Abstract
An autonomous synthetic programmable device that can diagnose a cell's state according to predefined markers and produce a corresponding therapeutic output may be the basis of future programmable drugs. Motivated to increase diagnosis precision, devices that integrate multiple disease markers have been implemented based on various molecular tools. As simplicity is key to future in-vivo applications, we sought a molecular device that a) integrates multiple inputs without requiring pairwise interactions, and b) harnesses only mechanisms that cells natively use. Here we show a synthetic NOR-based programmable device, operating via a biochemical obstructing approach rather than on a constructive approach, capable of differentiating between prokaryotic cell strains based on their unique expression profile. To demonstrate our system's strengths we further implemented the NOT, OR and AND gates. The device's programmability allows context-dependent selection of the inputs being sensed, and of the expressed output, thus, holding great promise in future biomedical applications.
Completion of the human genome sequence and technological advancements have made it possible to identify abnormal expression profiles in various diseases, including cancer1,2,3. Transcription Factors (TFs) are proteins that regulate the expression of genes by binding to specific DNA sequences. In various diseases, coordinated de-regulation of expression can be found underlying the development or maintenance of the diseased states. For example, cancer cells alter their expression profile to promote uncontrolled proliferation and suppress cell death mechanisms4. Expression-based targeting, in which a therapeutic gene is expressed under the control of an impaired transcription factor, expressed solely in the target cells, holds the promise for smart drugs capable of differentiating diseased cells from healthy ones, and affecting the latter accordingly5. Treatments based on single disease markers have been demonstrated by delivering a therapeutic gene under the control of a promoter that can be activated by transcription factors that are overexpressed and/or constitutively activated in cancer cells in numerous tumor types6,7,8,9,10.
However, diagnosis based on a single input may be error prone. Integration of multiple disease indicators, such as transcription factors, is advantageous over a single indicator since it increases diagnosis accuracy and decreases the probability of falsely classifying healthy versus diseased cells. For these reasons, systems integrating multiple inputs have been implemented11,12,13,14,15,16. These implementations are based on a constructive approach, in which the diagnostic computation is held in multiple steps. In the first step, each one of the disease markers controls a sub-component, such as a protein. In consecutive steps, sub-components repeatedly interact with each other to generate the final output, e.g., a reporter or a toxic protein, exclusively expressed in target diseased cells. Expanding these systems to a larger number of disease indicators requires addition of large number of sub-components which iteratively hold the sub-computations. Thus, to increase the diagnostic accuracy of these systems, multiple complex biochemical reactions are required, and therefore scaling them up may be difficult.
To overcome these constrains, we used an "obstructing" approach, similar to Tasmir et al.17. Here we show a NOR-gate based device that is capable of integrating multiple disease indicators without requiring pairwise interactions, harnessing only native cellular mechanisms to conduct computations. In accordance with NOR gate's logic, as can be seen in Figure 1a, we designed a single regulatory element that can serve as an integrator of several inputs and enables the expression of an output if and only if all inputs are absent (Fig 1b). The regulatory element is comprised of several potential binding regions, each corresponding to a specific pre-defined input (Fig 1b, balloon). One binding input is sufficient for inhibiting the expression of the output by physically blocking the transcription machinery. The binding regions are programmable and can utilize sequences of either prokaryotic TFs (such as lacI, which represses the expression of unnecessary proteins involved in the metabolism of lactose when the sugar is not available18) or eukaryotic TFs (such as p53, which binds the promoter of Survivin, an apoptosis inhibitor highly expressed in most human tumors, and therefore represses its expression19).
