Abstract
The title octadecatrienoic acid derivative, C18H26O4, was isolated from Silene maritima With. (Caryophyllaceae), the first time this natural compound has been found in the Caryophyllales order. This fatty acid has an 18-carbon backbone with three double bonds on trans (E) conformation and two carbonyl. In the crystal, molecules are linked via pairs of O—H⋯O hydrogen bonds, forming inversion dimers.
Related literature
For botanical information about Silene maritima With., see: Baker (1978 ▶); Bremer et al. (2009 ▶). For interactions between heavy-metals and Silene maritima With., see: Price & Abrahams (1994 ▶). For phytochemical investigation on Silene maritima With., see: Adrian-Romero et al. (1998 ▶). For previous descriptions of the title compound, see: Herz & Kulanthaivel (1984 ▶); Li et al. (2011 ▶). For lipoxygenase action on α-linoleic acid, see: Vellosillo et al. (2007 ▶). For environmental-stress-response involvement of oxylipines and their structure similarity with the title compound, see: Browse (2005 ▶); Schaller et al. (2004 ▶); Wasternack (2007 ▶). 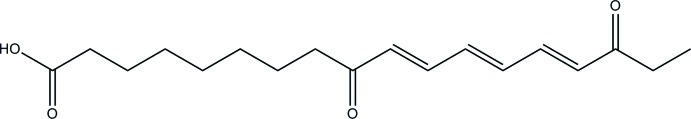
Experimental
Crystal data
C18H26O4
M r = 306.39
Triclinic,
a = 5.6859 (3) Å
b = 7.7535 (5) Å
c = 19.9045 (16) Å
α = 81.333 (4)°
β = 84.152 (4)°
γ = 87.660 (4)°
V = 862.68 (10) Å3
Z = 2
Mo Kα radiation
μ = 0.08 mm−1
T = 173 K
0.40 × 0.30 × 0.10 mm
Data collection
Nonius KappaCCD diffractometer
8217 measured reflections
3846 independent reflections
2648 reflections with I > 2σ(I)
R int = 0.063
Refinement
R[F 2 > 2σ(F 2)] = 0.065
wR(F 2) = 0.189
S = 1.06
3846 reflections
204 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.27 e Å−3
Δρmin = −0.24 e Å−3
Data collection: COLLECT (Nonius, 1998 ▶); cell refinement: DENZO (Otwinowski & Minor, 1997 ▶); data reduction: DENZO; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: PLATON (Spek, 2009 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812027870/zj2081sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812027870/zj2081Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812027870/zj2081Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O2—H2⋯O1i | 0.90 (4) | 1.78 (4) | 2.658 (2) | 167.0 (4) |
Symmetry code: (i) 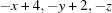 .
.
Acknowledgments
The authors thank Dr J. Suffert for his help in crystallizing the compound.
supplementary crystallographic information
Comment
Silene maritima With. belongs to the Caryophyllaceae family (Bremer et al., 2009) and is a perennial species found on cliffs and shingle beaches in coastal habitats (Baker, 1978). This species is known to be a heavy-metal indicator (Price & Abrahams, 1994), and the only phytochemical investigation previously carried out on its aerial parts has revealed the presence of glycinebetaine, a compound used by cells for protection against osmotic stress (Adrian-Romero et al., 1998).
This study is the first report of the presence of 9,16-dioxo-10E,12E,14E octadecatrienoic acid in the Caryophyllales order. This compound has previously been described only in the Asteraceae (Herz & Kulanthaivel, 1984) and Lamiaceae (Li et al., 2011) families.
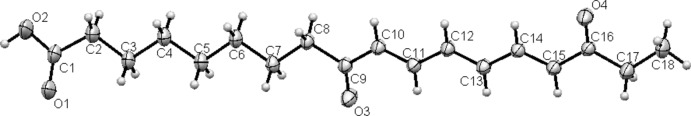
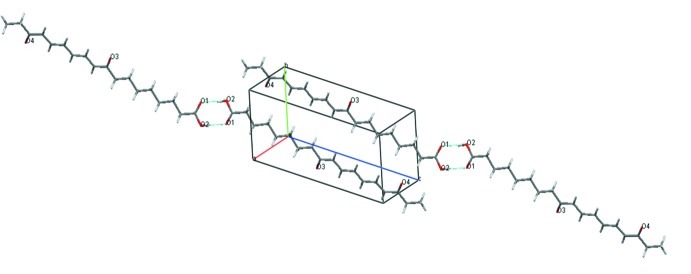
Its molecular structure contains an 18-carbon backbone with three double bonds in the trans conformation and two carbonyls (Fig. 1). The existence of intermolecular hydrogen interactions between two carboxylic functions was also observed (Fig. 2). The structure of this fatty acid might involve a lipoxygenase action on the α-linoleic acid (Vellosillo et al., 2007). Thus suggesting that it could belong to oxylipines, a class of compounds implicated in environmental stress responses (Browse, 2005; Schaller et al., 2004; Wasternack, 2007).
