Abstract
Colorectal tumors that are wild-type (WT) for KRAS are often sensitive to EGFR blockade, but almost always develop resistance within several months of initiating therapy1,2. The mechanisms underlying this acquired resistance to anti-EGFR antibodies are largely unknown. This situation stands in marked contrast to that of small molecule targeted agents, such as inhibitors of ABL, EGFR, BRAF, and MEK, in which mutations in the genes encoding the protein targets render the tumors resistant to the effects of the drugs3–6. The simplest hypothesis to account for the development of resistance to EGFR blockade are that rare cells with KRAS mutations pre-exist at low levels in tumors with ostensibly WT KRAS genes. Though this hypothesis would seem readily testable, there is no evidence in pre-clinical models to support it, nor is there data from patients. To test this hypothesis, we determined whether mutant KRAS DNA could be detected in the circulation of 28 patients receiving monotherapy with panitumumab, a therapeutic anti-EGFR antibody. We found that nine of 24 (38%) patients whose tumors were initially KRAS WT developed detectable mutations in KRAS in their sera, three of which developed multiple different KRAS mutations. The appearance of these mutations was very consistent, generally occurring between five to six months following treatment. Mathematical modeling indicated that the mutations were present in expanded subclones prior to the initiation of panitumumab. These results suggest that the emergence of KRAS mutations is a mediator of acquired resistance to EGFR blockade and that these mutations can be detected in a non-invasive manner. Moreover, they explain why solid tumors develop resistance to targeted therapies in a highly reproducible fashion.
One major barrier to testing any hypothesis about the nature of acquired resistance to anti-EGFR antibodies is limited access to post-treatment tumor tissue. Even when post-treatment tumor tissue is available, sampling bias confounds interpretation because only a small portion of one tumor is usually biopsied, precluding assessment of genetic heterogeneity within or among lesions. To circumvent the tissue access problem, we have examined circulating, cell-free DNA - a form of “liquid biopsy”. It has been previously shown that circulating tumor DNA (ctDNA) can be found in the majority of patients with metastatic colorectal cancers7–9. Analysis of ctDNA is informative because it not only can identify a specific mutant genotype but can also provide a measurement of the total tumor burden7. If tumors became resistant to anti-EGFR antibodies as a result of the emergence of KRAS mutations in their tumors, we expected that mutant KRAS genes would be released into the circulation in a time frame consistent with the emergence of resistance.
We retrospectively analyzed longitudinal serum samples from 28 patients with chemorefractory metastatic colorectal cancer (CRC) receiving single-agent therapy with panitumumab10. Four patients with KRAS mutant tumors, who never achieved disease control, were selected as controls. As expected, these four patients were found to have progressive disease at the time of first tumor assessment, 7 ± 2 weeks (mean ± 1 standard deviation) after initiating treatment with panitumumab (Supplementary Table 1)1,2. The other 24 patients with WT KRAS tumors achieved a partial response (n=8), had prolonged stable disease (n=14), or had retrospectively-determined progressive disease but remained on study for an extended period (n=2). These 24 patients developed clinically evident progressive disease 23 ± 10 weeks (mean ± 1 standard deviation) following initiation of treatment (Supplementary Table 1) as determined by radiographic imaging.
Serum samples obtained from patients prior to the initiation of therapy were evaluated for all common mutations at codons 12 and 13 of KRAS using a digital ligation assay with a detection limit of one mutant molecule per ml of serum (examples in Supplementary Fig. 1)11. Mutations were independently confirmed in a second aliquot of the same serum and the results quantified via a PCR assay that can digitally enumerate the fraction of rare variants in a complex mixture of DNA template molecules (examples in Supplementary Fig. 1 and Supplementary Table 2)12.
