Abstract
Insulators are genomic elements that regulate transcriptional activity by forming chromatin boundaries. Various DNA insulators have been identified or are postulated in many organisms, and the paradigmatic CTCF-dependent insulators are perhaps the best understood and most widespread in function. The diversity of DNA insulators is, however, understudied, especially in the context of embryonic development, when many new gene territories undergo transitions in functionality. Here we report the functional analysis of the arylsulfatase insulator (ArsI) derived from the sea urchin, which has conserved insulator activity throughout the many metazoans tested, but for which the molecular mechanism of function is unknown. Using a rapid in vivo assay system and a high-throughput mega-shift assay, we identified a minimal region in ArsI that is responsible for its insulator function. We discovered a small set of proteins specifically bound to the minimal ArsI region, including ISWI, a known chromatin-remodeling protein. During embryogenesis, ISWI was found to interact with select ArsI sites throughout the genome, and when inactivated led to misregulation of select gene expression, loss of insulator activity and aberrant morphogenesis. These studies reveal a mechanistic basis for ArsI function in the gene regulatory network of early development.
Keywords: Sea urchin, Insulator, ISWI, Embryogenesis, Gene regulation, Strongylocentrotus purpuratus
INTRODUCTION
The regulation of chromatin structure is a major mechanism of differential gene activity. Chromatin insulators are DNA sequences that function in the epigenetic regulation of gene expression, including three-dimensional structural alterations of the chromatin. Malfunction of insulators can cause maladies in humans, including diabetes, Angelman and Beckwith-Wiedemann syndromes (e.g. Steenman et al., 1994; Okamoto et al., 1997; Reik et al., 1995; Taniguchi et al., 1995) (reviewed by Herold et al., 2012). Although detailed explanations for developmental abnormalities resulting from insulator malfunction are not well understood, insulator-dependent defects are particularly pronounced during oogenesis and embryogenesis when major epigenetic events of reprogramming and imprinting occur. Identifying insulator function and mechanisms of action during embryogenesis will significantly impact our understanding of basic gene regulatory networks (GRNs) and lead to clinical treatments for polygenic diseases (reviewed by Ideraabdullah et al., 2008; Filippova, 2008; Emery, 2011).
Insulators are known to have several distinct chromatin activities, including enhancer-blocking and anti-silencing activities (Gaszer and Felsenfeld, 2006; Herold et al., 2012). Detailed classifications of insulator function are not well established, but insulators may confer one or a variety of these activities, suggesting some shared functionality (Brasset et al., 2010; Ghirlando et al., 2012). Enhancer-blocking activities include the repression of gene transcription as a result of the insulator being located between an enhancer and the promoter of that gene. One of the best-studied examples of this enhancer-blocking activity is the imprinting control region (ICR) of the human H19/IGF2 locus (Bell et al., 1999; Hark et al., 2000; Kanduri et al., 2000). Promoters for the H19 (which produces a long, non-coding RNA) and Igf2 (insulin-like growth factor 2) genes share a single enhancer. On the maternal allele, the ICR (insulator domain) is unmethylated and thus the CCCTC-binding factor (CTCF) insulator component is able to bind at that site, resulting in recruitment of the enhancer to the H19 promoter to the exclusion of its interaction with the Igf2 promoter. On the paternal allele the ICR is methylated, CTCF cannot bind to it, and the enhancer is instead recruited by three-dimensional looping to activate the Igf2 promoter. Thus, differential insulator activity leads to differences in gene activity, and malfunction of this insulator contributes to Beckwith-Wiedemann syndrome (prenatal overgrowth) (Prawitt et al., 2005), a paradigm of insulator malfunction.
Anti-silencing/barrier activities of an insulator function when a gene locus is flanked by homologous insulators. Upon insulator engagement with trans-factors, this activity isolates the locus from the adjacent cis-chromosomal environment, and thereby maintains differential gene activity in select genetic loci. Barrier elements may function as chain terminators by blocking a processive reaction, such as the histone acetyltransferase (HAT) and ATP-dependent nucleosome-remodeling complexes (Gaszer and Felsenfeld, 2006; Xie et al., 2007; Bi et al., 2004) (reviewed by Herold et al., 2012). Some enhancer-blocking insulators, such as chicken cHS4, are known to function also as barrier elements, with different proteins involved in each function (Pikaart et al., 1998; West et al., 2002). Barrier insulators are also capable of stabilizing transgene expression in many organisms (Recillas-Targa, 2006; Yajima et al., 2007).
The arylsulfatase insulator (ArsI) was originally discovered in the sea urchin Hemicentrotus pulcherrimus (Akasaka et al., 1999) through a process of cis-element analysis on the arylsulfatase locus. It is distinct in sequence from the CTCF-dependent insulator family and is functional in diverse organisms (Hino et al., 2006; Akasaka et al., 1999; Nagaya et al., 2001; Takada et al., 2000; Watanabe et al., 2006; Tajima et al., 2006). It is not known how this DNA element functions as an insulator, nor what proteins are associated with its activity. We sought to expand our understanding of diverse insulators and their functions and report here that the ArsI sequence is found throughout the genome and interacts with a small cohort of nuclear proteins responsible for its insulator activity. One of the ArsI-associated proteins discovered here, an ortholog of the chromatin-remodeling protein ISWI, was found to function in ArsI activities in vivo and to associate with ArsI sites differentially during the course of embryonic development. These results demonstrate in vivo important regulation of early embryonic development by a DNA insulator and document another essential element in our understanding of GRNs during embryonic development.
MATERIALS AND METHODS
Mega-shift assay, cloning and sequencing
A high-throughput binding assay called mega-shift (microarray evaluation of genomic aptamers by shift) was used to narrow down the exact location(s) of the ArsI sequence-protein complexes. For detailed methods, see Tantin et al. (Tantin et al., 2008). Briefly, a previously identified 573 bp region of ArsI sequence and its adjacent 1.4 kb sequence from the genomic loci of the Strongylocentrotus purpuratus arylsulfatase ortholog (Sp-Ars; accession number NW_001468620) were synthesized as a pool of tiled 30-mers with a common flanking primer sequence to amplify the entire pool. The common amplification primer sequence is 5′-AGAAAGCGAAAGGAG and 3′-TGTAGTTGCCGTCGT. This pool of over 2000 oligonucleotides was then assayed by iterative gel shifts for binding to 3 μg of nuclear proteins isolated from gastrulae of sea urchins. The nuclear extract was prepared by the method of Calzone et al. (Calzone et al., 1988) and each shifted oligonucleotide was identified by sequencing. For gel shift with a single probe, each test probe was amplified from cloned oligonucleotides in the presence of [32P]TTP.
In vivo functional assay
In vivo functional assays and the appropriate reporter constructs were as described previously (Yajima et al., 2007; Kurita et al., 2003). Mutated versions of constructs were produced by inserting restriction sites (BglI sites for M60 and NcoI sites for M70, created by Epoch Biolabs) into the ArsI sequence, followed by restriction enzyme digestion and self-ligation of the plasmid. Approximately 6 pl of 6 ng/μl GFP reporter constructs and 20 ng/μl genomic DNA as a carrier were injected into fertilized eggs with or without 6 pl of 0.25-1 mM test/control morpholinos. GFP reporter expression was observed at each developmental stage by fluorescence microscopy (Zeiss, Axioplan).
For quantitative analysis, 100 injected embryos were collected from each dish and RNA was extracted, cDNA was prepared using the TaqMan Reverse Transcription Reagents Kit (Applied Biosystems, Foster City, CA, USA), and quantitative (q) PCR was performed on the 7300 Real-Time PCR System (Applied Biosystems) with the SYBR Green PCR Master Mix Kit (Applied Biosystems). Experiments were performed in at least triplicate and the following primers were used for analysis (F, forward; R, reverse): GFP F, ATCAGACACAACATTGAGGA and R, TCGTTGGGATCTTTAGACAG; SM50 promoter and GFP F, TGGTAGTCGTGAATGCATC and R, GCCAGTGAACAGTTCCTC; Ars promoter and GFP F, AGCGTTCTCCCTGACAGGTTG and R, CGCCATCCAGTTCCACGAGA.
