Background: The function of multiple homologues of Atg4 in higher eukaryotes remains largely unknown.
Results: We demonstrated that the two Atg4 homologues in Caenorhabditis elegans have differential enzymatic activities in processing LGG-1/Atg8 and exhibit functional redundancy.
Conclusion: The two Atg4s contain redundant yet differential processing activities in the autophagy pathway.
Significance: We revealed physiological function of the two Atg4 homologues in the autophagy pathway.
Keywords: Autophagy, C. elegans, Protease, Protein Processing, Protein Turnover, atg4, atg8, Autophagosome
Abstract
The presence of multiple homologues of the same yeast Atg protein endows an additional layer of complexity on the autophagy pathway in higher eukaryotes. The physiological function of the individual genes, however, remains largely unknown. Here we investigated the role of the two Caenorhabditis elegans homologues of the cysteine protease Atg4 in the pathway responsible for degradation of protein aggregates. Loss of atg-4.1 activity causes defective degradation of a variety of protein aggregates, whereas atg-4.2 mutants remove these substrates normally. LGG-1 precursors accumulate in atg-4.1 mutants, but not atg-4.2 mutants. LGG-1 puncta, formation of which depends on lipidation of LGG-1, are present in atg-4.1 and atg-4.2 single mutants, but are completely absent in atg-4.1; atg-4.2 double mutants. In vitro enzymatic analysis revealed that ATG-4.1 processes LGG-1 precursors about 100-fold more efficiently than ATG-4.2. Expression of a mutant form LGG-1, which mimics the processed precursor, rescues the defective autophagic degradation of protein aggregates in atg-4.1 mutants and, to a lesser extent, in atg-4.1; atg-4.2 double mutants. Our study reveals that ATG-4.1 and ATG-4.2 are functionally redundant yet display differential LGG-1 processing and deconjugating activity in the aggrephagy pathway in C. elegans.
Introduction
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved lysosome-mediated degradation process (1). It can be dissected into a series of steps involving membrane dynamics (2); a cup-shaped membrane sac (known as the isolation membrane or phagophore) nucleates, elongates and seals off to form an enclosed double membrane vesicle (known as the autophagosome), which is then delivered to, and fuses with, lysosomes to form the autolysosome, where the sequestrated materials, together with the inner membrane of the autophagosome, are degraded (1, 2).
Genetic screens in yeast have identified about 19 autophagy-related (ATG)2 genes essential for autophagosome biogenesis (2). Among the core autophagic machinery, two ubiquitin-like systems, the Atg8 system and the Atg12 system, are essential for the expansion and closure of the autophagosome (1, 3). The Atg8 system involves the sequential action of processing, conjugation and deconjugation activities during autophagosome formation (4–6). The newly synthesized Atg8 precursor is cleaved by the cysteine protease Atg4 to expose its C-terminal glycine residue, which is activated by an E1-like activating enzyme, Atg7, and then transferred to an E2-like conjugating enzyme, Atg3, and finally conjugated to phosphatidylethanolamine (PE) via an amide bond (2–5). PE-conjugated Atg8 associates with the autophagosomal membrane (4, 5). During maturation of autophagosomes, Atg8-PE on the outer autophagosomal membrane is deconjugated by Atg4 for recycling, while Atg8-PE on the inner membrane is transported into the lysosome for degradation (4, 7). This reversible modification of Atg8 is crucial for the normal progression of autophagy (4, 8, 9).
The autophagy process in higher eukaryotes involves much more complicated membrane dynamics. One layer of complexity is conferred by the presence of multiple homologues of the same yeast Atg protein (10). The single Atg4 in yeast has four mammalian paralogues, Atg4A, Atg4B, Atg4C, and Atg4D (11–14). At least eight homologues of Atg8 have been identified in mammals (15, 16). The human (Hs) Atg4 paralogues exhibit differential processing activity toward the different Atg8 proteins (17). HsAtg4B processes all Atg8 proteins, including GATE-16, GABARAP, GABARAP-L1, and LC3 (13, 14). The activity of other Atg4 family members appears to be more specific toward individual Atg8 paralogues. HsAtg4A efficiently processes GATE-16, but is ineffective toward LC3 (11, 17). Despite the wide expression pattern of Atg4C, Atg4C is not essential for autophagy under normal conditions, but Atg4C−/− starved mice show decreased autophagic activity in the diaphragm (18). Mice deficient in Atg4B exhibit a reduction of basal- and starvation-induced autophagic flux in all tissues (19). The proteolytic cleavage of multiple Atg8 homologues is severely reduced but not totally impaired in Atg4B−/− cells (19, 20). Atg4C and Atg4D display minimal processing activity toward Atg8 substrates (17). Although mice deficient in Atg3, Atg5, Atg7, and Atg16L show severe defects in the autophagy pathway and die during the neonatal starvation period, mice deficient in Atg4C and Atg4B do not display any obvious abnormalities and survive the early neonatal period, suggesting that there are functional divergences and redundancies between mammalian Atg4 homologues (18–24). The physiological function of Atg4C and Atg4D and the contribution of each of the Atg4 paralogues during the autophagy process remains poorly understood.
Caenorhabditis elegans contains two Atg4 homologues, encoded by atg-4.1 and atg-4.2. Here we showed that genetic mutations in atg-4.1, but not in atg-4.2, cause defective autophagic degradation of a variety of protein aggregates during embryogenesis. In wild-type C. elegans, the nascent Atg8 homologue LGG-1 is rapidly processed by cleavage near the C terminus to give LGG-1-I. LGG-1-I can then be conjugated with PE to give LGG-1-II. Nascent LGG-1 precursors accumulate in atg-4.1 mutants but not in atg-4.2 mutants. The PE-conjugated form of LGG-1 is present in atg-4.1 and atg-4.2 single mutants, but absent in atg-4.1; atg-4.2 double mutants. An in vitro cleavage assay revealed that ATG-4.1 has much higher processing activity than ATG-4.2 toward LGG-1/Atg8. Overexpression of the processed form of LGG-1 rescues the defective degradation of protein aggregates in atg-4.1 mutants and, to a lesser extent, in atg-4.1; atg-4.2 double mutants. Our results show that atg-4.1 and atg-4.2 have differential processing activities and contribute differentially in the aggrephagy pathway in C. elegans.
EXPERIMENTAL PROCEDURES
Strains
The following strains were used in this work: atg-4.1(bp321), atg-4.1(bp451), atg-4.1(bp501), atg-4.1(bp504), atg-4.2(tm3948), atg-3(bp412), epg-1(bp414), epg-6(bp242), bpIs131(sepa-1::gfp), bpIs151(sqst-1::gfp), bpIs152(sqst-1::gfp), bpIs168(DFCP1::gfp). atg-4.1(bp501) was used for genetic analysis.
