Abstract
FK228 [systematic name: (1S,4S,7Z,10S,16E,21R)-7-ethylidene-4,21-di(propan-2-yl)-2-oxa-12,13-dithia-5,8,20,23-tetrazabicyclo[8.7.6]tricos-16-ene-3,6,9,19,22-pentone], C24H36N4O6S2, also known as FR901228, depsipeptide, NSC 630176, romidepsin, and marketed as Istodax by Celgene Corporation, is crystallized from ethyl acetate in P21 as compared to the absolute configuration of FK228, first crystallized from methanol in P212121 [Shigematsu et al. (1994 ▶). J. Antibiot. 47, 311–314]. A slight difference is observed between the absolute configuration of FK228 and the present structure. The molecular structure is stabilized by intramolecular N—H⋯O hydrogen bonds. In the crystal, molecules are linked via N—H⋯O hydrogen bonds.
Related literature
For diverse natural products, see: Nguyen et al. (2008 ▶); Knappe et al. (2008 ▶); Seyedsayamdost et al. (2010 ▶); Biggins et al. (2011 ▶); Klausmeyer et al. (2011 ▶); Wang et al. (2011 ▶, 2012 ▶). For large-scale genome sequencing, see: Yu et al. (2006 ▶); Mukhopadhyay et al. (2010 ▶); Zhuo et al. (2012 ▶). For the initial discovery of FK228 from Chromobacterium violaceum No. 968 and its crystal structure report, see: Shigematsu et al. (1994 ▶); Ueda, Nakajima, Hori, Fujita et al. (1994 ▶). For the biological activities and mode of action of FK228, see: Furumai et al. (2002 ▶); Ueda, Manda et al. (1994 ▶); Ueda, Nakajima, Hori, Goto & Okuhara (1994 ▶). For biosynthetic studies of FK228, see: Cheng et al. (2007 ▶); Potharla et al. (2011 ▶); Wesener et al. (2011 ▶). For clinical application of FK228, see: Robey et al. (2011 ▶); StatBite (2010 ▶).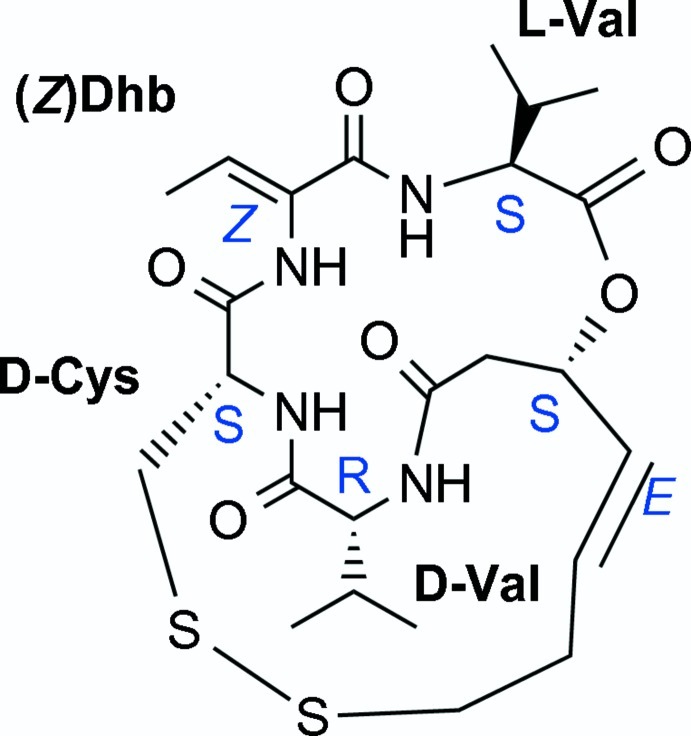
Experimental
Crystal data
C24H36N4O6S2
M r = 540.69
Monoclinic,
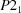
a = 9.1085 (2) Å
b = 16.2431 (4) Å
c = 9.4192 (2) Å
β = 92.096 (1)°
V = 1392.64 (5) Å3
Z = 2
Cu Kα radiation
μ = 2.10 mm−1
T = 100 K
0.26 × 0.14 × 0.07 mm
Data collection
Bruker SMART APEXII area-detector diffractometer
Absorption correction: analytical (SADABS; Sheldrick, 2003 ▶) T min = 0.611, T max = 0.867
22577 measured reflections
4918 independent reflections
4859 reflections with I > 2σ(I)
R int = 0.025
Refinement
R[F 2 > 2σ(F 2)] = 0.025
wR(F 2) = 0.065
S = 1.02
4918 reflections
343 parameters
1 restraint
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.29 e Å−3
Δρmin = −0.15 e Å−3
Absolute structure: Flack (1983 ▶), 2188 Friedel pairs
Flack parameter: 0.022 (9)
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXTL (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXTL; molecular graphics: SHELXTL and OLEX2 (Dolomanov et al., 2009 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S160053681203601X/jj2148sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681203601X/jj2148Isup2.hkl
Supplementary material file. DOI: 10.1107/S160053681203601X/jj2148Isup3.cdx
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H3⋯O5 | 0.84 (2) | 2.27 (2) | 3.0123 (18) | 147.1 (17) |
| N2—H4⋯O6 | 0.82 (2) | 2.05 (2) | 2.7867 (18) | 149.1 (18) |
| N3—H16⋯O3i | 0.87 (2) | 2.18 (2) | 3.0449 (17) | 175.9 (18) |
Symmetry code: (i) 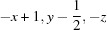 .
.
Acknowledgments
Support for this work was obtained from a Research Growth Initiative Award from the University of Wisconsin–Milwaukee and NIH/NCI grant R01 CA 152212 (both to YQC). The authors thank Lara C. Spencer and Ilia A. Guzei (University of Wisconsin–Madison, Department of Chemistry, Crystallography Facility) for collecting the crystallographic data.
supplementary crystallographic information
Comment
Diverse natural products, including thailandamides (Nguyen et al., 2008), capistruin (Knappe et al., 2008), bactobolin A—D (Seyedsayamdost et al., 2010), burkholdacs A—B (Biggins et al., 2011), spiruchostatin C (Klausmeyer et al., 2011), and thailandepsins (Wang et al., 2011, 2012), were discovered recently from Brukholderia thailandensis E264. In conjunction with large-scale genome sequencing (Yu et al., 2006; Mukhopadhyay et al., 2010; Zhuo et al., 2012), the Burkholderia thailandensis species have drawn much attention due to their capability to synthesize novel compounds with antibacterial, antitumor and antiviral activities.