Mass spectrometry
Biotinylated M70 double-stranded oligonucleotides were synthesized and bound to streptavidin magnetic beads (Invitrogen, catalog code 656.01) to produce an affinity probe. The nuclear extract retrieved by M70 probe beads with or without free M70 oligonucleotides (25-fold excess of the probe) was washed five times to remove unbound proteins and then run on an SDS-PAGE gel. Bands of interest were isolated, treated for in-gel tryptic digestion (Thermo Scientific, catalog code 89871) and analyzed by tandem mass spectrometry (MS/MS). Experiments were performed independently three times. Experimental MS and MS/MS spectra were matched against in silico tryptic digests of the entire GLEAN3 sea urchin database using the search programs ProteinPilot (Applied Biosystems) and Mascot (Matrix Science), and matched to candidate lists with the program ProID (Applied Biosystem USA). Results from ProID were filtered, condensed and compiled using the program ProGROUP (Applied Biosystem USA). Results from each lane with or without competitors were compared to minimize the background and extract proteins that bound specifically to the M70 sequence.
Immunostaining, morpholino design and detection of endogenous gene expression
Immunolabeling was performed as described previously (Yajima and Wessel, 2011a; Yajima and Wessel, 2011b). Briefly, embryos were fixed with 90% methanol, anti-Drosophila ISWI antibody (Abcam, Cambridge, MA, USA, ab10748) was used at 1:500, anti-rabbit Cy3 (Jackson ImmunoResearch, West Grove, PA, USA) was used as a secondary antibody at 1:300, and 10 mg/ml Hoechst was used at 1:1000 dilution.
Morpholinos against S.p.iswi (ISWI-MO, TTGCACGAAAGTAAGTGTGTTACGA) and S.p.sin3A (Sin3A-MO, TTGTAGCACCAACCAGAAGAAGAAT) were designed at the 5′-end of each coding region and produced by Gene Tools (Philomath, OR, USA); 6 pl of 0.25-1 mM morpholino was injected into fertilized eggs. For detection of endogenous expression of Sp-Ars-related genes, cDNA was prepared from 200 embryos either injected with or without 0.5 mm ISWI-MO, and the level of gene expression determined by qPCR. Primers were designed within a region specific to the Sp-Ars ortholog (Sp-Arsa_1 gene) containing the well-conserved ArsI site in its upstream region [NCBI NW_00146820, locus #582391 (Takagi et al., 2012)] (F, GCAGCACCAACCACGACAAGG and R, GTTCAAT CTG AGCAATCACTG) and regions specific to other Sp-Ars-related genes containing less conserved ArsI sites [Ars_1 (SPU_023353) F, GCAGCACCAACCACGACAAGG and R, GTTCAATCT GAGCAATCACTG; Ars_2 (SPU_006968) F, CAGTAGACCCCGATCTCCTTGAAC and R, GAAGCCAGGTTCCTGCGACGGGTG; Ars (SPU_013100) F, CTTCTTCTACTACTGCAAGGA and R, GTTGCAGTCGAAGTAGTCTTC]. Sp-ubiquitin was used as a control (F, CACAGGCAAGACCATCAC and R, GAGAGAGTGCGACCATCC). The 1/Ct value of each sample injected with ISWI-MO was then normalized to that of control samples. These experiments were performed independently three times.
Chromatin Immunoprecipitation (ChIP) and quantitative PCR
ChIP was performed as described previously (Alekseyenko et al., 2008). Briefly, 25000 embryos were collected for each chromatin sample preparation, ground in liquid nitrogen and fixed in 1% formaldehyde. Then, 5 μg of each antibody was used for ChIP reaction against 20 μg of each chromatin sample, 25 μl of Protein A beads (Invitrogen) was used for each pulldown, each final ChIP material was diluted in 50 μl of water and 1 μl used for the qPCR reaction. For qPCR primer design, individual ArsI loci were identified from the NCBI site as follows: S.p.arylsulfatase (Sp-Ars) ortholog locus, NW_00146820, locus #582891, annotated as Sp-Arsa_1; locus 1, NW_001463517, 59,169 bp from the 3′ side of myosin-RhoGAP protein; locus 2, NW_001330318, no known adjacent genes; locus 3, NW_001470764, 6386 bp from the 3′ side of 26S proteasome non-ATPase regulatory subunit 9; locus 4, NW_001292072, 11,549 bp from the 3′ side of proton/amino acid transporter 1; locus 5, NW_001357699, 1982 bp from the 3′ side of RNA pseudouridylate synthase domain containing 3. The following primers were used for qPCR. (1) Gene body primers were designed within the ORF of each test gene: Sp-nanos 2 F, GCAAGAACAACGGAGAGAGC and R, CCGCATAATGGACAGGTGTA; Sp-myosin IX F, GAGAGTACTACATTGTTGATAC and R, GCACAGGGCCTGCTCCTAGAAG; Sp-Prolactin-releasing peptide receptor (PrRP receptor) F, GAGCTTCACCGCTTTGGAATTC and R, GAATCACCCACCAATTCCATACG. (2) ArsI locus primers were designed to overhang ArsI M70 and adjacent sequences specific to each locus: Sp-Ars ortholog locus F, GTTCAGGGGCGTATCTTCTCAC and R, CATTCCGCATGCTCCTATTGTTATC; locus 1 M70 F, CATCTGACCGTAATTAAACGCC and R, CCCCAGAAGATCAAGGGTTTTAG; locus 2 F, CATCTGACCGTAATTAAACGCC and R, GACACTGATTGGAGTGAGGTCC; locus 3 F, CATCTGAC CGTAATTAAACGCC and R, ACACTGATTGGAGTGAGGTCC; locus 4 F, CGCCTAAAAACCCCTGAGCTT and R, GCACTAAAT TTTGGTCAAATGAG; locus 5 F, TTGCTCATCTGACCGTAATTA and R, GTATCAAGGGGCCTGCCCAAG. The 1/Ct value of each ISWI ChIP sample was normalized to that of control IgG ChIP samples. These experiments were performed independently three times.
