Abstract
Emerging evidence indicates that gene expression in higher organisms is regulated by RNA polymerase II stalling during early transcription elongation. To probe the mechanisms responsible for this regulation, we developed methods to isolate and characterize short RNAs derived from stalled RNA polymerase II in Drosophila cells. Significant levels of these short RNAs were generated from over one third of all genes, indicating that promoter-proximal stalling is a general feature of early polymerase elongation. Nucleotide composition of the initially transcribed sequence played an important role in promoting transcriptional stalling by rendering polymerase elongation complexes highly susceptible to backtracking and arrest. These results indicate that the intrinsic efficiency of early elongation can greatly affect gene expression.
Recent genome-wide studies of RNA polymerase II (Pol II) distribution have demonstrated that Pol II accumulates at promoters of many developmentally regulated and stimulus-responsive genes in their uninduced states (1–3). These findings challenge a common paradigm for gene regulation, which holds that recruiting the polymerase to a promoter is sufficient for gene activation, and indicate that regulation of many genes occurs after transcription initiates. An appealing model for such regulation involves promoter-proximal stalling, wherein an actively engaged polymerase pauses 25–50 nucleotides (nt) downstream of the Transcription Start Site (TSS) (4–6). Release of stalled Pol II into productive elongation is rate-limiting for the expression of several Drosophila and mammalian genes (4, 5, 7), and mounting evidence suggests that promoter-proximal stalling is a widespread strategy for governing transcription output (8). However, efforts to define the prevalence and mechanisms of Pol II stalling have been hampered by the lack of a high-resolution, high-throughput method to detect transcriptionally engaged polymerase.
To overcome this obstacle, we developed a strategy to map promoter-proximal Pol II in Drosophila on a genome-wide scale and with single-nucleotide resolution. We isolated RNAs derived from stalled polymerase, making use of their characteristic properties as previously delineated for heat shock (hsp) genes: short size (<100nt), nuclear localization, and presence of the 7-methylguanosine cap that is added to the 5’-end of nascent mRNA shortly after initiation (fig. S1) (6, 9–11). Short RNA libraries were prepared from two independent biological replicates and sequenced on an Illumina Genome Analyzer, yielding a combined total of 16.5 million uniquely mappable reads (Table S1) (12).
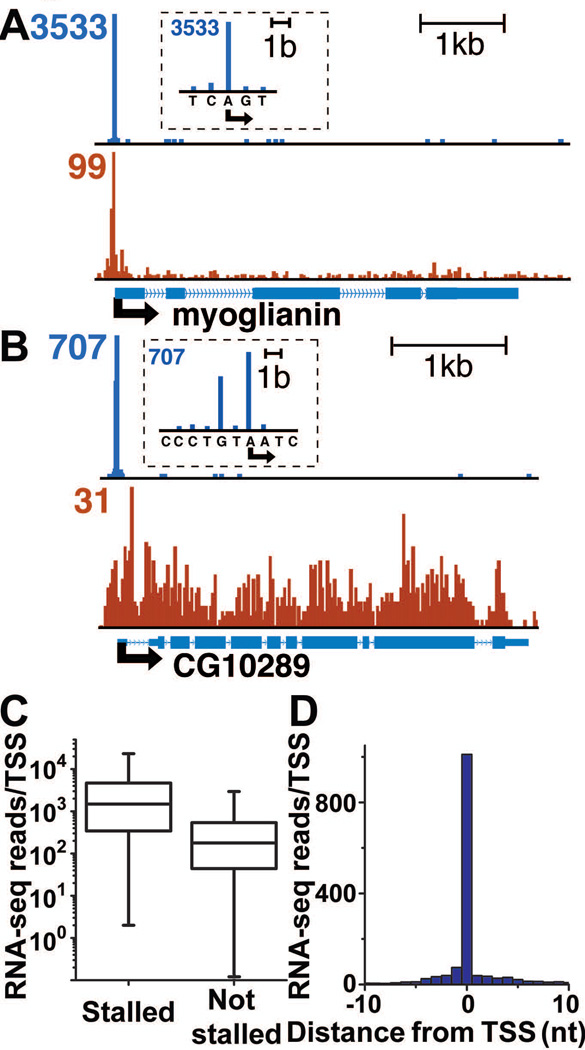
Our approach efficiently selected for Pol II transcripts: ~75% of the reads mapped within 200 bp of the annotated TSSs of mRNA genes (fig. S2). About 98% of short RNAs that mapped near promoters aligned with the sense DNA strand (fig. S2), presenting no evidence for significant divergent transcription in Drosophila. Statistically significant levels of short RNAs were observed from more than 7,400 TSSs (P<0.005) (fig. S3), including >93% of genes that were previously defined as possessing stalled polymerase in ChIP-chip studies (Fig. 1A; 1C) (1). Genes with stalled Pol II generated considerably more short RNAs than genes with Pol II that did not appear stalled (Fig. 1C; P<0.0001; fig. S4) (1). However, >85% of genes that were not considered stalled in prior work also produced short RNAs, presumably as transient intermediates on the pathway to productive transcription (Fig. 1B; 1C; fig. S4). Although the number of short RNAs was a poor predictor of gene expression level (fig. S5), it was highly correlated with Pol II ChIP-seq signal near the TSS (fig. S4). This finding agrees well with recent work indicating that much of the polymerase detected promoter-proximally is indeed engaged in early elongation (8).