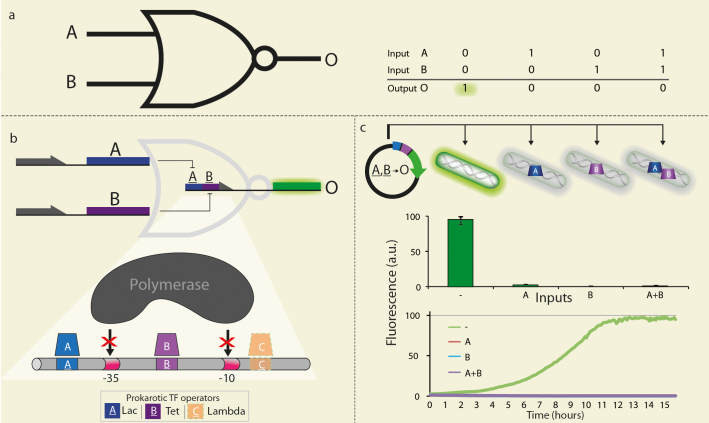
Figure 1. NOR-gate and its molecular implementation.
(a) The universal NOR-gate and it's truth table. (b) Molecular implementation of the NOR synthetic genetic circuit. A single regulatory element can be repressed by either one of several potential inputs. If and only if none are present, the RNA polymerase can attach to its binding site resulting in the GFP output's expression (A and B and A and B represent the TFs LacI and TetR and their corresponding potential binding regions Lac-Operator and Tet-Operator, respectively. O represents GFP.). The integrator is comprised of arbitrary regions, located downstream, upstream and in-between conserved regions, responsible for recruiting the transcription machinery (e.g., the RNA polymerase and its −35 −10 recruiting sequences). The arbitrary regions can be assigned with binding regions for TFs. This design applies for prokaryotic TFs (e.g., TetR, LacI, λ-Repressor, etc.) as well as for Eukaryotic TFs by principal (e.g., p53, E2F, FOXO, etc.). (c) The truth table of the four E. coli strains used to test the NOR synthetic genetic circuit, each genomically expressing one of the four possible input combinations. The NOR-gate plasmid was transformed into the four different strains. As can be seen, only the strain presenting none of the inputs resulted in a ‘1’ signal while the rest, presenting one or two inputs, resulted in a ‘0’ signal, in accordance to the NOR's truth table. Kinetics results are also shown, exhibiting efficient digital behavior over time - high signal strength while maintaining no signal leakage. Arbitrary unit (a.u.) is calculated as fluorescence/O.D2. Fluorescence values and their error bars are calculated as mean ± s.d. from three experiments.
Results
The NOR gate
We demonstrated this design in prokaryotic cells. Our integrator is capable of differentiating between four strains of E. coli, genomically expressing different logic combinations of two common TFs: NOR(A = 0, B = 0), XOR(A = 1, B = 0 or A = 0, B = 1) and AND(A = 1, B = 1). To test this ability we transformed the NOR-gate plasmid into the four different strains, as depicted in Fig. 1c. Only in strains expressing at least one of the TFs, the RNA polymerase is blocked from attaching to its binding site and the output protein is not expressed. All inputs and outputs are of the same type, i.e., TFs, allowing composition of logical circuits. The integrator controls the expression of another TF, which can serve as an input to another logic gate. To further test our NOR-gate in terms of robustness, efficiency and digital behavior, we've implemented three basic logic gates NOT, OR and AND (Figure 2).
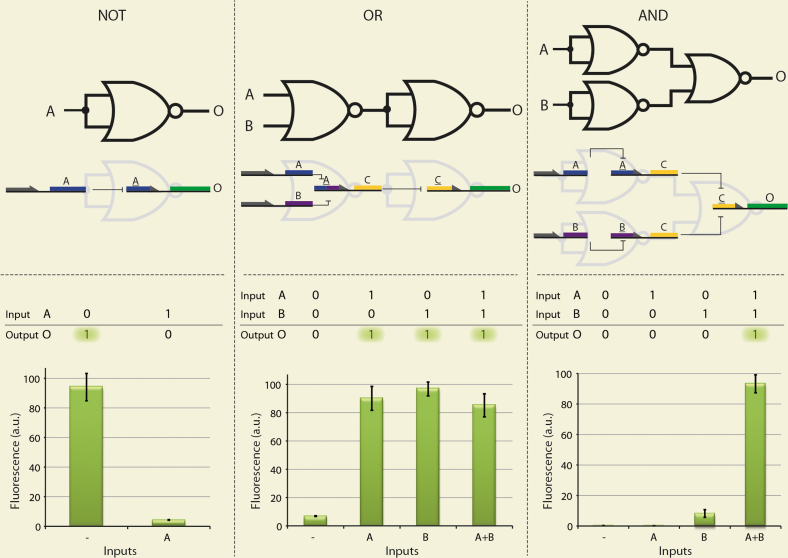
Figure 2. Molecular implementation of basic logic gates assembled by the NOR gate.
