Abstract
In the title compound, [Ir(C18H15P)2(CO)3]PF6·CH3OH, the IrI atom is coordinated by two triphenylphosphine ligands in axial sites and three carbonyl ligands in the equatorial plane of a fairly regular trigonal bipyramid: the equatorial C—Ir—C angles range from 115.45 (9) to 126.42 (10)°. The small deviations from the ideal tetrahedral geometry around the P atoms are illustrated by C—P—C angles ranging from 104.08 (9) to 106.46 (9)°. In the crystal, the molecules are linked by weak C—H⋯F, C—H⋯O and C—H⋯π interactions.
Related literature
For related complexes, see: Randall et al. (1991 ▶, 1994 ▶); Raper & McDonald (1973 ▶). For other P-donor ligands, see: Purcell et al. (1995 ▶); Otto & Roodt (2001 ▶); Otto et al. (2005 ▶); Muller et al. (2008 ▶). For their use in catalytic olefin transformation reactions, see: Haumann et al. (2004 ▶); Crous et al. (2005 ▶); Booyens et al. (2007 ▶); Ferreira et al. (2007 ▶).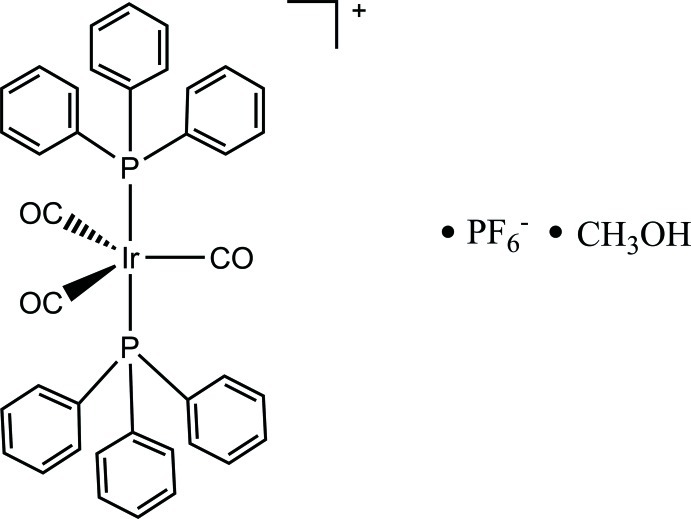
Experimental
Crystal data
[Ir(C18H15P)2(CO)3]PF6·CH4O
M r = 977.80
Monoclinic,
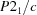
a = 16.487 (5) Å
b = 13.571 (4) Å
c = 20.903 (5) Å
β = 125.297 (5)°
V = 3817 (2) Å3
Z = 4
Mo Kα radiation
μ = 3.69 mm−1
T = 100 K
0.18 × 0.14 × 0.06 mm
Data collection
Bruker APEXII CCD diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2008 ▶) T min = 0.556, T max = 0.809
68611 measured reflections
9501 independent reflections
8699 reflections with I > 2σ(I)
R int = 0.037
Refinement
R[F 2 > 2σ(F 2)] = 0.019
wR(F 2) = 0.044
S = 1.03
9501 reflections
489 parameters
H-atom parameters constrained
Δρmax = 1.01 e Å−3
Δρmin = −0.75 e Å−3
Data collection: APEX2 (Bruker, 2011 ▶); cell refinement: SAINT-Plus (Bruker, 2008 ▶); data reduction: SAINT-Plus; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: DIAMOND (Brandenburg & Putz, 2005 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812035593/hb6932sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812035593/hb6932Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg1, Cg2 and Cg3 are the centroids of the C11–C16, C21–C26 and C41–C46 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C15—H15⋯F3i | 0.95 | 2.39 | 3.281 (3) | 157 |
| C16—H16⋯F6i | 0.95 | 2.53 | 3.319 (3) | 141 |
| C42—H42⋯F6i | 0.95 | 2.38 | 3.138 (3) | 136 |
| C43—H43⋯F2i | 0.95 | 2.49 | 3.386 (3) | 158 |
| C45—H45⋯O01ii | 0.95 | 2.50 | 3.281 (3) | 139 |
| C64—H64⋯F4iii | 0.95 | 2.47 | 3.200 (3) | 133 |
| O01—H01⋯F3i | 0.84 | 2.27 | 3.059 (3) | 157 |
| C53—H53⋯Cg1iv | 0.95 | 2.68 | 3.523 (2) | 148 |
| C35—H35⋯Cg3iv | 0.95 | 2.91 | 3.587 (2) | 129 |
| C13—H13⋯Cg2v | 0.95 | 2.97 | 3.744 (2) | 140 |
Symmetry codes: (i) 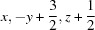 ; (ii)
; (ii) 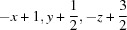 ; (iii)
; (iii) 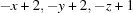 ; (iv)
; (iv) 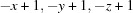 ; (v)
; (v) 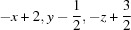 .
.
Acknowledgments
Financial assistance from the Department of Science and Technology (DST) of South Africa, the South African National Research Foundation (SA-NRF/THRIP), the DST–NRF centre of excellence (c*change), the University of the Free State and INKABA yeAfrica funding projects are gratefully acknowledged.
supplementary crystallographic information
Comment
P donor ligands (Muller et al., 2008; Purcell et al., 1995; Otto et al., 2005; Otto & Roodt, 2001) form part of ongoing research in different catalytic olefin transformation reactions such as hydroformylation (Haumann et al., 2004; Crous et al., 2005), metathesis (Booyens et al., 2007) and methoxycarbonylation (Ferreira et al., 2007). As part of our studies in this area, we now describe the structure of the title compound: all bond distances and angles fall within the range for similar complexes (Randall et al. 1991, 1994; Raper & McDonald, 1973).