Of the four cases whose archival tumors harbored KRAS mutations, three had detectable levels of mutant KRAS in the serum prior to treatment with panitumumab (Supplementary Table 2). In these three patients, the KRAS mutations found in the circulation were identical to those found in the patients’ tumor tissues even though the time of serum assessment was, on average, 88 weeks after the diagnosis of metastatic disease and even longer after the initial tumor excision (Supplementary Tables 1 and 2). No mutations in KRAS were detected in the pre-treatment serum DNA from patients whose archival tumor tissue was WT for KRAS (Supplementary Table 2).
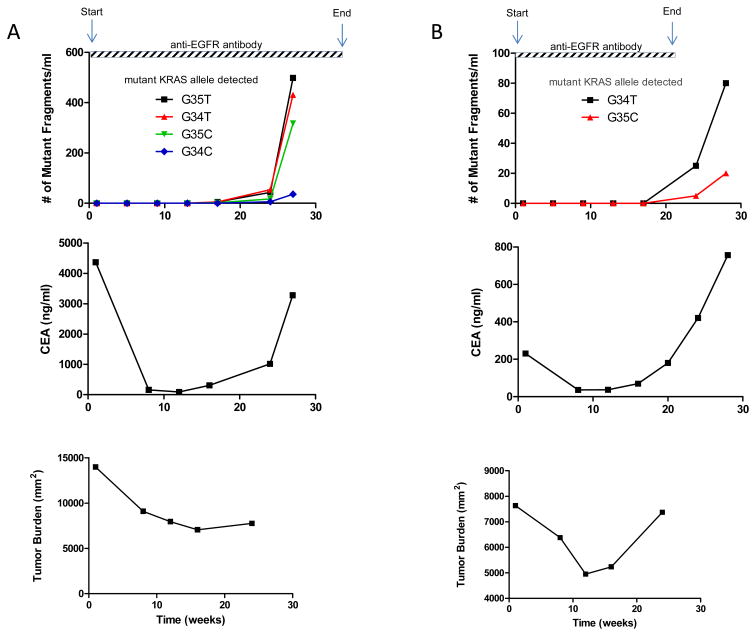
Next, we examined 169 serially acquired serum samples from the 28 patients for the presence of mutant KRAS fragments (Supplementary Fig. 2). These samples were collected at approximately four week intervals until disease progression (Supplementary Table 2). Serum was assessed for mutations at KRAS codons 12 and 13 as described above. When sufficient serum was available (23 of 28 patients), it was assessed for BRAF mutations at codons 600 and 601 using the identical assay. Of the 24 patients who did not have KRAS mutations at baseline, nine (38%) were found to develop KRAS mutations during the course of therapy (Supplementary Table 2) while none developed BRAF mutations. In three cases (Patients #1, 12, and 22), multiple KRAS mutations appeared in the circulation - two different mutants in one case and four different mutants in the two other cases (examples in Fig. 1). In each of these cases, the time to appearance of all mutations in the circulation was very similar (Fig. 2). Circulating mutant KRAS templates were identified prior to radiographic evidence of disease progression in three of the nine cases (Patients #1, 10 and 24). The lead time (i.e., the interval between detectable ctDNA and radiographic evidence of disease progression) averaged 21 weeks (Supplementary Table 2). The level of ctDNA generally paralleled that of CEA, the standard biomarker used for following disease progression in metastatic CRC (Supplementary Table 3 and Supplementary Fig. 2).
Fig. 1. Emergence of circulating mutant KRAS.
The time course of circulating mutant KRAS alleles, CEA and tumor burden are depicted in two patients where fragments of circulating DNA containing mutant KRAS were detected. (A) Demonstrates the emergence of four different mutant KRAS alleles in codon 12 (cDNA nt 35T, 34T, 35C and 34C) in the serum of Patient #1 and (B) demonstrates the emergence of two different mutant KRAS alleles in codon 12 (cDNA nt 34T and 35C) in the serum of Patient #12. Tumor burden refers to the aggregate cross-sectional diameter of the index lesions.