While exploring novel natural products from the culture broth of B. thailandensis MSMB43, the title compound FK228 was isolated in large quantity (~100 mg/L). The physicochemical properties (UV spectrum, IR spectrum, MS and NMR) of the FK228 from B. thailandensis MSMB43 are identical to those of the FK228 isolated in the early 1990s from Chromobacterium violaceum No. 968 (Ueda, Nakajima, Hori, Fujita et al., 1994; Shigematsu et al., 1994). The crystal structure of the newly isolated FK228, herein, is slightly different than that reported by (Ueda, Manda et al., 1994), due to different crystallization conditions employed.
FK228 is bicyclic depsipeptide and consists of five building blocks, D-cysteine (D-Cys), D-valine (D-Val), 4-amino-3-hydroxy-5-methylheptanoic acid (Ahhp, derived from an isoleucine and an acetate unit), L-valine (L-Val), Z-dehydrobutyrine(Z-Dhb) and 3-hydroxy-7-mercapto-4-heptenoic acid (Acyl, derived from a cysteine and two acetate units). The primary structure of FK228 is (L)Val-(Z)Dhb-(D)Cys-Ahhp-(D)Val. X-ray crystallographic analysis indicates that the skeleton of FK228 is a [8.7.6] 23-membered ring adopting an uncommon cage-shape including a 16-membered macrocyclic lactone and a 15-membered ring containing a signature disulfide bond.
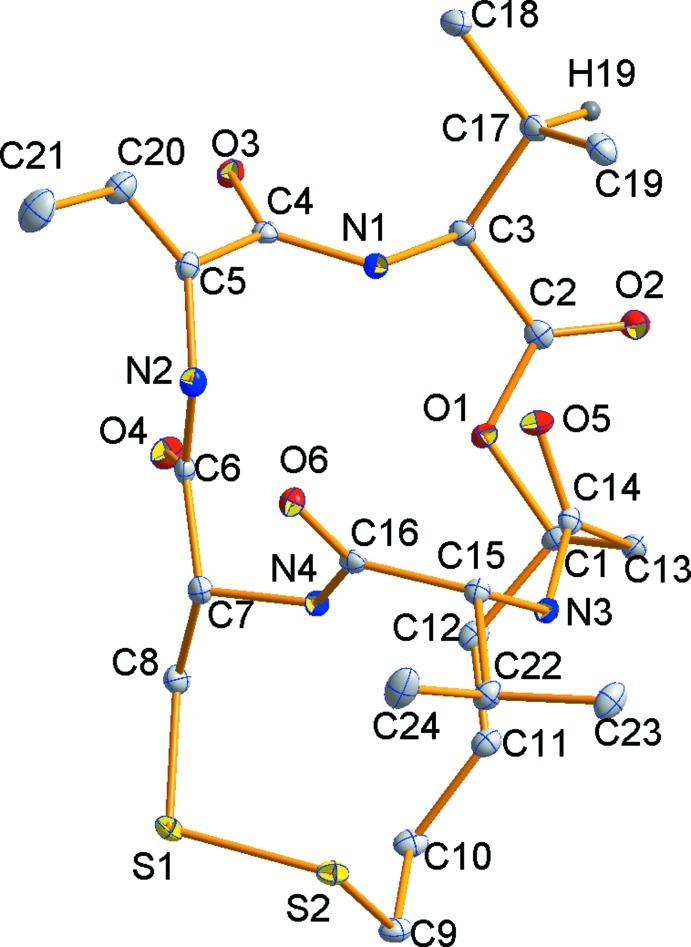
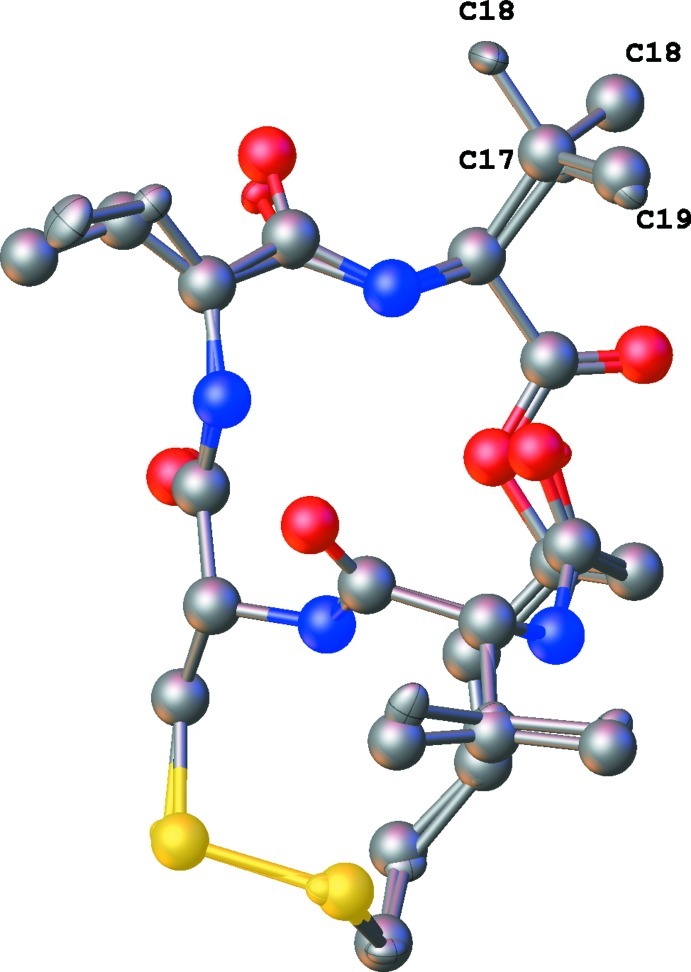
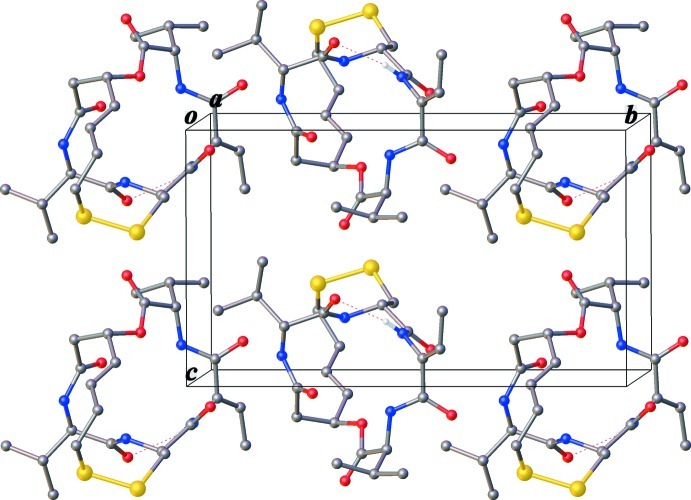
Under the different crystallization conditions presented, FK228 formed a different molecular arrangement than previously described (Fig. 1). The title crystal structure was obtained from ethyl acetate crystallizing in the P21 space group, while the provirus structure co-crystallized with one methanol in asymmetric unit in the P212121 space group. The absolute configurations of all the chiral centers in each of these two structures are the same: C1(S), C3(S), c7 (S) and C22 (R). The geometric configuration of the double bonds in the Acyl and Dhb components are all determined as E and Z respectively. In comparing the these two structures, the configuration of these skeletons are the same. But there is a slight difference at the end of L-Val group. Under the different crystallization conditions, C18 and H19 had the opposite positions in these two structures because of the free rotation of the single bond (Fig. 2). Hydrogen bonds N2—H4···O6 and the weak intermolecular interactions N1—H3···O5 and N3—H16···O3 are observed in the title crystal structure. (Table 1, Fig. 3).