Experimental
The sampling station is situated in the littoral zone of the western coast of Brittany (Brélès 29, France). Sampling was carried out in July 2008. The aerial parts of the plant were collected, air-dried, and grinded into a fine powder using a grinder (Retsch, ZM 200). Hydroalcoholic extract of aerial parts (1 kg) was prepared by soaking it at room temperature in 3 x 10 l of EtOH/H2O (6/4, v/v) during first 14 h, then 4 h and again 4 h, until exhaustion of raw materials. The extract was then filtered and dried under vacuum using a rotavapor. The amorphous solid, a black-brownish mass, was then dissolved in d-H2O and extracted sequentially with cyclohexane, CH2Cl2, AcOEt and n-BuOH. The CH2Cl2 extract (2.496 g) was fractionated on a silica gel column (SI60 0.050–0.16 mm in size, Merck) eluted successively with cyclohexane (500 ml), AcOEt (1170 ml) and MeOH (330 ml) to yield five main fractions. The third fraction (210 mg) was re-dissolved in MeOH and subjected to semi-preparative HPLC purification (Gilson, binary solvent system). The isolation was performed with a reverse phase Nucleodur C18 ec (250 mm x 21 mm, 5 µm) from Macherey-Nagel. Eluent A was H2O with 0.01% HCOOH, and eluent B was ACN. The flow rate was 10 ml/min and the injection volume was 400 µl at 40 mg ml-1. The elution conditions applied were: 0–5 min, linear gradient from 10% to 15% B; 5–55 min, 15% to 65% B; 55–60 min, 65% to 100% B; 60–70 min, 100% B isocratic. Simultaneous UV monitoring was set at 316 nm. This experimental procedure allowed us to isolate the title compound C18H26O4 at the retention time of 26 min. The pure compound (1 mg) was re-dissolved in 0.2 ml of MeOH/CHCl3 (2/1). The corresponding crystals of 9,16-dioxo-10E,12E,14E octadecatrienoic acid were grown thanks to a slow solubility decrease during two weeks at room temperature after addition of n-heptane (0.4 ml).
Refinement
The H atoms, except for the H-atom of the carboxyl group which was located from Fourier difference maps, were positioned geometrically and refined using a riding model, with C—H = 0.95–0.99 Å and with Uiso(H) = 1.2 (1.5 for methyl groups) times Ueq(C).
Figures
Fig. 1.
ORTEP representation of 9,16-dioxo-10E,12E,14E octadecatrienoic acid with 50% probability displacement ellipsoids for non-H atoms.
Fig. 2.
Packing diagram of four molecules of 9,16-dioxo-10E,12E,14E octadecatrienoic acid.