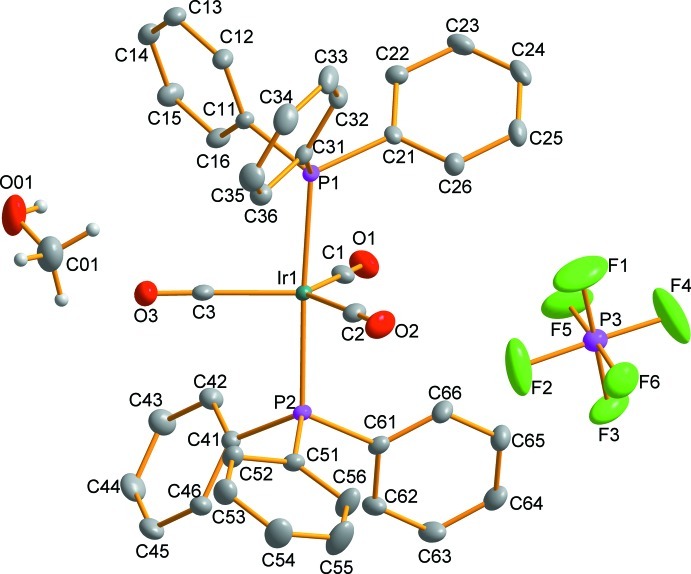
The main fragment of the crystal structure of the title compound, [Ir(CO)3(PPh3)2](PF6).MeOH, was originally reported by Randall et al., 1991, in the trigonal form, crystallizing in the space group R3 with hydrogen sulfate as counter ion. In this case, the Ir(I) complex (Figure 1) crystallizes with one hexafluoridophosphate anion and a methanol solvent molecule in the P21/c spacegroup. The trigonal bipyramidal complex consists of three carbonyl groups in the equatorial plane and two triphenylphosphine ligands in the axial plane.
Similar Ir—P distances (2.3620 (8) and 2.3599 (8) Å) and P1—Ir—P2 angle of 177.047 (18) ° make the phosphine ligands equally trans. Ir—C3 distance of 1.947 (2) Å is slightly longer than for Ir—C1 and Ir—C2 distances, both equal to 1.938 (2) Å. Ir—C—O angles are close to linear (175.6 (2) - 178.8 (2) °) and C—O distances range from 1.107 (3) - 1.135 (3) Å, with C3—O3 distance the shortest. Angles between the equatorial ligands show some distortion with C2—Ir1—C3 = 115.45 (9) ° and C1—Ir1—C3 = 118.13 (9) ° compared to C2—Ir1—C1 = 126.42 (10) °. C—P—C angles range from 104.07 (9) - 106.46 (9) ° illustrating the distorted tetrahedral geometry around the P atoms.
In the crystal, weak C—H···F, C—H···O and C—H···π, interactions link the molecules into a supra-molecular network (Table 1).
Experimental
CO was bubbled through a solution of [Ir(COD)(PPh3)2]PF6 (cod = 1,5-cyclooctadiene) (50.0 mg, 0.0515 mmol) in benzene while the mixture was vigorously stirred under gentle reflux. Rapid displacement of COD occurs after which all solvents were evaporated. The product was filtered after the addition of methanol and diethyl ether. Slow evaporation of methanol solution gave yellow blocks. (Yield: 40.1 mg, 82%)
Refinement
The methine and aromatic H atoms were placed in geometrically idealized positions at C—H = 1.00 and 0.95 Å, respectively and constrained to ride on their parent atoms, with Uiso(H) = 1.2Ueq(C). The highest peak is located 0.79 Å from Ir1 and the deepest hole is situated 0.79 Å from F4.
Figures
Fig. 1.
Molecular structure of the title compound. Displacement ellipsoids are drawn at the 50% probability level. Hydrogen atoms have been omitted for clarity.
Crystal data
| [Ir(C18H15P)2(CO)3]PF6·CH4O | F(000) = 1928 |
| Mr = 977.80 | Dx = 1.701 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71069 Å |
| Hall symbol: -P 2ybc | Cell parameters from 9817 reflections |
| a = 16.487 (5) Å | θ = 2.6–28.3° |
| b = 13.571 (4) Å | µ = 3.69 mm−1 |
| c = 20.903 (5) Å | T = 100 K |
| β = 125.297 (5)° | Block, yellow |
| V = 3817 (2) Å3 | 0.18 × 0.14 × 0.06 mm |
| Z = 4 |
Data collection
| Bruker APEXII CCD diffractometer | 8699 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.037 |
| φ and ω scans | θmax = 28.3°, θmin = 1.9° |
| Absorption correction: multi-scan (SADABS; Bruker, 2008) | h = −21→19 |
| Tmin = 0.556, Tmax = 0.809 | k = −18→15 |
| 68611 measured reflections | l = −27→27 |
| 9501 independent reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.019 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.044 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0153P)2 + 4.3801P] where P = (Fo2 + 2Fc2)/3 |
| 9501 reflections | (Δ/σ)max = 0.004 |
| 489 parameters | Δρmax = 1.01 e Å−3 |
| 0 restraints | Δρmin = −0.75 e Å−3 |
Special details
| Experimental. The intensity data were collected on a Bruker X8 ApexII 4 K Kappa CCD diffractometer using an exposure time of ?? s/frame. A total of ??? frames were collected with a frame width of 0.5\ % covering up to θ = 28.0 ° with 99.9% completeness accomplished.Spectroscopy data: 1H NMR (300 MHz, (CD3)2CO): δ = 7.5–7.8 (m, 30H). 31P NMR (121 MHz, (CD3)2CO): δ = -1.6 (s), -143.0 (m, PF6). ν(CO): 1989, 2008, 2025 cm-1. |