Fig. 1. Short capped RNAs are produced by promoter-proximal Pol II.
Distribution of RNA 5’-ends (blue) and Pol II ChIP-seq signal (red) on (A) myoglianin (CG1838) a gene with stalled Pol II and low expression and (B) CG10289, a gene with more uniform polymerase distribution and high expression. The peak number of RNA reads (1-nt windows) and Pol II ChIP-seq reads (25-bp windows) is indicated in the corresponding color. Insets contain magnified views of the TSSs. (C) The number of short RNAs that mapped to stalled genes (median reads/TSS=1,477) as compared to genes that were not considered stalled (median reads/TSS=178). Box plots represent the 25th, 50th and 75th percentiles, with whiskers denoting the 5th and 95th percentiles. (D) Metagene analysis of 5’-end reads aligned to observed TSSs.
The 5’-end reads around many TSSs mapped to a single nucleotide position (Fig. 1D), consistent with the idea that initiation in Drosophila is highly focused at most promoters (13). The observed 5’-end positions frequently differed from annotated TSSs, as suggested in earlier examinations of capped mRNA (fig. S6; Table S2) (14). Analysis of the sequences surrounding short RNA 5’-ends revealed a much better match to the consensus Initiator element (13) than sequences around the annotated TSSs (fig. S6), indicating that we have accurately identified TSSs. The TSSs observed from short RNAs were in good agreement with those observed from capped RNAs isolated without size restriction (fig. S7) (15), indicating that polymerases generating short RNAs initiated from the same TSSs as those that synthesized full-length mRNA.
Although our approach readily detected short RNAs derived from stalled polymerase, pinpointing Pol II location precisely required mapping the RNA 3’-ends. Since currently available high-throughput procedures allow sequencing of RNAs only from 5’-ends, we designed new RNA adapters to sequence RNAs directly from their 3’-ends. The resultant RNA libraries, prepared from the same RNA samples as above, remained fully compatible with commercial sequencing primers and chemistry (fig. S2, S3; Table S1).