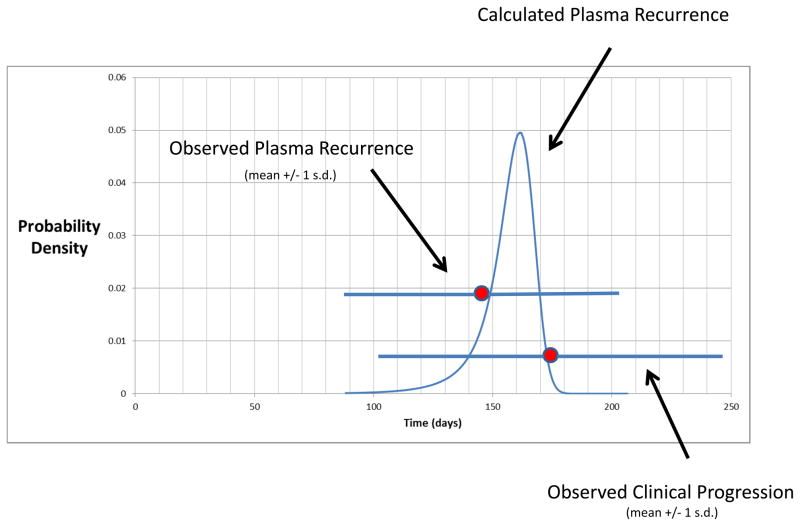
Fig. 2.
Predicted probability distribution of times from when treatment starts until resistance mutations become observable in circulating DNA. Overlaid is the observed time to detection of mutant KRAS fragments (23 ± 5 weeks, mean ± 1 standard deviation) and time to clinical progression (25 ± 10 weeks, mean ± 1 standard deviation) in the studied patients. Predictions were based on the Lea-Coulson model with death, as introduced by Dewanji et al.14, or equivalently, the branching process model of Iwasa et al14,16.
In the three patients (Patients #8, 20 and 28) with detectable mutant KRAS in their tumors as well as in their circulation, no new mutations in KRAS emerged (Supplementary Table 2).
The progression-free survival (Hazard ratio 0. 9; 95% CI 0.3366 to 2.453; p=0.85; Log-Rank Test) and overall survival (Hazard ratio 0.42; 95% CI 0.1599 to 1.144; p=0.09; Log-Rank Test) of patients was not significantly different whether they developed secondary KRAS mutations or not (Supplementary Fig. 3)
The availability of serially collected serum samples provided a unique opportunity to model the tumor evolutionary process in those patients that responded to panitumumab. The first question we addressed was whether KRAS mutations were likely to be present prior to the initiation of therapy with panitumumab. To estimate the average tumor growth rate, we used the ctDNA data (Supplementary Table 2) from the nine patients that developed KRAS mutations during therapy. The average tumor growth rate was found to be 0.069 - i.e., the number of tumor cells resistant to panitumumab doubled approximately every ten days (=ln2/0.069 days; see Supplementary Appendix). The growth rate represents the difference between the cell birth rate b and the cell death rate d, i.e., b – d. Previous studies13 have shown that b = ~0.25 for colorectal cancer cells, corresponding to one cell division every four days, yielding a value of 0.18 (=0.25-0.069) for d.
Using these data-derived estimates of b and d, a branching process model was used to test the null hypothesis that there were no cells with KRAS mutations in the tumors prior to the initiation of therapy. We calculated the probability that the number of mutant KRAS cells could have grown to at least the observed levels at the times they were measured, assuming this null hypothesis (see Supplementary Appendix). Using the known tumor burdens and pre-treatment ctDNA levels measured in Patients 8 and 20 who harbored KRAS mutations in their tumors prior to therapy, as well as data obtained on previously studied patients with metastatic disease7, we calculated that one mutant KRAS template per ml of serum corresponds to a tumor containing 44 million KRAS-mutant cells. We performed the statistical valuation separately for each patient. In the three patients that developed more than one circulating KRAS mutation, we assumed that each of the detected KRAS mutations originated from a different lesion (see below). Thus a total of 16 lesions from nine patients were analyzed. For each lesion, the possibility that the observed mutations were absent at the start of treatment could be rejected (thus confirming the presence of pre-existing KRAS mutations) with >95% confidence (in most cases, >99.9% confidence; Supplementary Table 4). 14,15 Furthermore, we varied the birth rate b around the previously calculated value of 0.25, allowing it to be as small as 0.15 or as large as 0.35, and found that this did not affect our conclusion that the mutations were present prior to therapy, though for one lesion the confidence interval fell below the 95% threshold as the birth rate was increased (Supplementary Table 4).