RESULTS
Proteins that interact with ArsI
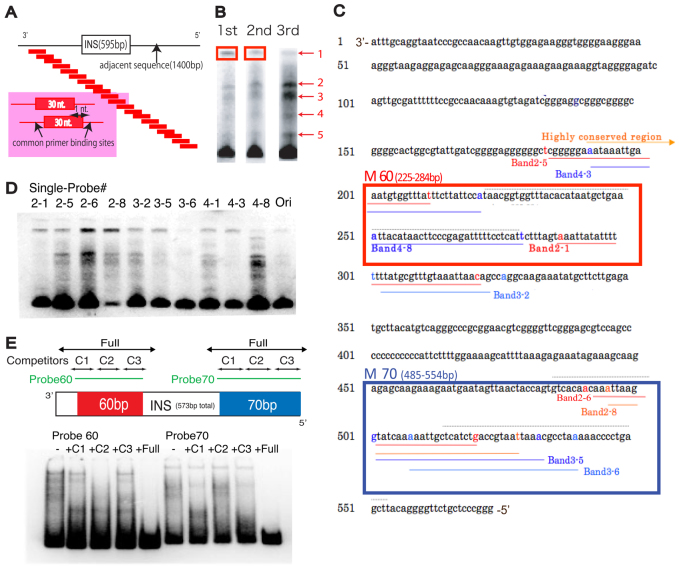
The original ArsI sequence from Hemicentrotus pulcherrimus was screened against the genome of another sea urchin, Strongylocentrotus purpuratus, and we found ∼120 sites that are ∼90% identical to 400 bp of the 3′-end of the ArsI sequence (supplementary material Fig. S1A). Previous work identified the minimal ArsI insulator element as a 574 bp region (Akasaka et al., 1999). Any further deletions of this element resulted in reduced insulator function. Here we mapped protein-binding elements within this insulator region by mega-shift, a high-throughput protein-DNA binding assay (Fig. 1A). This binding assay utilizes a scanning oligonucleotide library to identify DNA sequences bound to protein as detected using a gel-shift approach (Tantin et al., 2008; Ferraris et al., 2011). Proteins extracted from nuclei of the sea urchin embryo were incubated with a tiled 30-mer probe pool designed from a total of 2 kb of DNA sequence that included the entire ArsI region and an adjacent 1.4 kb sequence derived from the locus of the S. purpuratus arylsulfatase gene ortholog. The probe pool contains 2000 individual probes, each a 30 bp sequence from the Ars minimal insulator (and flanking amplification sites), with each probe differing from the next by sliding the 30 nucleotide segment a single nucleotide at a time. We then incubated the entire 2000 oligonucleotide library with the proteins extracted from nuclei, and following analysis on polyacrylamide DNA-shift gels, the subset of probes that formed DNA-protein complexes of reduced mobility were isolated, amplified (using their engineered flanking oligonucleotide sequences), and rerun following incubation with new nuclear extracts to sequentially enrich for tiled probe(s) of specific interaction. In each of the first three cycles of enrichment, DNA-protein complexes increased significantly and multiple complexes were resolved by gel mobility retardation (Fig. 1B). Remarkably, the reamplified mega-shifted complexes showed significant overlap; each oligonucleotide present in the complexes of differing mobility mapped to two regions of the ArsI sequence of less than 100 bases total within the 2 kb of DNA covered by the 2000 probe set (Fig. 1C; supplementary material Fig. S2). Thus, the specificity of binding within the probe set was pronounced, and tiles of significant protein binding were located both at the 5′-end (M-70, Fig. 1C) and in the middle region (M-60, Fig. 1C) of the minimal ArsI sequence.
Fig. 1.
The mega-shift assay identifies regions of protein binding. (A) Experimental design of the mega-shift assay. A tiled pool of 30-mer probes, each shifted by one nucleotide from the adjacent probe and containing common primer sites, were designed to cover 2 kb of total sequence flanking and including the ArsI site. (B) Probes extracted from the top band (boxed) were amplified and used as probes for the next round of mega-shift assay. Three rounds of mega-shift resulted in enriched pools of primers with five different but consistent complexes. Probes were extracted from each of the five bands and sequenced. (C) Probes sequenced from each band obtained in B were mapped to the ArsI element. Probes derived from diverse complexes were found to be overlapping in two regions of the ArsI sequence: M-60 and M-70. Each band number corresponds to the single probe #. (D) A single probe designed for each region labeled in C was used for a gel-shift assay and resulted in several bands, suggesting that ArsI-associated proteins might form a higher-order protein complex. (E) Competitors designed for each region (C1-C3) and the entire region (Full) within M-60 and M-70 of ArsI did not compete as effectively as the full-length competitor, indicating the existence of multiple protein binding sites.
To test whether ArsI-associated proteins formed complexes between different oligonucleotide tiles, a single probe from each band labeled in Fig. 1C (oligonucleotides 2-1, 2-5, 2-6, 2-8, 3-2, 3-5, 3-6, 4-1, 4-3, 4-8) was used for individual gel-shift analysis. Although each probe demonstrated a variety of enrichments in shifted bands, each of the single probes generated the same pattern of multiple bands (Fig. 1D), with significantly stronger probe shifting overall than with the original pooled probes (Ori in Fig. 1D). This suggests that ArsI-associated proteins form higher-order protein-oligonucleotide complexes with multivalent interactions. The ability to link two similar sequences in this assay is predicted if the complexes function in three-dimensional looping of chromatin within the genome.
To delineate the protein-binding regions and specificity within the M-60 and M-70 sequences, a gel-shift assay was performed with competitors (Fig. 1E). Double-stranded competitor probes that mask the entire M-60 or M-70 probe sequence diminished protein binding to the probes, yet none of the single competitors was sufficient to diminish the protein interaction, further suggesting that Ars-associated proteins might confer higher-order complexes and bind to multiple regions within the ArsI sequence, particularly to the M-60 and M-70 regions (Fig. 1E).
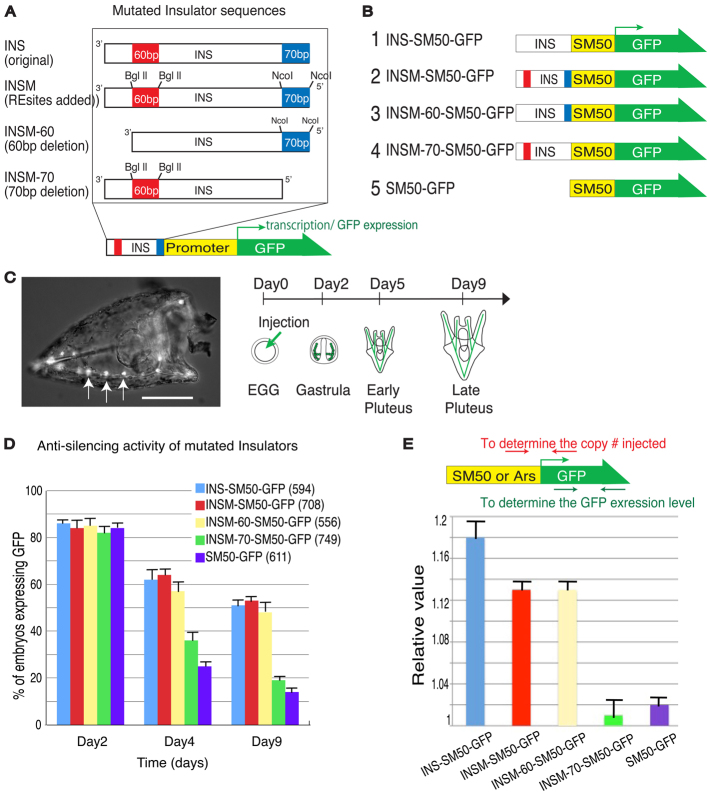
Minimal element(s) responsible for ArsI function in vivo
The minimal element sequences, 70 bp at the 5′-end (M70) and 60 bp at the 3′-end (M60), exhibit the most protein binding activity and these regions were evaluated further by in vivo functional assays. Mutated ArsI lacking either M70 (INSM-70) or M60 (INSM-60) were connected to a sea urchin general promoter driving GFP (Fig. 2A). These reporter constructs were injected into fertilized sea urchin eggs, and anti-silencing activity or enhancer blocking activity was surveyed both by GFP fluorescence and by qPCR for GFP mRNA. For assay of the anti-silencing activity, the sea urchin Spicule Matrix Protein 50 (SM50) promoter was flanked at its 5′-end either by ArsI (INS) or mutated versions of ArsI (INSM-60 or INSM-70) (Fig. 2B). The SM50 promoter functions specifically and consistently only within the skeletogenic cells (Fig. 2C, arrows) during embryogenesis. SM50 promoter function has been well characterized and was chosen here because expression in a single lineage is of great advantage in quantitating the numbers of embryos that exhibit GFP expression (Makabe et al., 1995; Wilt, 1999; Yajima et al., 2007) (Fig. 2C). Promoter activities without ArsI or with INSM-70 deletions decreased to only 10% of the original activity by day 9 postfertilization, whereas those with native ArsI, modified ArsI (INSM) or INSM-60 maintained activity within this timeframe at 50% (Fig. 2D). To quantify the transcript levels resulting from ArsI activities, embryos at day 4 were collected and subjected to qRT-PCR, and the results indicated that INSM-70 lacked significant anti-silencing activity (Fig. 2E). This suggests that the M-70 sequence is important both for protein binding and for anti-silencing activity in vivo.