Experimental
Bacteria and culture medium
Burkholderia thailandensis strain MSMB43 (obtained from the US Centers for Disease Control, CDC) was routinely activated on LB agar containing 50 mg ml-1 of apramycin (Am50) at 37°C for 1 to 2 days as a master plate. A single colony was then transferred into a 1 L flask containing 300 ml of LB medium and Am50, and the culture were growing at 37°C for 24 h as seed culture. For batch fermentation 250 ml of seed culture was transferred into a 20 L fermentor (BioFlo IV, New Brunswick Scientific Co., USA) containing 12 L of M8 medium (0.5% glucose, 0.5% peptone, 0.3% NaCl, 0.12% Na2HPO4, and 0.05% KH2PO4; pH 7), and fermented at 30°C for 96 hr under 20 L/min aeration and 200 rpm agitation; pH was maintained at 7.0 with 1 N NaCl or 1M HCl. For fed-batch fermentation, feeding of 3 L of 10X concentrated medium between 24 hr and 48 hr was performed on top of the batch fermentation.
Recovery of the crude extract
Bacteria cells were removed by centrifugation of cell broth at 3,800 rpm for 15 min. The supernatant was applied on a 2 L column (Φ 8.0 × 40 cm) packed with a mixture of Diaion HP-20 (Sigma-Aldrich, USA) resin and Amberlite XAD16 (Sigma-Aldrich, USA), which has been equilibrated in water at 5 ml/min. The resin was dried and then extracted with ethyl acetate for three times. The ethyl acetate extracts were combined and concentrated to dryness in vacuo at 35°C.
Isolation and purification of the title compound
The ethyl acetate extract was subjected to a two step isolation and purification protocol. Briefly, the ethyl acetate extract was mixed with 50 g silica gel (230–400 mesh, Whatman Purasil, USA). The mixture of silica gel was dried overnight and then applied to a 120 - g silica gel column (300 ml), which has been equilibrated with hexane. The column was eluted sequentially with 1 L hexane, 1 L hexane: ethyl acetate (3:1, v/v), 1 L hexane: ethyl acetate (1:1, v/v), 1 L ethyl acetate, 1 L ethyl acetate: acetone (1:1, v/v), and finally 2 L of acetone. The obtained fractions were applied on a flash chromatography equipped with a silica gel universal column (Yamazen Corporation, 23 ×123 mm, 16 g) connected to an injection column (Yamazen Corporation, 20 × 65 mm, 14 g). The column was eluted by a mixture of chloroform and acetone with an increasing of polarity according to the following concentrations of acetone, 1%, 5%, 10%, 15%, 20%, 25%, 30%, 35%, 65%, and 100%. Fractions by 15% and 20% acetone were concentrated into dryness in vacuo and dissolved in acetonitrile. The acetonitrile solution was applied on a preparative HPLC system equipped with an Agilent Prep-C18 column (21.2 × 250 mm, 10 µm, PN 410910–102, USA). The mobile phase was composed of acetonitrile and water (purified by NANO pure Diamond Life Science ultrapure water system). After loading the sample, the column was eluted by a linear gradient aqueous acetonitrile from 40% to 55% within 0–25 min, FK228 was collected at 21–25 min. The flow rate was 8 ml/min. The UV spectrum was monitored at 210 nm. The FK228 solution was concentrated into dryness in vacuo on a rotary evaporator at 35 °C. The dried FK228 was dissolved in ethyl acetate and the crystals were obtained at room temperature after 5–7 days.
Refinement
All hydrogen atoms attached to the carbon atoms were placed in geometrically idealized positions (C—H = 0.98, 0.99 and 1.00 Å on the primary, secondary and tertiary aliphatic C atoms respectively, 0.95 Å on aromatic C). The H atoms were refined as riding, with isotropic displacement coefficients of Uiso(H) = 1.5Ueq(C) for methyl groups or 1.2Ueq(C) otherwise. The hydrogen atoms attached to N and O were located in difference maps and refined independently with restraints and constraints. The H atoms on N atoms were restrained to have N—H distances of 0.880 (1) Å and their Uiso values were constrained as equal to 1.2 times the Ueq of their carrier atoms. The H atom on O was restrained to have an O—H distance of 0.840 (1) Å and the Uiso value was assigned as equal to 1.5 times the Ueq of the oxygen atom.
Figures
Fig. 1.
A molecular structure of FK228 with displacement ellipsoids shown at the 30% probability level. All hydrogen atoms were omitted for clarity except for H19.
Fig. 2.
Overlaid crystal structures of previous (isolated from Chromobacterium violaceum No. 968) (Balls & sticks) and newly (Ellipses & sticks) isolated FK228.
Fig. 3.
A packing diagram of FK228, viewed along the a axis. The dashed lines represent N—H···O hydrogen bonds.