Isolation and Identification of atg-4.1
Animals carrying gfp::pgl-1 or sepa-1::gfp reporters were used for EMS mutagenesis. Embryos derived from F2 animals were screened for accumulation of GFP::PGL-1 or SEPA-1::GFP aggregates. From ∼20,000 genomes screened (∼10,000 genomes for each reporter), a total of 96 mutant alleles were obtained (28 from the gfp::pgl-1 screen, 68 from the sepa-1::gfp screen). Subsequent genetic and molecular analysis revealed that we had isolated seven alleles of atg-4.1. bp410, bp418, and bp451 were identified from the gfp::pgl-1 screen. bp321, bp482, bp501, and bp504 were identified from the sepa-1::gfp screen.
Starvation Assay
Embryos collected by the synchronization method were incubated in deionized distilled water at 20 °C. An aliquot of each sample was placed on a plate seeded with OP50 at each time point. After 3 days, the number of animals surviving to L4 was counted. Differences in survival were analyzed with the Kaplan-Meier method and log-rank tests using SPSS version 20.0.
Indirect Immunofluorescence
Permeabilization of embryos was performed by freeze-cracking methods. Freeze-cracked slides were fixed, blocked, and incubated with diluted antibody at room temperature for 2–4 h. The worms were then washed three times and incubated with secondary antibodies. Slides were viewed using a confocal microscope (Zeiss LSM 510 Meta).
Reporter Construction
A cDNA encoding ATG-4.1 was cloned into pPD95.79 to create a reporter with GFP fused at the C terminus. The reporter contained the unc-54 3′-UTR and was driven by the atg-4.1 promoter (∼2 kb). The plasmid was co-injected with pRF4(rol-6) into wild-type animals. The reporter was functional in rescuing defective degradation of SEPA-1 aggregates in the null allele of atg-4.1, bp501.
Plasmid Construction
cDNAs encoding ATG-4.1 and ATG-4.2 were cloned into pET-28a for His tagging. cDNAs encoding LGG-1 and LGG-1(G116A) were cloned into pET-21b for His tagging at the C terminus. Genomic DNA containing the promoter, coding sequence and 3′-UTR of lgg-1 (c32d5, nt 34,945–37,706) was cloned into pBluescript II SK (+). This construct was further used to delete the C-terminal seven amino acids, directly exposing the conjugated glycine (Gly-116), and also to mutate glycine 116 to alanine.
Assay for in Vitro Cleavage
His-tagged ATG-4.1, ATG-4.2, and LGG-1 proteins were expressed in Escherichia coli BL21 strain and then purified using Ni-NTA beads (Qiagen). Purified His-ATG-4s (0.0125 mg/ml and 0.025 mg/ml) and LGG-1-His (0.5 mg/ml) were incubated together in reaction buffer (150 mm Nacl, 1 mm EDTA, 1 mm DTT, and 50 mm Tris-HCl, pH 7.2) in a volume of 20 μl at 37 °C for the given time. Laemmli sample buffer was added to stop the reactions. Proteins were then separated by 13% SDS-PAGE, followed by Coomassie Brilliant Blue (CBB) staining. The density of bands was measured by Image J (W. S. Rasband, ImageJ, U. S. National Institutes of Health, Bethesda, MD) to quantify the amount of products generated from the substrate. The percentage of substrate remaining was plotted against the reaction time and used to determine the optimal reaction time for further kinetic analysis of ATG-4.1 and ATG-4.2. Different concentrations of substrate (LGG-1-His6) were incubated with ATG-4.1 and ATG-4.2 for the set optimal reaction time. The initial velocity (V, y axis) was determined from the change in LGG-1 product concentration per second (mm/s). It was then plotted against substrate concentration (S (mm), x axis) for each reaction and the resulting curves were fitted by the non-linear regression method (GraphPad Prism version 5.00, GraphPad Software, San Diego CA). The values of Vmax (mol liter−1 s−1) and Km (mol/liter) were derived from the curves. The catalytic constant, kcat (s−1), was calculated for each reaction by dividing Vmax by the enzyme concentration.
RNA Isolation and Real-time RT-PCR
atg-4.1, atg-4.2 and wild-type animals were collected and total RNA was extracted with Trizol reagent (Sigma). Total isolated RNA (2 μg) from each sample was reverse transcribed using an Invitrogen Superscript III kit. Quantitative PCR reactions were carried out as described previously (25). atg-4.1 and atg-4.2 mRNA levels in mutants were normalized to the level of wild type worms, which was set to 1. Error bars indicate the standard deviation (S.D.) of three independent experiments.
Modeling
For constructing the HsAtg4B-LGG-1 complex model, Thr-118, Phe-119, and Thr-121 of LC3 were replaced with Val, Tyr, and Gly, respectively, using the program COOT while retaining the original configurations. These replacements did not cause any steric clash with the surrounding HsAtg4B residues. G258S and A263V mutations in HsAtg4B were also introduced in the same manner.
RESULTS
atg-4.1 Is Essential for Degradation of a Variety of Autophagy Substrates
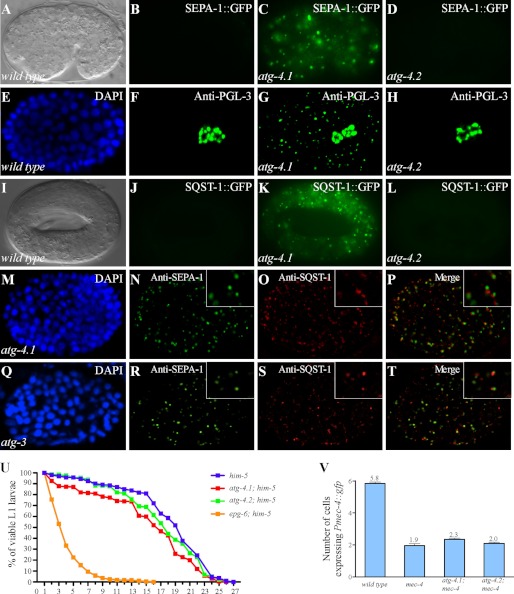
The germline P granule components PGL-1 and PGL-3 are selectively removed by autophagy in somatic cells during C. elegans embryogenesis (26, 27). Degradation of PGL-1 and PGL-3 requires the self-oligomerization protein SEPA-1, which functions as a receptor in recruiting PGL-1 and PGL-3 into aggregates, known as PGL granules (28). SEPA-1 is also removed by autophagy during embryogenesis (28). In wild type animals, SEPA-1 aggregates are present only in early stage embryos and become undetectable from the comma stage onwards. In autophagy mutants, PGL granules (containing PGL-1/PGL-3/SEPA-1) dramatically accumulate and persist throughout embryogenesis. In genetic screens to identify mutants with defective degradation of GFP::PGL-1 and SEPA-1::GFP, we isolated 7 alleles of atg-4.1 (Fig. 1, A–C and data not shown). Endogenous PGL-3 and SEPA-1, detected by specific antibodies, also failed to be removed and accumulated into a large number of aggregates in atg-4.1 mutants (Fig. 1, E–G and data not shown).
FIGURE 1.