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.85491 (15) | 0.69659 (15) | 0.67007 (13) | 0.0193 (4) | |
| C01 | 0.6389 (2) | 0.5239 (2) | 0.77777 (18) | 0.0440 (7) | |
| H01A | 0.6237 | 0.5943 | 0.7739 | 0.066* | |
| H01B | 0.5781 | 0.4873 | 0.74 | 0.066* | |
| H01C | 0.6881 | 0.5132 | 0.7663 | 0.066* | |
| C2 | 0.70334 (17) | 0.63622 (15) | 0.47318 (13) | 0.0203 (4) | |
| C3 | 0.64299 (15) | 0.57286 (14) | 0.59226 (12) | 0.0171 (4) | |
| C11 | 0.84576 (14) | 0.41711 (14) | 0.68487 (11) | 0.0147 (4) | |
| C12 | 0.85577 (16) | 0.31492 (15) | 0.68521 (12) | 0.0183 (4) | |
| H12 | 0.8498 | 0.2832 | 0.6422 | 0.022* | |
| C13 | 0.87431 (16) | 0.25962 (15) | 0.74810 (13) | 0.0209 (4) | |
| H13 | 0.8808 | 0.1901 | 0.7479 | 0.025* | |
| C14 | 0.88347 (16) | 0.30508 (16) | 0.81104 (13) | 0.0216 (4) | |
| H14 | 0.8963 | 0.2668 | 0.854 | 0.026* | |
| C15 | 0.87397 (17) | 0.40654 (17) | 0.81152 (13) | 0.0238 (5) | |
| H15 | 0.8806 | 0.4378 | 0.855 | 0.029* | |
| C16 | 0.85477 (16) | 0.46261 (15) | 0.74836 (12) | 0.0194 (4) | |
| H16 | 0.8478 | 0.5321 | 0.7486 | 0.023* | |
| C21 | 0.95198 (14) | 0.51021 (14) | 0.62908 (12) | 0.0143 (4) | |
| C22 | 1.03624 (15) | 0.46978 (15) | 0.69615 (13) | 0.0199 (4) | |
| H22 | 1.0308 | 0.4353 | 0.7331 | 0.024* | |
| C23 | 1.12827 (16) | 0.48030 (17) | 0.70857 (14) | 0.0257 (5) | |
| H23 | 1.1859 | 0.4531 | 0.7543 | 0.031* | |
| C24 | 1.13651 (16) | 0.52994 (16) | 0.65505 (14) | 0.0239 (5) | |
| H24 | 1.1995 | 0.5358 | 0.6637 | 0.029* | |
| C25 | 1.05332 (17) | 0.57136 (16) | 0.58874 (14) | 0.0227 (4) | |
| H25 | 1.0594 | 0.6059 | 0.5522 | 0.027* | |
| C26 | 0.96128 (16) | 0.56221 (15) | 0.57591 (12) | 0.0189 (4) | |
| H26 | 0.9044 | 0.5914 | 0.5309 | 0.023* | |
| C31 | 0.77070 (15) | 0.40644 (14) | 0.52184 (11) | 0.0153 (4) | |
| C32 | 0.82977 (17) | 0.34357 (15) | 0.51186 (13) | 0.0186 (4) | |
| H32 | 0.9001 | 0.348 | 0.5461 | 0.022* | |
| C33 | 0.78507 (18) | 0.27437 (16) | 0.45141 (14) | 0.0236 (5) | |
| H33 | 0.8251 | 0.2313 | 0.4448 | 0.028* | |
| C34 | 0.68332 (19) | 0.26832 (17) | 0.40143 (14) | 0.0273 (5) | |
| H34 | 0.6533 | 0.2209 | 0.3605 | 0.033* | |
| C35 | 0.62365 (18) | 0.33144 (17) | 0.41046 (13) | 0.0261 (5) | |
| H35 | 0.5533 | 0.3274 | 0.3755 | 0.031* | |
| C36 | 0.66749 (16) | 0.39985 (16) | 0.47064 (13) | 0.0207 (4) | |
| H36 | 0.627 | 0.4425 | 0.4771 | 0.025* | |
| C41 | 0.60501 (15) | 0.81216 (14) | 0.60849 (12) | 0.0162 (4) | |
| C42 | 0.66416 (16) | 0.78861 (16) | 0.68786 (12) | 0.0201 (4) | |
| H42 | 0.7243 | 0.7532 | 0.7092 | 0.024* | |
| C43 | 0.63557 (17) | 0.81664 (18) | 0.73602 (13) | 0.0254 (5) | |
| H43 | 0.6761 | 0.8006 | 0.7902 | 0.03* | |
| C44 | 0.54769 (18) | 0.86814 (17) | 0.70476 (15) | 0.0273 (5) | |
| H44 | 0.5286 | 0.8881 | 0.7379 | 0.033* | |
| C45 | 0.48746 (17) | 0.89072 (16) | 0.62554 (14) | 0.0250 (5) | |
| H45 | 0.4267 | 0.9249 | 0.6043 | 0.03* | |
| C46 | 0.51604 (16) | 0.86333 (15) | 0.57727 (13) | 0.0206 (4) | |
| H46 | 0.4752 | 0.8793 | 0.5231 | 0.025* | |
| C51 | 0.54265 (15) | 0.78888 (14) | 0.44876 (11) | 0.0160 (4) | |
| C52 | 0.46604 (15) | 0.72272 (16) | 0.42633 (13) | 0.0212 (4) | |
| H52 | 0.4704 | 0.6798 | 0.4641 | 0.025* | |
| C53 | 0.38338 (17) | 0.71896 (17) | 0.34916 (13) | 0.0253 (5) | |
| H53 | 0.3314 | 0.6735 | 0.334 | 0.03* | |
| C54 | 0.37697 (18) | 0.78165 (19) | 0.29442 (13) | 0.0298 (5) | |
| H54 | 0.3195 | 0.7806 | 0.2418 | 0.036* | |
| C55 | 0.4536 (2) | 0.8458 (2) | 0.31567 (14) | 0.0367 (6) | |
