxycholic acid
- UPLC
- ultraperformance liquid chromatography
- ωMCA
- omega-Muricholic acid
This work was supported by Instituto de Salud Carlos III of the Spanish Ministry of Science and Innovation (PI10/00923 and PI11/02942), by a Miguel Server contract (CP08/00125) from the Spanish Ministry of Science and Innovation/Instituto de Salud Carlos III. (A.L.), and by a predoctoral contract from the Vali+d program of the Conselleria d'Educació (Regional Valencian Ministry of Education) (J.C.G.-C.).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of seven figures and three tables.
REFERENCES
- 1.Botham K. M., Boyd G. S. 1983. The metabolism of chenodeoxycholic acid to beta-muricholic acid in rat liver. Eur. J. Biochem. 134: 191–196 [DOI] [PubMed] [Google Scholar]
- 2.Eyssen H., De Pauw G., Stragier J., Verhulst A. 1983. Cooperative formation of omega-muricholic acid by intestinal microorganisms. Appl. Environ. Microbiol. 45: 141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hofmann A. F., Hagey L. R. 2008. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell. Mol. Life Sci. 65: 2461–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang J. Y. 2009. Bile acids: regulation of synthesis. J. Lipid Res. 50: 1955–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hylemon P. B., Zhou H., Pandak W. M., Ren S., Gil G., Dent P. 2009. Bile acids as regulatory molecules. J. Lipid Res. 50: 1509–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan A. A., Chow E. C., Porte R. J., Pang K. S., Groothuis G. M. 2011. The role of lithocholic acid in the regulation of bile acid detoxication, synthesis, and transport proteins in rat and human intestine and liver slices. Toxicol. In Vitro. 25: 80–90 [DOI] [PubMed] [Google Scholar]
- 7.Pols T. W., Noriega L. G., Nomura M., Auwerx J., Schoonjans K. 2011. The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J. Hepatol. 54: 1263–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reschly E. J., Krasowski M. D. 2006. Evolution and function of the NR1I nuclear hormone receptor subfamily (VDR, PXR, and CAR) with respect to metabolism of xenobiotics and endogenous compounds. Curr. Drug Metab. 7: 349–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas C., Pellicciari R., Pruzanski M., Auwerx J., Schoonjans K. 2008. Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Discov. 7: 678–693 [DOI] [PubMed] [Google Scholar]
- 10.Trottier J., Bialek A., Caron P., Straka R. J., Milkiewicz P., Barbier O. 2011. Profiling circulating and urinary bile acids in patients with biliary obstruction before and after biliary stenting. PLoS ONE. 6: e22094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Want E. J., Coen M., Masson P., Keun H. C., Pearce J. T., Reily M. D., Robertson D. G., Rohde C. M., Holmes E., Lindon J. C., et al. 2010. Ultra performance liquid chromatography-mass spectrometry profiling of bile acid metabolites in biofluids: application to experimental toxicology studies. Anal. Chem. 82: 5282–5289 [DOI] [PubMed] [Google Scholar]
- 12.Yang L., Xiong A., He Y., Wang Z., Wang C., Li W., Hu Z. 2008. Bile acids metabonomic study on the CCl4- and alpha-naphthylisothiocyanate-induced animal models: quantitative analysis of 22 bile acids by ultraperformance liquid chromatography-mass spectrometry. Chem. Res. Toxicol. 21: 2280–2288 [DOI] [PubMed] [Google Scholar]
- 13.Trottier J., Bialek A., Caron P., Straka R. J., Heathcote J., Milkiewicz P., Barbier O. 2012. Metabolomic profiling of 17 bile acids in serum from patients with primary biliary cirrhosis and primary sclerosing cholangitis: a pilot study. Dig. Liver Dis. 44: 303–310 [DOI] [PubMed] [Google Scholar]