Mutations in atg-4.1 but not atg-4.2 cause defective degradation of autophagy substrates. A and B, SEPA-1::GFP aggregates are undetectable from the comma stage onwards in wild-type embryos. A, Nomarski image of the embryo in B. C, SEPA-1::GFP accumulates into a large number of aggregates in atg-4.1 mutants. D, expression level of SEPA-1::GFP in atg-4.2 mutants is identical to wild-type embryos. E–H, endogenous PGL-3, detected by anti-PGL-3 antibody, is restricted to germ precursor cells in a wild-type embryo (E and F), but ectopically accumulates in somatic cells in atg-4.1 mutant embryos (G). atg-4.2 mutants shows the same distribution of PGL-3 as wild-type embryos (H). E, DAPI image of the embryo shown in F. I–L, SQST-1::GFP is very weakly expressed and diffusely located in the cytoplasm in wild-type embryos (I and J). It accumulates into numerous aggregates in atg-4.1 mutants (K). The expression level of SQST-1::GFP reporter remains unchanged in atg-4.2 mutants (L). I, Nomarski image of the embryo in J. M–P, in atg-4.1(bp501) mutant embryos, SEPA-1 and SQST-1 aggregates, detected by specific antibodies, are largely separable. Q–T, SEPA-1 and SQST-1 aggregates accumulate and are separable in atg-3(bp412) mutant embryos. M and Q, DAPI images of the embryos shown in N–P and R–T, respectively. U, atg-4.1(bp501) mutants show a reduction in the survival of L1 larvae in the absence of food compared with wild type animals (log-rank test, p = 0.000). atg-4.2(tm3948) mutants show no defect in the L1 survival rate. At least 500 wild-type, atg-4.1(bp501) and atg-4.2(tm3948) L1 larvae were scored. V, number of neurons labeled by the touch neuron-specific reporter Pmec-4::gfp. atg-4.1(bp501); mec-4(u231) mutants have more Pmec-4::gfp-labeled neurons (p < 0.05) than mec-4(u231) single mutants. atg-4.2(tm3948); mec-4(u231) shows no significant increase in the neuron number.
The degradation of the C. elegans p62 homologue SQST-1 was next examined in atg-4.1 mutants. In wild type embryos, SQST-1 is very weakly expressed and diffusely localized in the cytoplasm (Fig. 1, I and J). In atg-4.1 mutants, SQST-1 was detected at dramatically elevated levels and accumulated into numerous aggregates (Fig. 1K). SEPA-1 and SQST-1 aggregates were round-shaped and largely separable in atg-4.1 mutants (Fig. 1, M–P), resembling those in embryos with loss of function mutations in other LGG-1 conjugation system genes, including atg-3, atg-7, and lgg-1 (Fig. 1Q-T). Thus, atg-4.1 is required for degradation of various autophagy substrates.
We further investigated whether atg-4.1 is involved in other autophagy-regulated processes in addition to aggrephagy (26). Optimal survival of L1 larvae under starvation conditions requires autophagy activity. In the absence of food, atg-4.1 mutant L1 larvae had a significantly reduced survival rate compared with wild type L1 larvae (Fig. 1U). The defect in atg-4.1 mutants, however, was much milder than epg-6 mutants. Autophagy activity also contributes to the necrosis-like degeneration of six touch receptor neurons caused by a toxic ion channel variant encoded by mec-4(u231) (26). Loss of atg-4.1 activity ameliorated the neurodegeneration in mec-4(u231) animals (Fig. 1V). In summary, atg-4.1 mutants show defects in multiple autophagy-regulated processes.
We constructed a translational fusion reporter for atg-4.1 with gfp inserted at its C terminus to determine its expression pattern. The atg-4.1::gfp reporter was functional in rescuing defective degradation of SEPA-1 aggregates in atg-4.1 mutants (supplemental Fig. S1). ATG-4.1::GFP was expressed in the cytoplasm of most cells during embryogenesis (supplemental Fig. S1).
Molecular Lesions in atg-4.1 Mutant Alleles
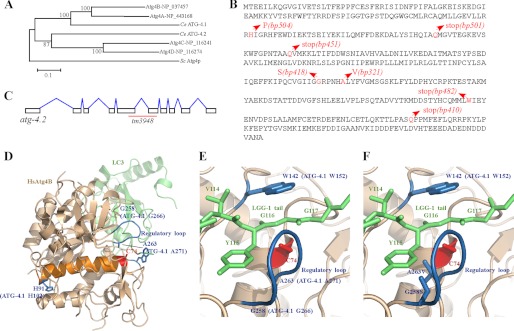
Phylogenetic analysis reveals that ATG-4.1 is closely related to human Atg4A and Atg4B (Fig. 2A). We identified molecular lesions in all seven alleles of atg-4.1 (Fig. 2B). In bp501, bp451, and bp410 mutants, glutamine residues are mutated to stop codons at positions 140, 160, and 385, respectively. The tryptophan at amino acid 347 is mutated to a stop codon in bp482. bp504 contains a histidine to proline mutation at codon 102. The glycine residue at amino acid 266 and the alanine residue at amino acid 271 are mutated to serine and valine in bp418 and bp321 mutants, respectively. atg-4.1(bp501) is likely to be null and was used in this study.
FIGURE 2.
Molecular lesions in atg-4.1 mutant alleles. A, phylogenetic tree of Atg4 family proteins. MEGA version 5.05 was used to construct a Neighbor-Joining phylogenetic tree. Bootstrap values are shown in percentages at nodes. The 0.1 scale bar represents 10% change. B, protein sequence of ATG-4.1. Glutamine residues are mutated to stop codons at amino acids 140, 160, and 385 in bp501, bp451 and bp410 mutants, respectively. The tryptophan at amino acid 347 is mutated to a stop codon in bp482. bp504 contains a histidine to proline mutation at amino acid 102. The glycine residue at amino acid 266 and the alanine residue at amino acid 271 are mutated to serine and valine in bp418 and bp321 mutants, respectively. C, schematic structure of atg-4.2. tm3948 deletes 498 nucleotides, introducing an early stop codon and creating a truncated protein with 266 amino acids. Intron-exon boundaries were confirmed by cDNA sequencing. D, crystal structure of the HsAtg4B-LC3 complex. The residues that are mutated in bp504, bp418, and bp321 are labeled. Although HsAtg4B has C74S mutation, Ser-74 is labeled as “C74” in Fig. 2, D–F. E, modeled structure of the HsAtg4B-LGG-1 complex. The LGG-1 tail (green) is recognized by the regulatory loop (blue) and W142 (blue) of HsAtg4B. F, G258S and A263V mutations, found in bp418 and bp321, respectively, disrupt the interaction between Atg4B and LGG-1.