| H55 | 0.4495 | 0.8874 | 0.2774 | 0.044* | |
| C56 | 0.53717 (18) | 0.84969 (17) | 0.39324 (13) | 0.0273 (5) | |
| H56 | 0.5899 | 0.8938 | 0.4078 | 0.033* | |
| C61 | 0.72295 (15) | 0.89687 (14) | 0.56249 (11) | 0.0160 (4) | |
| C62 | 0.69580 (16) | 0.98774 (15) | 0.57625 (13) | 0.0212 (4) | |
| H62 | 0.641 | 0.9916 | 0.5798 | 0.025* | |
| C63 | 0.74831 (16) | 1.07195 (16) | 0.58478 (13) | 0.0247 (5) | |
| H63 | 0.7293 | 1.1336 | 0.5938 | 0.03* | |
| C64 | 0.82869 (17) | 1.06647 (16) | 0.58012 (13) | 0.0241 (5) | |
| H64 | 0.8649 | 1.1244 | 0.5862 | 0.029* | |
| C65 | 0.85644 (17) | 0.97694 (16) | 0.56662 (13) | 0.0232 (5) | |
| H65 | 0.9117 | 0.9733 | 0.5637 | 0.028* | |
| C66 | 0.80312 (17) | 0.89215 (15) | 0.55739 (13) | 0.0207 (4) | |
| H66 | 0.8217 | 0.8308 | 0.5475 | 0.025* | |
| O1 | 0.92145 (12) | 0.73141 (12) | 0.72421 (10) | 0.0309 (4) | |
| O01 | 0.67752 (15) | 0.49056 (17) | 0.85393 (12) | 0.0471 (5) | |
| H01 | 0.7328 | 0.5175 | 0.8858 | 0.071* | |
| O2 | 0.67796 (14) | 0.63197 (12) | 0.40991 (10) | 0.0315 (4) | |
| O3 | 0.58781 (12) | 0.53708 (12) | 0.59882 (10) | 0.0289 (4) | |
| F1 | 0.9118 (2) | 0.70324 (13) | 0.40752 (13) | 0.0768 (7) | |
| F2 | 0.80028 (14) | 0.79216 (18) | 0.41110 (11) | 0.0678 (6) | |
| F3 | 0.88792 (15) | 0.92851 (11) | 0.43228 (10) | 0.0497 (5) | |
| F4 | 0.99908 (15) | 0.8406 (2) | 0.42799 (15) | 0.0823 (8) | |
| F5 | 0.95832 (15) | 0.79933 (13) | 0.51178 (9) | 0.0612 (6) | |
| F6 | 0.83935 (13) | 0.83280 (13) | 0.32776 (9) | 0.0436 (4) | |
| P1 | 0.82838 (4) | 0.48994 (3) | 0.60498 (3) | 0.01235 (9) | |
| P2 | 0.65123 (4) | 0.78758 (4) | 0.54975 (3) | 0.01324 (10) | |
| P3 | 0.90070 (4) | 0.81519 (4) | 0.41996 (3) | 0.02208 (12) | |
| Ir1 | 0.737840 (5) | 0.637207 (5) | 0.578891 (4) | 0.01155 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0173 (10) | 0.0155 (9) | 0.0248 (11) | 0.0027 (8) | 0.0119 (9) | −0.0008 (8) |
| C01 | 0.0380 (16) | 0.0500 (17) | 0.0486 (17) | −0.0059 (13) | 0.0276 (14) | 0.0050 (14) |
| C2 | 0.0247 (11) | 0.0148 (9) | 0.0231 (11) | 0.0036 (8) | 0.0148 (9) | 0.0028 (8) |
| C3 | 0.0192 (10) | 0.0129 (9) | 0.0193 (10) | 0.0051 (7) | 0.0111 (8) | 0.0033 (7) |
| C11 | 0.0152 (9) | 0.0134 (9) | 0.0157 (9) | 0.0012 (7) | 0.0089 (8) | 0.0029 (7) |
| C12 | 0.0227 (10) | 0.0138 (9) | 0.0221 (10) | 0.0004 (8) | 0.0151 (9) | 0.0000 (8) |
| C13 | 0.0227 (11) | 0.0145 (9) | 0.0261 (11) | 0.0026 (8) | 0.0144 (9) | 0.0042 (8) |
| C14 | 0.0220 (11) | 0.0228 (11) | 0.0193 (10) | 0.0028 (8) | 0.0116 (9) | 0.0074 (8) |
| C15 | 0.0323 (12) | 0.0226 (11) | 0.0175 (10) | 0.0031 (9) | 0.0151 (9) | 0.0012 (8) |
| C16 | 0.0252 (11) | 0.0141 (9) | 0.0188 (10) | 0.0016 (8) | 0.0126 (9) | 0.0006 (8) |
| C21 | 0.0156 (9) | 0.0112 (8) | 0.0191 (9) | −0.0007 (7) | 0.0119 (8) | −0.0032 (7) |
| C22 | 0.0193 (10) | 0.0158 (9) | 0.0231 (10) | 0.0014 (8) | 0.0113 (9) | 0.0012 (8) |
| C23 | 0.0162 (10) | 0.0230 (11) | 0.0312 (12) | 0.0028 (8) | 0.0097 (9) | −0.0021 (9) |
| C24 | 0.0184 (10) | 0.0218 (11) | 0.0369 (13) | −0.0046 (8) | 0.0190 (10) | −0.0099 (9) |
| C25 | 0.0256 (11) | 0.0220 (10) | 0.0297 (11) | −0.0068 (8) | 0.0213 (10) | −0.0062 (9) |
| C26 | 0.0197 (10) | 0.0189 (10) | 0.0199 (10) | −0.0020 (8) | 0.0124 (9) | −0.0009 (8) |
| C31 | 0.0203 (10) | 0.0116 (8) | 0.0170 (9) | −0.0027 (7) | 0.0125 (8) | −0.0007 (7) |
| C32 | 0.0229 (11) | 0.0163 (9) | 0.0223 (10) | −0.0030 (8) | 0.0164 (9) | −0.0026 (8) |
| C33 | 0.0354 (13) | 0.0198 (10) | 0.0290 (11) | −0.0067 (9) | 0.0264 (11) | −0.0075 (9) |
| C34 | 0.0379 (13) | 0.0244 (11) | 0.0253 (11) | −0.0109 (10) | 0.0215 (11) | −0.0102 (9) |
| C35 | 0.0236 (11) | 0.0272 (11) | 0.0225 (11) | −0.0077 (9) | 0.0104 (10) | −0.0061 (9) |