Next, we estimated how many mutations gave rise to resistance to panitumumab in patients in vivo. The total number of lesions large enough to detect by imaging averaged 7.0 lesions/patient in the 24 patients studied with WT KRAS tumors (Supplementary Table 1). Assuming that each individual KRAS mutation detected in the serum emanated from a single metastatic lesion, the 16 lesions responsible for contributing ctDNA (Supplementary Table 2) accounted for 9.6% of the total 167 lesions. The other 90% of lesions presumably had developed genetic alterations that were not assessed in our study. As we assessed four different mutated nucleotides (nt) of KRAS (at codons 12 and 13, Supplementary Table 5), this analysis suggests that a total of ~42 (= 4 × 167/16) nt gave rise to resistance in our 24 patients. These remaining 38 nt (42 minus 4) would represent a maximum of 38 genes (one potential mutant nt per gene), or ~ 10 genes if, like KRAS, there was an average of four nt per gene that could give rise to resistance when mutated.
Would at least one of these 42 mutant nt be expected to be present in a metastatic lesion initiated by a single cell that was WT at all 42 nt? We analyzed this question using a Luria-Delbruck model generalized to incorporate cell death14–16. If we conservatively assume that cancer cells have the same mutation rate as normal cells and the same birth rate described above (0.25)15,17, then a tumor detectable by CT-scanning (one billion cells) is almost certain (probability > 1 × 10−32) to contain at least one cell with a mutation at one of these 42 nt. The expected number of such cells is ~3,200 (Supplementary Appendix). 16,17 These 3,200 cells are distributed among various clones that arose during the growth of the metastatic lesion prior to therapy with panitumumab. The largest clonal subpopulation is in most cases the progeny of the subpopulation containing the first resistance mutation to arise and survive stochastic drift. Using the equations described in the Supplementary Appendix, we calculated the expected size of this clonal subpopulation as 2,200 cells; thus, 69% (2,200/3,200) of the resistant cells should be derived from a single clone. As described in the Supplementary Appendix, varying the number of possible mutations conferring resistance from 4 to 100 did not change this expectation: there are hundreds to thousands of cells with resistance mutations in each metastasis prior to the initiation of panitumumab therapy and more than half of these resistant cells are expected to be derived from a single clone.
Our mathematical model also accounts for the very similar times at which circulating KRAS fragments were observed across patients and lesions. We calculated the probability distribution for the times at which circulating KRAS fragments should become detectable (Supplementary Appendix). As shown in Fig. 2, the mean of this distribution was 22 weeks with a 95% confidence interval ranging from 18 to 25 weeks. These predicted times are strikingly similar to those actually observed in the patients analyzed in this study (23 ± 5 weeks, mean ± one standard deviation) and when clinical progression was observed (25 ± 10 weeks, mean ± one standard deviation). Finally, we performed simulations to further validate these analytical findings. The results were similar: mutant KRAS fragments were predicted to become evident at 22 ± 1.5 weeks (mean ± one standard deviation) following treatment (Supplementary Appendix).
