Background: The role of microRNAs in cutaneous squamous cell carcinoma (cSCC) is not well understood.
Results: cSCC has a unique miRNAome. MicroRNA-125b is down-regulated in human cSCC and suppresses growth and motility of cSCC cells through targeting Matrix Metallopeptidase 13.
Conclusion: MicroRNA-125b may play a tumor suppressive role in cSCC.
Significance: This study suggests a role for microRNAs in cSCC pathogenesis.
Keywords: Cancer, Epidermis, Gene Regulation, Matrix Metalloproteinase (MMP), MicroRNA, Tumor Metastases
Abstract
Cutaneous squamous cell carcinoma (cSCC) is the second most common human cancer. Although dysregulation of microRNAs (miRNAs) is known to be involved in a variety of cancers, the role of miRNAs in cSCC is unclear. In this study, we aimed to identify tumor suppressive and oncogenic miRNAs involved in the pathogenesis of cSCC. MiRNA expression profiles in healthy skins (n = 4) and cSCCs (n = 4) were analyzed using MicroRNA Low Density Array. MiR-125b expression was analyzed by quantitative real-time PCR and in situ hybridization in skin biopsies from 40 healthy donors, 13 actinic keratosis, and 74 cSCC patients. The effect of miR-125b was analyzed in wound closure, colony formation, migration, and invasion assays in two cSCC cell lines, UT-SCC-7 and A431. The genes regulated by miR-125b in cSCC were identified by microarray analysis and its direct target was validated by luciferase reporter assay. Comparing cSCC with healthy skin, we identified four up-regulated miRNAs (miR-31, miR-135b, miR-21, and miR-223) and 54 down-regulated miRNAs, including miR-125b, whose function was further examined. We found that miR-125b suppressed proliferation, colony formation, migratory, and invasive capacity of cSCC cells. Matrix metallopeptidase 13 (MMP13) was identified as a direct target suppressed by miR-125b, and there was an inverse relationship between the expression of miR-125b and MMP13 in cSCC. Knockdown of MMP13 expression phenocopied the effects of miR-125b overexpression. These findings provide a novel molecular mechanism by which MMP13 is up-regulated in cSCCs and indicate that miR-125b plays a tumor suppressive role in cSCC.
Introduction
Cutaneous squamous cell carcinoma (cSCC)5 is an epidermal keratinocyte derived skin tumor, which is the second most common human cancer with a yearly incidence of about 250,000 cases in the United States alone, posing a significant threat for the public health (1). The incidence of cSCC has been steadily increasing during the past decades (2). The most important risk factors for cSCC are solar radiation and immunosuppression (3). While most sporadic cSCCs are cured by surgery and/or radiotherapy, cSCCs in individuals on life-long immunosuppressive therapy (e.g. organ transplant recipients) represent a particular clinical problem. These tumor are often multiple and aggressive, with increased recurrences and metastasis (4). Metastatic cSCC often raise therapeutic problems, since chemotherapy is not consistently efficient (5). Although patients with primary cSCC have a favorable prognosis, for those with metastatic disease, the long-term prognosis is extremely poor with a disease-specific survival at 1 year of 44–56% (4). Thus, there is a great need for more effective therapeutic strategies.
MicroRNAs (miRNAs) are a class of small (22∼24 nt), non-coding RNAs that can regulate gene expression at the post-transcriptional level by binding to the 3′-untranslated region (3′-UTR) of target genes and suppressing target gene expression (6). miRNAs are proposed to regulate ∼60% of all protein-coding genes in humans and participate in the regulation of almost every cellular process investigated to date (7). Accordingly, deregulation of miRNA expression has been shown to contribute to a variety of human diseases, including cancer. Tumors frequently overexpress oncogenic miRNAs (such as miR-21, miR-155), while down-regulate tumor suppressive miRNAs (such as miR-16, let-7), which allow their growth and metastasis (8, 9). By regulating the multiple target genes simultaneously, miRNAs may function as the critical control nodes in the existing tumor signaling network, which make them become promising targets for cancer treatment (10).
Herein we identify the miRNA expression profile in cSCC. MiR-125b, one of the top down-regulated miRNAs in cSCC compared with healthy skin, is found to suppress cell proliferation, migration and invasion. Moreover, we identify a novel molecular mechanism, miR-125b-mediated regulation of MMP13, which may account for the over-expression of MMP13 in cSCC. Together our data indicate that miR-125b plays a tumor suppressive role in cSCC.
EXPERIMENTAL PROCEDURES
Clinical Samples
4 mm punch biopsies were taken, after informed consent, from skin of healthy donors (n = 44), actinic keratosis (AK, n = 13), and cSCC patients (n = 23) at the Dermatology and Venerology Unit, Karolinska University Hospital, Stockholm, Sweden and at the Department of Dermatology, Heinrich Heine University, Düsseldorf, Germany. The clinical diagnosis was made by a dermatologist and confirmed by histopathological evaluation. The formalin-fixed, paraffin embedded cSCC biopsies (n = 55) used in Fig. 1C was obtained from Karolinska University Hospital Biobank. The study was approved by the Regional Ethics Committees and conducted according to the Declaration of Helsinki Principles. RNA was extracted from frozen biopsies as described previously (11) or from formalin-fixed, paraffin-embedded tissue sections using miRNeasy FFPE Kit (Qiagen).
FIGURE 1.
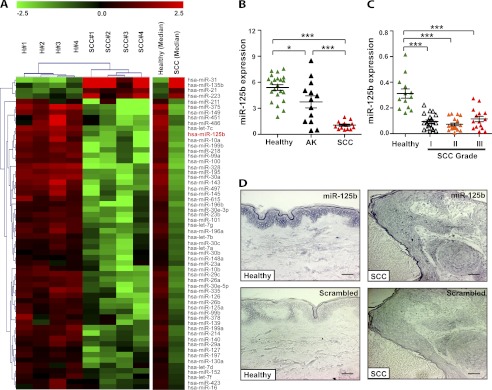
MiR-125b is down-regulated in cSCC. A, unsupervised hierarchical clustering was performed on a subset of 58 genes that were differentially expressed between healthy skin (H) and squamous cell carcinomas (SCC) as determined by SAM analysis. Heatmap colors represent relative miRNA expression. A median expression value equal to 1 was designated black; red, increased expression; green, reduced expression. Note that the color scale is logarithmic. B, MiR-125b expression was analyzed in healthy skin biopsies (n = 21), actinic keratoses (AK, n = 13) and cSCCs (n = 14) using qRT-PCR. *, p < 0.05, ***, p < 0.001, Mann-Whitney test. C, MiR-125b expression was analyzed in healthy skin samples (n = 13), well-differentiated cSCCs (grade I, n = 20), moderately differentiated SCCs (grade II, n = 20), and poorly differentiated cSCCs (grade III, n = 15). ***, p < 0.001, Mann-Whitney test. D, in situ hybridization was performed on healthy skin samples (n = 6) and cSCC sections (n = 5) using miR-125b-specific locked nucleic acid (LNA) probe or scrambled probe. Blue-purple color indicates miR-125b expression. Bar, 50 μm.