A previously published x-ray crystallography study of the HsAtg4B-LC3 complex showed that Cys-74, Asp-278, and His-280 of HsAtg4B form the catalytic triad (29). The catalytic cleft of HsAtg4B is masked by a regulatory loop (residues 259 to 262) that is lifted by Phe-119 of LC3 (29, 30). HsAtg4B recognizes the Phe-119 and Gly-120 residues of LC3 using the regulatory loop and Trp-142, respectively, and this recognition is essential for the processing of LC3 by HsAtg4B (Fig. 2D). The catalytic triad, Trp-142 and the regulatory loop are conserved in ATG-4.1 (supplemental Fig. S2). His-102, Gly-266, and Ala-271 of ATG4.1, which are mutated in bp504, bp418, and bp321 mutants, correspond to H91, G258 and Ala-263 of HsAtg4B, respectively (Fig. 2D). Gly-258 and Ala-263 of HsAtg4B are located in the regulatory loop of Atg4, and both are involved in direct recognition of the side-chain of LC3 Phe-119 in the HsAtg4B-LC3 complex (29). Using the crystal structure of the HsAtg4B-LC3 complex (PDB ID 2ZZP), the C-terminal tail residues of LC3 were replaced with those of LGG-1. In this new version, ATG-4.1 will recognize Y115 (corresponding to LC3 Phe-119) and Gly-116 (corresponding to LC3 Gly-120) of LGG-1 using the regulatory loop and Trp-152 (corresponding to HsAtg4B W142) (Fig. 2E). The side-chains of Ser-266 in ATG-4.1(bp418) and Val-271 in ATG-4.1(bp321) interfere with the side-chain of Tyr-115 (Fig. 2F), destroying the proper interaction between ATG-4.1 and LGG-1 and resulting in severely impaired activity of ATG-4.1. The Pro-102 in ATG-4.1(bp504) is located in the same helix (orange) as the catalytic cysteine (Cys-74) (Fig. 2D). This mutation is likely to induce a large conformational change of the helix and thus affect the conformation of the catalytic cysteine. Alternatively, the H102P mutation may destroy the overall structure of ATG-4.1.
atg-4.2 Mutants Exhibit No Defects in Degradation of Autophagy Substrates
atg-4.2 encodes a second C. elegans Atg4 homologue, which is more closely related to human Atg4C and Atg4D (Fig. 2A). ATG-4.1 and ATG-4.2 display 22% overall amino acid identity and 44% similarity. The catalytic sites and the regulatory loop are conserved in ATG-4.2 (supplemental Fig. S2). Mutations in atg-4.2 were not recovered in our genetic screens. We obtained a mutant allele of atg-4.2, tm3948, which has a deletion of 498 nt that affects exons 5 and 6 (Fig. 2C). cDNA sequencing revealed the presence of an early stop codon, resulting in a truncated protein with 266 amino acids. In atg-4.2 mutants, there were no changes in the expression levels of GFP reporters for PGL-1, SEPA-1 and SQST-1 or the corresponding endogenous proteins (Fig. 1, D, H, L), indicating that atg-4.2 is not essential for autophagic degradation of protein aggregates during embryogenesis. Loss of function of atg-4.2 had no effect on the survival of L1 larvae under starvation conditions (Fig. 1U). The degeneration of touch neurons in mec-4(u231) mutants was also not affected by the atg-4.2 mutation (Fig. 1V).
We crossed the atg-4.1::gfp transgene into atg-4.2(tm3948) mutants and found that the fluorescent intensity remained unchanged (supplemental Fig. S1). Consistent with this, quantitative real-time PCR showed that the atg-4.1 mRNA level in atg-4.2 mutants was identical to that in wild-type worms. Similarly, the atg-4.2 mRNA level showed no difference to wild type in atg-4.1-null mutants. Thus, loss of function of one atg-4 paralogue does not cause compensatory up-regulation of the other.
atg-4.1 and atg-4.2 Function Redundantly in Mediating LGG-1 Conjugation
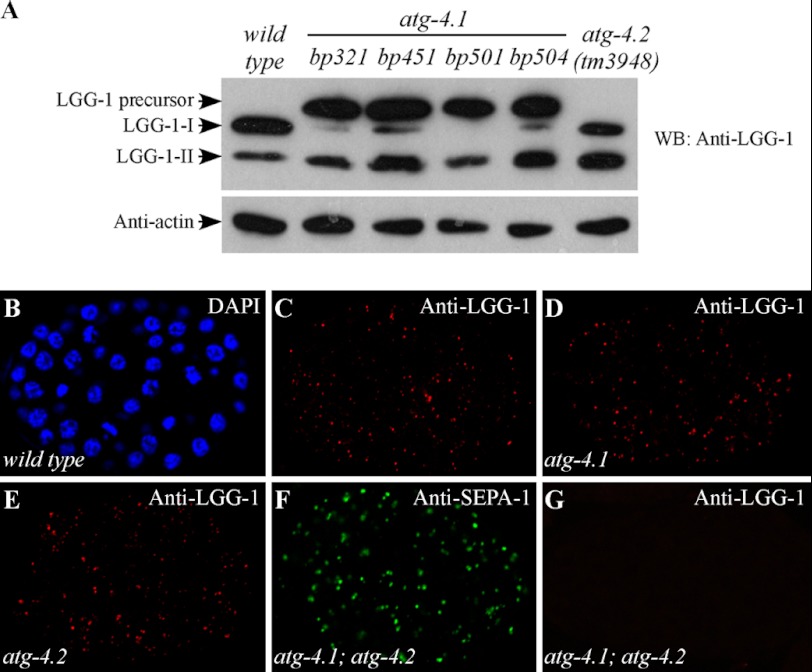
To determine whether ATG-4.1 and ATG-4.2 cleave Atg8, we examined endogenous levels of LGG-1/Atg8 in both atg-4.1 and atg-4.2 mutants. In wild-type animals, nascent LGG-1 is cleaved at a conserved glycine residue (Gly-116), releasing the seven C-terminal amino acids to give the processed cytoplasmic form LGG-1-I. This can then be lipidated to give LGG-1-II. LGG-1 precursors were absent in wild type animals, but dramatically accumulated in atg-4.1 mutants (Fig. 3A). LGG-1-II was present at a level similar to that in wild type animals, suggesting that the defective autophagic process in atg-4.1 mutants is not due to lack of lipidated LGG-1-II. LGG-1-I, however, was very weakly detected in bp321, bp451, and bp504 mutants and absent in bp501 mutants. In atg-4.2 mutants, LGG-1 was properly processed and no LGG-1 precursors were found. Compared with wild type animals, we consistently detected more LGG-1-II than LGG-1-I in atg-4.2 mutants. These results indicate that atg-4.1, but not atg-4.2, plays an essential role in processing LGG-1 precursors.
FIGURE 3.
atg-4.1 and atg-4.2 function redundantly in mediating conjugation of LGG-1. A, LGG-1/Atg8 levels in atg-4.1 and atg-4.2 mutants. Western analysis shows accumulation of LGG-1 precursors and also LGG-1-II in atg-4.1 mutants. LGG-1-I is very weakly detected in bp321, bp451, and bp504 mutants and is absent in bp501 mutants. In atg-4.2 mutants, no LGG-1 precursors are detected. LGG-1-II accumulates at a higher level than in wild type animals. B–E, atg-4.1- and atg-4.2-null mutants exhibit the wild-type dynamic immunostaining pattern of LGG-1 during embryogenesis. B, DAPI image of the embryo in C. F, in atg-4.1(bp501); atg-4.2(tm3948) double mutants, SEPA-1 aggregates accumulate at the comma stage. G, no LGG-1 puncta are detected in atg-4.1(bp501); atg-4.2(tm3948) double mutants.