| C36 | 0.0211 (10) | 0.0175 (10) | 0.0235 (10) | −0.0013 (8) | 0.0129 (9) | −0.0007 (8) |
| C41 | 0.0182 (10) | 0.0128 (9) | 0.0200 (10) | −0.0001 (7) | 0.0124 (8) | −0.0003 (7) |
| C42 | 0.0201 (10) | 0.0214 (10) | 0.0183 (10) | 0.0020 (8) | 0.0107 (9) | −0.0013 (8) |
| C43 | 0.0271 (12) | 0.0296 (12) | 0.0188 (10) | 0.0013 (9) | 0.0129 (9) | −0.0038 (9) |
| C44 | 0.0315 (13) | 0.0272 (12) | 0.0326 (13) | −0.0019 (9) | 0.0240 (11) | −0.0093 (10) |
| C45 | 0.0235 (11) | 0.0203 (10) | 0.0352 (13) | 0.0038 (8) | 0.0193 (10) | −0.0017 (9) |
| C46 | 0.0193 (10) | 0.0183 (10) | 0.0249 (11) | 0.0033 (8) | 0.0131 (9) | 0.0034 (8) |
| C51 | 0.0154 (9) | 0.0142 (9) | 0.0148 (9) | 0.0017 (7) | 0.0067 (8) | 0.0010 (7) |
| C52 | 0.0179 (10) | 0.0217 (10) | 0.0216 (10) | 0.0002 (8) | 0.0100 (9) | 0.0052 (8) |
| C53 | 0.0187 (11) | 0.0272 (11) | 0.0245 (11) | −0.0036 (9) | 0.0094 (9) | 0.0008 (9) |
| C54 | 0.0238 (12) | 0.0374 (13) | 0.0170 (10) | −0.0045 (10) | 0.0054 (9) | 0.0017 (9) |
| C55 | 0.0366 (14) | 0.0411 (15) | 0.0195 (11) | −0.0104 (11) | 0.0088 (11) | 0.0101 (10) |
| C56 | 0.0272 (12) | 0.0264 (11) | 0.0191 (11) | −0.0097 (9) | 0.0081 (10) | 0.0021 (9) |
| C61 | 0.0175 (10) | 0.0123 (9) | 0.0144 (9) | −0.0004 (7) | 0.0070 (8) | 0.0014 (7) |
| C62 | 0.0168 (10) | 0.0151 (9) | 0.0277 (11) | 0.0006 (8) | 0.0105 (9) | −0.0013 (8) |
| C63 | 0.0221 (11) | 0.0147 (10) | 0.0304 (12) | −0.0002 (8) | 0.0111 (10) | −0.0029 (8) |
| C64 | 0.0260 (11) | 0.0158 (10) | 0.0270 (11) | −0.0059 (8) | 0.0132 (10) | −0.0004 (8) |
| C65 | 0.0271 (11) | 0.0198 (10) | 0.0287 (11) | −0.0054 (9) | 0.0196 (10) | −0.0020 (9) |
| C66 | 0.0283 (11) | 0.0134 (9) | 0.0264 (11) | −0.0018 (8) | 0.0191 (10) | −0.0006 (8) |
| O1 | 0.0211 (8) | 0.0299 (9) | 0.0331 (9) | −0.0048 (7) | 0.0107 (7) | −0.0090 (7) |
| O01 | 0.0427 (12) | 0.0584 (14) | 0.0467 (12) | −0.0160 (10) | 0.0296 (10) | 0.0019 (10) |
| O2 | 0.0462 (11) | 0.0303 (9) | 0.0228 (9) | 0.0040 (8) | 0.0226 (8) | 0.0037 (7) |
| O3 | 0.0280 (9) | 0.0250 (8) | 0.0375 (10) | 0.0001 (7) | 0.0211 (8) | 0.0052 (7) |
| F1 | 0.136 (2) | 0.0262 (9) | 0.0671 (13) | 0.0119 (11) | 0.0580 (14) | −0.0049 (9) |
| F2 | 0.0527 (11) | 0.1141 (18) | 0.0538 (11) | −0.0414 (12) | 0.0407 (10) | −0.0299 (12) |
| F3 | 0.0909 (14) | 0.0237 (8) | 0.0456 (10) | 0.0033 (8) | 0.0458 (10) | 0.0005 (7) |
| F4 | 0.0368 (11) | 0.129 (2) | 0.0927 (17) | −0.0237 (12) | 0.0442 (12) | −0.0429 (15) |
| F5 | 0.0807 (14) | 0.0453 (10) | 0.0234 (8) | 0.0142 (9) | 0.0104 (9) | 0.0076 (7) |
| F6 | 0.0566 (11) | 0.0478 (9) | 0.0238 (8) | −0.0030 (8) | 0.0216 (8) | 0.0000 (7) |
| P1 | 0.0147 (2) | 0.0100 (2) | 0.0142 (2) | 0.00026 (17) | 0.00941 (19) | 0.00037 (17) |
| P2 | 0.0144 (2) | 0.0102 (2) | 0.0147 (2) | 0.00088 (17) | 0.0081 (2) | 0.00118 (18) |
| P3 | 0.0224 (3) | 0.0202 (3) | 0.0206 (3) | −0.0001 (2) | 0.0107 (2) | −0.0002 (2) |
| Ir1 | 0.01286 (4) | 0.00918 (4) | 0.01346 (4) | 0.00053 (3) | 0.00809 (3) | 0.00089 (3) |
Geometric parameters (Å, º)
| Ir1—C1 | 1.938 (2) | C35—H35 | 0.95 |
| Ir1—C2 | 1.938 (2) | C36—H36 | 0.95 |
| Ir1—C3 | 1.947 (2) | C41—C42 | 1.391 (3) |
| Ir1—P1 | 2.3620 (8) | C41—C46 | 1.398 (3) |
| Ir1—P2 | 2.3599 (8) | C41—P2 | 1.811 (2) |
| C1—O1 | 1.128 (3) | C42—C43 | 1.389 (3) |
| C01—O01 | 1.405 (3) | C42—H42 | 0.95 |
| C01—H01A | 0.98 | C43—C44 | 1.386 (3) |
| C01—H01B | 0.98 | C43—H43 | 0.95 |
| C01—H01C | 0.98 | C44—C45 | 1.386 (3) |
| C2—O2 | 1.135 (3) | C44—H44 | 0.95 |
| C3—O3 | 1.107 (3) | C45—C46 | 1.388 (3) |
| C11—C16 | 1.392 (3) | C45—H45 | 0.95 |
| C11—C12 | 1.396 (3) | C46—H46 | 0.95 |