Fig. 2.
In vivo functional assay for anti-silencing activity of ArsI. (A) ArsI mutation constructs. ArsI is indicated as INS in this diagram. (B) Mutation constructs designed to test the anti-silencing activity of ArsI. SM50 indicates the SM50 promoter. (C) Injected SM50-GFP construct drives specific GFP expression (green in schematic) in skeletogenic cells during S. purpuratus development. A day-3 sea urchin larva injected with SM50-GFP shows GFP expression in skeletogenic cells (arrows). Scale bar: 50 μm. (D) Each reporter construct was injected into fertilized eggs and the number of GFP-expressing larvae was counted at each developmental point. The total number of embryos injected is indicated in parentheses. (E) For a quantitative assay, day-3 larvae injected with each construct were subjected to qRT-PCR and the level of GFP expression normalized to the copy number of injected constructs. The location of primers is indicated above. Error bars indicate variations among three independent experiments.
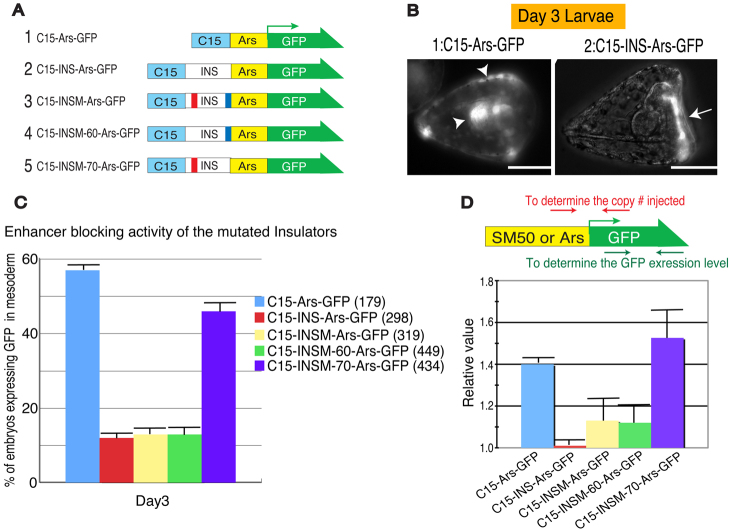
To delineate the essential elements of enhancer-blocking activity in vivo, ArsI or mutated ArsI elements were placed between the sea urchin C15 enhancer and the Ars promoter (Fig. 3A) (Akasaka et al., 1999; Kurita et al., 2003). The Ars promoter drives expression specifically in oral ectoderm (Fig. 3B, arrow), but when under the regulation of the C15 enhancer Ars gene expression is strongly active in both the mesoderm and endoderm, in addition to the ectoderm (Yang et al., 1989; Kurita et al., 2003) (Fig. 3B, arrowheads). As ArsI effectively knocks down Ars promoter activity when it is functional, these constructs work as an effective tool to test whether mutated ArsI elements are still capable of blocking enhancer activity. Reporter constructs without ArsI or with INSM-70 showed higher expression in mesoderm, indicating a loss of ArsI function. A minor difference in the activities of these two constructs implies a possible contribution of another element for the enhancer-blocking activity. Constructs with ArsI and INSM-60 showed little mesoderm or endoderm expression (Fig. 3C), indicating little contribution of the M-60 sequence on its own to insulator activity. A quantitative analysis was performed to measure the overall level of overexpression induced by the lack of ArsI activity in embryos, and the results consistently demonstrated that the M-70 region is important for the enhancer-blocking activity of ArsI (Fig. 3D).
Fig. 3.
In vivo functional assay for enhancer-blocking activity of ArsI. (A) Mutation constructs designed to test the enhancer-blocking activity of ArsI. C15 indicates C15 enhancer, INS indicates ArsI, and Ars indicates the arylsulfatase promoter. (B) Injected C15-Ars-GFP drives ectopic expression in endoderm/mesoderm in addition to ectoderm (arrowheads), whereas INS (ArsI) prevents C15 enhancer activity and thus C15-INS-Ars-GFP drives specific GFP expression in oral ectoderm (arrow), of day-3 S. purpuratus larvae. Scale bars: 50 μm. (C) Each reporter construct was injected into fertilized eggs and the number of larvae expressing GFP ectopically in endoderm/mesoderm was counted at day 3. The total number of embryos injected is indicated in parentheses. (D) For a quantitative assay, day-2 larvae injected with each construct were subjected to qRT-PCR and the level of GFP expression was normalized to the copy number of injected constructs. The location of primers is indicated above. Error bars indicate variations among three independent experiments.
Overall, in both anti-silencing and enhancer-blocking activities, constructs lacking the M-70 region failed to show insulator function, whereas insulator sequences lacking the M-60 region showed normal function. These results suggest that the M-70 sequence is important for protein interaction and in vivo function for both anti-silencing and enhancer-blocking activities.
Proteins that confer insulator function to ArsI
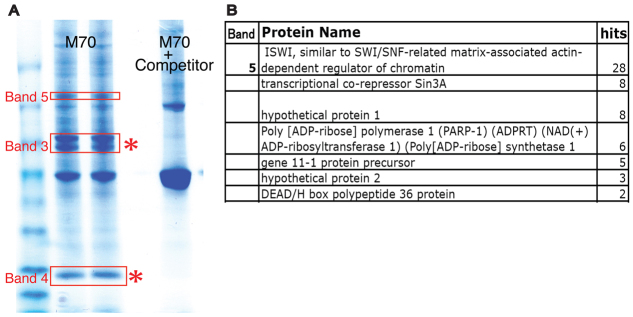
To identify proteins that bind to the minimal ArsI insulator sequence, we used tandem mass spectrometry (MS). Results from mega-shift assays and in vivo analyses suggested that M-70 is essential for ArsI function, and thus proteins bound to M-70 were prioritized. The M70 beads were incubated with sea urchin nuclear extract, with or without competitors (free M70 sequence), and bound proteins were resolved on an SDS-PAGE gel. Bands that were competed off with specific competitors (Fig. 4A, bands 3, 4 and 5) were excised and analyzed by tandem MS. As a result, 23 significant peptide hits were found (Fig. 4B). Peptide significance in this experiment was set at three or more independent peptide hits from a single protein per sample compared with each complementary blank band and with reproducibility in each of the three experiments.
Fig. 4.
Identifying proteins bound to ArsI. (A) Nuclear proteins enriched by M-70 sequence were run on an SDS-PAGE gel with or without competitors. Bands competed away with competitors were isolated and subjected to mass spectrometry. (B) Summary of a series of proteins identified from band 5. Proteins identified from bands 3 and 4 (asterisks) are not shown in this paper.
In vivo functional assays
From 23 candidate proteins, SWI/SNF-related matrix-associated protein (ISWI) showed the greatest number of hits and was one of the candidate proteins selected for further analysis. ISWI is a chromatin-remodeling protein and is considered to be required for regulation of higher-order chromatin structure in several organisms (Deuring et al., 2000; Varga-Weisz, 2005; Erdel and Rippe, 2011). Further, one report in Drosophila documented that ISWI is enriched in the nucleus as expected and functions in self-renewal of germline stem cells, perhaps through its chromatin-remodeling activities (Xi and Xie, 2005). In sea urchins, ISWI is highly expressed during embryogenesis (Wei et al., 2006), although no functional analysis has been reported.