Crystal data
| C24H36N4O6S2 | F(000) = 576 |
| Mr = 540.69 | Dx = 1.289 Mg m−3 |
| Monoclinic, P21 | Cu Kα radiation, λ = 1.54178 Å |
| Hall symbol: P 2yb | Cell parameters from 999 reflections |
| a = 9.1085 (2) Å | θ = 4.7–69.8° |
| b = 16.2431 (4) Å | µ = 2.10 mm−1 |
| c = 9.4192 (2) Å | T = 100 K |
| β = 92.096 (1)° | Block, colourless |
| V = 1392.64 (5) Å3 | 0.26 × 0.14 × 0.07 mm |
| Z = 2 |
Data collection
| Bruker SMART APEXII area-detector diffractometer | 4918 independent reflections |
| Radiation source: fine-focus sealed tube | 4859 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.025 |
| 0.50° ω and 0.5 ° φ scans | θmax = 69.8°, θmin = 4.7° |
| Absorption correction: analytical (SADABS; Sheldrick, 2003) | h = −10→11 |
| Tmin = 0.611, Tmax = 0.867 | k = −19→19 |
| 22577 measured reflections | l = −11→11 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.025 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.065 | w = 1/[σ2(Fo2) + (0.0447P)2 + 0.2408P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.02 | (Δ/σ)max = 0.001 |
| 4918 reflections | Δρmax = 0.29 e Å−3 |
| 343 parameters | Δρmin = −0.15 e Å−3 |
| 1 restraint | Absolute structure: Flack (1983), 2188 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: 0.022 (9) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.21741 (4) | 0.87614 (3) | 0.41260 (4) | 0.02391 (9) | |
| S2 | 0.21966 (4) | 0.75561 (3) | 0.35057 (4) | 0.02177 (9) | |
| O1 | 0.47759 (11) | 0.86900 (7) | −0.17912 (11) | 0.0166 (2) | |
| O2 | 0.54893 (12) | 0.83426 (7) | −0.39718 (11) | 0.0228 (2) | |
| O3 | 0.73140 (12) | 1.08712 (7) | −0.12864 (11) | 0.0188 (2) | |
| O4 | 0.48889 (12) | 1.02648 (7) | 0.11054 (12) | 0.0234 (2) | |
| O5 | 0.68455 (11) | 0.76827 (7) | −0.04745 (11) | 0.0206 (2) | |
| O6 | 0.74839 (11) | 0.82368 (7) | 0.32442 (11) | 0.0175 (2) | |
| N1 | 0.72350 (14) | 0.94804 (8) | −0.11503 (13) | 0.0155 (2) | |
| H3 | 0.724 (2) | 0.9060 (13) | −0.063 (2) | 0.019* | |
| N2 | 0.70812 (14) | 0.97489 (8) | 0.18875 (13) | 0.0170 (3) | |
| H4 | 0.750 (2) | 0.9394 (13) | 0.237 (2) | 0.020* | |
| N3 | 0.54236 (14) | 0.69120 (8) | 0.09041 (13) | 0.0153 (2) | |
| H16 | 0.467 (2) | 0.6595 (13) | 0.099 (2) | 0.018* | |
| N4 | 0.51449 (14) | 0.83271 (8) | 0.23715 (14) | 0.0159 (3) | |
| H18 | 0.454 (2) | 0.8103 (13) | 0.195 (2) | 0.019* | |
| C1 | 0.37318 (16) | 0.80077 (9) | −0.18177 (16) | 0.0171 (3) | |
| H1 | 0.3247 | 0.7979 | −0.2786 | 0.021* | |
| C2 | 0.56600 (16) | 0.87327 (10) | −0.28962 (14) | 0.0168 (3) | |
| C3 | 0.68786 (16) | 0.93675 (10) | −0.26594 (15) | 0.0167 (3) | |
| H2 | 0.6488 | 0.9905 | −0.3027 | 0.020* | |
| C4 | 0.74312 (15) | 1.02321 (10) | −0.05887 (15) | 0.0157 (3) | |
| C5 | 0.78862 (16) | 1.02503 (9) | 0.09680 (15) | 0.0172 (3) | |
| C6 | 0.55899 (17) | 0.97821 (9) | 0.18570 (15) | 0.0169 (3) | |
| C7 | 0.48646 (16) | 0.91621 (10) | 0.28501 (15) | 0.0171 (3) | |
| H5A | 0.5281 | 0.9234 | 0.3839 | 0.021* | |
| C8 | 0.32175 (17) | 0.93220 (10) | 0.28278 (16) | 0.0200 (3) | |
| H6 | 0.2808 | 0.9185 | 0.1869 | 0.024* | |
| H7 | 0.3061 | 0.9918 | 0.2977 | 0.024* | |
| C9 | 0.05712 (16) | 0.74578 (10) | 0.23338 (17) | 0.0216 (3) | |
| H8 | 0.0435 | 0.6869 | 0.2092 | 0.026* | |
| H9 | −0.0295 | 0.7635 | 0.2860 | 0.026* | |
| C10 | 0.06058 (16) | 0.79491 (11) | 0.09558 (17) | 0.0214 (3) | |
| H10 | −0.0326 | 0.7857 | 0.0404 | 0.026* | |
| H11 | 0.0667 | 0.8543 | 0.1189 | 0.026* | |
| C11 | 0.18736 (16) | 0.77249 (10) | 0.00426 (16) | 0.0190 (3) | |
| H12 | 0.2182 | 0.7166 | 0.0052 | 0.023* | |
| C12 | 0.25928 (16) | 0.82409 (10) | −0.07712 (17) | 0.0190 (3) | |
| H13 | 0.2369 | 0.8810 | −0.0687 | 0.023* | |
| C13 | 0.45645 (17) | 0.71982 (10) | −0.15341 (15) | 0.0172 (3) | |
| H14 | 0.3854 | 0.6764 | −0.1291 | 0.021* | |
| H15 | 0.5048 | 0.7027 | −0.2411 | 0.021* | |
| C14 | 0.57163 (16) | 0.72764 (9) | −0.03355 (15) | 0.0162 (3) | |