| C11—P1 | 1.815 (2) | C51—C56 | 1.384 (3) |
| C12—C13 | 1.386 (3) | C51—C52 | 1.392 (3) |
| C12—H12 | 0.95 | C51—P2 | 1.815 (2) |
| C13—C14 | 1.380 (3) | C52—C53 | 1.386 (3) |
| C13—H13 | 0.95 | C52—H52 | 0.95 |
| C14—C15 | 1.387 (3) | C53—C54 | 1.380 (3) |
| C14—H14 | 0.95 | C53—H53 | 0.95 |
| C15—C16 | 1.393 (3) | C54—C55 | 1.379 (3) |
| C15—H15 | 0.95 | C54—H54 | 0.95 |
| C16—H16 | 0.95 | C55—C56 | 1.396 (3) |
| C21—C22 | 1.395 (3) | C55—H55 | 0.95 |
| C21—C26 | 1.398 (3) | C56—H56 | 0.95 |
| C21—P1 | 1.815 (2) | C61—C66 | 1.388 (3) |
| C22—C23 | 1.391 (3) | C61—C62 | 1.397 (3) |
| C22—H22 | 0.95 | C61—P2 | 1.818 (2) |
| C23—C24 | 1.378 (3) | C62—C63 | 1.382 (3) |
| C23—H23 | 0.95 | C62—H62 | 0.95 |
| C24—C25 | 1.386 (3) | C63—C64 | 1.386 (3) |
| C24—H24 | 0.95 | C63—H63 | 0.95 |
| C25—C26 | 1.386 (3) | C64—C65 | 1.384 (3) |
| C25—H25 | 0.95 | C64—H64 | 0.95 |
| C26—H26 | 0.95 | C65—C66 | 1.392 (3) |
| C31—C36 | 1.394 (3) | C65—H65 | 0.95 |
| C31—C32 | 1.398 (3) | C66—H66 | 0.95 |
| C31—P1 | 1.816 (2) | O01—H01 | 0.84 |
| C32—C33 | 1.395 (3) | P3—F1 | 1.5698 (19) |
| C32—H32 | 0.95 | P3—F2 | 1.5870 (19) |
| C33—C34 | 1.374 (3) | P3—F3 | 1.5933 (17) |
| C33—H33 | 0.95 | P3—F4 | 1.569 (2) |
| C34—C35 | 1.397 (3) | P3—F5 | 1.5890 (17) |
| C34—H34 | 0.95 | P3—F6 | 1.5943 (16) |
| C35—C36 | 1.384 (3) | ||
| O1—C1—Ir1 | 177.8 (2) | C45—C46—C41 | 119.9 (2) |
| O01—C01—H01A | 109.5 | C45—C46—H46 | 120 |
| O01—C01—H01B | 109.5 | C41—C46—H46 | 120 |
| H01A—C01—H01B | 109.5 | C56—C51—C52 | 119.69 (19) |
| O01—C01—H01C | 109.5 | C56—C51—P2 | 121.59 (16) |
| H01A—C01—H01C | 109.5 | C52—C51—P2 | 118.61 (16) |
| H01B—C01—H01C | 109.5 | C53—C52—C51 | 120.4 (2) |
| O2—C2—Ir1 | 175.6 (2) | C53—C52—H52 | 119.8 |
| O3—C3—Ir1 | 178.8 (2) | C51—C52—H52 | 119.8 |
| C16—C11—C12 | 119.26 (18) | C54—C53—C52 | 119.7 (2) |
| C16—C11—P1 | 120.58 (15) | C54—C53—H53 | 120.2 |
| C12—C11—P1 | 120.06 (15) | C52—C53—H53 | 120.2 |
| C13—C12—C11 | 120.23 (19) | C55—C54—C53 | 120.3 (2) |
| C13—C12—H12 | 119.9 | C55—C54—H54 | 119.8 |
| C11—C12—H12 | 119.9 | C53—C54—H54 | 119.8 |
| C14—C13—C12 | 120.3 (2) | C54—C55—C56 | 120.3 (2) |
| C14—C13—H13 | 119.8 | C54—C55—H55 | 119.9 |
| C12—C13—H13 | 119.8 | C56—C55—H55 | 119.9 |
| C13—C14—C15 | 120.0 (2) | C51—C56—C55 | 119.6 (2) |
| C13—C14—H14 | 120 | C51—C56—H56 | 120.2 |
| C15—C14—H14 | 120 | C55—C56—H56 | 120.2 |
| C14—C15—C16 | 120.0 (2) | C66—C61—C62 | 119.29 (19) |
| C14—C15—H15 | 120 | C66—C61—P2 | 121.26 (16) |
| C16—C15—H15 | 120 | C62—C61—P2 | 119.44 (16) |
| C11—C16—C15 | 120.14 (19) | C63—C62—C61 | 120.3 (2) |
| C11—C16—H16 | 119.9 | C63—C62—H62 | 119.8 |
| C15—C16—H16 | 119.9 | C61—C62—H62 | 119.8 |
| C22—C21—C26 | 119.65 (19) | C62—C63—C64 | 120.0 (2) |
| C22—C21—P1 | 121.71 (16) | C62—C63—H63 | 120 |
| C26—C21—P1 | 118.50 (15) | C64—C63—H63 | 120 |
| C23—C22—C21 | 119.5 (2) | C65—C64—C63 | 120.2 (2) |
| C23—C22—H22 | 120.2 | C65—C64—H64 | 119.9 |
| C21—C22—H22 | 120.2 | C63—C64—H64 | 119.9 |
| C24—C23—C22 | 120.5 (2) | C64—C65—C66 | 119.8 (2) |
| C24—C23—H23 | 119.8 | C64—C65—H65 | 120.1 |
| C22—C23—H23 | 119.8 | C66—C65—H65 | 120.1 |
| C23—C24—C25 | 120.4 (2) | C61—C66—C65 | 120.3 (2) |
| C23—C24—H24 | 119.8 | C61—C66—H66 | 119.9 |
| C25—C24—H24 | 119.8 | C65—C66—H66 | 119.9 |
| C26—C25—C24 | 119.8 (2) | C01—O01—H01 | 109.5 |
| C26—C25—H25 | 120.1 | C11—P1—C21 | 105.79 (9) |
| C24—C25—H25 | 120.1 | C11—P1—C31 | 104.93 (9) |
| C25—C26—C21 | 120.2 (2) | C21—P1—C31 | 104.07 (9) |
| C25—C26—H26 | 119.9 | C11—P1—Ir1 | 114.58 (7) |
| C21—C26—H26 | 119.9 | C21—P1—Ir1 | 113.22 (7) |
| C36—C31—C32 | 119.51 (19) | C31—P1—Ir1 | 113.28 (7) |
| C36—C31—P1 | 120.43 (16) | C41—P2—C51 | 105.55 (10) |