As the ISWI protein sequence is highly conserved among metazoans (supplementary material Fig. S3A), we utilized the Drosophila ISWI antibody (from Abcam) to observe ISWI localization during embryogenesis. As expected, ISWI was present in the nuclei of every cell throughout early development (Fig. 5A; supplementary material Fig. S3B). We next designed a morpholino antisense oligonucleotide (MO) against ISWI to specifically knockdown translation of endogenous ISWI; the ISWI-MO reduced ISWI accumulation in nuclei by ∼90% (Fig. 5A). This also indicated that the conservation of ISWI and the reagents used to detect it are indeed specific in detecting endogenous ISWI in the sea urchin. A higher dose (1 mM stock solution) of ISWI-MO injection was lethal, suggesting that the ISWI ortholog is essential for development, and thus a 0.5 mM stock ISWI-MO solution was selected for further studies as it was the most effective dosage per egg, yet was calibrated to allow embryos to develop successfully. These ISWI-MO-injected embryos demonstrated consistent atypical morphologies to the late gastrula stage, with a rounded shape yet with all the major body parts present, such as endoderm, mesoderm and ectoderm (Fig. 5B).
Fig. 5.
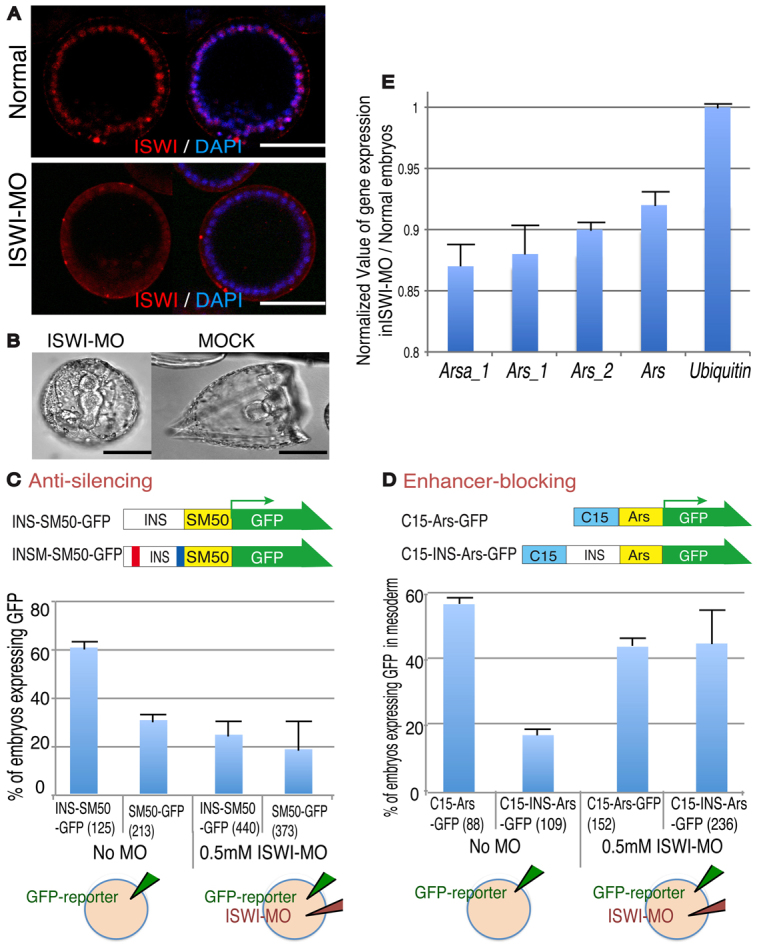
ISWI is an ArsI-associated protein. (A) The specific localization of ISWI in the nucleus of each cell in the S. purpuratus blastula is severely diminished by treatment with ISWI-MO. Scale bars: 50 μm (B) ISWI-MO-injected S. purpuratus embryos demonstrate developmental defects yet successfully form the three germ layers, a blastocoel, skeleton and cilia. (C,D) In vivo functional assay for anti-silencing and enhancer-blocking activities of ArsI. The promoter activity of each reporter construct was tested by GFP expression with or without ISWI-MO. The total number of embryos injected is indicated in parentheses. (E) Endogenous Sp-Ars-related gene expression was measured by qPCR in the presence or absence of ISWI. Gene expression from embryos injected with ISWI-MO was normalized to that from normal embryos. Error bars indicate variations among three independent experiments.
To test whether ISWI functions in insulator activity in vivo, we examined ArsI function by measuring reporter activities under the presence/reduction of ISWI protein, by co-injecting reporters and ISWI-MO (Fig. 5C,D). To test the anti-silencing activity, INS-SM50-GFP or SM50-GFP was injected into embryos and the level of GFP expression was compared between these two constructs containing/lacking the ArsI element at day 4 in the presence/absence of ISWI-MO (Fig. 5C). In the presence of ISWI-MO, reporter activity was significantly decreased compared with controls, suggesting that ArsI activity was downregulated in the absence of ISWI. To test enhancer-blocking activity, C15-INS-Ars-GFP or C15-Ars-GFP was injected with or without ISWI-MO (Fig. 5D). In the presence of ISWI-MO, reporter expression was as high as that of controls on day 3 (C15-Ars-GFP with and without MO), suggesting that ArsI enhancer-blocking activity was abolished in the absence of ISWI.
In Drosophila it is known that ISWI physically interacts with the Sin3A/Rpd3 protein complex (Burgio et al., 2008). Since both ISWI and Sin3A were identified by tandem MS in the oligonucleotide affinity isolation, we also tested whether Sin3A has a similar functional contribution to ArsI activity in vivo or even functions coordinately with ISWI. We injected each reporter construct with 0.5 mM Sin3A-MO alone, or with a half dose (0.25 mM) of Sin3A-MO and ISWI-MO together. Sin3A-MO at 0.5 mM caused similar developmental defects as ISWI-MO, implying a similar general function in the context of chromatin (supplementary material Fig. S4A). In the presence of 0.5 mM Sin3A-MO the anti-silencing activity of ArsI was inhibited, whereas its enhancer-blocking activity was less affected compared with the ISWI knockdown. In the presence of both Sin3A-MO and ISWI-MO, the anti-silencing activity was additively blocked whereas a minimal effect was seen on enhancer-blocking activity, implying that ISWI and Sin3A have overlapping functions for the anti-silencing activity but not for the enhancer-blocking activity of ArsI (supplementary material Fig. S4B,C).
We next tested whether ISWI makes a functional contribution to the endogenous expression of S.p.arylsulfatase (Sp-Ars)-related genes that are orthologous to the original H.p.arylsulfatase (Takagi et al., 2012). The Sp-Ars family includes more than 22 genes and we selected members that are associated with a well-conserved ArsI sequence in the upstream region (locus #582391 or Sp-Arsa_1 locus; supplementary material Fig. S4D). Several other genes containing less well conserved ArsI sequence (Sp-Ars_1, Sp-Ars_2 and Sp-Ars), and Sp-ubiquitin, which has no conserved ArsI sequence adjacent to its gene locus, were used as negative controls. Embryos injected with or without 0.5 mM ISWI-MO were collected at mid-gastrula stage and subjected to qRT-PCR. The level of expression of each gene in ISWI-MO embryos was then normalized to that in control embryos (Fig. 5E). Expression of Sp-Arsa_1 (which contains a conserved ArsI upstream sequence) was largely inhibited, whereas that of other Sp-Ars-related genes was less affected, and Sp-ubiquitin showed no affect, implicating that the effect of ISWI on gene activity is highly specific.
Taken together, these results support the conclusion that not only does ISWI interact within a complex that includes ArsI essential sequence, but that ISWI is necessary for ArsI activity in vivo, contributes to endogenous gene regulation and might function as an insulator complex protein with another ArsI-associated protein, Sin3A.