Cell Culture and Transfections
UT-SCC-7 cells (a kind gift from Professor Veli-Matti Kähäri, Turku, Finland) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mm 4-(2-hydroxyethyl)-1- piperazineethanesulfonic acid (HEPES), 100 μm MEM Non-Essential Amino Acids Solution, and 100 units/ml penicillin/streptomycin (PEST); A431 cells were cultured in DMEM with 10% FBS and 1% PEST (Invitrogen). All cells were cultured at 37 °C in 5% CO2. Cells were transfected with 10 nm miR-125b miRNA precursor (Pre-miR-125b) or miRNA precursor negative control #1 (Pre-miR-Ctrl) (Ambion) and with 30 nm Silencer® select pre-designed siRNA for MMP13 or siRNA negative control #1 (Ambion) using Lipofectamine 2000 (Invitrogen) following the manufacturers' instruction. Overexpression of the mature, biologically active form of miR-125b was confirmed by real time PCR after transfection (data not shown).
TaqMan MicroRNA Low Density Array
MiRNAs were reverse transcribed and amplified using the multiplex RT TaqMan MicroRNA Low Density Array (TLDA) (Applied Biosystems). Global miRNA profiling for 365 human miRNAs was performed by using the TLDA Human microRNA Panel v1.0 (Applied Biosystems). TLDAs were run on an ABI7900 HT analyzer with TLDA upgrade and analyzed by RQ Manager software (Applied Biosystems). All quality control tests were validated: blanks and reproducibility (standard deviation of cycle threshold (Ct) < 1) of the two small nucleolar house-keeping RNAs, RNU48 (SNORD48) and RNU44 (SNORD44). The amount of RNA from each sample was calibrated to the more stable (between the different arrays) small nucleolar housekeeping RNA, RNU48. To find consistently differentially expressed genes, the data were subjected to significance analysis of microarrays (SAM) as described (12). Genes showing at least 1.7-fold regulation and a q value less than 2.5% were considered to be differentially expressed.
Quantitative Real Time PCR
Quantification of miRNAs by TaqMan® Real-Time PCR was carried out as previously described (11). Target gene expression was normalized between different samples based on the values of U48 small nucleolar RNA expression. Quantification of mRNAs was carried out according to standard procedures (13). Specific primers and probes were purchased from Applied Biosystems. Target gene expression was normalized based on the values of the expression of 18S RNA (18S-F: 5′-CGG CTA CCA CAT CCA AGG AA-3′, 18S-R: 5′-GCT GGA ATT ACC GCG GCT-3′, 18S TaqMan probe: 5′-FAM-TGC TGG CAC CAG ACT TGC CCT C-TAMRA -3′).
In Situ Hybridization
In situ hybridization was performed on formalin-fixed paraffin-embedded sections (10 μm thickness) of skin biopsy specimens as previously described (11). Briefly, after dewaxation, sections were treated with proteinase K (2 μg/ml) at 37 °C for 15 min, washed, and prehybridized for 1 h at 49 °C. Hybridization with digoxygenin (DIG) -labeled miRCURY LNA probes (Exiqon) was performed overnight at 49 °C. Slides were then washed at 49 °C and incubated with alkaline phosphatase-conjugated sheep anti-DIG Fab fragments (1:1500 (Roche)) for 1 h at room temperature. The staining was visualized by adding BM purple AP substrate (Roche) according to the manufacturer's instructions.
Cell Cycle Analysis
EdU was added at a 10 μm final concentration to the transfected cells 2 h before harvesting. Click-iT™ EdU Flow Cytometry Assay (Invitrogen) was carried out according to the manufacturer's instructions and analyzed on a CyAnTM ADP Analyzer (Beckman Coulter) to determine EdU-positive cells and the cell cycle distribution.
Colony Formation Assay
Cells were seeded into 12-well-plate with a density of 200 cells/well 48 h after transfection. Medium was changed every third day. After 8 days colonies containing at least 50 cells formed which were stained with crystal violet (Sigma-Aldrich) and counted.
Scratch-Wound Assay
Transfected cells were grown to confluence and a scratch was made with a sterile 200 μl pipette tip. The cells were kept in medium containing 1% FBS and photographed at different time points until the scratch was closed. Migration rate at each time point equals to (area of the wound at 0 h − area of the wound at this time point)/area of the wound at 0 h.
Transwell Migration and Invasion Assay
Transwell migration and invasion assay was performed using the BD BioCoat Matrigel Invasion Chamber (BD Bioscience). Briefly, 2.5 × 104 transfected cells in FBS-free medium were placed into the upper chamber of the insert with bare polyethylene terephthalate (PET) base membrane (for migration assay) or coated with Matrigel (for invasion assay). Medium containing 10% FBS was added into the lower chamber. After 24 or 48 h, the cells remaining on the upper membrane were removed by cotton buds, whereas the cells migrating or invading through the membrane were stained with crystal violet and counted under microscope.
Gene Expression Microarray
Expression profiling of UT-SCC-7 cells transfected with 10 nm Pre-miR-125b or Pre-miR-Ctrl for 48 h (in triplicates) was performed using Affymetrix Genechip system at the Microarray core facility of Karolinska Institute. In brief, total RNA was extracted using the miRNeasy Mini Kit (Qiagen) and RNA quality and quantity were determined using Agilent 2100 Bioanalyzer and Nanodrop ND-1000. 100 ng of total RNA were used to prepare cDNA following the Affymetrix 3′IVT Express Kit labeling protocol. Standardized array processing procedures recommended by Affymetrix included hybridization, fluidics processing, and scanning were used. The functional annotations of resulting gene lists were performed using the NIH web-based tool DAVID (Database for Annotation, Visualization and Integrated Discovery) (14). Genes showing at least 1.2-fold regulation and a p value less than 0.05 were considered to be differentially expressed.