| C15 | 0.64392 (16) | 0.70259 (9) | 0.21230 (15) | 0.0154 (3) | |
| H17 | 0.7451 | 0.6916 | 0.1792 | 0.018* | |
| C16 | 0.64070 (16) | 0.79203 (9) | 0.26422 (14) | 0.0153 (3) | |
| C17 | 0.82380 (17) | 0.91592 (10) | −0.35219 (16) | 0.0199 (3) | |
| H19 | 0.7900 | 0.9098 | −0.4539 | 0.024* | |
| C18 | 0.93314 (19) | 0.98732 (11) | −0.34389 (17) | 0.0274 (4) | |
| H22 | 0.8836 | 1.0383 | −0.3742 | 0.041* | |
| H21 | 1.0145 | 0.9761 | −0.4063 | 0.041* | |
| H20 | 0.9714 | 0.9934 | −0.2459 | 0.041* | |
| C19 | 0.89729 (19) | 0.83572 (11) | −0.30681 (18) | 0.0257 (4) | |
| H24 | 0.9370 | 0.8410 | −0.2091 | 0.039* | |
| H25 | 0.9773 | 0.8234 | −0.3703 | 0.039* | |
| H23 | 0.8250 | 0.7910 | −0.3118 | 0.039* | |
| C20 | 0.90281 (18) | 1.07072 (10) | 0.14031 (17) | 0.0230 (3) | |
| H26 | 0.9536 | 1.1000 | 0.0699 | 0.028* | |
| C21 | 0.9578 (2) | 1.07966 (13) | 0.2911 (2) | 0.0358 (4) | |
| H27 | 0.8802 | 1.0637 | 0.3550 | 0.054* | |
| H29 | 0.9856 | 1.1371 | 0.3093 | 0.054* | |
| H28 | 1.0436 | 1.0441 | 0.3079 | 0.054* | |
| C22 | 0.61246 (18) | 0.63859 (10) | 0.32827 (17) | 0.0201 (3) | |
| H30 | 0.5052 | 0.6402 | 0.3475 | 0.024* | |
| C23 | 0.65029 (19) | 0.55289 (10) | 0.27385 (19) | 0.0253 (3) | |
| H31 | 0.5935 | 0.5418 | 0.1855 | 0.038* | |
| H32 | 0.6262 | 0.5117 | 0.3453 | 0.038* | |
| H33 | 0.7555 | 0.5502 | 0.2558 | 0.038* | |
| C24 | 0.6992 (2) | 0.65666 (11) | 0.46668 (18) | 0.0301 (4) | |
| H35 | 0.8045 | 0.6571 | 0.4488 | 0.045* | |
| H34 | 0.6786 | 0.6140 | 0.5368 | 0.045* | |
| H36 | 0.6700 | 0.7105 | 0.5033 | 0.045* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.02086 (19) | 0.0278 (2) | 0.02363 (18) | 0.00080 (16) | 0.00810 (13) | −0.00407 (16) |
| S2 | 0.01743 (17) | 0.02238 (19) | 0.02566 (19) | 0.00194 (15) | 0.00309 (13) | 0.00550 (15) |
| O1 | 0.0174 (5) | 0.0153 (5) | 0.0171 (5) | −0.0017 (4) | 0.0011 (4) | 0.0016 (4) |
| O2 | 0.0244 (6) | 0.0277 (6) | 0.0161 (5) | −0.0058 (5) | −0.0005 (4) | −0.0013 (5) |
| O3 | 0.0221 (5) | 0.0140 (5) | 0.0204 (5) | −0.0002 (4) | 0.0005 (4) | 0.0020 (4) |
| O4 | 0.0225 (6) | 0.0182 (6) | 0.0295 (6) | 0.0029 (4) | −0.0013 (5) | 0.0046 (5) |
| O5 | 0.0159 (5) | 0.0223 (6) | 0.0236 (5) | −0.0028 (4) | 0.0004 (4) | 0.0032 (4) |
| O6 | 0.0171 (5) | 0.0176 (5) | 0.0175 (5) | 0.0001 (4) | −0.0034 (4) | −0.0004 (4) |
| N1 | 0.0191 (6) | 0.0142 (6) | 0.0132 (6) | −0.0004 (5) | 0.0006 (5) | 0.0013 (5) |
| N2 | 0.0199 (6) | 0.0152 (6) | 0.0157 (6) | 0.0017 (5) | −0.0022 (5) | 0.0012 (5) |
| N3 | 0.0141 (6) | 0.0140 (6) | 0.0179 (6) | −0.0013 (5) | 0.0000 (5) | −0.0006 (5) |
| N4 | 0.0139 (6) | 0.0150 (6) | 0.0187 (6) | −0.0002 (5) | −0.0011 (5) | −0.0017 (5) |
| C1 | 0.0168 (7) | 0.0170 (8) | 0.0174 (7) | −0.0040 (6) | −0.0028 (5) | 0.0020 (6) |
| C2 | 0.0189 (7) | 0.0183 (7) | 0.0130 (6) | 0.0034 (6) | −0.0021 (5) | 0.0040 (6) |
| C3 | 0.0181 (7) | 0.0177 (7) | 0.0144 (6) | −0.0005 (6) | −0.0001 (5) | 0.0015 (6) |
| C4 | 0.0111 (6) | 0.0171 (7) | 0.0190 (7) | −0.0007 (5) | 0.0030 (5) | 0.0001 (6) |
| C5 | 0.0182 (7) | 0.0141 (7) | 0.0192 (7) | 0.0018 (6) | 0.0004 (5) | −0.0011 (6) |
| C6 | 0.0206 (7) | 0.0128 (7) | 0.0172 (7) | 0.0004 (6) | 0.0010 (6) | −0.0048 (6) |
| C7 | 0.0184 (7) | 0.0171 (7) | 0.0159 (7) | 0.0006 (6) | 0.0010 (5) | −0.0021 (6) |
| C8 | 0.0196 (7) | 0.0167 (7) | 0.0238 (7) | 0.0023 (6) | 0.0033 (6) | −0.0011 (6) |
| C9 | 0.0143 (7) | 0.0229 (9) | 0.0278 (8) | −0.0014 (6) | 0.0033 (6) | 0.0025 (7) |
| C10 | 0.0146 (7) | 0.0220 (8) | 0.0275 (8) | 0.0015 (6) | −0.0007 (6) | 0.0041 (7) |
| C11 | 0.0163 (7) | 0.0173 (8) | 0.0231 (7) | 0.0001 (6) | −0.0024 (6) | 0.0012 (6) |
| C12 | 0.0159 (7) | 0.0161 (7) | 0.0246 (8) | 0.0016 (6) | −0.0038 (6) | 0.0022 (6) |
| C13 | 0.0189 (7) | 0.0170 (7) | 0.0157 (7) | −0.0014 (6) | 0.0013 (5) | −0.0001 (6) |
| C14 | 0.0166 (7) | 0.0135 (7) | 0.0185 (7) | 0.0027 (6) | 0.0010 (5) | −0.0012 (6) |