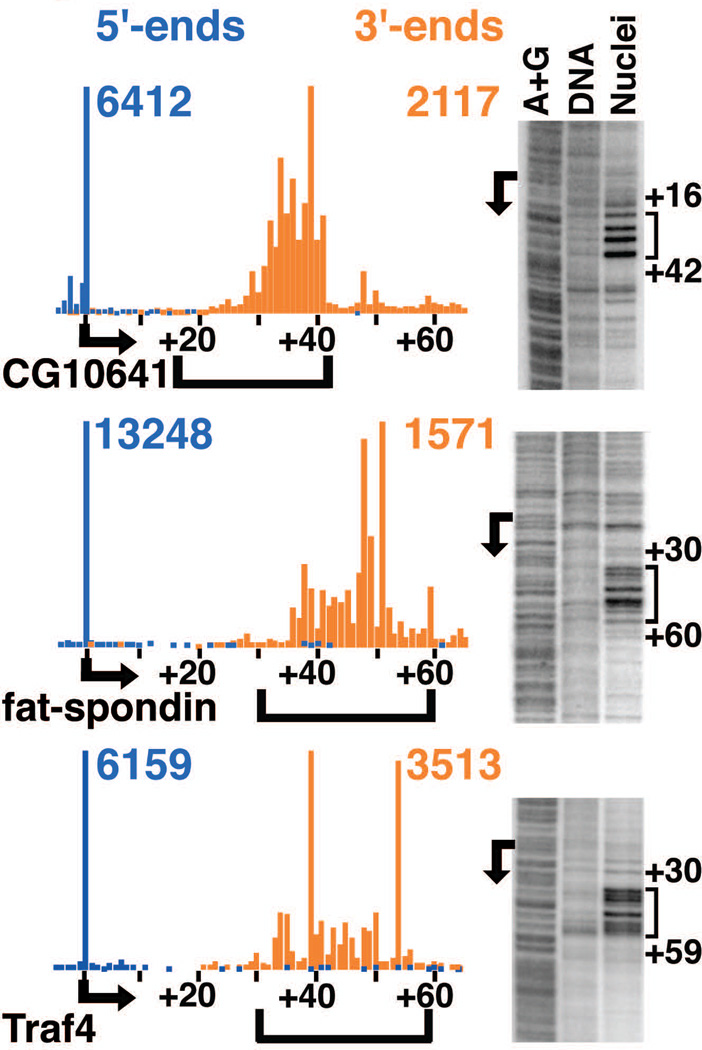
We confirmed that the 3’-ends of short RNAs accurately defined locations of Pol II stalling by comparing their distribution to positions of engaged Pol II independently determined by permanganate probing. Permanganate reacts with single-stranded thymines on DNA, such as those in an open transcription bubble, and is currently the most authoritative means to detect stalled polymerase in vivo (16, 17). We found a remarkable correspondence between locations of short RNA 3’-ends and regions of permanganate reactivity at newly identified genes (Fig. 2) and in published studies (fig. S8) (1, 2, 17, 18). Metagene analysis showed that the 3’-ends were distributed primarily between +25 and +60 relative to the TSS (Fig. 3A; ~6,500 genes), which agreed well with prior analysis of permanganate reactivity on ~60 genes (17).
Fig. 2. 3’-ends of short RNAs identify sites of polymerase stalling.
5’-end (blue) and 3’-end (orange) RNA reads are shown for example genes, designated by Flybase gene names, with observed TSSs indicated by arrows. The number of 5’- and 3’-end reads at the peak locations is given in the corresponding colors. Permanganate footprints from nuclei are shown for each gene, alongside purified DNA and the A+G DNA ladder. Numbers in black indicate positions relative to the TSS, and brackets show areas of permanganate reactivity.
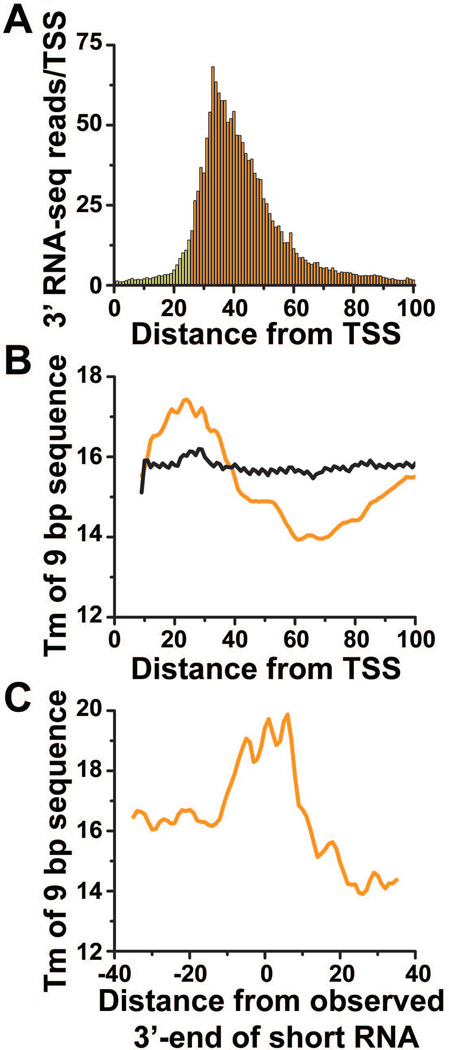
Fig. 3. Promoter-proximal sequences impact Pol II stalling.