The distribution of LGG-1 was then examined in atg-4.1 and atg-4.2 mutants. In wild type embryos, LGG-1 forms distinct punctate structures, mostly around the ∼100 to ∼200 cell stage (Fig. 3, B and C). The number of LGG-1 puncta decreases as development proceeds and only a few could be detected during late embryonic stages. LGG-1-II (lipidated LGG-1) is required for the formation of LGG-1 puncta, as they are absent in atg-3 and atg-7 mutants (31). Consistent with the presence of the lipidated form of LGG-1, the temporal pattern of formation of LGG-1 puncta was identical to wild type in atg-4.1 and atg-4.2 mutant embryos (Fig. 3, D and E).
We next determined whether ATG-4.1 and ATG-4.2 function redundantly in processing LGG-1 precursors. atg-4.1(bp501); atg-4.2(tm3948) double mutants were homozygous lethal. The double mutants survived into the young adult stage but failed to lay viable embryos. In embryos lacking both atg-4.1 and atg-4.2, derived from atg-4.1(bp501)/+; atg-4.2(tm3948) hermaphrodites, SEPA-1 and SQST-1 ectopically accumulated (Fig. 3F and data not shown). No LGG-1 puncta were detected in atg-4.1(bp501); atg-4.2(tm3948) mutant embryos (Fig. 3G), suggesting an absence of lipidated LGG-1. Thus, atg-4.1 and atg-4.2 function redundantly in LGG-1 processing.
ATG-4.1 and ATG-4.2 Show Different Processing Activity toward LGG-1
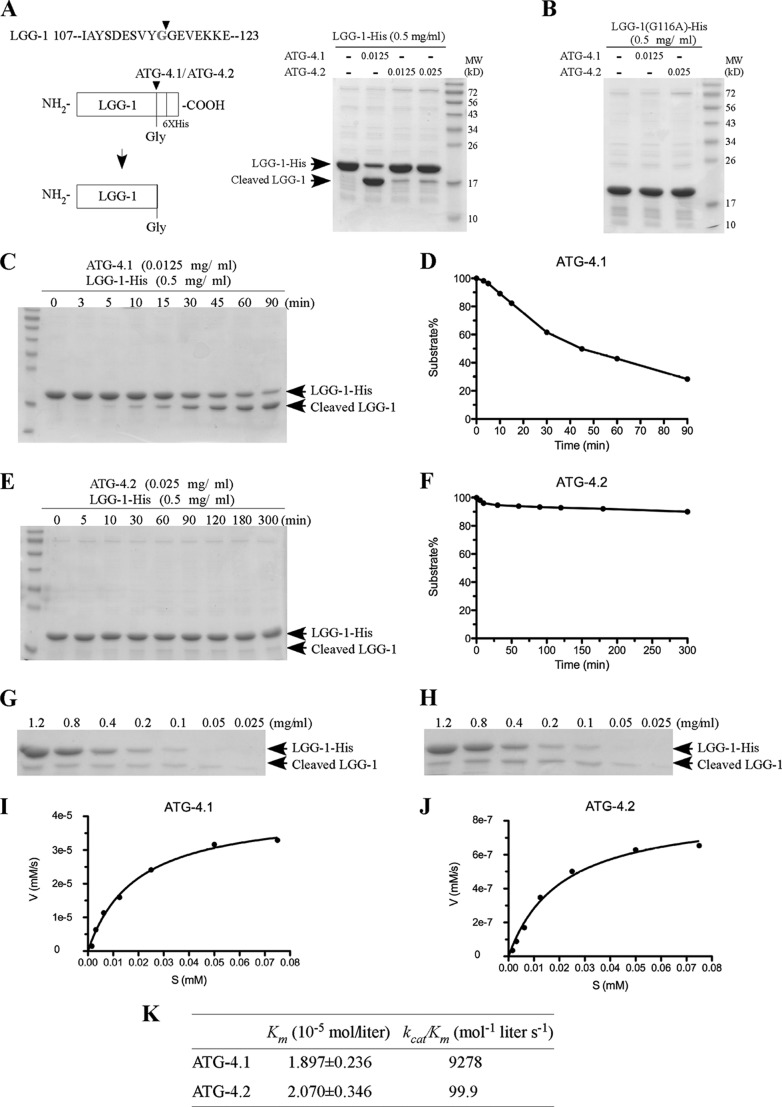
We further performed in vitro cleavage assays to determine the processing activity of ATG-4.1 and ATG-4.2 toward LGG-1. Wild type His-tagged ATG-4.1 and ATG-4.2 were expressed in bacteria and affinity-purified on Ni-NTA beads. The purified proteins were incubated with recombinant LGG-1-His6 in reaction buffer. The cleaved form of LGG-1 (LGG-1-I), which lacks the final seven residues at its C terminus, was separated from the full-length LGG-1 precursor by SDS-PAGE. We found that both ATG-4.1 and ATG-4.2 were able to cleave full-length LGG-1, generating the shorter LGG-1-I in the assay (Fig. 4A). ATG-4.1 showed much higher LGG-1 processing activity than ATG-4.2. The LGG-1(G116A) mutant protein, in which the conserved glycine at amino acid 116 was mutated to alanine, failed to be cleaved by ATG-4.1 and ATG-4.2 (Fig. 4B).
FIGURE 4.
ATG-4.1 and ATG-4. 2 show different processing activity toward LGG-1/Atg8. A, schematic diagram showing cleavage of LGG-1-His6 by ATG-4.1 and ATG-4.2. The cleaved form of LGG-1 lacks the final seven residues as well as the His-tag at its C terminus, resulting in a lower molecular weight than LGG-1-His6 when separated by SDS-PAGE. Bacterially expressed His-tagged LGG-1, ATG-4.1, and ATG-4.2 were purified with Ni-NTA. LGG-1-His6 substrate (0.5 mg/ml) was incubated with vehicle control (first lane), ATG-4.1 (0.0125 mg/ml, second lane), or ATG-4.2 (0.0125 mg/ml and 0.025 mg/ml, third and fourth lanes), in a volume of 20 μl for 1.5 h at 37 °C. The mixture was resolved using SDS-PAGE followed by CBB staining after the reaction was stopped. B, LGG-1(G116tA)-His6 substrate (0.5 mg/ml), in which the conserved glycine is mutated to alanine, was incubated with vehicle control (first lane), ATG-4.1 (0.0125 mg/ml, second lane), or ATG-4.2 (0.025 mg/ml, third lane), under the same reaction conditions as in Fig. 4A. C, ATG-4.1 (0.0125 mg/ml) was incubated with the substrate LGG-1-His6 (0.5 mg/ml) in a volume of 20 μl for the indicated time. The mixture was resolved using SDS-PAGE followed by CBB staining after the reaction was stopped. D, gel was analyzed by Image J and the densities of both substrate and product were measured. About 50% LGG-1-His substrate was cleaved by ATG-4.1 in 45 min. E, ATG-4.2 (0.025 mg/ml) was incubated with the substrate LGG-1-His6 (0.5 mg/ml) in a volume of 20 μl for the indicated time. The mixture was resolved using SDS-PAGE followed by CBB staining. F, gel was analyzed by Image J and the densities of both substrate and product were measured. G–J, decreasing concentrations of the substrate LGG-1-His6 were incubated with fixed amounts of ATG-4.1 (0.0125 mg/ml) (G) and ATG-4.2 (0.025 mg/ml) (H) for 5 min and 300 min, respectively. The mixture was resolved using SDS-PAGE followed by CBB staining. The initial velocity (V, y axis (mm/s), i.e. the change in LGG-1 product concentration/s) was plotted against each substrate concentration (S (mm), x axis) for each reaction. The curves were fitted by the non-linear regression method (GraphPad Prism 5.00). K, kinetic parameters of ATG-4.1 and ATG-4.2 toward LGG-1. The values of Vmax (mol liter−1 s−1) and Km (mol/liter) were derived from curves fitted by the non-linear regression method (GraphPad Prism 5.00). The catalytic constant, kcat (s−1), was calculated by dividing Vmax by the enzyme concentration of each reaction.