- 14.Palmeira C. M., Rolo A. P. 2004. Mitochondrially-mediated toxicity of bile acids. Toxicology. 203: 1–15 [DOI] [PubMed] [Google Scholar]
- 15.Reddy B. S., Watanabe K., Weisburger J. H., Wynder E. L. 1977. Promoting effect of bile acids in colon carcinogenesis in germ-free and conventional F344 rats. Cancer Res. 37: 3238–3242 [PubMed] [Google Scholar]
- 16.Monte M. J., Martinez-Diez M. C., El-Mir M. Y., Mendoza M. E., Bravo P., Bachs O., Marin J. J. 2002. Changes in the pool of bile acids in hepatocyte nuclei during rat liver regeneration. J. Hepatol. 36: 534–542 [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Canaveras J. C., Donato M. T., Castell J. V., Lahoz A. 2011. A comprehensive untargeted metabonomic analysis of human steatotic liver tissue by RP and HILIC chromatography coupled to mass spectrometry reveals important metabolic alterations. J. Proteome Res. 10: 4825–4834 [DOI] [PubMed] [Google Scholar]
- 18.Legido-Quigley C., McDermott L., Vilca-Melendez H., Murphy G. M., Heaton N., Lindon J. C., Nicholson J. K., Holmes E. 2011. Bile UPLC-MS fingerprinting and bile acid fluxes during human liver transplantation. Electrophoresis. 32: 2063–2070 [DOI] [PubMed] [Google Scholar]
- 19.Griffiths W. J., Sjovall J. 2010. Bile acids: analysis in biological fluids and tissues. J. Lipid Res. 51: 23–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bobeldijk I., Hekman M., de Vries-van der Weij J., Coulier L., Ramaker R., Kleemann R., Kooistra T., Rubingh C., Freidig A., Verheij E. 2008. Quantitative profiling of bile acids in biofluids and tissues based on accurate mass high resolution LC-FT-MS: compound class targeting in a metabolomics workflow. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 871: 306–313 [DOI] [PubMed] [Google Scholar]
- 21.Steiner C., von Eckardstein A., Rentsch K. M. 2010. Quantification of the 15 major human bile acids and their precursor 7alpha-hydroxy-4-cholesten-3-one in serum by liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 878: 2870–2880 [DOI] [PubMed] [Google Scholar]
- 22.Huang J., Bathena S. P., Csanaky I. L., Alnouti Y. 2011. Simultaneous characterization of bile acids and their sulfate metabolites in mouse liver, plasma, bile, and urine using LC-MS/MS. J. Pharm. Biomed. Anal. 55: 1111–1119 [DOI] [PubMed] [Google Scholar]
- 23.Alnouti Y., Csanaky I. L., Klaassen C. D. 2008. Quantitative-profiling of bile acids and their conjugates in mouse liver, bile, plasma, and urine using LC-MS/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 873: 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ando M., Kaneko T., Watanabe R., Kikuchi S., Goto T., Iida T., Hishinuma T., Mano N., Goto J. 2006. High sensitive analysis of rat serum bile acids by liquid chromatography/electrospray ionization tandem mass spectrometry. J. Pharm. Biomed. Anal. 40: 1179–1186 [DOI] [PubMed] [Google Scholar]
- 25.Honda A., Yamashita K., Numazawa M., Ikegami T., Doy M., Matsuzaki Y., Miyazaki H. 2007. Highly sensitive quantification of 7alpha-hydroxy-4-cholesten-3-one in human serum by LC-ESI-MS/MS. J. Lipid Res. 48: 458–464 [DOI] [PubMed] [Google Scholar]
- 26.Nakashima T., Sakamoto Y., Inaba K., Mitsuyoshi H., Ishikawa H., Nakajima Y., Sakai M., Shima T., Kashima K. 1999. A paucity of unusual trihydroxy bile acids in the urine of patients with severe liver diseases. Hepatology. 29: 1518–1522 [DOI] [PubMed] [Google Scholar]
- 27.FDA. 2001. Guidance for industry: bioanalytical method validation. FDA, Washington, DC.