(A) 3’-end reads were aligned relative to observed TSSs. RNAs shorter than 26 nt are not efficiently mapped to the genome (colored in yellow). (B) Metagene analysis of melting temperatures (°C) for 9-bp sequences across the initially transcribed regions for genes that generate significant numbers of short RNAs (orange) and all other genes (black). (C) Tm analysis of 9-bp sequences surrounding the primary 3’-end location for a subset of genes with focused 3’-end positions (N=434).
Since Pol II stalls within the same promoter-proximal interval globally, we wondered whether the initially transcribed sequence might contribute to stalling. Given that the stability of the 9-bp RNA-DNA hybrid in the elongation complex greatly influences the efficiency of elongation (19, 20), we calculated the melting temperature (Tm) of each 9-bp sequence across the initially transcribed region for genes shown in Fig. 3A and for control genes that did not generate significant short RNAs (Fig. 3B). There were clear differences between these profiles: whereas the profile of the control group was essentially flat, genes that produced short RNAs exhibited a peak of Tm between positions +20 and +35 that corresponded to the primary sites of Pol II stalling. This peak was followed by a decline in Tm which would serve to progressively destabilize the elongation complex (Fig. 3B) (19, 20). These observations support a two-step model of stalling wherein the elongating polymerase first pauses transiently within the downstream region of weak RNA-DNA hybrid stability, and then slides backwards along DNA to a site with high thermodynamic stability (21–23). To verify that the position of stalling coincides with the peak in hybrid stability, we repeated Tm analysis on a subset of genes that displayed one predominant 3’-end position, and found that it aligned with the region of highest Tm (Fig. 3C; fig. S9).
Although there were clear differences in the Tm profiles between genes that produced short RNAs and those that did not, some genes that lacked short RNAs in S2 cells nonetheless displayed an elevated Tm within the region from +20 and +35 (Fig. 3B). To test whether these genes might be stalled in another cell type, we isolated short RNAs from 0–16h Drosophila embryos and found >1,500 genes that did not generate short RNAs in S2 cells but did so during embryo development. The Tm within the promoter-proximal region (+20 to +35) of these new genes was significantly higher than for genes without short RNAs (fig. S10; P<0.0001), and calculation of a Tm profile around the embryo-derived 3’-ends confirmed that they mapped within a peak in melting temperature. These results indicated that sequence composition of the initially transcribed region predisposes the polymerase to stall, and could be used to predict genes that are most likely to possess stalled Pol II under different conditions.
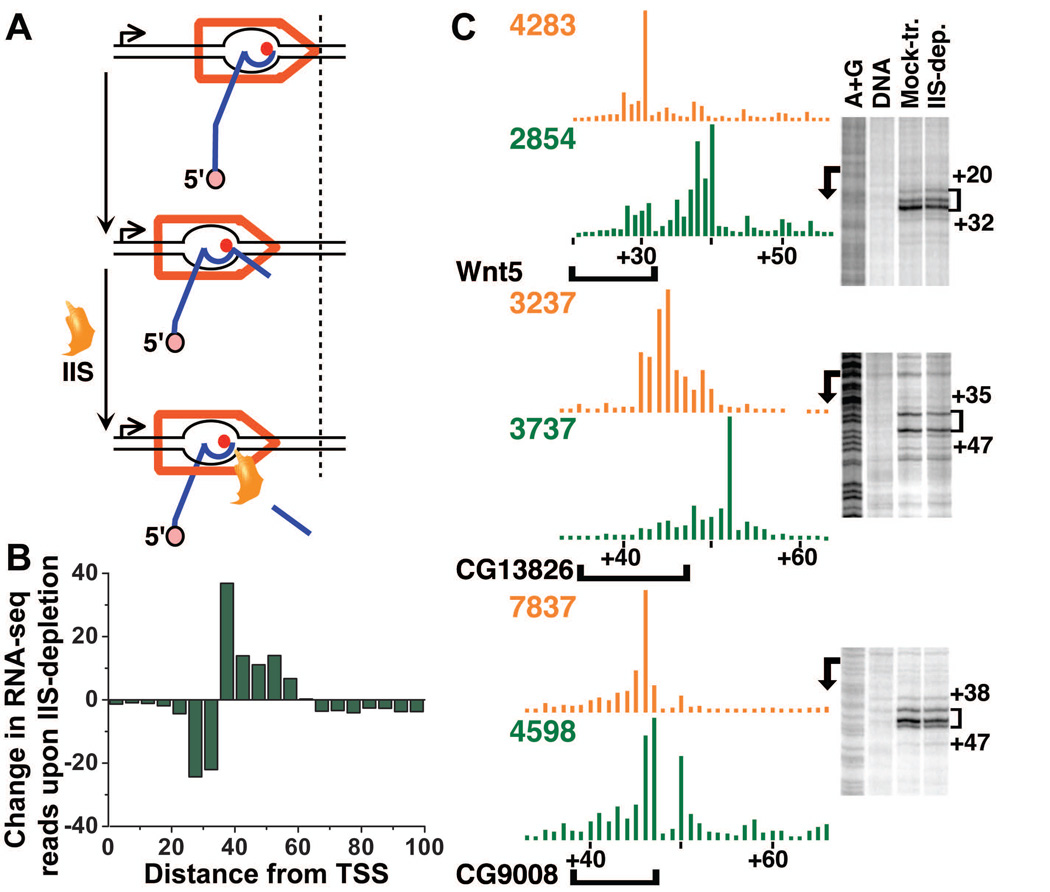