ATG-4.1 Possesses Higher Catalytic Activity than ATG-4.2
To further characterize the processing activity of ATG-4.1 and ATG-4.2, we determined their enzymatic kinetics by measuring levels of the Coomassie Brilliant Blue (CBB)-stained cleaved form of LGG-1. LGG-1-His and the cleaved form of LGG-1 were separated by SDS-PAGE and then stained with CBB. The density of bands was measured to determine the amount of products generated from the substrate. This assay has been used previously to determine the kinetic differences among mammalian Atg4 homologues (17).
We first determined the optimal time for the kinetic analysis of ATG-4.1 and ATG-4.2. Under the given experimental conditions, about 50% of the LGG-1-His substrate was cleaved by ATG-4.1 in 45 min (Fig. 4C). The cleavage reaction became linear after 5 min, so this was the time point we selected for further experiments on ATG-4.1 (Fig. 4D). The catalytic protease reaction of ATG-4.2 was much slower than ATG-4.1. The substrate processing of ATG-4.2 occurred at very low levels, but persisted even up to 300 min (Fig. 4, E and F). For further kinetic analysis, we determined the activity of ATG-4.2 at 300 min.
We next incubated decreasing concentrations of the LGG-1-His substrate with fixed amounts of ATG-4.1 and ATG-4.2 at the chosen reaction time (Fig. 4, G and H). The amount of product generated from the substrate was measured, and the reaction velocity for each substrate concentration point was calculated. The values of Km and kcat/Km were then determined (Fig. 4, I and J). ATG-4.1 and ATG-4.2 bound to LGG-1/Atg8 with similar affinities (1/Km) (Fig. 4K). However, ATG-4.1 displayed about 100-fold higher catalytic efficiency (kcat/Km) in cleaving the LGG-1 precursor compared with ATG-4.2 (Fig. 4K). The catalytic efficiency of ATG-4.1 for LGG-1 was comparable with mammalian Atg4A toward its most highly preferred substrate GATE-16 (17). The catalytic efficiency of ATG-4.2 was in the same range as mammalian Atg4C and Atg4D (17). ATG-4.2 did not reduce the catalytic efficiency of ATG-4.1 in the mixture reaction (supplemental Fig. S3).
Overexpression of the Processed Form of LGG-1 Rescues Defective Degradation of Protein Aggregates in atg-4.1 Mutants
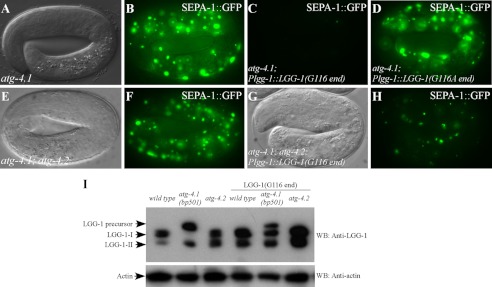
To determine whether expression of the processed form of LGG-1 rescues the defect in atg-4.1(bp501) mutants, we created a Plgg-1::LGG-1(G116 end) transgene, in which LGG-1 ends with the glycine at position 116. SEPA-1::GFP aggregates accumulated in atg-4.1(bp501) mutants at late embryonic stages (Fig. 5, A and B). atg-4.1(bp501) embryos carrying the Plgg-1::LGG-1(G116 end) transgene had a dramatically reduced number of SEPA-1::GFP aggregates (Fig. 5C), indicating that expression of the processed form of LGG-1 rescued defective degradation of protein aggregates in atg-4.1 mutants. In contrast, a transgene expressing a mutant form of LGG-1 with Gly-116 mutated to alanine did not rescue atg-4.1(bp501) (Fig. 5D). Immunoblotting assays showed that wild type and atg-4.2 mutant animals carrying the pLGG-1(G116) transgene had increased levels of LGG-1-II (Fig. 5I). atg-4.1(bp501) mutants carrying this transgene also showed accumulation of LGG-1-I (unlipidated form) (Fig. 5I).
FIGURE 5.
The processed form of LGG-1 rescues the defect in atg-4. 1 mutants. A and B, SEPA-1 aggregates, labeled by a gfp reporter, accumulate in 4-fold stage atg-4.1(bp501) mutant embryos. A, Nomarski image of the embryo in B. C, number of SEPA-1::GFP aggregates is dramatically decreased in atg-4.1(bp501) embryos carrying the Plgg-1::LGG-1(G116 end) transgene. D, numerous SEPA-1::GFP aggregates exist in atg-4.1(bp501) embryos carrying the Plgg-1::LGG-1(A116 end) transgene. E and F, SEPA-1::GFP aggregates ectopically accumulate in atg-4.1(bp501); atg-4.2(tm3948) double mutant embryos at the 4-fold stage. E, Nomarski image of the embryo in F. G and H, accumulation of SEPA-1::GFP aggregates is greatly reduced in atg-4.1(bp501); atg-4.2(tm3948); Plgg-1::LGG-1(G116 end) embryos. G, Nomarski image of the embryo in H. I, expression of LGG-1 in wild type, atg-4.1 and atg-4.2 mutants carrying the Plgg-1::LGG-1(G116 end) transgene. LGG-1-II accumulates in wild type and atg-4.2 mutant animals carrying the Plgg-1::LGG-1(G116 end) transgene. In atg-4.1(bp501) mutants carrying this transgene, in addition to LGG-1 precursors and LGG-1-II, the unlipidated form of LGG-1-I also accumulates.
The Plgg-1::LGG-1(G116 end) transgene was crossed into atg-4.1(bp501); atg-4.2(tm3948) mutants. The lethal phenotype of atg-4.1; atg-4.2 double mutants could not be rescued by this transgene. However, accumulation of SEPA-1 aggregates was significantly reduced in atg-4.1(bp501); atg-4.2(tm3948); Plgg-1::LGG-1(G116 end) embryos compared with that in atg-4.1; atg-4.2 mutants (Fig. 5, E–H), indicating that the defect is partially rescued.