- 28.Ye L., Liu S., Wang M., Shao Y., Ding M. 2007. High-performance liquid chromatography-tandem mass spectrometry for the analysis of bile acid profiles in serum of women with intrahepatic cholestasis of pregnancy. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 860: 10–17 [DOI] [PubMed] [Google Scholar]
- 29.McRae M., Rezk N. L., Bridges A. S., Corbett A. H., Tien H. C., Brouwer K. L., Kashuba A. D. 2010. Plasma bile acid concentrations in patients with human immunodeficiency virus infection receiving protease inhibitor therapy: possible implications for hepatotoxicity. Pharmacotherapy. 30: 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burkard I., von Eckardstein A., Rentsch K. M. 2005. Differentiated quantification of human bile acids in serum by high-performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 826: 147–159 [DOI] [PubMed] [Google Scholar]
- 31.Aoki M., Konya Y., Takagaki T., Umemura K., Sogame Y., Katsumata T., Komuro S. 2011. Metabolomic investigation of cholestasis in a rat model using ultra-performance liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 25: 1847–1852 [DOI] [PubMed] [Google Scholar]
- 32.Xiang X., Han Y., Neuvonen M., Laitila J., Neuvonen P. J., Niemi M. 2010. High performance liquid chromatography-tandem mass spectrometry for the determination of bile acid concentrations in human plasma. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 878: 51–60 [DOI] [PubMed] [Google Scholar]
- 33.Tessier E., Neirinck L., Zhu Z. 2003. High-performance liquid chromatographic mass spectrometric method for the determination of ursodeoxycholic acid and its glycine and taurine conjugates in human plasma. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 798: 295–302 [DOI] [PubMed] [Google Scholar]
- 34.Hagio M., Matsumoto M., Fukushima M., Hara H., Ishizuka S. 2009. Improved analysis of bile acids in tissues and intestinal contents of rats using LC/ESI-MS. J. Lipid Res. 50: 173–180 [DOI] [PubMed] [Google Scholar]
- 35.Scherer M., Gnewuch C., Schmitz G., Liebisch G. 2009. Rapid quantification of bile acids and their conjugates in serum by liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 3920–3925 [DOI] [PubMed] [Google Scholar]
- 36.Tagliacozzi D., Mozzi A. F., Casetta B., Bertucci P., Bernardini S., Di Ilio C., Urbani A., Federici G. 2003. Quantitative analysis of bile acids in human plasma by liquid chromatography-electrospray tandem mass spectrometry: a simple and rapid one-step method. Clin. Chem. Lab. Med. 41: 1633–1641 [DOI] [PubMed] [Google Scholar]
- 37.Mendoza M. E., Monte M. J., El-Mir M. Y., Badia M. D., Marin J. J. 2002. Changes in the pattern of bile acids in the nuclei of rat liver cells during hepatocarcinogenesis. Clin. Sci. (Lond.). 102: 143–150 [PubMed] [Google Scholar]
- 38.Batta A. K., Arora R., Salen G., Tint G. S., Eskreis D., Katz S. 1989. Characterization of serum and urinary bile acids in patients with primary biliary cirrhosis by gas-liquid chromatography-mass spectrometry: effect of ursodeoxycholic acid treatment. J. Lipid Res. 30: 1953–1962 [PubMed] [Google Scholar]
- 39.Nakashima T., Sano A., Seto Y., Nakajima T., Shima T., Sakamoto Y., Okuno T., Kashima K., Hasegawa T. 1990. Unusual trihydroxy bile acids in the urine of patients treated with chenodeoxycholate, ursodeoxycholate or rifampicin and those with cirrhosis. Hepatology. 11: 255–260 [DOI] [PubMed] [Google Scholar]
- 40.Hofmann A. F., Hagey L. R., Krasowski M. D. 2010. Bile salts of vertebrates: structural variation and possible evolutionary significance. J. Lipid Res. 51: 226–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.