Our data suggested that stalled elongation complexes have undergone backtracking following transient pausing. The transcript cleavage factor TFIIS (IIS) has been shown to reactivate Pol II in backtracked and arrested elongation complexes (Fig. 4A), including stalled Pol II at the Drosophila hsp70 gene (24) (fig. S11). If backtracking is a general feature of early polymerase elongation, then we should be able to detect evidence for IIS-mediated cleavage and shortening of promoter-proximal RNAs by comparing RNA profiles in mock-treated and IIS-depleted cells. Indeed, RNAi-depletion of IIS caused a global increase in RNA lengths (Fig. 4B; P <10−15). Moreover, RNAs between 35–60 nt long were specifically enriched in IIS-depleted cells, indicating that they are the primary targets of IIS-induced cleavage (Fig. 4B). A concomitant reduction in 20–35 nt RNAs in IIS-deficient cells suggested that these RNAs are produced in part by IIS. Genes with stalled Pol II were significantly more likely to exhibit RNA lengthening upon IIS-depletion than genes lacking stalled polymerase (fig. S12; P<10−4), indicating that polymerase stalling generally involves backtracking. Comparison of short RNA and permanganate reactivity profiles on individual genes showed that, unlike the RNA 3’-ends, which were clearly shifted downstream in IIS-depleted cells, the location of the transcription bubble did not change (Fig. 4C). These results indicated that the primary location of the stalled elongation complex at steady-state reflected the position to which the polymerase has backtracked.
Fig. 4. Stalled polymerase complexes are predominantly backtracked.
(A) The role of transcription cleavage factor IIS in rescuing arrested Pol II elongation complexes. Following transcriptional pausing (top panel) the polymerase can backtrack along DNA (middle panel), which dislodges the RNA 3’-end from the polymerase active site (shown as red dot) and blocks further transcription. IIS induces internal cleavage of nascent RNA (bottom panel), re-aligning the 3’-end with the active site, such that Pol II can resume transcription. (B) Depletion of IIS leads to an increase in RNA length within the promoter-proximal region. Shown is the difference between the normalized number of reads in IIS-depleted and mock-treated samples, binned in 5-nt windows. (C) IIS-depletion affects RNA 3’-end positions but not permanganate reactivity profiles. RNA 3’-ends from mock-treated samples are shown in orange and IIS-depleted cells in green. Brackets denote regions of permanganate reactivity.
Recent work has underscored the importance of early transcription elongation and its regulation in vivo (1–3, 25, 26). Our data reveal that fluctuations in RNA-DNA hybrid stability in the initially transcribed sequence make the polymerase susceptible to pausing and backtracking. This tendency is likely amplified by the presence of downstream nucleosomes and the reported absence of secondary structures within these short RNAs that would inhibit backtracking (17, 27). Furthermore, elongation factors specifically target promoter-proximal Pol II to regulate the duration of stalling. For example, the negative transcription elongation factor NELF has been shown biochemically to both enhance the duration of intrinsic pauses and to inhibit IIS activity (28, 29).