Our study indicates that the resistance mutations in KRAS and other genes were highly likely to be present in a clonal subpopulation within the tumors prior to the initiation of panitumumab therapy. Though we base this conclusion on new data and mathematical analyses, it is consistent with prior experimental data on other targeted agents 18–21 as well as theoretical predictions22–24. As noted in several previous studies of targeted therapeutic agents 18–21, most lesions recur at approximately the same time following therapy. This was also true in our selected group of patients where disease progression occurred at the same time in patients who did or did not develop circulating KRAS mutations (Fig. 2).
Our study suggests that only a limited number of nucleotide mutations and genes are likely to be able to exert a resistance phenotype. If there were many more genes with this capacity, then it would be unlikely that such a high fraction of patients (38%) and lesions (9.6%) would develop mutations in a single gene (KRAS). This finding is consistent with prior studies in that only a small number of genes has been found to be expressed differentially in resistant tumors and an even smaller number to be mutated - none as frequently as KRAS18,25,26. That only a small number of genes can confer resistance is encouraging; if there were a very large number of genes (and nucleotides) that had the capacity to produce resistance to panitumumab, there would be little hope of combining this drug with others to circumvent resistance.
In sum, our results suggest the following scenario for the development of resistance to panitumumab. Each relatively large metastatic lesion is expected to contain a subclone containing hundreds or thousands of cells with one of ~42 mutations conferring resistance to the antibody. Resistance is therefore a fait accompli - the time to recurrence is simply the interval required for the subclone to re-populate the lesion. This generally takes five to six months (Fig. 2) and is due to the rapid expansion of the resistant subclone immediately following treatment initiation. To make these remissions last longer than five to six months, combination therapies targeting at least two different pathways will be required.
METHODS
Patients and specimens
Patients with chemorefractory metastatic colorectal carcinoma were enrolled into one of two panitumumab monotherapy studies (NCT00089635 and NCT00083616)10. The study protocols were approved by the institutional review boards and all patients signed a written consent form. A subset of patients was selected for this analysis from a total of 388 patients. Patients received panitumumab 6mg/kg every two weeks until disease progression. Tumor scans were read centrally by a panel of at least two blinded independent radiologists using a modification of the WHO criteria. Assessments were performed at four week intervals through week 28 and every three months thereafter until progression of disease. Responses were confirmed at least four weeks after response criteria were first met. KRAS mutational status in the tissue was predetermined using the DxS assay (Qiagen).
DNA purification
DNA was purified from of 0.2–1mL of banked serum using the QIAamp circulating nucleic acid kit (Qiagen, cat. # 55114) following the manufacturer’s recommendations. Amplifiable DNA was quantified with qPCR, employing primers and conditions as previously described27.
Assessments of circulating KRAS-mutant DNA
The ligation assays were performed as previously described11 using the primers and probes indicated in Supplementary Table 5. In brief, KRAS fragments containing codons 12 and 13 were amplified with primers designed to yield a small PCR product (106 bp) to accommodate the degraded DNA found in serum28. Note that some of the probes contained locked-nucleic acid (LNA) linkages (Exiqon). To confirm and further quantify samples containing KRAS mutations, BEAMing assays were employed12 using the primers and probes listed in Supplementary Table 5. The number of mutant fragments per ml serum was determined from the fraction of alleles containing the mutant allele (determined by BEAMing7) and the number of alleles assessed per ml of plasma (determined by qPCR27 and reported in Supplementary Table 6).
Supplementary Material
Acknowledgments
The authors thank J. Schaeffer, J. Ptak, N. Silliman, and L. Dobbyn for technical assistance and Mark Ekdahl for operational assistance. This work was supported by The Virginia and D.K. Ludwig Fund for Cancer Research, the National Colorectal Cancer Research Alliance, and NIH grants CA129825, CA43460, CA57345, CA62924, and CA095103 and NCI contract N01-CN-43309. Simulations were performed on the Orchestra cluster supported by the Harvard Medical School Research Information Technology Group. ERC Start grant (279307: Graph Games) and FWF NFN Grant No S11407-N23 (Rise).