A, B and C and A, B and C represent the TFs LacI, TetR and λ-Repressor and their corresponding potential binding regions Lac-Operator, Tet-Operator and λ-Operator, respectively. O represents GFP. (a) NOT gate. If and only if (IFF) A is present, its corresponding promoter which controls the expression of the output protein is blocked, resulting in a ‘0’ output signal. (b) OR gate. IFF both A and B are absent, the expression of C is enabled, which in turn represses its promoter that controls the expression of the output protein, resulting in a ‘0’ output signal. (c) AND gate. Both A and B are needed to repress C, which in turn controls the expression of the output protein. Thus, IFF both input signals are ‘1’ the output signal is ‘1’. Arbitrary unit (a.u.) is calculated as fluorescence\O.D2. Fluorescence values and their error bars are calculated as mean ± s.d. from three experiments.
NOT gate
The NOT gate is based upon a rather straight-forward signal inverter. If and only if input A's signal is ‘1’, i.e. repressor TF that represents input A is present, its corresponding promoter which controls the expression of the output protein is blocked, resulting in a ‘0’ output signal. As seen in Figure 2a, the output protein was expressed only in strains lacking input A, corresponding to a NOT-gate's logic.
OR gate
The OR gate plasmid was derived from the previously constructed NOR gate, in which the output protein was replaced with an intermediate repressor, C. The resulting plasmid is comprised of a promoter incorporating the binding regions of inputs A and B, and controls the expression of C in a NOR fashion. Based on the abstract digital logical representation, in which the OR gate is formed by inverting the NOR gate's signal, an additional element was added, in which the output protein is controlled by the inverting repressor, C. If and only if both A and B are absent, repressor C is expressed and the output protein is blocked from expression. As seen in Figure 2b, the output protein was expressed in strains containing either input A, input B, or both – corresponding to an OR-gate's logic.
AND gate
In order to implement the AND gate, the intermediate repressor C was placed under the control of both inputs, A and B, in an independent manner. The output protein was placed under the control of the C repressor. If and only if repressor C is absent, the output protein is expressed. Repressor C's absence is dependent on both input A and input B's presence. Overall, as seen in Figure 2c, the output protein was expressed only in strains containing both input A and input B – corresponding to an AND gate's logic.
As can be seen all gates maintained robust and digital behavior, exhibiting very low signal leakage and keeping a high signal yield and strength (control experiments, including kinetics of the system can be found in supplementary Fig. S1 and Fig. S2).
Discussion
In this work we implemented a dual-repressed promoter, serving as a NOR gate, along with a complete set of Boolean gates (NOT, OR & AND) in prokaryotic cells. Our system is modular and programmable by design – any repressing TF can be used as its input, and any gene of interest can be set as the expressed output. This is in line with the systems of Elowitz20 and Gardner21 who pioneered the field of synthetic gene circuits. Their systems are also based on the utilization of TFs, in which the inputs and outputs are of the same type, allowing direct and easy composition of basic logic gates into cascadable circuits, unlike systems based on tRNA22, aptamers or RNA alternative splicing11, and microRNAs and RNA interference15,23,24. A system possessing these features—input and output modularity, programmability and cascadability—allows accurate targeting of desired cells without falsely targeting other cells.