| C32—C31—P1 | 119.92 (16) | C41—P2—C61 | 104.08 (9) |
| C33—C32—C31 | 119.8 (2) | C51—P2—C61 | 106.46 (9) |
| C33—C32—H32 | 120.1 | C41—P2—Ir1 | 114.40 (7) |
| C31—C32—H32 | 120.1 | C51—P2—Ir1 | 110.59 (7) |
| C34—C33—C32 | 120.2 (2) | C61—P2—Ir1 | 114.98 (7) |
| C34—C33—H33 | 119.9 | F4—P3—F1 | 91.02 (14) |
| C32—C33—H33 | 119.9 | F4—P3—F2 | 178.61 (15) |
| C33—C34—C35 | 120.4 (2) | F1—P3—F2 | 90.22 (14) |
| C33—C34—H34 | 119.8 | F4—P3—F5 | 92.35 (13) |
| C35—C34—H34 | 119.8 | F1—P3—F5 | 91.35 (11) |
| C36—C35—C34 | 119.7 (2) | F2—P3—F5 | 88.26 (12) |
| C36—C35—H35 | 120.2 | F4—P3—F3 | 89.76 (12) |
| C34—C35—H35 | 120.2 | F1—P3—F3 | 179.21 (13) |
| C35—C36—C31 | 120.4 (2) | F2—P3—F3 | 88.99 (12) |
| C35—C36—H36 | 119.8 | F5—P3—F3 | 88.74 (9) |
| C31—C36—H36 | 119.8 | F4—P3—F6 | 89.48 (12) |
| C42—C41—C46 | 119.6 (2) | F1—P3—F6 | 89.89 (11) |
| C42—C41—P2 | 119.11 (16) | F2—P3—F6 | 89.89 (10) |
| C46—C41—P2 | 120.93 (16) | F5—P3—F6 | 177.78 (11) |
| C43—C42—C41 | 120.3 (2) | F3—P3—F6 | 90.00 (9) |
| C43—C42—H42 | 119.9 | C2—Ir1—C1 | 126.42 (10) |
| C41—C42—H42 | 119.9 | C2—Ir1—C3 | 115.45 (9) |
| C44—C43—C42 | 119.8 (2) | C1—Ir1—C3 | 118.13 (9) |
| C44—C43—H43 | 120.1 | C2—Ir1—P2 | 88.65 (6) |
| C42—C43—H43 | 120.1 | C1—Ir1—P2 | 90.26 (6) |
| C43—C44—C45 | 120.4 (2) | C3—Ir1—P2 | 90.49 (6) |
| C43—C44—H44 | 119.8 | C2—Ir1—P1 | 89.28 (6) |
| C45—C44—H44 | 119.8 | C1—Ir1—P1 | 89.27 (6) |
| C44—C45—C46 | 120.0 (2) | C3—Ir1—P1 | 92.30 (6) |
| C44—C45—H45 | 120 | P2—Ir1—P1 | 177.047 (18) |
| C46—C45—H45 | 120 | ||
| C16—C11—C12—C13 | 0.1 (3) | C12—C11—P1—C31 | 26.42 (19) |
| P1—C11—C12—C13 | 176.57 (16) | C16—C11—P1—Ir1 | −32.28 (19) |
| C11—C12—C13—C14 | −0.3 (3) | C12—C11—P1—Ir1 | 151.30 (14) |
| C12—C13—C14—C15 | 0.1 (3) | C22—C21—P1—C11 | 2.27 (19) |
| C13—C14—C15—C16 | 0.3 (3) | C26—C21—P1—C11 | 177.92 (16) |
| C12—C11—C16—C15 | 0.3 (3) | C22—C21—P1—C31 | −108.02 (17) |
| P1—C11—C16—C15 | −176.18 (17) | C26—C21—P1—C31 | 67.63 (17) |
| C14—C15—C16—C11 | −0.5 (3) | C22—C21—P1—Ir1 | 128.54 (15) |
| C26—C21—C22—C23 | −1.1 (3) | C26—C21—P1—Ir1 | −55.80 (17) |
| P1—C21—C22—C23 | 174.49 (16) | C36—C31—P1—C11 | 91.01 (18) |
| C21—C22—C23—C24 | −0.2 (3) | C32—C31—P1—C11 | −84.74 (18) |
| C22—C23—C24—C25 | 1.0 (3) | C36—C31—P1—C21 | −158.07 (17) |
| C23—C24—C25—C26 | −0.5 (3) | C32—C31—P1—C21 | 26.19 (19) |
| C24—C25—C26—C21 | −0.9 (3) | C36—C31—P1—Ir1 | −34.68 (18) |
| C22—C21—C26—C25 | 1.7 (3) | C32—C31—P1—Ir1 | 149.57 (14) |
| P1—C21—C26—C25 | −174.07 (16) | C42—C41—P2—C51 | 161.74 (16) |
| C36—C31—C32—C33 | −0.6 (3) | C46—C41—P2—C51 | −24.91 (19) |
| P1—C31—C32—C33 | 175.17 (16) | C42—C41—P2—C61 | −86.35 (18) |
| C31—C32—C33—C34 | 0.4 (3) | C46—C41—P2—C61 | 86.99 (18) |
| C32—C33—C34—C35 | 0.2 (4) | C42—C41—P2—Ir1 | 39.93 (18) |
| C33—C34—C35—C36 | −0.6 (4) | C46—C41—P2—Ir1 | −146.72 (15) |
| C34—C35—C36—C31 | 0.4 (3) | C56—C51—P2—C41 | 124.9 (2) |
| C32—C31—C36—C35 | 0.2 (3) | C52—C51—P2—C41 | −58.94 (19) |
| P1—C31—C36—C35 | −175.57 (17) | C56—C51—P2—C61 | 14.6 (2) |
| C46—C41—C42—C43 | −0.7 (3) | C52—C51—P2—C61 | −169.15 (17) |
| P2—C41—C42—C43 | 172.75 (17) | C56—C51—P2—Ir1 | −110.90 (19) |
| C41—C42—C43—C44 | 0.1 (3) | C52—C51—P2—Ir1 | 65.30 (18) |
| C42—C43—C44—C45 | 0.9 (4) | C66—C61—P2—C41 | 151.24 (17) |
| C43—C44—C45—C46 | −1.2 (4) | C62—C61—P2—C41 | −29.95 (19) |
| C44—C45—C46—C41 | 0.6 (3) | C66—C61—P2—C51 | −97.51 (18) |
| C42—C41—C46—C45 | 0.3 (3) | C62—C61—P2—C51 | 81.29 (18) |
| P2—C41—C46—C45 | −173.00 (17) | C66—C61—P2—Ir1 | 25.32 (19) |
| C56—C51—C52—C53 | −1.5 (3) | C62—C61—P2—Ir1 | −155.87 (15) |
| P2—C51—C52—C53 | −177.75 (18) | C41—P2—Ir1—C2 | 154.66 (10) |