Footnotes
AUTHOR CONTRIBUTIONS
L.D., K.O. and B.V. designed and performed experiments, analyzed data and wrote the paper. I.K. and J.W. performed experiments, analyzed data and provided input to the manuscript. R.W., J.B. and R.H. provided critical materials, reagents, analyzed data and provided input to the manuscript. B.A., I.B., J.R. and M.N. analyzed data, performed the mathematical modeling and provided input to the manuscript.
Supplementary Information is available online.
References
- 1.Amado RG, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 2.Karapetis CS, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 5.Corcoran RB, et al. BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci Signal. 2010;3:ra84. doi: 10.1126/scisignal.2001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diehl F, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diehl F, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holdhoff M, Schmidt K, Donehower R, Diaz LA., Jr Analysis of circulating tumor DNA to confirm somatic KRAS mutations. J Natl Cancer Inst. 2009;101:1284–1285. doi: 10.1093/jnci/djp240. [DOI] [PubMed] [Google Scholar]
- 10.Hecht JR, et al. Lack of correlation between epidermal growth factor receptor status and response to Panitumumab monotherapy in metastatic colorectal cancer. Clin Cancer Res. 2010;16:2205–2213. doi: 10.1158/1078-0432.CCR-09-2017. [DOI] [PubMed] [Google Scholar]
- 11.Wu J, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med. 2011;3:92ra66. doi: 10.1126/scitranslmed.3002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diehl F, et al. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006;3:551–559. doi: 10.1038/nmeth898. [DOI] [PubMed] [Google Scholar]
- 13.Jones S, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dewanji A, Luebeck EG, Moolgavkar SH. A generalized Luria-Delbruck model. Math Biosci. 2005;197:140–152. doi: 10.1016/j.mbs.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwasa Y, Nowak MA, Michor F. Evolution of resistance during clonal expansion. Genetics. 2006;172:2557–2566. doi: 10.1534/genetics.105.049791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomasetti C, Levy D. Role of symmetric and asymmetric division of stem cells in developing drug resistance. Proc Natl Acad Sci U S A. 2010;107:16766–16771. doi: 10.1073/pnas.1007726107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montagut C, et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nat Med. 2012;18:221–223. doi: 10.1038/nm.2609. [DOI] [PubMed] [Google Scholar]
- 19.Maheswaran S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turke AB, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leder K, et al. Fitness conferred by BCR-ABL kinase domain mutations determines the risk of pre-existing resistance in chronic myeloid leukemia. PLoS One. 2011;6:e27682. doi: 10.1371/journal.pone.0027682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durrett R, Moseley S. Evolution of resistance and progression to disease during clonal expansion of cancer. Theor Popul Biol. 2010;77:42–48. doi: 10.1016/j.tpb.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Lenaerts T, Pacheco JM, Traulsen A, Dingli D. Tyrosine kinase inhibitor therapy can cure chronic myeloid leukemia without hitting leukemic stem cells. Haematologica. 2010;95:900–907. doi: 10.3324/haematol.2009.015271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komarova NL, Wodarz D. Drug resistance in cancer: principles of emergence and prevention. Proc Natl Acad Sci U S A. 2005;102:9714–9719. doi: 10.1073/pnas.0501870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Y, et al. Epidermal growth factor receptor (EGFR) ubiquitination as a mechanism of acquired resistance escaping treatment by the anti-EGFR monoclonal antibody cetuximab. Cancer Res. 2007;67:8240–8247. doi: 10.1158/0008-5472.CAN-07-0589. [DOI] [PubMed] [Google Scholar]
- 26.Yonesaka K, et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med. 2011;3:99ra86. doi: 10.1126/scitranslmed.3002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rago C, et al. Serial assessment of human tumor burdens in mice by the analysis of circulating DNA. Cancer Res. 2007;67:9364–9370. doi: 10.1158/0008-5472.CAN-07-0605. [DOI] [PubMed] [Google Scholar]
- 28.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proceedings of the National Academy of Sciences. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.