3′-UTR Luciferase Reporter Assays
Firefly luciferase reporter plasmids containing 3′-UTR of the MMP13 gene and empty luciferase vector were obtained from SwitchGear Genomics. The mutations were generated with the predicted target site of MMP13 3′-UTR using the QuickChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. UT-SCC-7 cells were co-transfected with the luciferase reporters (50 ng per well) together with 10 nm Pre-miR-125b or Pre-miR-Ctrl using Lipofectamine 2000 (Invitrogen). Luciferase activity was analyzed 24 h post-transfection using Dual Luciferase® Reporter Assay System (Promega) following the manufacturer's instructions.
Western Blot Analysis
MMP13 protein expression was analyzed by Western blotting with mouse anti-human MMP13 antibody (1:300) (Calbiochem). Protein levels were visualized by ECL (GE Healthcare) using horseradish peroxidise (HRP)-conjugated anti-mouse antibody (1:2000) (DakoCytomation). As loading controls, the blots were re-probed using a mouse anti-human β-actin antibody (1:5000) (Abcam) and secondary HRP-conjugated anti-mouse antibody (1:2000) (DakoCytomation).
Statistics
Statistical significance for experiments was determined by Mann-Whitney U Test or Student's t test. Correlation between the expressions of different genes in the same samples was made using Pearson's correlation test on log-transformed data. p values < 0.05 were considered to be statistically significant.
RESULTS
A Distinct miRNAome in Cutaneous Squamous Cell Carcinoma
To identify miRNAs deregulated in cSCC, we compared the expression of 365 miRNAs in cSCC (n = 4) and healthy skin (n = 4) by TaqMan MicroRNA Low Density Array (TLDA) (Fig. 1A and supplemental Tables S1-S2). Analysis of the array data using the Significance Analysis of Microarrays (SAM) algorithm led to the identification of 58 miRNAs that were significantly differentially expressed in cSCC relative to healthy skin. Interestingly, many of these deregulated miRNAs could be classified into several gene families sharing 5′ seed sequences (the highly conserved 7- or 8-mer sequence within a miRNA that establishes target specificity) (supplemental Table S2). We found that different members of the deregulated miRNA gene families tended to be co-expressed in cSCC even when they are located on different chromosomal sites, which indicates that dysregulation of miRNA expression in cSCC occurs in a regulated manner. Unsupervised hierarchical clustering based on miRNA expression clearly separated cSCC tumor samples from healthy skin. Four miRNAs were found to be up-regulated in cSCC, that is, miR-31, miR-135b, miR-21, and miR-223. The majority of miRNAs with significantly changed expression (54 out of 58) were down-regulated in cSCC, such as miR-375, miR-125a/b family, let-7a/b/c/d/f/g family, miR-99a/b/100 family, miR-143, and miR-101, many of which have been implicated in various cancers.
One of the top down-regulated miRNAs was miR-125b, a miRNA previously shown to be deregulated in multiple cancers (15–19) as well as in psoriasis lesions, a benign skin disease characterized by hyperproliferation of keratinocytes (11). To validate our array results and to explore the expression of miR-125b in pre-cancerous skin lesions (actinic keratosis), we performed quantitative real-time PCR (qRT-PCR) on a set of RNA isolated from 21 healthy skin biopsies, 13 actinic keratosis, and 14 cSCCs (Fig. 1B). Interestingly, the results showed that miR-125b expression was slightly, but significantly decreased in actinic keratosis (1.4-fold down-regulation, p = 0.03) and decreased further in cSCC (5.0-fold down-regulation, p = 9.0 × 10−12) compared with healthy skin. In addition, we examined miR-125b expression in an independent set of samples, including 20 well differentiated cSCCs (grade I), 20 moderately differentiated cSCCs (grade II), 15 poorly differentiated cSCCs (grade III) and 13 healthy skin biopsies by qRT-PCR (Fig. 1C). MiR-125b was significantly down-regulated in Grade I cSCCs (3.3-fold down-regulation, p = 4.3 × 10−5) and its level was comparable in Grade II (4.3-fold down-regulation, p = 1.7 × 10−5) and Grade III (2.7-fold down-regulation, p = 4.5 × 10−5) cSCCs. These results indicate that loss of miR-125b expression may be an early step during the pathogenesis of cSCC. To examine whether the loss of miR-125b in cSCC is due to changes in a small subset of tumor cells, we performed in situ hybridization in healthy skin (n = 6) and cSCC (n = 5) specimens. The results showed that miR-125b was expressed throughout the epidermis in healthy skin but absent in cSCC cells (Fig. 1D).
MiR-125b Suppresses Proliferation and Colony Formation Ability of cSCC
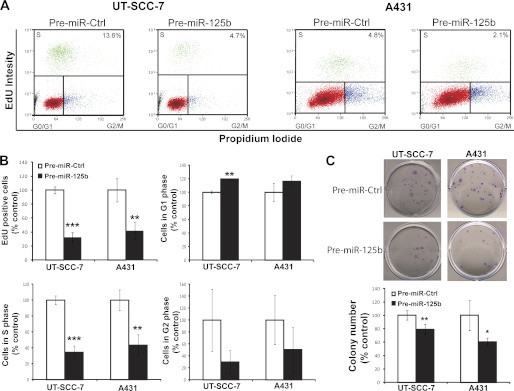
To determine whether miR-125b affects the proliferative capacity of cSCC cells, we performed cell cycle progression assay in two human SCC cell lines, UT-SCC-7 (20), and A431. MiR-125b significantly decreased the percentage of cells that underwent cell division in UT-SCC-7 cells (3-fold reduction, p = 0.0003) and A431 cells (2.4-fold reduction, p = 0.01) when compared with cells over-expressing scrambled miRNA (Fig. 2, A and B). Further analysis revealed that miR-125b increased the percentage of SCC cells arrested in G1 phase and decreased the percentage of cells in S phase, indicating that miR-125b inhibits G1/S-phase transition (Fig. 2B).
FIGURE 2.
Overexpression of miR-125b suppresses the growth and colony formation ability of cSCC cells. UT-SCC-7 cells and A431 cells were transfected with miR-125b precursor RNAs (Pre-miR-125b) or miRNA precursor control (Pre-miR-Ctrl). Cell proliferation and cell cycle progression were measured 72 h post-transfection by EdU Alexa Fluor 647 labeling and subsequent cell cycle analysis by flow cytometry. A, in each representative flow cytometry plot, the percentage of EdU+ cells (depicted as green dots) is listed in the up-right corner. B, bars depict mean ± S.D. of triplicates in one representative experiment out of three independent experiments. C, colonies formed by the transfected cells were stained with crystal violet and counted 10 days after transfection. A representative experiment performed in quintuplicate is shown and this experiment was repeated three times. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Student's t test.