| C15 | 0.0127 (7) | 0.0160 (7) | 0.0173 (7) | 0.0008 (5) | −0.0007 (5) | −0.0003 (6) |
| C16 | 0.0164 (7) | 0.0181 (7) | 0.0114 (6) | −0.0009 (6) | 0.0014 (5) | 0.0014 (6) |
| C17 | 0.0196 (7) | 0.0263 (8) | 0.0141 (6) | −0.0025 (7) | 0.0017 (5) | −0.0025 (6) |
| C18 | 0.0245 (8) | 0.0333 (10) | 0.0249 (8) | −0.0088 (7) | 0.0087 (7) | −0.0060 (7) |
| C19 | 0.0219 (8) | 0.0319 (9) | 0.0235 (8) | 0.0026 (7) | 0.0035 (6) | −0.0028 (7) |
| C20 | 0.0228 (8) | 0.0198 (8) | 0.0259 (8) | −0.0017 (6) | −0.0038 (6) | 0.0025 (6) |
| C21 | 0.0375 (10) | 0.0349 (11) | 0.0338 (10) | −0.0069 (8) | −0.0157 (8) | −0.0019 (8) |
| C22 | 0.0214 (8) | 0.0171 (8) | 0.0215 (7) | −0.0006 (6) | −0.0021 (6) | 0.0021 (6) |
| C23 | 0.0248 (8) | 0.0166 (8) | 0.0340 (9) | 0.0021 (6) | −0.0072 (7) | 0.0032 (7) |
| C24 | 0.0458 (11) | 0.0210 (9) | 0.0226 (8) | −0.0021 (8) | −0.0097 (7) | 0.0059 (7) |
Geometric parameters (Å, º)
| S1—C8 | 1.8200 (16) | C9—H9 | 0.9900 |
| S1—S2 | 2.0434 (6) | C10—C11 | 1.510 (2) |
| S2—C9 | 1.8211 (15) | C10—H10 | 0.9900 |
| O1—C2 | 1.3408 (17) | C10—H11 | 0.9900 |
| O1—C1 | 1.4599 (18) | C11—C12 | 1.325 (2) |
| O2—C2 | 1.2003 (19) | C11—H12 | 0.9500 |
| O3—C4 | 1.2311 (19) | C12—H13 | 0.9500 |
| O4—C6 | 1.2208 (19) | C13—C14 | 1.518 (2) |
| O5—C14 | 1.2331 (19) | C13—H14 | 0.9900 |
| O6—C16 | 1.2277 (19) | C13—H15 | 0.9900 |
| N1—C4 | 1.340 (2) | C15—C16 | 1.534 (2) |
| N1—C3 | 1.4580 (18) | C15—C22 | 1.542 (2) |
| N1—H3 | 0.84 (2) | C15—H17 | 1.0000 |
| N2—C6 | 1.359 (2) | C17—C19 | 1.519 (2) |
| N2—C5 | 1.413 (2) | C17—C18 | 1.529 (2) |
| N2—H4 | 0.82 (2) | C17—H19 | 1.0000 |
| N3—C14 | 1.344 (2) | C18—H22 | 0.9800 |
| N3—C15 | 1.4596 (18) | C18—H21 | 0.9800 |
| N3—H16 | 0.87 (2) | C18—H20 | 0.9800 |
| N4—C16 | 1.342 (2) | C19—H24 | 0.9800 |
| N4—C7 | 1.455 (2) | C19—H25 | 0.9800 |
| N4—H18 | 0.76 (2) | C19—H23 | 0.9800 |
| C1—C12 | 1.505 (2) | C20—C21 | 1.496 (2) |
| C1—C13 | 1.537 (2) | C20—H26 | 0.9500 |
| C1—H1 | 1.0000 | C21—H27 | 0.9800 |
| C2—C3 | 1.525 (2) | C21—H29 | 0.9800 |
| C3—C17 | 1.543 (2) | C21—H28 | 0.9800 |
| C3—H2 | 1.0000 | C22—C23 | 1.527 (2) |
| C4—C5 | 1.509 (2) | C22—C24 | 1.528 (2) |
| C5—C20 | 1.330 (2) | C22—H30 | 1.0000 |
| C6—C7 | 1.540 (2) | C23—H31 | 0.9800 |
| C7—C8 | 1.522 (2) | C23—H32 | 0.9800 |
| C7—H5A | 1.0000 | C23—H33 | 0.9800 |
| C8—H6 | 0.9900 | C24—H35 | 0.9800 |
| C8—H7 | 0.9900 | C24—H34 | 0.9800 |
| C9—C10 | 1.525 (2) | C24—H36 | 0.9800 |
| C9—H8 | 0.9900 | ||
| C8—S1—S2 | 106.04 (5) | C11—C12—C1 | 125.93 (15) |
| C9—S2—S1 | 103.94 (6) | C11—C12—H13 | 117.0 |
| C2—O1—C1 | 115.81 (12) | C1—C12—H13 | 117.0 |
| C4—N1—C3 | 121.41 (13) | C14—C13—C1 | 112.39 (13) |
| C4—N1—H3 | 120.8 (13) | C14—C13—H14 | 109.1 |
| C3—N1—H3 | 117.6 (13) | C1—C13—H14 | 109.1 |
| C6—N2—C5 | 120.32 (13) | C14—C13—H15 | 109.1 |
| C6—N2—H4 | 118.8 (14) | C1—C13—H15 | 109.1 |
| C5—N2—H4 | 120.4 (14) | H14—C13—H15 | 107.9 |
| C14—N3—C15 | 119.16 (13) | O5—C14—N3 | 121.37 (13) |
| C14—N3—H16 | 122.0 (13) | O5—C14—C13 | 121.48 (13) |
| C15—N3—H16 | 118.8 (13) | N3—C14—C13 | 117.09 (13) |
| C16—N4—C7 | 123.97 (13) | N3—C15—C16 | 110.61 (12) |
| C16—N4—H18 | 117.8 (15) | N3—C15—C22 | 110.05 (12) |
| C7—N4—H18 | 118.2 (15) | C16—C15—C22 | 113.98 (12) |
| O1—C1—C12 | 105.19 (12) | N3—C15—H17 | 107.3 |
| O1—C1—C13 | 109.24 (12) | C16—C15—H17 | 107.3 |
| C12—C1—C13 | 116.69 (13) | C22—C15—H17 | 107.3 |
| O1—C1—H1 | 108.5 | O6—C16—N4 | 123.09 (14) |
| C12—C1—H1 | 108.5 | O6—C16—C15 | 121.31 (14) |
| C13—C1—H1 | 108.5 | N4—C16—C15 | 115.59 (13) |
| O2—C2—O1 | 124.40 (14) | C19—C17—C18 | 110.89 (14) |
| O2—C2—C3 | 123.47 (13) | C19—C17—C3 | 113.19 (13) |
| O1—C2—C3 | 112.08 (12) | C18—C17—C3 | 109.91 (13) |
| N1—C3—C2 | 111.25 (12) | C19—C17—H19 | 107.5 |
| N1—C3—C17 | 112.74 (12) | C18—C17—H19 | 107.5 |
| C2—C3—C17 | 111.62 (13) | C3—C17—H19 | 107.5 |
| N1—C3—H2 | 106.9 | C17—C18—H22 | 109.5 |
| C2—C3—H2 | 106.9 | C17—C18—H21 | 109.5 |