Crystal data
| C18H26O4 | Z = 2 |
| Mr = 306.39 | F(000) = 332 |
| Triclinic, P1 | Dx = 1.180 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 5.6859 (3) Å | Cell parameters from 12739 reflections |
| b = 7.7535 (5) Å | θ = 1.0–27.5° |
| c = 19.9045 (16) Å | µ = 0.08 mm−1 |
| α = 81.333 (4)° | T = 173 K |
| β = 84.152 (4)° | Plate, colorless |
| γ = 87.660 (4)° | 0.40 × 0.30 × 0.10 mm |
| V = 862.68 (10) Å3 |
Data collection
| Nonius KappaCCD diffractometer | 2648 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.063 |
| Graphite monochromator | θmax = 27.5°, θmin = 1.0° |
| phi and ω scans | h = −7→7 |
| 8217 measured reflections | k = −10→10 |
| 3846 independent reflections | l = −25→25 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.065 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.189 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0932P)2 + 0.1679P] where P = (Fo2 + 2Fc2)/3 |
| 3846 reflections | (Δ/σ)max < 0.001 |
| 204 parameters | Δρmax = 0.27 e Å−3 |
| 0 restraints | Δρmin = −0.24 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 1.7412 (3) | 0.9602 (2) | 0.07113 (9) | 0.0370 (4) | |
| C2 | 1.5281 (3) | 0.9351 (3) | 0.12198 (10) | 0.0426 (5) | |
| H2A | 1.3914 | 0.9132 | 0.0977 | 0.051* | |
| H2B | 1.4935 | 1.0453 | 0.1410 | 0.051* | |
| C3 | 1.5485 (3) | 0.7882 (2) | 0.18074 (9) | 0.0355 (4) | |
| H3A | 1.6700 | 0.8163 | 0.2094 | 0.043* | |
| H3B | 1.5999 | 0.6793 | 0.1625 | 0.043* | |
| C4 | 1.3134 (3) | 0.7601 (2) | 0.22433 (9) | 0.0371 (4) | |
| H4A | 1.2613 | 0.8705 | 0.2413 | 0.044* | |
| H4B | 1.1935 | 0.7311 | 0.1954 | 0.044* | |
| C5 | 1.3238 (3) | 0.6162 (2) | 0.28491 (9) | 0.0349 (4) | |
| H5A | 1.4352 | 0.6489 | 0.3157 | 0.042* | |
| H5B | 1.3859 | 0.5074 | 0.2684 | 0.042* | |
| C6 | 1.0834 (3) | 0.5817 (2) | 0.32511 (9) | 0.0344 (4) | |
| H6A | 1.0212 | 0.6904 | 0.3417 | 0.041* | |
| H6B | 0.9720 | 0.5489 | 0.2944 | 0.041* | |
| C7 | 1.0945 (3) | 0.4374 (2) | 0.38581 (9) | 0.0332 (4) | |
| H7A | 1.2019 | 0.4722 | 0.4173 | 0.040* | |
| H7B | 1.1622 | 0.3300 | 0.3693 | 0.040* | |
| C8 | 0.8539 (3) | 0.3977 (2) | 0.42492 (8) | 0.0310 (4) | |
| H8A | 0.7810 | 0.5074 | 0.4381 | 0.037* | |
| H8B | 0.7506 | 0.3545 | 0.3942 | 0.037* | |
| C9 | 0.8631 (3) | 0.2648 (2) | 0.48833 (9) | 0.0295 (4) | |
| C10 | 0.6343 (3) | 0.2012 (2) | 0.52376 (8) | 0.0300 (4) | |
| H10 | 0.4917 | 0.2397 | 0.5045 | 0.036* | |
| C11 | 0.6245 (3) | 0.0905 (2) | 0.58252 (9) | 0.0299 (4) | |
| H11 | 0.7696 | 0.0515 | 0.6002 | 0.036* | |
| C12 | 0.4081 (3) | 0.0265 (2) | 0.62080 (9) | 0.0298 (4) | |
| H12 | 0.2623 | 0.0608 | 0.6027 | 0.036* | |
| C13 | 0.4038 (3) | −0.0797 (2) | 0.68114 (9) | 0.0303 (4) | |
| H13 | 0.5503 | −0.1168 | 0.6984 | 0.036* | |
| C14 | 0.1889 (3) | −0.1398 (2) | 0.72083 (8) | 0.0291 (4) | |