Next, we performed colony formation assay using both UT-SCC-7 and A431 cells (Fig. 2C). MiR-125b significantly reduced the number of colonies formed by UT-SCC-7 cells (1.3-fold reduction, p = 0.002) and A431 cells (1.7-fold reduction, p = 0.015) compared with cells transfected with scrambled controls. These data indicate that loss of miR-125b in cSCC may, at least partially, contribute to tumor growth.
MiR-125b Suppresses Migration and Invasion of SCC Cells
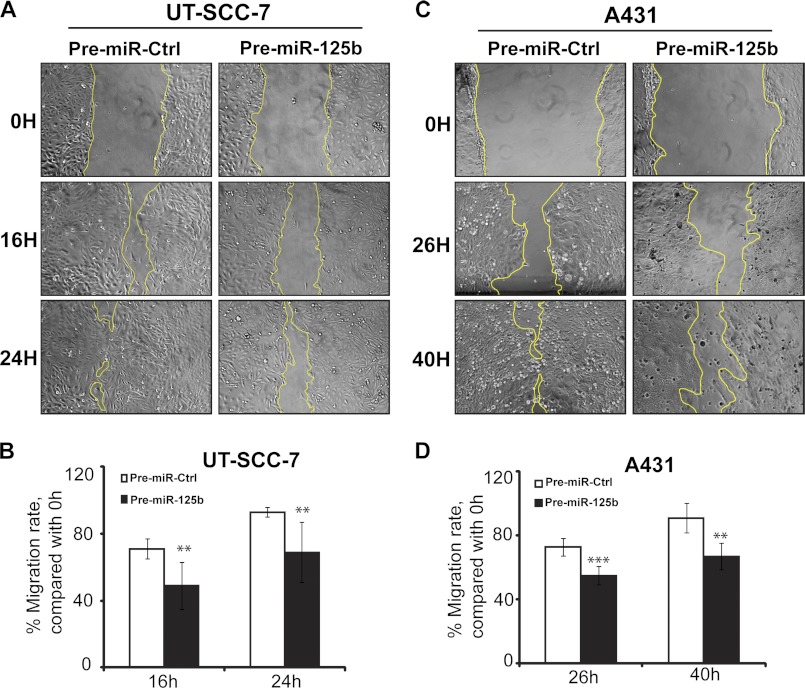
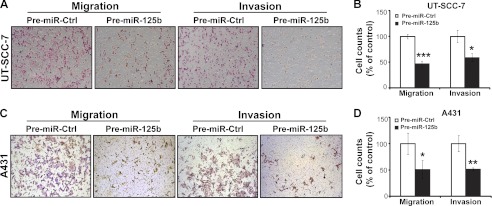
Migration and invasion are two key elements of tumor metastasis. As shown in Fig. 3, miR-125b suppressed cell migration in both UT-SCC-7 (1.4-fold reduction (p = 0.008) at 16 h; 1.3-fold reduction (p = 0.0098) at 24 h) and A431 cells (1.3-fold reduction (p = 0.0003) at 26 h; 1.4-fold reduction (p = 0.0014) at 40 h), as measured by scratch-wound assay. In line with this, miR-125b suppressed transwell cell migration of both UT-SCC-7 (2.1-fold reduction, p = 0.003) and A431 cells (2.0-fold reduction, p = 0.038) significantly (Fig. 4). Moreover, using transwell cell invasion assay, we showed that miR-125b decreased the number of UT-SCC-7 (1.7-fold reduction, p = 0.012) and A431 cells (2.0-fold reduction, p = 0.005) invading through the Matrigel basement membrane (Fig. 4). Taken together, our data suggest that miR-125b can suppress the mobility and invasive capacity of cSCC cells.
FIGURE 3.
MiR-125b suppresses the mobility of cSCC cells. Scratch-wound assay was performed to assess the migration rate of UT-SCC-7 (A) and A431cells (C) transfected with Pre-miR-125b or Pre-miR-Ctrl. Photographs were taken at indicated time points after scratch injury. The migration rates of UT-SCC-7 (B) and A431 (D) were quantified by measuring the area of the injured region. Data of one representative experiment performed with six replicates are shown and this experiment was repeated three times. **, p < 0.01, ***, p < 0.001. Student's t test.
FIGURE 4.
MiR-125b inhibits transwell migration and invasion of cSCC cells. Representative photomicrographs of transwell results for UT-SCC-7 (A) and A431 cells (C) transfected with Pre-miR-125b or Pre-miR-Ctrl were taken under ×100 original magnification. The number of UT-SCC-7 (B) and A431 cells (D) passing through the PET membrane (migration) and Matrigel (invasion) were counted. Data of one representative experiment out of three independent experiments are shown. *, p < 0.05, **, p < 0.01, ***, p < 0.001. Student's t test.
Identification of the Gene Network Regulated by miR-125b in cSCC Cells
Our data strongly suggest that miR-125b has tumor suppressing properties in the context of cSCC. To determine the mechanism of miR-125b's action in cSCC, we aimed to identify the genes regulated by this miRNA in cSCC cells. To this end, we performed a global transcriptome analysis of UT-SCC-7 cells upon over-expression of miR-125b using Affymetrix arrays and identified 250 genes whose levels were significantly regulated by miR-125b in cSCC cells (supplemental Table S3). Notably, according to DIANA-mirExTra algorithm (21), miR-125b seed sequence is the most overrepresented, conserved hexamer motif among the seeds of all the miRNAs found in the 3′-UTRs of the down-regulated genes (supplemental Fig. S1), indicating that the microarray analysis is specific and sensitive for detecting miR-125b direct target genes.
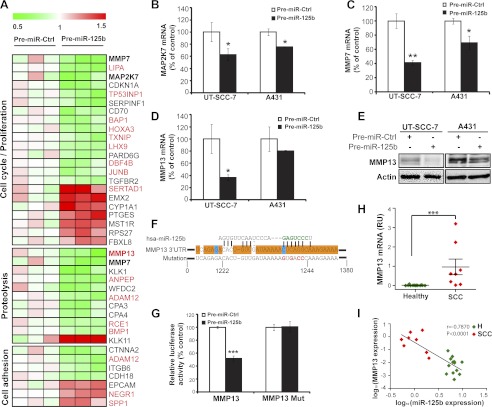
Gene ontology analysis identified several groups of genes involved in biological processes important for tumor growth and metastasis, such as cell proliferation, proteolysis and cell adhesion (Fig. 5A). Interestingly, two matrix metallopeptidases, MMP7 and MMP13 as well as mitogen-activated protein kinase kinase 7 (MAP2K7), genes with known functions in cSCC (22–24), were also down-regulated by miR-125b. To validate the microarray results, we performed qRT-PCR on an independent set of RNA samples isolated from miR-125b overexpressing cSCC cells (Fig. 5, B--D). All three genes were confirmed to be down-regulated by miR-125b in both UT-SCC-7 and A431 cells, which may partially explain the suppressive effects of miR-125b on proliferation, migration, and invasion of cSCC cells.