| C17—C3—H2 | 106.9 | H22—C18—H21 | 109.5 |
| O3—C4—N1 | 123.36 (13) | C17—C18—H20 | 109.5 |
| O3—C4—C5 | 121.16 (14) | H22—C18—H20 | 109.5 |
| N1—C4—C5 | 115.42 (13) | H21—C18—H20 | 109.5 |
| C20—C5—N2 | 123.37 (14) | C17—C19—H24 | 109.5 |
| C20—C5—C4 | 119.61 (14) | C17—C19—H25 | 109.5 |
| N2—C5—C4 | 116.99 (13) | H24—C19—H25 | 109.5 |
| O4—C6—N2 | 122.62 (14) | C17—C19—H23 | 109.5 |
| O4—C6—C7 | 123.05 (14) | H24—C19—H23 | 109.5 |
| N2—C6—C7 | 114.33 (13) | H25—C19—H23 | 109.5 |
| N4—C7—C8 | 109.81 (13) | C5—C20—C21 | 125.28 (16) |
| N4—C7—C6 | 109.72 (11) | C5—C20—H26 | 117.4 |
| C8—C7—C6 | 108.94 (12) | C21—C20—H26 | 117.4 |
| N4—C7—H5A | 109.4 | C20—C21—H27 | 109.5 |
| C8—C7—H5A | 109.4 | C20—C21—H29 | 109.5 |
| C6—C7—H5A | 109.4 | H27—C21—H29 | 109.5 |
| C7—C8—S1 | 116.36 (11) | C20—C21—H28 | 109.5 |
| C7—C8—H6 | 108.2 | H27—C21—H28 | 109.5 |
| S1—C8—H6 | 108.2 | H29—C21—H28 | 109.5 |
| C7—C8—H7 | 108.2 | C23—C22—C24 | 110.22 (14) |
| S1—C8—H7 | 108.2 | C23—C22—C15 | 109.06 (13) |
| H6—C8—H7 | 107.4 | C24—C22—C15 | 111.79 (13) |
| C10—C9—S2 | 115.36 (11) | C23—C22—H30 | 108.6 |
| C10—C9—H8 | 108.4 | C24—C22—H30 | 108.6 |
| S2—C9—H8 | 108.4 | C15—C22—H30 | 108.6 |
| C10—C9—H9 | 108.4 | C22—C23—H31 | 109.5 |
| S2—C9—H9 | 108.4 | C22—C23—H32 | 109.5 |
| H8—C9—H9 | 107.5 | H31—C23—H32 | 109.5 |
| C11—C10—C9 | 113.50 (13) | C22—C23—H33 | 109.5 |
| C11—C10—H10 | 108.9 | H31—C23—H33 | 109.5 |
| C9—C10—H10 | 108.9 | H32—C23—H33 | 109.5 |
| C11—C10—H11 | 108.9 | C22—C24—H35 | 109.5 |
| C9—C10—H11 | 108.9 | C22—C24—H34 | 109.5 |
| H10—C10—H11 | 107.7 | H35—C24—H34 | 109.5 |
| C12—C11—C10 | 125.57 (15) | C22—C24—H36 | 109.5 |
| C12—C11—H12 | 117.2 | H35—C24—H36 | 109.5 |
| C10—C11—H12 | 117.2 | H34—C24—H36 | 109.5 |
| C8—S1—S2—C9 | −90.18 (7) | S1—S2—C9—C10 | 65.73 (12) |
| C2—O1—C1—C12 | −162.59 (12) | S2—C9—C10—C11 | 59.05 (17) |
| C2—O1—C1—C13 | 71.42 (15) | C9—C10—C11—C12 | −145.87 (15) |
| C1—O1—C2—O2 | 12.5 (2) | C10—C11—C12—C1 | −172.08 (15) |
| C1—O1—C2—C3 | −170.18 (12) | O1—C1—C12—C11 | −146.55 (14) |
| C4—N1—C3—C2 | −135.83 (14) | C13—C1—C12—C11 | −25.3 (2) |
| C4—N1—C3—C17 | 97.90 (17) | O1—C1—C13—C14 | 43.45 (16) |
| O2—C2—C3—N1 | −155.37 (14) | C12—C1—C13—C14 | −75.63 (16) |
| O1—C2—C3—N1 | 27.23 (17) | C15—N3—C14—O5 | 2.3 (2) |
| O2—C2—C3—C17 | −28.5 (2) | C15—N3—C14—C13 | −175.03 (12) |
| O1—C2—C3—C17 | 154.12 (12) | C1—C13—C14—O5 | −70.20 (18) |
| C3—N1—C4—O3 | 0.7 (2) | C1—C13—C14—N3 | 107.10 (15) |
| C3—N1—C4—C5 | −176.38 (12) | C14—N3—C15—C16 | 68.10 (16) |
| C6—N2—C5—C20 | 131.38 (16) | C14—N3—C15—C22 | −165.07 (13) |
| C6—N2—C5—C4 | −50.66 (19) | C7—N4—C16—O6 | −4.9 (2) |
| O3—C4—C5—C20 | −46.7 (2) | C7—N4—C16—C15 | 176.20 (12) |
| N1—C4—C5—C20 | 130.44 (16) | N3—C15—C16—O6 | −152.01 (13) |
| O3—C4—C5—N2 | 135.23 (15) | C22—C15—C16—O6 | 83.37 (17) |
| N1—C4—C5—N2 | −47.60 (18) | N3—C15—C16—N4 | 26.90 (17) |
| C5—N2—C6—O4 | −3.8 (2) | C22—C15—C16—N4 | −97.72 (15) |
| C5—N2—C6—C7 | 176.31 (12) | N1—C3—C17—C19 | 61.93 (18) |
| C16—N4—C7—C8 | −160.22 (13) | C2—C3—C17—C19 | −64.14 (16) |
| C16—N4—C7—C6 | 80.06 (16) | N1—C3—C17—C18 | −62.66 (17) |
| O4—C6—C7—N4 | 114.64 (16) | C2—C3—C17—C18 | 171.27 (12) |
| N2—C6—C7—N4 | −65.46 (16) | N2—C5—C20—C21 | −3.5 (3) |
| O4—C6—C7—C8 | −5.61 (19) | C4—C5—C20—C21 | 178.62 (16) |
| N2—C6—C7—C8 | 174.30 (12) | N3—C15—C22—C23 | 66.98 (16) |
| N4—C7—C8—S1 | 68.66 (14) | C16—C15—C22—C23 | −168.10 (12) |
| C6—C7—C8—S1 | −171.15 (10) | N3—C15—C22—C24 | −170.88 (13) |
| S2—S1—C8—C7 | −69.44 (12) | C16—C15—C22—C24 | −45.97 (19) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H3···O5 | 0.84 (2) | 2.27 (2) | 3.0123 (18) | 147.1 (17) |
| N2—H4···O6 | 0.82 (2) | 2.05 (2) | 2.7867 (18) | 149.1 (18) |
| N3—H16···O3i | 0.87 (2) | 2.18 (2) | 3.0449 (17) | 175.9 (18) |
Symmetry code: (i) −x+1, y−1/2, −z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: JJ2148).