| H14 | 0.0433 | −0.1026 | 0.7031 | 0.035* | |
| C15 | 0.1797 (3) | −0.2443 (2) | 0.78105 (9) | 0.0318 (4) | |
| H15 | 0.3235 | −0.2847 | 0.7992 | 0.038* | |
| C16 | −0.0463 (3) | −0.2989 (2) | 0.82020 (9) | 0.0295 (4) | |
| C17 | −0.0310 (3) | −0.4111 (3) | 0.88816 (10) | 0.0420 (5) | |
| H17A | 0.0712 | −0.3535 | 0.9150 | 0.050* | |
| H17B | 0.0468 | −0.5239 | 0.8804 | 0.050* | |
| C18 | −0.2653 (4) | −0.4474 (3) | 0.93005 (10) | 0.0502 (5) | |
| H18A | −0.3392 | −0.3374 | 0.9411 | 0.075* | |
| H18B | −0.2393 | −0.5251 | 0.9724 | 0.075* | |
| H18C | −0.3693 | −0.5032 | 0.9039 | 0.075* | |
| O1 | 1.9060 (2) | 0.85667 (18) | 0.06800 (7) | 0.0537 (4) | |
| O2 | 1.7321 (3) | 1.10716 (19) | 0.02820 (8) | 0.0565 (4) | |
| O3 | 1.0497 (2) | 0.21570 (18) | 0.51052 (7) | 0.0464 (4) | |
| O4 | −0.2355 (2) | −0.25612 (17) | 0.79837 (6) | 0.0434 (4) | |
| H2 | 1.868 (7) | 1.118 (5) | 0.001 (2) | 0.140 (14)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0422 (10) | 0.0350 (9) | 0.0294 (9) | −0.0042 (8) | 0.0027 (8) | 0.0061 (7) |
| C2 | 0.0401 (10) | 0.0458 (11) | 0.0351 (10) | −0.0011 (8) | 0.0066 (8) | 0.0093 (8) |
| C3 | 0.0367 (9) | 0.0362 (9) | 0.0293 (9) | −0.0033 (7) | 0.0052 (7) | 0.0037 (7) |
| C4 | 0.0362 (10) | 0.0399 (10) | 0.0306 (9) | −0.0047 (8) | 0.0049 (7) | 0.0050 (8) |
| C5 | 0.0352 (9) | 0.0362 (9) | 0.0299 (9) | −0.0051 (7) | 0.0042 (7) | 0.0021 (7) |
| C6 | 0.0346 (9) | 0.0356 (9) | 0.0293 (9) | −0.0053 (7) | 0.0039 (7) | 0.0041 (7) |
| C7 | 0.0310 (9) | 0.0343 (9) | 0.0308 (9) | −0.0053 (7) | 0.0030 (7) | 0.0031 (7) |
| C8 | 0.0296 (9) | 0.0340 (9) | 0.0267 (9) | −0.0050 (7) | −0.0010 (7) | 0.0041 (7) |
| C9 | 0.0275 (8) | 0.0321 (9) | 0.0274 (8) | −0.0046 (7) | −0.0020 (7) | 0.0009 (7) |
| C10 | 0.0265 (8) | 0.0342 (9) | 0.0277 (9) | −0.0034 (7) | −0.0034 (7) | 0.0023 (7) |
| C11 | 0.0253 (8) | 0.0324 (9) | 0.0303 (9) | −0.0029 (7) | −0.0032 (7) | 0.0015 (7) |
| C12 | 0.0275 (8) | 0.0322 (9) | 0.0278 (9) | −0.0026 (7) | −0.0025 (7) | 0.0027 (7) |
| C13 | 0.0259 (8) | 0.0328 (9) | 0.0299 (9) | −0.0028 (7) | −0.0028 (7) | 0.0030 (7) |
| C14 | 0.0260 (8) | 0.0311 (8) | 0.0285 (9) | −0.0011 (7) | −0.0032 (7) | 0.0017 (7) |
| C15 | 0.0253 (8) | 0.0345 (9) | 0.0324 (9) | −0.0019 (7) | −0.0035 (7) | 0.0059 (7) |
| C16 | 0.0276 (8) | 0.0291 (8) | 0.0293 (9) | −0.0010 (7) | −0.0030 (7) | 0.0034 (7) |
| C17 | 0.0349 (9) | 0.0471 (11) | 0.0369 (10) | −0.0026 (8) | −0.0018 (8) | 0.0157 (8) |
| C18 | 0.0468 (12) | 0.0550 (12) | 0.0389 (11) | −0.0009 (9) | 0.0072 (9) | 0.0176 (9) |
| O1 | 0.0502 (8) | 0.0507 (8) | 0.0466 (9) | 0.0085 (7) | 0.0175 (6) | 0.0193 (7) |
| O2 | 0.0603 (10) | 0.0444 (8) | 0.0501 (9) | 0.0071 (7) | 0.0212 (7) | 0.0200 (7) |
| O3 | 0.0292 (7) | 0.0591 (9) | 0.0442 (8) | −0.0057 (6) | −0.0048 (6) | 0.0166 (6) |