FIGURE 5.
Identification of genes regulated by miR-125b in cSCC cells. A, microarray analysis performed in independent biological triplicates for UT-SCC-7 cells transfected with either Pre-miR-125b or Pre-miR-Ctrl. Heat map showing the relative expression of miR-125b regulated genes involved in proliferation, proteolysis and cell adhesion. Putative miR-125b targets predicted by miRWalk are labeled in red. B, miR-125b mediated regulation for MAP2K7 (B), MMP7 (C), and MMP13 (D) was verified in both cSCC cell lines by qRT-PCR. *, p < 0.05, **, p < 0.01. Student's t test. E, cSCC cells were transfected with Pre-miR-125b or Pre-miR-Ctrl for 48 h and MMP13 protein was detected by Western blotting. β-actin was detected on the same blot as loading control. F, nucleotide resolution of the predicted miR-125b target site in 3′-UTR of MMP13 mRNA: the seed sequence (green letters); the evolutionarily conserved regions in at least 12 species (orange boxes) or in 6 species (blue boxes); the mutant miR-125b binding sites (red letters). G, UT-SCC-7 cells were transfected with luciferase reporter construct containing MMP13 3′UTR together with Pre-miR-125b or Pre-miR-Ctrl. ***, p < 0.001. Student's t test. H, expression of MMP13 mRNA was analyzed in healthy skin (n = 13) and cSCC biopsies (n = 8) using qRT-PCR. ***, p < 0.001, Mann-Whitney test. I, correlation of miR-125b with MMP13 expression in cSCC and healthy skin. Spearman-Correlation on log-transformed values.
MMP13 Is a Direct Target Gene Down-regulated by miR-125b in cSCC
Using target prediction algorithm miRWalk (25), we found that MMP13 contains an evolutionarily conserved putative binding site of miR-125b in the 3′-UTR (Fig. 5F). To determine whether MMP13 is a bona fide target of miR-125b, we performed 3′-UTR luciferase reporter assays with the MMP13–3′UTR-luciferase reporter gene constructs in UT-SCC-7 cells (Fig. 5G). Overexpression of miR-125b suppressed MMP13–3′-UTR-luciferase activity by 48% (p = 0.0005). Mutation of four nucleotides within the seed-matching sequence of the predicted miR-125b binding site (Fig. 5F) abolished the inhibitory effect of miR-125b on luciferase activity (Fig. 5G). These results demonstrated that miR-125b directly regulates MMP13 expression by binding to the 3′-UTR of MMP13 mRNA. In accordance with this result, overexpression of miR-125b in UT-SCC-7 and A431 cells suppressed the expression of MMP13 at both mRNA (Fig. 5D) and protein levels (Fig. 5E), as shown by qRT-PCR and Western blotting respectively. MMP13 expression was significantly higher in cSCC compared with healthy skin (Fig. 5H), which is in line with previous reports (26). Furthermore, we found an inverse correlation between the expression levels of miR-125b and MMP13 mRNA (r = −0.7870, p < 0.0001) (Fig. 5I), supporting the regulation of MMP13 by miR-125b in cSCC tumors in vivo.
Knockdown of MMP13 Expression Phenocopies the Effects of miR-125b Overexpression in cSCC Cells
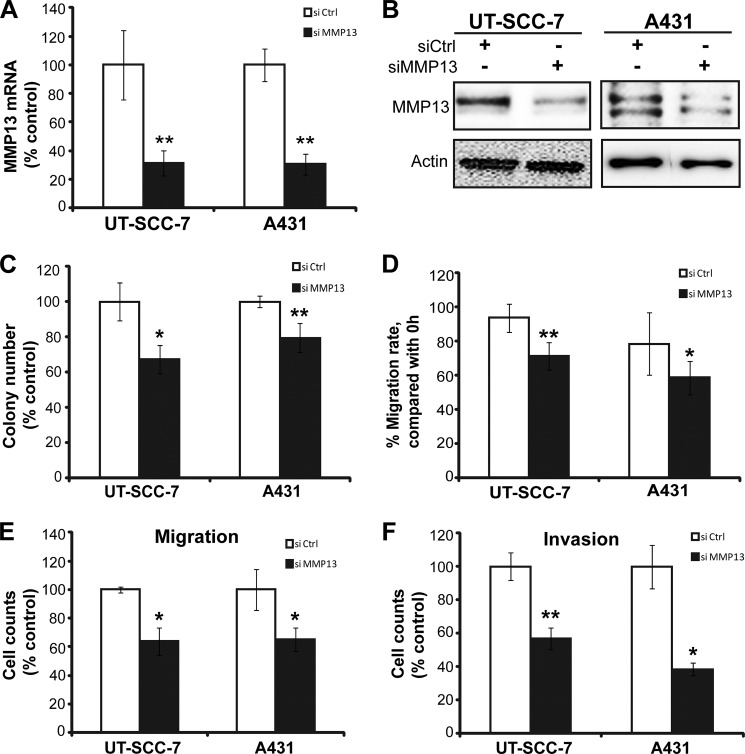
To determine whether regulation of MMP13 contributes to the actions of miR-125b in cSCC, we silenced the expression of MMP13 using siRNA and studied its effect on growth, migration, and invasion of cSCC cells (Fig. 6). The efficacy of knockdown in UT-SCC-7 and A431 cells was confirmed at both mRNA (Fig. 6A) and protein levels (Fig. 6B). Knockdown of MMP13 expression suppressed the cell growth (colony formation assay in Fig. 6C), migration (scratch wound assay in Fig. 6D and transwell migration assay in Fig. 6E), and invasion (Matrigel transwell invasion assay in Fig. 6F) in both cell lines. Thus, knockdown of MMP13 can phenocopy the effects of overexpression of miR-125b in cSCC cells, indicating that MMP13 is a key mediator of the tumor suppressive effects of miR-125b in cSCC.
FIGURE 6.