ISWI interacts with multiple endogenous ArsI sites on chromatin throughout development
We expect that insulators in general are differentially regulated through early development, and identifying binding sites of ISWI during development might indicate which loci are regulated via interaction with ISWI. To indicate ISWI binding sites in endogenous genetic loci, ChIP-qPCR was performed with anti-ISWI at three different developmental stages: mid-gastrula, late gastrula and pluteus stages.
ChIP materials prepared from each developmental stage were subjected to qPCR to measure the specificity of interactions between ISWI and the Sp-Ars orthologous locus (locus #582391, Sp-Arsa_1) or five different ArsI genomic loci. These five loci were identified and selected from the sea urchin genome database (NCBI BLAST and SpBase.org) based on sequence identity to ArsI. Primers for qPCR were designed to overhang the M-70 sequence and its adjacent sequence specific to each locus, or designed as negative controls within gene bodies where insulator proteins are unlikely to bind with insulator activity (Fig. 6A). Each ArsI site randomly selected from Sp-Ars orthologous loci demonstrated a distinct range of interactions with ISWI during development, yet each activity was always higher than that of the negative controls (nanos, myosin IX, PrRP) (Fig. 6B). These results suggest that ISWI might function as a core member of ArsI-associated complexes throughout the genome, yet regulate aspects of development by changing its interaction during embryogenesis.
Fig. 6.
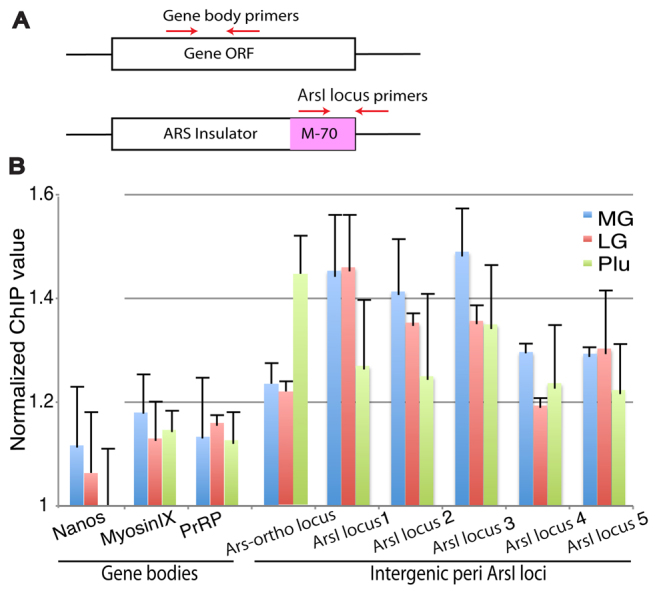
A differential level of interaction between ISWI and ArsI during development as detected by ChIP-qPCR. (A) Location of qPCR primers. Gene body primers include nanos, myosin IX and PrRP, and ArsI locus primers include the S.p.arylsulfatase ortholog locus and the random intergenic peri-ArsI loci in the genome. (B) ChIP-qPCR results from each developmental stage at mid-gastrula (MG), late gastrula (LG) and pluteus (Plu) stages. y-axis indicates values relative to IgG controls. Error bars indicate variations among three independent experiments.
DISCUSSION
Understanding the function of diverse DNA insulators is essential for our understanding of epigenetic regulation mechanisms, especially in early embryos. The ability to compare and contrast different insulator types from a range of organisms is required to reveal principles of insulator impact on GRNs. The CTCF insulator and its functional mechanism with partner proteins Cohesin and p68 is paradigmatic in the field (Wendt et al., 2008; Parelho et al., 2008; Yao et al., 2010). The number of CTCF binding sites in the human genome is substantial (between 14,000 and 20,000) and, given that a CTCF pair-defined domain (CPD) is on average 210 kb in length, with each domain containing 2.5 genes on average, this means that nearly all the genes in the human genome might come under the control of this insulator at least at one point in the life cycle (Kim et al., 2007; Barski et al., 2007; Xie et al., 2007). The importance of this insulator cannot be overestimated and interest now focuses on how CTCF domains may be differentially regulated. Therefore, understanding CTCF-dependent insulator function has a marked impact on our understanding of epigenetics in general. However, CTCF orthologs are absent in C. elegans, yeast and plants, organisms with robust epigenetic modifications that are used for differential gene expression during development. In sea urchins, the mRNA of the CTCF ortholog is present abundantly until after fertilization and then remains expressed at low level during embryogenesis. Its functional contribution to embryonic development has not been reported. Furthermore, CTCF-independent insulators have been found among a wide range of organisms, from mammals to yeast and plants (Heger et al., 2009; Ong and Corces, 2009), suggesting the biological importance of various CTCF-independent insulators.
The ArsI DNA insulator functions both independently of CTCF and of interaction with the nuclear matrix (Hino et al., 2006), making it mechanistically distinct from the CTCF paradigm. Around 120 ArsI sites have ∼90% sequence identity within the genome of the sea urchin S. purpuratus, and we predict that more cryptic sites exist that are partially conserved and that might be involved in gene activities that lead to fate decisions in early embryos. Some of these activities might be in response to environmental conditions (other cells, growth factors or stress conditions of the environment) and be functionally conserved among many organisms (Hino et al., 2006; Nagaya et al., 2001; Takada et al., 2000; Watanabe et al., 2006; Tajima et al., 2006). In this report, we revealed that 400 bp of the 5′ ArsI sequence is highly conserved between H. pulcherrimus and S. purpuratus and, in particular, that 70 bp of the 5′-end (M70 region) is largely responsible for ArsI activity as a binding site for protein complexes and for in vivo function. This functional region is also the most conserved among the ArsI elements (∼70 different sites) of the other sea urchin species examined (Strongylocentrotus franciscanus, Allocentrotus fragilis and Lytechinus variegatus; partial genomic BLAST database was obtained from SpBase.org) and this phylogenomic footprint independently suggests its functional importance. Although these sequence-dependent and CTCF-independent features are distinct from typical insulators, the fundamental mechanisms of ArsI function might be similar to those of CTCF-dependent insulators. CTCF binding at insulators is methylation sensitive, and this insulator protects transgene expression by preventing methylation of its adjacent promoter sequence (Steenman et al., 1994; Pikaart et al., 1998; Hark et al., 2000). As we previously identified that the anti-silencing activity of ArsI is dependent on methylation (Yajima et al., 2007), it would be intriguing to test whether the binding of ArsI-associated proteins, including ISWI and Sin3A, is also methylation sensitive in a similar manner to other insulator-associated proteins.
Several candidate proteins were identified within the oligonucleotide complexes seen in the mega-shift analysis, including the chromatin-modifying protein ISWI. Although ISWI may contribute to many insulator sites and chromatin-modifying mechanisms, the identification of ISWI in ArsI function broadens the perspective for conserved trans-elements in diverse insulator function of many organisms. Notably, the protein complex identified here on the ArsI sequence lacked cohesins and CTCF, two important proteins for CTCF-dependent insulator function. Although a negative result and as such not necessarily conclusive, it does support the premise that the ArsI core machinery functions differently from the CTCF insulator. ISWI was also recently reported to function in several locus-specific insulators in Drosophila (Li et al., 2010), suggesting that ISWI could be another highly conserved insulator protein, like Cohesin. Although a detailed functional test of ISWI in insulator regulation is awaited, recent reports demonstrated that ISWI may regulate gene transcription indirectly by altering chromatin to enable transcription factors to bind targets directly. For example, Isw2, an ISWI homolog in yeast, was found to bind transiently on chromatin and to scan the genome for its targets (Gelbart et al., 2005). Also, Drosophila ISWI appears to bind genes near their promoters, causing specific alterations in nucleosome positioning at the transcription start site (Sala et al., 2011). This premise may be transposed to insulator function as well, i.e. ISWI and its associated factors might function to prepare an enhanced state for insulator activity, which in turn would regulate gene activities. Furthermore, ISWI has been reported to function in the germ line and in multipotent cell divisions (Demeret et al., 2002; Xi and Xie, 2005; Yokoyama et al., 2009; Cherry and Matunis, 2010). We found that the ISWI protein accumulates in nuclei of every blastomere of the sea urchin embryo but becomes restricted to the multipotent germ line stem cells at later stages of larval development (data not shown). Cells of the early sea urchin embryo have broad developmental potentiality and, as such (Ransick and Davidson, 1993), ISWI might be functioning in blastomeres as a regulator of potency in cells. Further, in the ISWI knockdown approaches we found significant developmental abnormalities and, although we cannot exclude the possibility that ISWI also regulates promoter activity directly, the reporter for the ArsI insulator minimally reveals and quantifies insulator function independently of any other functions that the protein might play.