| O4 | 0.0275 (6) | 0.0587 (9) | 0.0382 (7) | −0.0020 (6) | −0.0040 (5) | 0.0127 (6) |
Geometric parameters (Å, º)
| C1—O1 | 1.211 (2) | C9—O3 | 1.215 (2) |
| C1—O2 | 1.320 (2) | C9—C10 | 1.481 (2) |
| C1—C2 | 1.497 (3) | C10—C11 | 1.340 (2) |
| C2—C3 | 1.515 (2) | C10—H10 | 0.9500 |
| C2—H2A | 0.9900 | C11—C12 | 1.444 (2) |
| C2—H2B | 0.9900 | C11—H11 | 0.9500 |
| C3—C4 | 1.522 (2) | C12—C13 | 1.349 (2) |
| C3—H3A | 0.9900 | C12—H12 | 0.9500 |
| C3—H3B | 0.9900 | C13—C14 | 1.440 (2) |
| C4—C5 | 1.519 (2) | C13—H13 | 0.9500 |
| C4—H4A | 0.9900 | C14—C15 | 1.339 (2) |
| C4—H4B | 0.9900 | C14—H14 | 0.9500 |
| C5—C6 | 1.524 (2) | C15—C16 | 1.476 (2) |
| C5—H5A | 0.9900 | C15—H15 | 0.9500 |
| C5—H5B | 0.9900 | C16—O4 | 1.2161 (19) |
| C6—C7 | 1.522 (2) | C16—C17 | 1.502 (2) |
| C6—H6A | 0.9900 | C17—C18 | 1.511 (3) |
| C6—H6B | 0.9900 | C17—H17A | 0.9900 |
| C7—C8 | 1.523 (2) | C17—H17B | 0.9900 |
| C7—H7A | 0.9900 | C18—H18A | 0.9800 |
| C7—H7B | 0.9900 | C18—H18B | 0.9800 |
| C8—C9 | 1.508 (2) | C18—H18C | 0.9800 |
| C8—H8A | 0.9900 | O2—H2 | 0.90 (4) |
| C8—H8B | 0.9900 | ||
| O1—C1—O2 | 122.25 (17) | C7—C8—H8A | 108.8 |
| O1—C1—C2 | 124.48 (16) | C9—C8—H8B | 108.8 |
| O2—C1—C2 | 113.26 (16) | C7—C8—H8B | 108.8 |
| C1—C2—C3 | 115.59 (15) | H8A—C8—H8B | 107.7 |
| C1—C2—H2A | 108.4 | O3—C9—C10 | 121.41 (15) |
| C3—C2—H2A | 108.4 | O3—C9—C8 | 121.50 (15) |
| C1—C2—H2B | 108.4 | C10—C9—C8 | 117.06 (14) |
| C3—C2—H2B | 108.4 | C11—C10—C9 | 121.25 (15) |
| H2A—C2—H2B | 107.4 | C11—C10—H10 | 119.4 |
| C2—C3—C4 | 111.17 (15) | C9—C10—H10 | 119.4 |
| C2—C3—H3A | 109.4 | C10—C11—C12 | 124.38 (15) |
| C4—C3—H3A | 109.4 | C10—C11—H11 | 117.8 |
| C2—C3—H3B | 109.4 | C12—C11—H11 | 117.8 |
| C4—C3—H3B | 109.4 | C13—C12—C11 | 122.93 (15) |
| H3A—C3—H3B | 108.0 | C13—C12—H12 | 118.5 |
| C5—C4—C3 | 113.63 (15) | C11—C12—H12 | 118.5 |
| C5—C4—H4A | 108.8 | C12—C13—C14 | 123.43 (15) |
| C3—C4—H4A | 108.8 | C12—C13—H13 | 118.3 |
| C5—C4—H4B | 108.8 | C14—C13—H13 | 118.3 |
| C3—C4—H4B | 108.8 | C15—C14—C13 | 124.62 (15) |
| H4A—C4—H4B | 107.7 | C15—C14—H14 | 117.7 |
| C4—C5—C6 | 112.83 (15) | C13—C14—H14 | 117.7 |
| C4—C5—H5A | 109.0 | C14—C15—C16 | 122.26 (15) |
| C6—C5—H5A | 109.0 | C14—C15—H15 | 118.9 |
| C4—C5—H5B | 109.0 | C16—C15—H15 | 118.9 |
| C6—C5—H5B | 109.0 | O4—C16—C15 | 121.69 (15) |
| H5A—C5—H5B | 107.8 | O4—C16—C17 | 121.60 (15) |
| C7—C6—C5 | 112.67 (15) | C15—C16—C17 | 116.71 (14) |
| C7—C6—H6A | 109.1 | C16—C17—C18 | 115.05 (15) |
| C5—C6—H6A | 109.1 | C16—C17—H17A | 108.5 |
| C7—C6—H6B | 109.1 | C18—C17—H17A | 108.5 |
| C5—C6—H6B | 109.1 | C16—C17—H17B | 108.5 |
| H6A—C6—H6B | 107.8 | C18—C17—H17B | 108.5 |
| C6—C7—C8 | 113.09 (14) | H17A—C17—H17B | 107.5 |
| C6—C7—H7A | 109.0 | C17—C18—H18A | 109.5 |
| C8—C7—H7A | 109.0 | C17—C18—H18B | 109.5 |