Knockdown of MMP13 suppresses growth, migration, and invasion of cSCC cells. UT-SCC-7 and A431 cells were transfected with siRNA for MMP13 (si MMP13) or siRNA-negative control (si Ctrl). MMP13 mRNA and protein were analyzed by qRT-PCR (A) and Western blotting (B). Colony formation assay (C), scratch-wound assay (D), transwell migration (E), and invasion assay (F) were performed with transfected cells. *, p < 0.05, **, p < 0.01. Student's t test.
DISCUSSION
In this study, we described for the first time the miRNAome of one of the most common human malignancies, cSCC. The miRNA expression profile of cSCC shares similarities with that of head-and-neck SCC (HNSCC), including the up-regulation of miR-31, miR-21 and miR-223 and down-regulation of miR-99a/100, miR-375 and miR-125b (27). The oncogenic activity of some miRNAs up-regulated in cSCC has been previously reported: miR-21 is an oncomiR due to its overexpression and oncogenic activity in the vast majority of cancer types (reviewed in Ref. 28), including cSCC (29); miR-31 increases the oncogenic potential of HNSCC both in vitro and in vivo (30); miR-223 is up-regulated and promotes tumor progression in esophageal SCC (31). Moreover, tumor suppressive activities were reported for down-regulated miRNAs in cSCC, for example, let-7 family members in many tumor types (reviewed in Ref. 32); miR-375 (33), miR-99a/b/100 family members (27), miR-143 (34), and miR-101 in HNSCC (35).
Here we show that miR-125b, one of the top down-regulated miRNAs in cSCC, suppresses proliferation, migration and invasion of cSCC cells. Although miR-125b has been implicated in various cancers, its role in diseases is not completely clear. Because in some cell types it seemingly plays oncogenic role, while in others tumor suppressive role. For example, miR-125b expression is up-regulated in prostate cancer, and it stimulates androgen-independent growth of prostate cells (18). In contrast, miR-125b is down-regulated in breast cancer (19), oral SCC (15), osteosarcoma (17), and bladder cancer (16) and suppresses tumor growth in vitro and in vivo (15–17, 19). In addition, we previously showed that miR-125b was down-regulated in the keratinocytes of psoriasis, which is an inflammatory skin disease characterized by nonmalignant hyperproliferation of keratinocytes and showed that miR-125b inhibits cell proliferation in human primary keratinocytes (11). The seemingly paradoxical findings with regard to miR-125b in the context of different tumors indicate that the biological function of miR-125b is complex and highly cell-type dependent, which may result from the varied expression context of miR-125b target genes in each tumor. Our results establish miR-125b as a negative regulator of tumor growth in cutaneous SCC.
We found that many of the genes regulated by miR-125b in cSCC cells are involved in the biological processes important for tumor growth and metastasis. MMP7 and MMP13 belong to the Matrix Metalloproteinase family, which play important roles in extracellular matrix turnover, cancer cell migration, cell growth, inflammation, and angiogenesis (reviewed in Ref. 36). Both MMP7 and MMP13 are highly expressed in cSCC (26, 37). Knockdown of MMP13 in cSCC cells resulted in impaired growth, migration and invasion of cSCC (Fig. 6) in line with earlier observations obtained with ribozyme-mediated inhibition of MMP13 expression (22). We showed that miR-125b suppressed MAP2K7, a major upstream activator of c-Jun-NH2 kinase (JNK). MAP2K7 is activated in the majority of cSCCs and its inhibition abolished invasive human epidermal neoplasia, indicating its oncogenic potency (24). Thus, the down-regulation of multiple pro-tumorigenic genes, MMP7, MMP13, and MAP2K7, in cSCC by miR-125b may explain its suppressive effects on proliferation, migration, and invasion of cSCC cells (Fig. 7).
FIGURE 7.
A simplified model of the interactions of miR-125b and its target genes in the context of cutaneous SCC. The down-regulation of miR-125b contributes to the increased expression of MMP13 (a direct target of miR-125b), MMP7 and MAP2K7 in cSCC cells, which promotes proliferation, migration, and invasion of cSCC cells.
The regulatory mechanisms of MMP13 expression have been well studied in cancer. Our result showing that miR-125b can down-regulate MMP13 expression through binding to its 3′-UTR of mRNA in cSCC cells adds a new layer to the existing regulatory network of MMP13 expression. This novel regulation mechanism may also contribute to the up-regulation of MMP13 expression in other cancer types with decreased miR-125b level, such as breast cancer (38), oral SCC (39), bladder cancer (40), and osteosarcoma (41). One of the factors that have been implicated in the regulation of MMP13 in cSCC is transforming growth factor-β (TGF-β) (42). Interestingly, our gene expression profiling showed that miR-125b decreased the expression of TGF-β receptor II (TGFBR2). Down-regulation of TGFBR2 by miR-125b may damp the activation effect of TGF-β on MMP13 expression, suggesting that miR-125b can regulate MMP13 both by directly binding to the 3′-UTR of MMP13 mRNA and regulating the TGF-β pathway. Additionally, MMP13 has recently been reported to be the direct target of miR-143 in osteosarcoma (43). MiR-143 is down-regulated also in cSCC, which suggests that MMP13 may be regulated by multiple miRNAs simultaneously in cSCC.
In conclusion, our study suggests that miR-125b is a negative regulator of tumor growth in cSCC, which is lost at early steps of tumorigenesis. MiR-125b suppresses the growth, migration, and invasion of cSCC cells through the regulation of network of pro-tumorigenic genes, including MMP7, MMP13, MAP2K7, and many others with yet unknown functions in cSCC (Fig. 7). The therapeutic potential of specific targeting of MMP13 has been previously reported (22), which raises the question whether overexpression of miR-125b in cSCC cells could be a therapeutic option in cSCC. MiRNA-based therapy is expected to be more efficient than the traditional single target therapy, since miRNAs regulate multiple target genes simultaneously. Thus, the chance for tumor cells developing resistance by accumulating mutations is less. Investigation of miRNA-mRNA gene networks in cSCC will increase our understanding about the molecular mechanisms of this common cancer and may lead to the development of miRNA-based treatments for cutaneous SCC.
Supplementary Material
Acknowledgments
We thank Anna-Lena Kastman for excellent technical support, Mari-Anne Hedblad for assistance of clinical sample collection, and Professor Veli-Matti Kähäri for the fruitful discussions and suggestions. We also would like to thank the Microarray core facility at Novum, BEA, Bioinformatics and Expression Analysis, which is supported by the board of research at Karolinska Institute and the research committee at the Karolinska hospital.