Epistasis Analysis between atg-4.1 and Other Autophagy Mutants
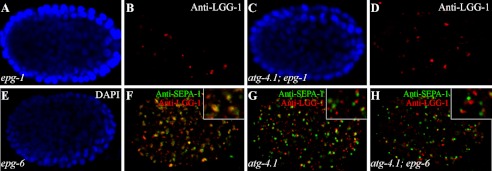
By analyzing a series of genetic phenotypes, including the formation, morphology and distribution of LGG-1 puncta and SEPA-1 aggregates, autophagy genes have been placed in a hierarchical order in the aggrephagy pathway in C. elegans (31, 32). We thus performed epistasis analysis between atg-4.1 and other autophagy mutants. In epg-1/Atg13 mutants, LGG-1 forms large aggregates in a few cells but is absent in most embryonic cells (Fig. 6, A and B) (31, 33). We constructed atg-4.1; epg-1 double mutants and found that LGG-1 accumulated into large puncta, resembling epg-1 single mutants (Fig. 6, C and D). We also examined phenotypes in atg-4.1; epg-6 double mutants. epg-6 is involved in the progression of omegasomes to autophagosomes (32). In epg-6 mutants, SEPA-1 aggregates and LGG-1 puncta accumulate and form clusters and are largely colocalized (Fig. 6, E and F, supplemental Fig. S4). In atg-4.1 mutants, SEPA-1 aggregates and LGG-1 puncta are small and spherical shaped and do not colocalize (Fig. 6G, supplemental Fig. S4). In atg-4.1; epg-6 double mutants, SEPA-1 formed small, spherical aggregates that were separable from LGG-1 puncta, resembling those in atg-4.1 mutants (Fig. 6H, supplemental Fig. S4). These results indicate that atg-4.1 acts upstream of epg-6 in the autophagic pathway by which PGL granules are degraded.
FIGURE 6.
Genetic interactions of atg-4.1 with other autophagy mutants. A–D, in epg-1/Atg13 mutants, LGG-1 forms large puncta in a few cells but is absent in most embryonic cells (B). The morphology and distribution of LGG-1 puncta in atg-4.1; epg-1 double mutants (D) resemble those in epg-1 single mutants. A and C, DAPI images of the embryos shown in B and D, respectively. E–H, SEPA-1 aggregates and LGG-1 puncta form clusters and are largely colocalized in epg-6 mutants. In atg-4.1; epg-6 double mutants, SEPA-1 aggregates are small and are separable from LGG-1 puncta (H), resembling those in atg-4.1 single mutants. E, DAPI image of the embryo in F. Separate images for Fig. 6, F–H are shown in supplemental Fig. S3.
DISCUSSION
The physiological function of the four mammalian homologues of the single yeast Atg4 protein remains poorly understood. The multiple homologues might act redundantly, function in a tissue-specific fashion, or be expressed at different developmental stages; alternatively they may have evolved to accomplish other functions distinct from autophagy. The substrate preferences of four mammalian Atg4s toward different Atg8s have been characterized in vitro (11, 13, 14, 17). HsAtg4B cleaves all Atg8s (13–15). HsAtg4A efficiently cleaves GATE-16, but not other Atg8s, while HsAtg4C and HsAtg4D both have minimal activity (11, 17). HsAtg4B is likely to be the principle Atg4 homologue in processing and deconjugation of Atg8s (15, 28). Targeted deletion of Atg3, Atg5, Atg7, and Atg16L in mice causes lethal phenotypes during the neonatal starvation period (21–24). Tissue-specific knock-out of Atg5 and Atg7 has been shown to lead to neurodegeneration, hepatomegaly, and many other defects (22, 23). Atg4C- and Atg4B- deficient mice exhibit largely normal development, indicating that Atg4s act redundantly in processing and deconjugation of Atg8s (18–20). Although HsAtg4D is a poor processing and deconjugation enzyme, truncation of the N-terminal 63 amino acids by caspase-3 increases its activity against GABARAP-L1, thus linking mitochondrial dysfunction with apoptosis (34).
C. elegans has two Atg4 orthologues, atg-4.1 and atg-4.2. ATG-4.1 is closer to HsAtg4B and HsAtg4A, while ATG-4.2 is more related to HsAtg4C and HsAtg4B. Here we showed that mutations in atg-4.1, but not atg-4.2, cause defects in degradation of various autophagy substrates. LGG-1 precursors are absent in wild type and atg-4.2 embryos but dramatically accumulate in atg-4.1 mutants. In vitro cleavage assays showed that ATG-4.1 has ∼100-fold higher LGG-1 processing activity than ATG-4.2. The LGG-1 puncta formed by the conjugated form of LGG-1 are detected in atg-4.1 and atg-4.2 single mutants, but are absent in the atg-4.1; atg-4.2 double mutants. In atg-4.1 single mutants, the lipidated form of LGG-1 (LGG-1-II) accumulates at a level comparable to that in wild type embryos, indicating that ATG-4.2 processes LGG-1 precursors in vivo. However, in atg-4.1 mutants, unlipidated LGG-1 (LGG-1-I) is present at very low levels, suggesting that ATG-4.2 possesses a very weak deconjugating activity toward LGG-1-PE, or that ATG-4.2 is sequestrated by nascent LGG-1 precursors because it processes LGG-1 inefficiently. The normal lipidated LGG-1 level in atg-4.1 mutants could also be resulted from the accumulation of nonfunctional autophagosomes that fail to mature, subsequently affecting the LGG-1 turnover. We consistently detected more LGG-1-II in atg-4.2 mutants. One explanation for this could that in wild type embryos, LGG-1-I is bound by ATG-4.2 and is unavailable for lipidation.
Both the processing and deconjugating activities of Atg4 are required for normal progression of autophagy in yeast (4). A recent study showed that yeast Atg4 also recycles Atg8-PE produced on inappropriate membranes to maintain a cytoplasmic pool of unlipidated Atg8 that can undergo lipidation and participate in autophagosome formation at the pre-autophagosomal structure (PAS) (35). The mutant form of LGG-1, in which the conjugated glycine residue is directly exposed, rescues defective degradation of protein aggregates in atg-4.1 mutants. atg-4.1 mutant animals expressing this mutant form of LGG-1 show no significant changes in levels of LGG-1-II, but accumulate unlipidated LGG-1, which is absent in atg-4.1 mutants, implying that maintenance of an unlipidated pool of LGG-1 is essential for autophagy flux. The LGG-1(G116 end) transgene also rescues, although not completely, defective degradation of protein aggregates in atg-4.1; atg-4.2 double mutants. In Δatg4Δatg8 yeast cells, expressing Atg8G116 also partially suppresses the defect in autophagy (35). Thus, the deconjugation activity is not completely required for autophagic flux. Our results demonstrate that ATG-4.1 and ATG-4.2 act redundantly, but ATG-4.1 plays a principle role in processing and deconjugation of LGG-1 in the autophagy pathway.