From our ChIP-qPCR results, ISWI appears to interact with multiple ArsI loci throughout the genome, yet the extent of interaction differs through development, suggesting that ISWI might function as a general/basic insulator protein that contributes to ArsI activity by manipulating the level of interactions dynamically to regulate the GRN of this embryo. Insulator function has been studied extensively in cultured cells, which may be subject to a variety of technical manipulations, whereas the dynamic function of insulators during development, as seen here and recently for example in flies (reviewed by Herold et al., 2012), may provide an important perspective for comparative insulator analysis with strong physiological relevance to gene regulation within the context of developmental change. Further genome-wide analysis and detailed functional assays of ArsI-protein complex(es), especially those involving Sin3A, which appears from this report to function together with ISWI in the anti-silencing activity of ArsI, will likely open the window to new functional perspectives of these important chromatin regulators within a GRN.
Supplementary Material
Acknowledgments
We thank Mr Matthew Gemberling for generous support with mega-shift assays; Drs Sorin Istrail and Ryan Tarpine for assistance in the whole-genome analysis of ArsI distribution; and Drs Erica Larschan and Judith Bender for guidance and critical reading of the manuscript; and technical assistance from Lifespan Rhode Island Hospital and the Division of Biology and Medicine, Brown University.
Footnotes
Funding
This work was supported by grants from the National Institutes of Health [2R01HD028152] and National Science Foundation [NSF IOS-1120972] to G.M.W., by a Human Frontier Science Program long-term fellowship to M.Y. by the Genomics Core Facility at Brown University, which has received support from the National Institutes of Health [NIGMS P30GM103410, NCRR P30RR031153, P20RR018728 and S10RR02763] and by National Science Foundation [EPSCoR 0554548 to W.G.F.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.081828/-/DC1
References
- Akasaka K., Nishimura A., Takata K., Mitsunaga K., Mibuka F., Ueda H., Hirose S., Tsutsui K., Shimada H. (1999). Upstream element of the sea urchin arylsulfatase gene serves as an insulator. Cell. Mol. Biol. 45, 555–565 [PubMed] [Google Scholar]
- Alekseyenko A. A., Peng S., Larschan E., Gorchakov A. A., Lee O. K., Kharchenko P., McGrath S. D., Wang C. I., Mardis E. R., Park P. J., et al. (2008). A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 134, 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- Bell A. C., West A. G., Felsenfeld G. (1999). The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98, 387–396 [DOI] [PubMed] [Google Scholar]
- Bi X., Yu Q., Sandmeier J. J., Zou Y. (2004). Formation of boundaries of transcriptionally silent chromatin by nucleosome-excluding structures. Mol. Cell. Biol. 24, 2118–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasset E., Hermant C., Jensen S., Vaury C. (2010). The Idefix enhancer-blocking insulator also harbors barrier activity. Gene 450, 25–31 [DOI] [PubMed] [Google Scholar]
- Burgio G., La Rocca G., Sala A., Arancio W., Di Gesù D., Collesano M., Sperling A. S., Armstrong J. A., van Heeringen S. J., Logie, et al. (2008). Genetic identification of a network of factors that functionally interact with the nucleosome remodeling ATPase ISWI. PLoS Genet. 4, e1000089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzone F. J., Thézé N., Thiebaud P., Hill R. L., Britten R. J., Davidson E. H. (1988). Developmental appearance of factors that bind specifically to cis-regulatory sequences of a gene expressed in the sea urchin embryo. Genes Dev. 2, 1074–1088 [DOI] [PubMed] [Google Scholar]
- Cherry C. M., Matunis E. L. (2010). Epigenetic regulation of stem cell maintenance in the Drosophila testis via the nucleosome-remodeling factor NURF. Cell Stem Cell 6, 557–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeret C., Bocquet S., Lemaítre J. M., Françon P., Méchali M. (2002). Expression of ISWI and its binding to chromatin during the cell cycle and early development. J. Struct. Biol. 140, 57–66 [DOI] [PubMed] [Google Scholar]
- Deuring R., Fanti L., Armstrong J. A., Sarte M., Papoulas O., Prestel M., Daubresse G., Verardo M., Moseley S. L., Berloco M., et al. (2000). The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5, 355–365 [DOI] [PubMed] [Google Scholar]
- Emery D. W. (2011). The use of chromatin insulators to improve the expression and safety of integrating gene transfer vectors. Hum. Gene Ther. 22, 761–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F., Rippe K. (2011). Chromatin remodelling in mammalian cells by ISWI-type complexes – where, when and why? FEBS J. 278, 3608–3618 [DOI] [PubMed] [Google Scholar]
- Ferraris L., Stewart A. P., Gemberling M. P., Reid D. C., Lapadula M. J., Thompson W. A., Fairbrother W. G. (2011). High-throughput mapping of protein occupancy identifies functional elements without the restriction of a candidate factor approach. Nucleic Acids Res. 39, e33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova G. N. (2008). Genetics and epigenetics of the multifunctional protein CTCF. Curr. Top. Dev. Biol. 80, 337–360 [DOI] [PubMed] [Google Scholar]
- Gaszner M., Felsenfeld G. (2006). Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7, 703–713 [DOI] [PubMed] [Google Scholar]
- Gelbart M. E., Bachman N., Delrow J., Boeke J. D., Tsukiyama T. (2005). Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 19, 942–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirlando R., Giles K., Gowher H., Xiao T., Xu Z., Yao H., Felsenfeld G. (2012). Chromatin domains, insulators, and the regulation of gene expression. Biochim. Biophys. Acta 1819, 644–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark A. T., Schoenherr C. J., Katz D. J., Ingram R. S., Levorse J. M., Tilghman S. M. (2000). CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405, 486–489 [DOI] [PubMed] [Google Scholar]
- Heger P., Marin B., Schierenberg E. (2009). Loss of the insulator protein CTCF during nematode evolution. BMC Mol. Biol. 10, 84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold M., Bartkuhn M., Renkawitz R. (2012). CTCF: insights into insulator function during development. Development 139, 1045–1057 [DOI] [PubMed] [Google Scholar]
- Hino S., Akasaka K., Matsuoka M. (2006). Sea urchin arylsulfatase insulator exerts its anti-silencing effect without interacting with the nuclear matrix. J. Mol. Biol. 357, 18–27 [DOI] [PubMed] [Google Scholar]
- Ideraabdullah F. Y., Vigneau S., Bartolomei M. S. (2008). Genomic imprinting mechanisms in mammals. Mutat. Res. 647, 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri C., Holmgren C., Pilartz M., Franklin G., Kanduri M., Liu L., Ginjala V., Ullerås E., Mattsson R., Ohlsson R. (2000). The 5′ flank of mouse H19 in an unusual chromatin conformation unidirectionally blocks enhancer-promoter communication. Curr. Biol. 10, 449–457 [DOI] [PubMed] [Google Scholar]