Our NOR-based design can be scaled to multiple inputs while maintaining a simple molecular implementation by forsaking pairwise interaction of the different individual inputs. Unlike AND-gate based systems13, which require pairwise interactions of inputs through iterative sub-computations (as depicted in Supplementary Fig. 3), our NOR-based design is based on the direct integration of different inputs, where each input directly and independently controls the output gene, in parallel with the other inputs. In addition, the system is based on an obstructive approach, e.g., repressing TFs that interfere with the regular regulatory machinery by steric blockage, similar to Tasmir et al.17, rather than a constructive approach, e.g., protein–protein interactions which is not easy to scale. Tasmir et al.17 recently demonstrated a genetic NOR gate based on the concatenation of two potentially repressible tandem promoters in E. coli. Either promoter, if in an unrepressed state, can solely suffice to drive the expression of a downstream repressor, which in turn can repress its corresponding downstream output gene. In terms of scalability, given that promoters are large entities, only a small number can be concatenated, since each added promoter will have to be farther from the transcriptional start site. This is particularly relevant for future medical applications given that mammalian cells' promoters are of much greater magnitude. In contrast, the repression operators (approximately 20 bases) are significantly smaller than promoters and therefore many can be concatenated within one promoter. Additionally, in the system of Tasmir et al.17, the inputs are two chemical external inducers incubated in the culture tubes together with the bacteria. These inducers can bind and inhibit the two TFs repressors that repress the two tandem promoters. If and only if the two external inducers are absent the output gene was expressed. External inputs accommodated Tasmir et al.17 goal of interconnecting individual E. coli colonies via chemical components functioning as the ‘wires’. However, the changes and anomalies underlining various diseases start and subside with endogenous intra-cellular changes4 (such as the deregulation of TFs levels). Therefore, for the goal of cell-state diagnosis computing we chose to use internal inputs. Delivery of the NOR circuit using traditional methods (such as transfection25) into all cells (target and normal) will allow the circuit to sense and analyze these intra-cellular inputs present inside the cell. Accordingly, we designed an integrator that accepts innate TFs as inputs and computes NOR-based logic gates with them. Together, these features offer an advance over previous approaches as they simplify the biochemical reactions underlying the computation and increase the feasibility to operate in a biological environment.
We have demonstrated our system's abilities in prokaryotic cells which are far less complex than mammalian cells. However, we believe its true potential is for diagnosis of disease indicators in mammalian cells as it is based on: a native cellular machinery; a destructive approach; and, can analyze both over-expressed TFs (such as oncogenes), and under-expressed TFs (such as tumor suppressors). When detecting the absence of tumor suppressors, it suffices for one tumor suppressor (which normally should be present) to directly attach onto its corresponding potential binding region and inhibit the expression of a protein which induces apoptotic cell death, as shown in Supplementary Figure 4a. When detecting the presence of oncogenic TFs, the over-expressed oncogenes converge to inhibit the expression of an intermediate repressor which in turn inhibits the expression of the output protein. One normally absent oncogene suffices to inhibit the expression of the output protein, as shown in Supplementary Figure 4b. Thus, in accordance with the NOR gate truth table, if and only if all inputs are aberrantly expressed, i.e., all tumor suppressors are absent and all oncogenes are present, the output is expressed. The system presented in this work demonstrates how the NOR gate can analyze TF inputs based on their digital presence or absence (as opposed to being able to analyze any analog or gradual level of expression). Although analog gradual de-regulation is more common than digital exclusive presence or absence, it is the last that holds the promise for cancer-specific gene therapies1. Digital, i.e., unique and distinct markers, enable greater specificity and optimized target versus non-target cells discrimination. And indeed, cancer-specific gene therapies1 based on this digital absence or presence principle, have already been clinically tested in numerous cancer types6,7,8,9,10. In these transcriptionally targeted gene therapies, a digital TF4 exclusively present in target cells, while absent in normal cells, solely controls the expression of a therapeutic gene. Thus, corresponding exclusive expression in target cells and not in normal cells is achieved. Scaling up the number of sensed inputs, while sensing both aberrantly present (e.g., oncogenes) and aberrantly absent (e.g., tumor suppressors) TFs, vastly broadens the repertoire of potential markers that can be analyzed. A mammalian system based on this design may allow analyzing the presence or absence of numerous cancer-related TFs and the induction of cells death if all TFs were aberrantly expressed, and therefore may have important future biological and medical applications.