Our data indicate that stalled polymerase complexes do not efficiently escape into productive elongation even after rescue by IIS-induced cleavage. In fact, many rounds of pausing, backtracking and cleavage, or perhaps even termination, may ensue before a positive signal such as the activity of the PTEF-b kinase releases the stalled Pol II from the promoter-proximal region (28, 29). In addition, our data suggest that transient stalling of polymerase is a general feature of early elongation, even at highly active genes, since we observe short RNAs with similar distributions arising from nearly all active genes (fig. S13). Thus, understanding how the duration of stalling is regulated under various conditions is of considerable interest, and our RNA-based approach opens a possibility for detailed dissection of this process on a genome-wide scale.
Supplementary Material
Acknowledgments
We thank P. Wade, T. Kunkel, and members of the Adelman lab for insightful discussions. We acknowledge S. Dai and J. Grovenstein for computational support. Sequence data are in the GEO database under accession number GSE18643. This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01 ES101987).
References and notes
- 1.Muse GW, et al. Nat Genet. 2007;39:1507. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeitlinger J, et al. Nat Genet. 2007;39:1512. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. Cell. 2007;130:77. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krumm A, Hickey LB, Groudine M. Genes Dev. 1995;9:559. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- 5.Rougvie AE, Lis JT. Mol Cell Biol. 1990;10:6041. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rasmussen EB, Lis JT. J Mol Biol. 1995;252:522. doi: 10.1006/jmbi.1995.0517. [DOI] [PubMed] [Google Scholar]
- 7.Aida M, et al. Mol Cell Biol. 2006;26:6094. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Core LJ, Waterfall JJ, Lis JT. Science. 2008;322:1845. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasmussen EB, Lis JT. Proc Natl Acad Sci U S A. 1993;90:7923. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coppola JA, Field AS, Luse DS. Proc Natl Acad Sci U S A. 1983;80:1251. doi: 10.1073/pnas.80.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jove R, Manley JL. J Biol Chem. 1984;259:8513. [PubMed] [Google Scholar]
- 12.Materials and Methods are available as supporting material on Science Online.
- 13.Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. Curr Opin Cell Biol. 2008;20:253. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahsan B, et al. Nucleic Acids Res. 2009;37:D49. doi: 10.1093/nar/gkn694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fejes-Toth K, et al. Nature. 2009;457:1028. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kainz M, Roberts J. Science. 1992;255:838. doi: 10.1126/science.1536008. [DOI] [PubMed] [Google Scholar]
- 17.Lee C, et al. Mol Cell Biol. 2008;28:3290. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilchrist DA, et al. Genes Dev. 2008;22:1921. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tadigotla VR, et al. Proc Natl Acad Sci U S A. 2006;103:4439. doi: 10.1073/pnas.0600508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palangat M, Landick R. J Mol Biol. 2001;311:265. doi: 10.1006/jmbi.2001.4842. [DOI] [PubMed] [Google Scholar]
- 21.Komissarova N, Kashlev M. J Biol Chem. 1997;272:15329. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 22.Reeder TC, Hawley DK. Cell. 1996;87:767. doi: 10.1016/s0092-8674(00)81395-1. [DOI] [PubMed] [Google Scholar]
- 23.Fish RN, Kane CM. Biochim Biophys Acta. 2002;1577:287. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- 24.Adelman K, et al. Mol Cell. 2005;17:103. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 25.Kephart DD, Marshall NF, Price DH. Mol Cell Biol. 1992;12:2067. doi: 10.1128/mcb.12.5.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pal M, McKean D, Luse DS. Mol Cell Biol. 2001;21:5815. doi: 10.1128/MCB.21.17.5815-5825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kireeva ML, et al. Mol Cell. 2005;18:97. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 28.Renner DB, Yamaguchi Y, Wada T, Handa H, Price DH. J Biol Chem. 2001;276:42601. doi: 10.1074/jbc.M104967200. [DOI] [PubMed] [Google Scholar]
- 29.Palangat M, Renner DB, Price DH, Landick R. Proc Natl Acad Sci U S A. 2005;102:15036. doi: 10.1073/pnas.0409405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.