- Kim T. H., Abdullaev Z. K., Smith A. D., Ching K. A., Loukinov D. I., Green R. D., Zhang M. Q., Lobanenkov V. V., Ren B. (2007). Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128, 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita M., Kondoh H., Mitsunaga-Nakatsubo K., Shimotori T., Sakamoto N., Yamamoto T., Shimada H., Takata K., Akasaka K. (2003). Utilization of a particle gun DNA introduction system for the analysis of cis-regulatory elements controlling the spatial expression pattern of the arylsulfatase gene (HpArs) in sea urchin embryos. Dev. Genes Evol. 213, 44–49 [DOI] [PubMed] [Google Scholar]
- Li M., Belozerov V. E., Cai H. N. (2010). Modulation of chromatin boundary activities by nucleosome-remodeling activities in Drosophila melanogaster. Mol. Cell. Biol. 30, 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makabe K. W., Kirchhamer C. V., Britten R. J., Davidson E. H. (1995). Cis-regulatory control of the SM50 gene, an early marker of skeletogenic lineage specification in the sea urchin embryo. Development 121, 1957–1970 [DOI] [PubMed] [Google Scholar]
- Nagaya S., Yoshida K., Kato K., Akasaka K., Shinmyo A. (2001). An insulator element from the sea urchin Hemicentrotus pulcherrimus suppresses variation in transgene expression in cultured tobacco cells. Mol. Genet. Genomics 265, 405–413 [DOI] [PubMed] [Google Scholar]
- Okamoto K., Morison I. M., Taniguchi T., Reeve A. E. (1997). Epigenetic changes at the insulin-like growth factor II/H19 locus in developing kidney is an early event in Wilms tumorigenesis. Proc. Natl. Acad. Sci. USA 94, 5367–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C. T., Corces V. G. (2009). Insulators as mediators of intra- and inter-chromosomal interactions: a common evolutionary theme. J. Biol. 8, 73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H. C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T., et al. (2008). Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132, 422–433 [DOI] [PubMed] [Google Scholar]
- Pikaart M. J., Recillas-Targa F., Felsenfeld G. (1998). Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 12, 2852–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prawitt D., Enklaar T., Gärtner-Rupprecht B., Spangenberg C., Lausch E., Reutzel D., Fees S., Korzon M., Brozek I., Limon J., et al. (2005). Microdeletion and IGF2 loss of imprinting in a cascade causing Beckwith-Wiedemann syndrome with Wilms’ tumor. Nat. Genet. 37, 785–786 [DOI] [PubMed] [Google Scholar]
- Ransick A., Davidson E. H. (1993). A complete second gut induced by transplanted micromeres in the sea urchin embryo. Science 259, 1134–1138 [DOI] [PubMed] [Google Scholar]
- Recillas-Targa F. (2006). Multiple strategies for gene transfer, expression, knockdown, and chromatin influence in mammalian cell lines and transgenic animals. Mol. Biotechnol. 34, 337–354 [DOI] [PubMed] [Google Scholar]
- Reik W., Brown K. W., Schneid H., Le Bouc Y., Bickmore W., Maher E. R. (1995). Imprinting mutations in the Beckwith-Wiedemann syndrome suggested by altered imprinting pattern in the IGF2-H19 domain. Hum. Mol. Genet. 4, 2379–2385 [DOI] [PubMed] [Google Scholar]
- Sala A., Toto M., Pinello L., Gabriele A., Di Benedetto V., Ingrassia A. M., Lo Bosco G., Di Gesù V., Giancarlo R., Corona D. F. (2011). Genome-wide characterization of chromatin binding and nucleosome spacing activity of the nucleosome remodelling ATPase ISWI. EMBO J. 30, 1766–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenman M. J., Rainier S., Dobry C. J., Grundy P., Horon I. L., Feinberg A. P. (1994). Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms’ tumour. Nat. Genet. 7, 433–439 [DOI] [PubMed] [Google Scholar]
- Tajima S., Shinohara K., Fukumoto M., Zaitsu R., Miyagawa J., Hino S., Fan J., Akasaka K., Matsuoka M. (2006). Ars insulator identified in sea urchin possesses an activity to ensure the transgene expression in mouse cells. J. Biochem. 139, 705–714 [DOI] [PubMed] [Google Scholar]
- Takada T., Iida K., Akasaka K., Yasue H., Torii R., Tsujimoto G., Taira M., Kimura H. (2000). Evaluation of heterologous insulator function with regard to chromosomal position effect in the mouse blastocyst and fetus. Mol. Reprod. Dev. 57, 232–237 [DOI] [PubMed] [Google Scholar]
- Takagi H., Inai Y., Watanabe S., Tatemoto S., Yajima M., Akasaka K., Yamamoto T., Sakamoto N. (2012). Nucleosome exclusion from the interspecies-conserved central AT-rich region of the Ars insulator. J. Biochem. 151, 75–87 [DOI] [PubMed] [Google Scholar]
- Taniguchi T., Sullivan M. J., Ogawa O., Reeve A. E. (1995). Epigenetic changes encompassing the IGF2/H19 locus associated with relaxation of IGF2 imprinting and silencing of H19 in Wilms tumor. Proc. Natl. Acad. Sci. USA 92, 2159–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantin D., Gemberling M., Callister C., Fairbrother W. G. (2008). High-throughput biochemical analysis of in vivo location data reveals novel distinct classes of POU5F1(Oct4)/DNA complexes. Genome Res. 18, 631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Weisz P. (2005). Chromatin remodeling factors and DNA replication. Prog.Mol. Subcell. Biol. 38, 1–30 [DOI] [PubMed] [Google Scholar]
- Watanabe S., Watanabe S., Sakamoto N., Sato M., Akasaka K. (2006). Functional analysis of the sea urchin-derived arylsulfatase (Ars)-element in mammalian cells. Genes Cells 11, 1009–1021 [DOI] [PubMed] [Google Scholar]
- Wei Z., Angerer R. C., Angerer L. M. (2006). A database of mRNA expression patterns for the sea urchin embryo. Dev. Biol. 300, 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt K. S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., et al. (2008). Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796–801 [DOI] [PubMed] [Google Scholar]
- West A. G., Gaszner M., Felsenfeld G. (2002). Insulators: many functions, many mechanisms. Genes Dev. 16, 271–288 [DOI] [PubMed] [Google Scholar]
- Wilt F. H. (1999). Matrix and mineral in the sea urchin larval skeleton. J. Struct. Biol. 126, 216–226 [DOI] [PubMed] [Google Scholar]
- Xi R., Xie T. (2005). Stem cell self-renewal controlled by chromatin remodeling factors. Science 310, 1487–1489 [DOI] [PubMed] [Google Scholar]
- Xie X., Mikkelsen T. S., Gnirke A., Lindblad-Toh K., Kellis M., Lander E. S. (2007). Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl. Acad. Sci. USA 104, 7145–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M., Wessel G. M. (2011a). The DEAD-box RNA helicase Vasa functions in embryonic mitotic progression in the sea urchin. Development 138, 2217–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M., Wessel G. M. (2011b). Small micromeres contribute to the germline in the sea urchin. Development 138, 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M., Kiyomoto M., Akasaka K. (2007). Ars insulator protects transgenes from long-term silencing in sea urchin larva. Dev. Genes Evol. 217, 331–336 [DOI] [PubMed] [Google Scholar]
- Yang Q., Angerer L. M., Angerer R. C. (1989). Structure and tissue-specific developmental expression of a sea urchin arylsulfatase gene. Dev. Biol. 135, 53–65 [DOI] [PubMed] [Google Scholar]
- Yao H., Brick K., Evrard Y., Xiao T., Camerini-Otero R. D., Felsenfeld G. (2010). Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 24, 2543–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama H., Rybina S., Santarella-Mellwig R., Mattaj I. W., Karsenti E. (2009). ISWI is a RanGTP-dependent MAP required for chromosome segregation. J. Cell Biol. 187, 813–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.