| C51—C52—C53—C54 | −0.3 (4) | C51—P2—Ir1—C2 | 35.65 (10) |
| C52—C53—C54—C55 | 1.9 (4) | C61—P2—Ir1—C2 | −84.94 (10) |
| C53—C54—C55—C56 | −1.7 (4) | C41—P2—Ir1—C1 | −78.91 (10) |
| C52—C51—C56—C55 | 1.6 (4) | C51—P2—Ir1—C1 | 162.08 (10) |
| P2—C51—C56—C55 | 177.8 (2) | C61—P2—Ir1—C1 | 41.48 (10) |
| C54—C55—C56—C51 | 0.0 (4) | C41—P2—Ir1—C3 | 39.22 (10) |
| C66—C61—C62—C63 | −0.1 (3) | C51—P2—Ir1—C3 | −79.79 (10) |
| P2—C61—C62—C63 | −178.91 (17) | C61—P2—Ir1—C3 | 159.61 (9) |
| C61—C62—C63—C64 | −0.3 (3) | C11—P1—Ir1—C2 | −156.99 (10) |
| C62—C63—C64—C65 | 0.3 (4) | C21—P1—Ir1—C2 | 81.56 (10) |
| C63—C64—C65—C66 | 0.3 (4) | C31—P1—Ir1—C2 | −36.64 (10) |
| C62—C61—C66—C65 | 0.6 (3) | C11—P1—Ir1—C1 | 76.58 (10) |
| P2—C61—C66—C65 | 179.41 (17) | C21—P1—Ir1—C1 | −44.87 (10) |
| C64—C65—C66—C61 | −0.7 (3) | C31—P1—Ir1—C1 | −163.08 (10) |
| C16—C11—P1—C21 | 93.15 (18) | C11—P1—Ir1—C3 | −41.55 (9) |
| C12—C11—P1—C21 | −83.26 (18) | C21—P1—Ir1—C3 | −163.00 (9) |
| C16—C11—P1—C31 | −157.16 (17) | C31—P1—Ir1—C3 | 78.80 (10) |
Hydrogen-bond geometry (Å, º)
Cg1, Cg2 and Cg3 are the centroids of the C11–C16, C21–C26 and C41–C46 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C15—H15···F3i | 0.95 | 2.39 | 3.281 (3) | 157 |
| C16—H16···F6i | 0.95 | 2.53 | 3.319 (3) | 141 |
| C42—H42···F6i | 0.95 | 2.38 | 3.138 (3) | 136 |
| C43—H43···F2i | 0.95 | 2.49 | 3.386 (3) | 158 |
| C45—H45···O01ii | 0.95 | 2.50 | 3.281 (3) | 139 |
| C64—H64···F4iii | 0.95 | 2.47 | 3.200 (3) | 133 |
| O01—H01···F3i | 0.84 | 2.27 | 3.059 (3) | 157 |
| C53—H53···Cg1iv | 0.95 | 2.68 | 3.523 (2) | 148 |
| C35—H35···Cg3iv | 0.95 | 2.91 | 3.587 (2) | 129 |
| C13—H13···Cg2v | 0.95 | 2.97 | 3.744 (2) | 140 |
Symmetry codes: (i) x, −y+3/2, z+1/2; (ii) −x+1, y+1/2, −z+3/2; (iii) −x+2, −y+2, −z+1; (iv) −x+1, −y+1, −z+1; (v) −x+2, y−1/2, −z+3/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HB6932).
References
- Booyens, S., Roodt, A. & Wendt, O. F. (2007). J. Organomet. Chem. 692, 5508–5512.
- Brandenburg, K. & Putz, H. (2005). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Bruker (2008). SAINT-Plus and SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2011). APEX2 Bruker AXS Inc., Madison, Wisconsin, USA.
- Crous, R., Datt, M., Foster, D., Bennie, L., Steenkamp, C., Huyser, J., Kirsten, L., Steyl, G. & Roodt, A. (2005). Dalton Trans. pp. 1108–1116. [DOI] [PubMed]
- Farrugia, L. J. (1999). J. Appl. Cryst. 32, 837–838.
- Ferreira, A. C., Crous, R., Bennie, L., Meij, A. M. M., Blann, K., Bezuidenhoudt, B. C. B., Young, D. A., Green, M. J. & Roodt, A. (2007). Angew. Chem. Int. Ed. 46, 2273–2275. [DOI] [PubMed]
- Haumann, M., Meijboom, R., Moss, J. R. & Roodt, A. (2004). Dalton Trans. pp. 1679–1686. [DOI] [PubMed]
- Muller, A., Otto, S. & Roodt, A. (2008). Dalton Trans. pp. 650–657. [DOI] [PubMed]
- Otto, S., Ionescu, A. & Roodt, A. (2005). J. Organomet. Chem. 690, 4337–4342.
- Otto, S. & Roodt, A. (2001). Inorg. Chem. Commun. 4, 49–52.
- Purcell, W., Basson, S. S., Leipoldt, J. G., Roodt, A. & Preston, H. (1995). Inorg. Chim. Acta, 234, 153–156.
- Randall, S. L., Miller, C. A., See, R. F., Churchill, M. R., Janik, T. S., Lake, C. H. & Atwood, J. D. (1994). Organometallics, 13, 5088–5095.
- Randall, S. L., Thompson, J. S., Buttrey, L. A., Ziller, J. W., Churchill, M. R. & Atwood, J. D. (1991). Organometallics, 10, 683–688.
- Raper, G. & McDonald, W. S. (1973). Acta Cryst. B29, 2013–2014.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536812035593/hb6932sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812035593/hb6932Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report