This work was supported by the Swedish Research Council (K2012-67X-22042-01-3), Karolinska Institutet, the Welander Finsens Foundations, Tore Nilsons Foundation, Konsul ThC Bergh Foundations, and the Stockholm County Council.

This article contains supplemental Table S1–S3 and Fig. S1.
- cSCC
- cutaneous squamous cell carcinoma
- miRNA and miR
- microRNA
- 3′-UTR
- 3′-untranslated region.
REFERENCES
- 1. Ratushny V., Gober M. D., Hick R., Ridky T. W., Seykora J. T. (2012) From keratinocyte to cancer: the pathogenesis and modeling of cutaneous squamous cell carcinoma. J. Clin. Invest. 122, 464–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hussain S. K., Sundquist J., Hemminki K. (2010) Incidence trends of squamous cell and rare skin cancers in the Swedish national cancer registry point to calendar year and age-dependent increases. J. Invest. Dermatol. 130, 1323–1328 [DOI] [PubMed] [Google Scholar]
- 3. Lauth M., Unden A. B., Toftgård R. (2004) Non-melanoma skin cancer: pathogenesis and mechanisms. Drug Discovery Today 1, 267–272 [Google Scholar]
- 4. Wells J. L., Shirai K. (2011) Systemic therapy for squamous cell carcinoma of the skin in organ transplant recipients. Am. J. Clin. Oncol., in press [DOI] [PubMed] [Google Scholar]
- 5. Cranmer L. D., Engelhardt C., Morgan S. S. (2010) Treatment of unresectable and metastatic cutaneous squamous cell carcinoma. Oncologist 15, 1320–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 7. Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., Downing J. R., Jacks T., Horvitz H. R., Golub T. R. (2005) MicroRNA expression profiles classify human cancers. Nature 435, 834–838 [DOI] [PubMed] [Google Scholar]
- 9. Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., Prueitt R. L., Yanaihara N., Lanza G., Scarpa A., Vecchione A., Negrini M., Harris C. C., Croce C. M. (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. U.S.A. 103, 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Valastyan S., Weinberg R. A. (2009) MicroRNAs: Crucial multi-tasking components in the complex circuitry of tumor metastasis. Cell Cycle 8, 3506–3512 [DOI] [PubMed] [Google Scholar]
- 11. Xu N., Brodin P., Wei T., Meisgen F., Eidsmo L., Nagy N., Kemeny L., Ståhle M., Sonkoly E., Pivarcsi A. (2011) MiR-125b, a microRNA downregulated in psoriasis, modulates keratinocyte proliferation by targeting FGFR2. J. Invest. Dermatol. 131, 1521–1529 [DOI] [PubMed] [Google Scholar]
- 12. Sonkoly E., Wei T., Janson P. C., Sääf A., Lundeberg L., Tengvall-Linder M., Norstedt G., Alenius H., Homey B., Scheynius A., Ståhle M., Pivarcsi A. (2007) MicroRNAs: novel regulators involved in the pathogenesis of Psoriasis? PLoS ONE 2, e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pivarcsi A., Müller A., Hippe A., Rieker J., van Lierop A., Steinhoff M., Seeliger S., Kubitza R., Pippirs U., Meller S., Gerber P. A., Liersch R., Buenemann E., Sonkoly E., Wiesner U., Hoffmann T. K., Schneider L., Piekorz R., Enderlein E., Reifenberger J., Rohr U. P., Haas R., Boukamp P., Haase I., Nürnberg B., Ruzicka T., Zlotnik A., Homey B. (2007) Tumor immune escape by the loss of homeostatic chemokine expression. Proc. Natl. Acad. Sci. U.S.A. 104, 19055–19060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 15. Henson B. J., Bhattacharjee S., O'Dee D. M., Feingold E., Gollin S. M. (2009) Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Genes Chromosomes Cancer 48, 569–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang L., Luo J., Cai Q., Pan Q., Zeng H., Guo Z., Dong W., Huang J., Lin T. (2010) MicroRNA-125b suppresses the development of bladder cancer by Targeting E2F3. Int. J. Cancer 128, 1758–1769 [DOI] [PubMed] [Google Scholar]
- 17. Liu L. H., Li H., Li J. P., Zhong H., Zhang H. C., Chen J., Xiao T. (2011) miR-125b suppresses the proliferation and migration of osteosarcoma cells through down-regulation of STAT3. Biochem. Biophys. Res. Commun. 416, 31–38 [DOI] [PubMed] [Google Scholar]
- 18. Shi X. B., Xue L., Yang J., Ma A. H., Zhao J., Xu M., Tepper C. G., Evans C. P., Kung H. J., deVere White R. W. (2007) An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc. Natl. Acad. Sci. U.S.A. 104, 19983–19988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y., Yan L. X., Wu Q. N., Du Z. M., Chen J., Liao D. Z., Huang M. Y., Hou J. H., Wu Q. L., Zeng M. S., Huang W. L., Zeng Y. X., Shao J. Y. (2011) miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 71, 3552–3562 [DOI] [PubMed] [Google Scholar]
- 20. Johansson N., Airola K., Grénman R., Kariniemi A. L., Saarialho-Kere U., Kähäri V. M. (1997) Expression of collagenase-3 (matrix metalloproteinase-13) in squamous cell carcinomas of the head and neck. Am. J. Pathol. 151, 499–508 [PMC free article] [PubMed] [Google Scholar]
- 21. Alexiou P., Maragkakis M., Papadopoulos G. L., Simmosis V. A., Zhang L., Hatzigeorgiou A. G. (2010) The DIANA-mirExTra web server: from gene expression data to microRNA function. PLoS One 5, e9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ala-aho R., Ahonen M., George S. J., Heikkilä J., Grénman R., Kallajoki M., Kähäri V. M. (2004) Targeted inhibition of human collagenase-3 (MMP-13) expression inhibits squamous cell carcinoma growth in vivo. Oncogene 23, 5111–5123 [DOI] [PubMed] [Google Scholar]
- 23. Kivisaari A. K., Kallajoki M., Ala-aho R., McGrath J. A., Bauer J. W., Königová R., Medvecz M., Beckert W., Grénman R., Kähäri V. M. (2010) Matrix metalloproteinase-7 activates heparin-binding epidermal growth factor-like growth factor in cutaneous squamous cell carcinoma. Br. J. Dermatol. 163, 726–735 [DOI] [PubMed] [Google Scholar]