Methods
Strains
All studies were performed using four different DH5α E. coli strains, genomically expressing the four inputs combinations, none, LacI, TetR and LacI and TetR, termed DH5α, DH5αZi, DH5αZr and DH5αZ1, respectively. DH5αZr (chromosomal TetR integration) was achived as follows: DH5αZr was prepared via chromosomal integration procedure as follows: The TetR gene was integrated in a DH5α E. coli strain that carries in its chromosome the attB site via Int mediated site specific recombination. For this, plasmid pZS4Int-tetR together with pIntAssist were used. pIntAssist carries a temperature sensitive origin of replication and upon heat treatment was lost after the integration procedure, i.e. the resulting strain carries a Spectinomycin resistance cassette in the chromosome only. A respective protocol can be found by the supplier of the pZ system (more details can be found on the website http://expressys.com/).
Media
Lysogeny broth (LB) plates with appropriate antibiotics were obtained from the bacteriology services (Weizmann Institute) and prepared as described26. Strains were grown in Lysogeny broth (LB) medium supplied by the Weizmann institute bacteriology unit and were grown overnight at 37°C with 250 rpm shaking. The cultures were diluted 1:100 into 200 μl of medium in a 96-well plate with different combinations of antibiotics and/or inducers; 34 μg/ml chloramphenicol and/or 50 μg/ml kanamycin and/or 100 μg/ml Ampicillin and/or 50 μg/ml Spectomycin and/or IPTG 1mM and/or anhydrotetracycline 100 ng/ml.
Plasmids
All plasmids are based on the components of the pZ Expression System and its nomenclature is as follows: The letter (E, A, S, S*) denotes the origin of replication. The first number indicates the resistance marker (1 to 5). The second number (1 to 5) defines the promoter controlling the transcription of the gene of interest. The MCS or the description of the gene of interest, e.g. GFP, follows this code as exemplified. The nomenclature can be found in Supplementary Table 1, and the derivative plasmids and their nomenclature used in our paper can be found in Supplementary table 2. We wish to thank the kind members of Uri Alon's and Michael Elowitz laboratories for sharing their wisdom and plasmids.
Liquid handling and measurements
Assembly, execution and readout of the experiments, i.e., liquid handling, orbital shaking, growth in stable 37°C temperature, were done on a Tecan Freedom® 2000 robot controlled by in-house developed software. Fluorescence signals were read by a Tecan Infinite® 200 microplate-reader: GFP (Excitation Wavelength: 497 nm. Emission Wavelength: 535 nm). mCherry (Excitation Wavelength: 587 nm. Emission Wavelength: 614 nm). Reaction's components: *) LB. *) Bacteria strain, expressing one of the four desired input combinations (none, LacI, TetR and LacI and TetR), and transformed with one or more of the plasmids implementing desired gates. *) Appropriate antibiotics according to Supplementary Table 1 and Supplementary Table 2.
Author Contributions
E.S. supervised the project. T.R. conceived the idea of this study and its corresponding experimental design. T.R., Y.D. and L.M. performed the experiments, discussed the results and their implications, and suggested technical and conceptual advice accordingly. T.R., Y.D. and E.S. wrote the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We thank R. Adar and M. Oren for their assistance and advice. In addition, we thank A. Mishali and K. Katzav for their help in preparation of the figures. This research was supported by the European Union FP7-ERC-AdG. E. Shapiro is the Incumbent of the Harry Weinrebe Professorial Chair of Computer Science and Biology.