References
- Biggins, J. B., Gleber, C. D. & Brady, S. F. (2011). Org. Lett. 13, 1536–1539. [DOI] [PMC free article] [PubMed]
- Bruker (2007). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Cheng, Y. Q., Yang, M. & Matter, A. M. (2007). Appl. Environ. Microbiol. 73, 3460–3469. [DOI] [PMC free article] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Furumai, R., Matsuyama, A., Kobashi, N., Lee, K. H., Nishiyama, M., Nakajima, H., Tanaka, A., Komatsu, Y., Nishino, N., Yoshida, M. & Horinouchi, S. (2002). Cancer Res. 62, 4916–4921. [PubMed]
- Klausmeyer, P., Shipley, S. M., Zuck, K. M. & McCloud, T. G. (2011). J. Nat. Prod. 74, 2039–2044. [DOI] [PMC free article] [PubMed]
- Knappe, T. A., Linne, U., Zirah, S., Rebuffat, S., Xie, X. & Marahiel, M. A. (2008). J. Am. Chem. Soc. 130, 11446–11454. [DOI] [PubMed]
- Mukhopadhyay, S., Thomason, M. K., Lentz, S., Nolan, N., Willner, K., Gee, J. E., Glass, M. B., Inglis, T. J., Merritt, A., Levy, A., Sozhamannan, S., Mateczun, A. & Read, T. D. (2010). J. Bacteriol. 192, 6313–6314. [DOI] [PMC free article] [PubMed]
- Nguyen, T., Ishida, K., Jenke-Kodama, H., Dittmann, E., Gurgui, C., Hochmuth, T., Taudien, S., Platzer, M., Hertweck, C. & Piel, J. (2008). Nat. Biotechnol. 26, 225–233. [DOI] [PubMed]
- Potharla, V. Y., Wesener, S. R. & Cheng, Y. Q. (2011). Appl. Environ. Microbiol. 77, 1508–1511. [DOI] [PMC free article] [PubMed]
- Robey, R. W., Chakraborty, A. R., Basseville, A., Luchenko, V., Bahr, J., Zhan, Z. & Bates, S. E. (2011). Mol. Pharm. 8, 2021–2031. [DOI] [PMC free article] [PubMed]
- Seyedsayamdost, M. R., Chandler, J. R., Blodgett, J. A., Lima, P. S., Duerkop, B. A., Oinuma, K., Greenberg, E. P. & Clardy, J. (2010). Org. Lett. 12, 716–719. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2003). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shigematsu, N., Ueda, H., Takase, S., Tanaka, H., Yamamoto, K. & Tada, T. (1994). J. Antibiot. 47, 311–314. [DOI] [PubMed]
- StatBite (2010). J. Natl Cancer Inst. 102, 219–230.
- Ueda, H., Manda, T., Matsumoto, S., Mukumoto, S., Nishigaki, F., Kawamura, I. & Shimomura, K. (1994). J. Antibiot. 47, 315–323. [DOI] [PubMed]
- Ueda, H., Nakajima, H., Hori, Y., Fujita, T., Nishimura, M., Goto, T. & Okuhara, M. (1994). J. Antibiot. 47, 301–310. [DOI] [PubMed]
- Ueda, H., Nakajima, H., Hori, Y., Goto, T. & Okuhara, M. (1994). Biosci. Biotechnol. Biochem. 58, 1579–1583. [DOI] [PubMed]
- Wang, C., Flemming, C. J. & Cheng, Y.-Q. (2012). MedChemComm, 3, 976–981. [DOI] [PMC free article] [PubMed]
- Wang, C., Henkes, L. M., Doughty, L. B., He, M., Wang, D., Meyer-Almes, F. J. & Cheng, Y. Q. (2011). J. Nat. Prod. 74, 2031–2038. [DOI] [PMC free article] [PubMed]
- Wesener, S. R., Potharla, V. Y. & Cheng, Y. Q. (2011). Appl. Environ. Microbiol. 77, 1501–1507. [DOI] [PMC free article] [PubMed]
- Yu, Y., Kim, H. S., Chua, H. H., Lin, C. H., Sim, S. H., Lin, D., Derr, A., Engels, R., DeShazer, D., Birren, B., Nierman, W. C. & Tan, P. (2006). BMC Microbiol. 6, 46. [DOI] [PMC free article] [PubMed]
- Zhuo, Y., Wang, Q., Liu, L., Liu, X., Ren, B., Liu, M., Cheng, Y.-Q. & Zhang, L. (2012). J. Bacteriol. 194, 4749–4750. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S160053681203601X/jj2148sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S160053681203601X/jj2148Isup2.hkl
Supplementary material file. DOI: 10.1107/S160053681203601X/jj2148Isup3.cdx
Additional supplementary materials: crystallographic information; 3D view; checkCIF report