- 24. Zhang J. Y., Adams A. E., Ridky T. W., Tao S., Khavari P. A. (2007) Tumor necrosis factor receptor 1/c-Jun-NH2-kinase signaling promotes human neoplasia. Cancer Res. 67, 3827–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dweep H., Sticht C., Pandey P., Gretz N. (2011) miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 44, 839–847 [DOI] [PubMed] [Google Scholar]
- 26. Airola K., Johansson N., Kariniemi A. L., Kähäri V. M., Saarialho-Kere U. K. (1997) Human collagenase-3 is expressed in malignant squamous epithelium of the skin. J. Invest. Dermatol. 109, 225–231 [DOI] [PubMed] [Google Scholar]
- 27. Chen Z., Jin Y., Yu D., Wang A., Mahjabeen I., Wang C., Liu X., Zhou X. (2012) Down-regulation of the microRNA-99 family members in head and neck squamous cell carcinoma. Oral. Oncol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Selcuklu S. D., Donoghue M. T., Spillane C. (2009) miR-21 as a key regulator of oncogenic processes. Biochem. Soc. Trans. 37, 918–925 [DOI] [PubMed] [Google Scholar]
- 29. Darido C., Georgy S. R., Wilanowski T., Dworkin S., Auden A., Zhao Q., Rank G., Srivastava S., Finlay M. J., Papenfuss A. T., Pandolfi P. P., Pearson R. B., Jane S. M. (2011) Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell 20, 635–648 [DOI] [PubMed] [Google Scholar]
- 30. Liu C. J., Tsai M. M., Hung P. S., Kao S. Y., Liu T. Y., Wu K. J., Chiou S. H., Lin S. C., Chang K. W. (2010) miR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Res. 70, 1635–1644 [DOI] [PubMed] [Google Scholar]
- 31. Kurashige J., Watanabe M., Iwatsuki M., Kinoshita K., Saito S., Hiyoshi Y., Kamohara H., Baba Y., Mimori K., Baba H. (2012) Overexpression of microRNA-223 regulates the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. Br. J. Cancer 106, 182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boyerinas B., Park S. M., Hau A., Murmann A. E., Peter M. E. (2010) The role of let-7 in cell differentiation and cancer. Endocr. Relat. Cancer 17, F19–36 [DOI] [PubMed] [Google Scholar]
- 33. Nohata N., Hanazawa T., Kikkawa N., Mutallip M., Sakurai D., Fujimura L., Kawakami K., Chiyomaru T., Yoshino H., Enokida H., Nakagawa M., Okamoto Y., Seki N. (2011) Tumor suppressive microRNA-375 regulates oncogene AEG-1/MTDH in head and neck squamous cell carcinoma (HNSCC). J. Hum. Genet. 56, 595–601 [DOI] [PubMed] [Google Scholar]
- 34. Peschiaroli A., Giacobbe A., Formosa A., Markert E. K., Bongiorno-Borbone L., Levine A. J., Candi E., D'Alessandro A., Zolla L., Finazzi Agro A., Melino G. (2012) miR-143 regulates hexokinase 2 expression in cancer cells. Oncogene [DOI] [PubMed] [Google Scholar]
- 35. Banerjee R., Mani R. S., Russo N., Scanlon C. S., Tsodikov A., Jing X., Cao Q., Palanisamy N., Metwally T., Inglehart R. C., Tomlins S., Bradford C., Carey T., Wolf G., Kalyana-Sundaram S., Chinnaiyan A. M., Varambally S., D'Silva N. J. (2011) The tumor suppressor gene rap1GAP is silenced by miR-101-mediated EZH2 overexpression in invasive squamous cell carcinoma. Oncogene 30, 4339–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kessenbrock K., Plaks V., Werb Z. (2010) Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141, 52–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kivisaari A. K., Kallajoki M., Mirtti T., McGrath J. A., Bauer J. W., Weber F., Königová R., Sawamura D., Sato-Matsumura K. C., Shimizu H., Csikós M., Sinemus K., Beckert W., Kähäri V. M. (2008) Transformation-specific matrix metalloproteinases (MMP)-7 and MMP-13 are expressed by tumour cells in epidermolysis bullosa-associated squamous cell carcinomas. Br. J. Dermatol. 158, 778–785 [DOI] [PubMed] [Google Scholar]
- 38. Chang H. J., Yang M. J., Yang Y. H., Hou M. F., Hsueh E. J., Lin S. R. (2009) MMP13 is potentially a new tumor marker for breast cancer diagnosis. Oncol. Rep. 22, 1119–1127 [DOI] [PubMed] [Google Scholar]
- 39. Impola U., Uitto V. J., Hietanen J., Hakkinen L., Zhang L., Larjava H., Isaka K., Saarialho-Kere U. (2004) Differential expression of matrilysin-1 (MMP-7), 92 kD gelatinase (MMP-9), and metalloelastase (MMP-12) in oral verrucous and squamous cell cancer. J. Pathol. 202, 14–22 [DOI] [PubMed] [Google Scholar]
- 40. Boström P. J., Ravanti L., Reunanen N., Aaltonen V., Söderström K. O., Kähäri V. M., Laato M. (2000) Expression of collagenase-3 (matrix metalloproteinase-13) in transitional-cell carcinoma of the urinary bladder. Int. J. Cancer 88, 417–423 [PubMed] [Google Scholar]
- 41. Tuckermann J. P., Vallon R., Gack S., Grigoriadis A. E., Porte D., Lutz A., Wagner E. F., Schmidt J., Angel P. (2001) Expression of collagenase-3 (MMP-13) in c-fos-induced osteosarcomas and chondrosarcomas is restricted to a subset of cells of the osteo-/chondrogenic lineage. Differentiation 69, 49–57 [DOI] [PubMed] [Google Scholar]
- 42. Johansson N., Ala-aho R., Uitto V., Grénman R., Fusenig N. E., López-Otín C., Káhári V. M. (2000) Expression of collagenase-3 (MMP-13) and collagenase-1 (MMP-1) by transformed keratinocytes is dependent on the activity of p38 mitogen-activated protein kinase. J. Cell Sci. 113, 227–235 [DOI] [PubMed] [Google Scholar]
- 43. Osaki M., Takeshita F., Sugimoto Y., Kosaka N., Yamamoto Y., Yoshioka Y., Kobayashi E., Yamada T., Kawai A., Inoue T., Ito H., Oshimura M., Ochiya T. (2011) MicroRNA-143 regulates human osteosarcoma metastasis by regulating matrix metalloprotease-13 expression. Mol. Ther. 19, 1123–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.