Supplementary Material
Acknowledgments
We thank members of Dr. Zhang's laboratory for helpful comments on the manuscript and Dr. Isabel Hanson for editing work. We thank Dr. Shohei Matani for the atg-4.2(tm3948) allele.
This work was supported by the National Basic Research Program of China (2011CB910100, 2010CB835201) (to H. Z.) and Grant-in-Aid for Young Scientists (A) (23687012) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to N. N. N).

This article contains supplemental Figs. S1–S4.
- ATG
- autophagy-related
- PE
- phosphatidylethanolamine
- CBB
- Coomassie Brilliant Blue
- PAS
- pre-autophagosomal structure.
REFERENCES
- 1. Xie Z., Klionsky D. J. (2007) Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 2. Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 [DOI] [PubMed] [Google Scholar]
- 3. Ohsumi Y. (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2, 211–216 [DOI] [PubMed] [Google Scholar]
- 4. Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., Ohsumi M., Takao T., Noda T., Ohsumi Y. (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yoshimori T., Noda T. (2008) Toward unraveling membrane biogenesis in mammalian autophagy. Curr. Opin. Cell Biol. 20, 401–407 [DOI] [PubMed] [Google Scholar]
- 6. Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. (1999) Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Geng J., Klionsky D. J. (2008) The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep. 9, 859–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ichimura Y., Imamura Y., Emoto K., Umeda M., Noda T., Ohsumi Y. (2004) In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J. Biol. Chem. 279, 40584–40592 [DOI] [PubMed] [Google Scholar]
- 9. Nakatogawa H., Ichimura Y., Ohsumi Y. (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178 [DOI] [PubMed] [Google Scholar]
- 10. Levine B., Klionsky D. J. (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463–477 [DOI] [PubMed] [Google Scholar]
- 11. Scherz-Shouval R., Sagiv Y., Shorer H., Elazar Z. (2003) The COOH terminus of GATE-16, an intra-Golgi transport modulator, is cleaved by the human cysteine protease HsApg4A. J. Biol. Chem. 278, 14053–14058 [DOI] [PubMed] [Google Scholar]
- 12. Mariño G., Uría J. A., Puente X. S., Quesada V., Bordallo J., López-Otín C. (2003) Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J. Biol. Chem. 278, 3671–3678 [DOI] [PubMed] [Google Scholar]
- 13. Hemelaar J., Lelyveld V. S., Kessler B. M., Ploegh H. L. (2003) A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J. Biol. Chem. 278, 51841–51850 [DOI] [PubMed] [Google Scholar]
- 14. Tanida I., Sou Y. S., Ezaki J., Minematsu-Ikeguchi N., Ueno T., Kominami E. (2004) HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J. Biol. Chem. 279, 36268–36276 [DOI] [PubMed] [Google Scholar]
- 15. Kabeya Y., Mizushima N., Yamamoto A., Oshitani-Okamoto S., Ohsumi Y., Yoshimori T. (2004) LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117, 2805–2812 [DOI] [PubMed] [Google Scholar]
- 16. Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., Elazar Z. (2010) LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29, 1792–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li M., Hou Y., Wang J., Chen X., Shao Z. M., Yin X. M. (2011) Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J. Biol. Chem. 286, 7327–7338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mariño G., Salvador-Montoliu N., Fueyo A., Knecht E., Mizushima N., López-Otín C. (2007) Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J. Biol. Chem. 282, 18573–18583 [DOI] [PubMed] [Google Scholar]
- 19. Cabrera S., Mariño G., Fernández A. F., López-Otín C. (2010) Autophagy, proteases and the sense of balance. Autophagy 6, 961–963 [DOI] [PubMed] [Google Scholar]
- 20. Mariño G., Fernández A. F., Cabrera S., Lundberg Y. W., Cabanillas R., Rodríguez F., Salvador-Montoliu N., Vega J. A., Germanà A., Fueyo A., Freije J. M., López-Otín C. (2010) Autophagy is essential for mouse sense of balance. J. Clin. Invest. 120, 2331–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sou Y. S., Waguri S., Iwata J., Ueno T., Fujimura T., Hara T., Sawada N., Yamada A., Mizushima N., Uchiyama Y., Kominami E., Tanaka K., Komatsu M. (2008) The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell 19, 4762–4775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 23. Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Kominami E., Tanaka K., Chiba T. (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cadwell K., Liu J. Y., Brown S. L., Miyoshi H., Loh J., Lennerz J. K., Kishi C., Kc W., Carrero J. A., Hunt S., Stone C. D., Brunt E. M., Xavier R. J., Sleckman B. P., Li E., Mizushima N., Stappenbeck T. S., Virgin H. W., 4th. (2008) A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang X., Zhang H., Zhang H. (2011) The zinc-finger protein SEA-2 regulates larval developmental timing and adult lifespan in C. elegans. Development 138, 2059–2068 [DOI] [PubMed] [Google Scholar]
- 26. Kovacs A. L., Zhang H. (2010) Role of autophagy in Caenorhabditis elegans. FEBS Lett. 584, 1335–1341 [DOI] [PubMed] [Google Scholar]
- 27. Zhao Y., Tian E., Zhang H. (2009) Selective autophagic degradation of maternally-loaded germline P granule components in somatic cells during C. elegans embryogenesis. Autophagy 5, 717–719 [DOI] [PubMed] [Google Scholar]
- 28. Zhang Y., Yan L., Zhou Z., Yang P., Tian E., Zhang K., Zhao Y., Li Z., Song B., Han J., Miao L., Zhang H. (2009) SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell 136, 308–321 [DOI] [PubMed] [Google Scholar]
- 29. Kumanomidou T., Mizushima T., Komatsu M., Suzuki A., Tanida I., Sou Y. S., Ueno T., Kominami E., Tanaka K., Yamane T. (2006) The crystal structure of human Atg4b, a processing and de-conjugating enzyme for autophagosome-forming modifiers. J. Mol. Biol. 355, 612–618 [DOI] [PubMed] [Google Scholar]
- 30. Satoo K., Noda N. N., Kumeta H., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. (2009) The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 28, 1341–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tian Y., Li Z., Hu W., Ren H., Tian E., Zhao Y., Lu Q., Huang X., Yang P., Li X., Wang X., Kovács A. L., Yu L., Zhang H. (2010) C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 141, 1042–1055 [DOI] [PubMed] [Google Scholar]
- 32. Lu Q., Yang P., Huang X., Hu W., Guo B., Wu F., Lin L., Kovács A. L., Yu L., Zhang H. (2011) The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev. Cell 21, 343–357 [DOI] [PubMed] [Google Scholar]
- 33. Tian E., Wang F., Han J., Zhang H. (2009) epg-1 functions in autophagy-regulated processes and may encode a highly divergent Atg13 homolog in C. elegans. Autophagy 5, 608–615 [DOI] [PubMed] [Google Scholar]
- 34. Betin V. M., Lane J. D. (2009) Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J. Cell Sci. 122, 2554–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nakatogawa H., Ishii J., Asai E., Ohsumi Y. (2012) Atg4 recycles inappropriately lipidated Atg8 to promote autophagosome biogenesis. Autophagy 8, 177–186 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.