References
- Lo H. W., Day C. P. & Hung M. C. Cancer-specific gene therapy. Advances in genetics 54, 235–255 (2005). [DOI] [PubMed] [Google Scholar]
- Ramaswamy S. et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proceedings of the National Academy of Sciences of the United States of America 98, 15149–15154 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. H., Bernard P. S. & Perou C. M. Molecular portraits and the family tree of cancer. Nature genetics 32 Suppl 533–540 (2002). [DOI] [PubMed] [Google Scholar]
- Vogelstein B. & Kinzler K. W. Cancer genes and the pathways they control. Nature Medicine 10, 789–799 (2004). [DOI] [PubMed] [Google Scholar]
- DeFatta R. J., Li Y. & De Benedetti A. Selective killing of cancer cells based on translational control of a suicide gene. Cancer gene therapy 9, 573–578 (2002). [DOI] [PubMed] [Google Scholar]
- Venkataraman S., Dirks R. M., Ueda C. T. & Pierce N. A. Selective cell death mediated by small conditional RNAs. Proceedings of the National Academy of Sciences of the United States of America 107, 16777–16782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pandha H. S. et al. Genetic prodrug activation therapy for breast cancer: A phase I clinical trial of erbB-2-directed suicide gene expression. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 17, 2180–2189 (1999). [DOI] [PubMed] [Google Scholar]
- Terazaki Y. et al. An optimal therapeutic expression level is crucial for suicide gene therapy for hepatic metastatic cancer in mice. Hepatology 37, 155–163 (2003). [DOI] [PubMed] [Google Scholar]
- Song J. S. & Kim H. P. Adenovirus-mediated HSV-TK gene therapy using the human telomerase promoter induced apoptosis of small cell lung cancer cell line. Oncology reports 12, 443–447 (2004). [PubMed] [Google Scholar]
- Song J. S. et al. Adenovirus-mediated suicide gene therapy using the human telomerase catalytic subunit (hTERT) gene promoter induced apoptosis of ovarian cancer cell line. Bioscience, Biotechnology and Biochemistry 67, 2344–2350 (2003). [DOI] [PubMed] [Google Scholar]
- Culler S. J., Hoff K. G. & Smolke C. D. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science 330, 1251–1255 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim L., Beatus T. & Bar-Ziv R. An autonomous system for identifying and governing a cell's state in yeast. Physical biology 4, 154–163 (2007). [DOI] [PubMed] [Google Scholar]
- Nissim L. & Bar-Ziv R. H. A tunable dual-promoter integrator for targeting of cancer cells. Molecular systems biology 6, 444 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaudo K. et al. A universal RNAi-based logic evaluator that operates in mammalian cells. Nature Biotechnology 25, 795–801 (2007). [DOI] [PubMed] [Google Scholar]
- Leisner M., Bleris L., Lohmueller J., Xie Z. & Benenson Y. Rationally designed logic integration of regulatory signals in mammalian cells. Nature nanotechnology 5, 666–670 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Wroblewska L., Prochazka L., Weiss R. & Benenson Y. Multi-Input RNAi-Based Logic Circuit for Identification of Specific Cancer Cells. Science 333, 1307–1311 (2011). [DOI] [PubMed] [Google Scholar]
- Tamsir A., Tabor J. J. & Voigt C. A. Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires’. Nature 469, 212–215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M. The lac repressor. Comptes Rendus Biologies 328, 521–548 (2005). [DOI] [PubMed] [Google Scholar]
- Hoffman W. H., Biade S., Zilfou J. T., Chen J. & Murphy M. Transcriptional Repression of the Anti-apoptoticsurvivin Gene by Wild Type p53. Journal of Biological Chemistry 277, 3247–3257 (2002). [DOI] [PubMed] [Google Scholar]
- Elowitz M. B. & Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000). [DOI] [PubMed] [Google Scholar]
- Gardner T. S., Cantor C. R. & Collins J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000). [DOI] [PubMed] [Google Scholar]
- Anderson J. C., Voigt C. A. & Arkin A. P. Environmental signal integration by a modular AND gate. Molecular systems biology 3, 133 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaudo K. et al. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat Biotechnol 25, 795–801 (2007). [DOI] [PubMed] [Google Scholar]
- Deans T. L., Cantor C. R. & Collins J. J. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell 130, 363–372 (2007). [DOI] [PubMed] [Google Scholar]
- Patil S. D., Rhodes D. G. & Burgess D. J. DNA-based therapeutics and DNA delivery systems: a comprehensive review. Aaps J 7, E61–77 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J R. D. Molecular Cloning: A laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, New York (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information