| C6—C7—H7B | 109.0 | H18A—C18—H18B | 109.5 |
| C8—C7—H7B | 109.0 | C17—C18—H18C | 109.5 |
| H7A—C7—H7B | 107.8 | H18A—C18—H18C | 109.5 |
| C9—C8—C7 | 113.98 (14) | H18B—C18—H18C | 109.5 |
| C9—C8—H8A | 108.8 | C1—O2—H2 | 109 (2) |
| O1—C1—C2—C3 | 12.6 (3) | C8—C9—C10—C11 | −176.26 (15) |
| O2—C1—C2—C3 | −168.51 (16) | C9—C10—C11—C12 | 178.37 (15) |
| C1—C2—C3—C4 | −173.08 (16) | C10—C11—C12—C13 | −177.39 (16) |
| C2—C3—C4—C5 | −179.00 (15) | C11—C12—C13—C14 | 177.91 (15) |
| C3—C4—C5—C6 | −176.13 (15) | C12—C13—C14—C15 | −179.56 (16) |
| C4—C5—C6—C7 | −179.98 (14) | C13—C14—C15—C16 | 178.72 (15) |
| C5—C6—C7—C8 | −178.05 (14) | C14—C15—C16—O4 | 2.6 (3) |
| C6—C7—C8—C9 | −175.62 (14) | C14—C15—C16—C17 | −177.86 (16) |
| C7—C8—C9—O3 | 10.3 (3) | O4—C16—C17—C18 | −7.0 (3) |
| C7—C8—C9—C10 | −171.76 (14) | C15—C16—C17—C18 | 173.45 (16) |
| O3—C9—C10—C11 | 1.7 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O2—H2···O1i | 0.90 (4) | 1.78 (4) | 2.658 (2) | 167.0 (4) |
Symmetry code: (i) −x+4, −y+2, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: ZJ2081).
References
- Adrian-Romero, M., Wilson, S. J., Gerald, B., Yang, M.-H., Carabot-Cuervo, A. & Bashir, A. K. (1998). Biochem. Syst. Ecol. 26, 535–543.
- Baker, A. J. M. (1978). New Phytol. 81, 321–330.
- Bremer, B., Bremer, K., Chase, M. W., Fay, M. F., Reveal, J. L., Soltis, D. E., Soltis, P. S., Stevens, P. F., Anderberg, A. A., Moore, M. J., Olmstead, R. G., Rudall, P. J., Sytsma, K. J., Tank, D. C., Wurdack, K., Xiang, J. Q. Y. & Zmarzty, S. (2009). Bot. J. Linn. Soc. 161, 105–121.
- Browse, J. (2005). Vitam. Horm. (N.Y.), 72, 431–456. [DOI] [PubMed]
- Herz, W. & Kulanthaivel, P. (1984). Phytochemistry, 23, 1453–1459.
- Li, L. M., Pu, J. X., Xiao, W. L. & Sun, H. D. (2011). Arch. Pharmacal. Res. 34, 875–879. [DOI] [PubMed]
- Nonius (1998). COLLECT Nonius BV, Delft, The Netherlands.
- Otwinowski, Z. & Minor, W. (1997). Methods in Enzymology, Vol. 276, Macromolecular Crystallography, Part A, edited by C. W. Carter Jr & R. M. Sweet, pp. 307–326. New York: Academic Press.
- Price, G. C. & Abrahams, W. P. (1994). Environ. Geochem. Health, 16, 27–31. [DOI] [PubMed]
- Schaller, F., Schaller, A. & Stintzi, A. (2004). J. Plant Growth Regul. 23, 179–199.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Vellosillo, T., Martinez, M., Lopez, M. A., Vicente, J., Cascon, T., Dolan, L., Hamberg, M. & Castresana, C. (2007). Plant Cell, 19, 831–846. [DOI] [PMC free article] [PubMed]
- Wasternack, C. (2007). Ann. Bot. (Oxford, U.K.), 100, 681–697. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536812027870/zj2081sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536812027870/zj2081Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536812027870/zj2081Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report