Background: AT-1 is an ER membrane transporter that regulates the influx of acetyl-CoA into the ER lumen.
Results: IRE1/XBP1 controls the induction of autophagy by regulating AT-1 expression levels and Atg9A acetylation.
Conclusion: AT-1 acts downstream of the UPR to control ERAD(II).
Significance: Close regulation of the acetylation status of the ER is essential for cell viability during the UPR.
Keywords: Acetyl Coenzyme A, Endoplasmic Reticulum (ER), ERAD, Membrane Transport, Post-translational Modification, Protein Degradation, Unfolded Protein Response, AT-1, Lysine Acetylation
Abstract
One of the main functions of the unfolded protein response is to ensure disposal of large protein aggregates that accumulate in the lumen of the endoplasmic reticulum (ER) whereas avoiding, at least under nonlethal levels of ER stress, cell death. When tightly controlled, autophagy-dependent ER-associated degradation (ERAD(II)) allows the cell to recover from the transient accumulation of protein aggregates; however, when unchecked, it can be detrimental and cause autophagic cell death/type 2 cell death. Here we show that IRE1/XBP1 controls the induction of autophagy/ERAD(II) during the unfolded protein response by activating the ER membrane transporter SLC33A1/AT-1, which ensures continuous supply of acetyl-CoA into the lumen of the ER. Failure to induce AT-1 leads to widespread autophagic cell death. Mechanistically, the regulation of the autophagic process involves Nϵ-lysine acetylation of Atg9A.
Introduction
One of the essential functions of the endoplasmic reticulum (ER)2 is to ensure that proteins that reside within and transit along the secretory pathway are properly folded (1, 2). Proteins that fail quality control are retained in the ER and directed toward the ER-associated degradation (ERAD). In contrast to incorrectly folded polypeptides, which are typically retro-translocated to the cytosol and sent to the proteasome, large protein aggregates are mostly dealt with by expanding the ER and activating autophagy (3). This process is still part of ERAD, and the terms ERAD(I) and ERAD(II) have also been used to differentiate the proteasome-dependent from the autophagy-dependent process, respectively. When tightly controlled, autophagy can allow the cell to recover from the transient accumulation of protein aggregates; however, when unchecked, it can be detrimental and cause autophagic cell death (also referred to as Type 2 programmed cell death) (4, 5). Therefore, “check points” must be in place to keep autophagy/ERAD(II) under control.
Both ERAD(I) and ERAD(II) are under the control of the unfolded protein response (UPR) signaling pathway, which in mammalian cells consists of three main branches: the inositol-requiring protein-1 (IRE1), the activating transcription factor-6 (ATF6), and the protein kinase RNA (PKR)-like ER kinase (PERK). Together they signal to the nucleus to correct possible imbalances resulting in ER stress (1, 6–8). A recent study has revealed that down-regulation of X-box-binding-1 protein (XBP1), which acts immediately downstream of IRE1, results in uncontrolled autophagy (9, 10), suggesting that IRE1/XBP1 signaling might serve as the regulatory check point for the induction of autophagy during the UPR. However, the downstream targets responsible for such a regulatory function are unknown. Also unknown is the possible involvement, if any, of ATF6 and PERK.
Our group has recently reported that mammalian cells are able to acetylate the Nϵ-lysine residue of nascent membrane proteins in the lumen of the ER (11–14). This event was initially discovered while studying the mechanisms that regulate the levels of β-site amyloid precursor protein (APP)-cleaving enzyme 1 (BACE1), a type I membrane protein that is involved in the pathogenesis of Alzheimer disease (11). However, it is now clear that this process is not limited to BACE1. In fact, other membrane and secreted proteins as well as ER-resident proteins that are involved with synthesis and folding of nascent proteins in the ER lumen are also acetylated (12, 14–16). Therefore, it is likely that the ER acetylation machinery exerts a more global function that is essential for the homeostasis of the organelle.
The import of acetyl-CoA into the ER lumen by the ER membrane transporter SLC33A1/AT-1 is the key step for the intraluminal acetylation of ER-resident and -transiting proteins. In fact, acetyl-CoA serves as the donor of the acetyl group in the reaction of Nϵ-lysine acetylation; as a result, changes in acetyl-CoA influx dramatically affect the acetylation status of the ER (14).
AT-1 appears to be essential for cell viability. In fact, down-regulation of AT-1 in the zebrafish causes embryonic lethality and severe morphological defects as well as defective neuronal outgrowth in the surviving animals (17), whereas down-regulation of AT-1 in cultured cells causes engulfment and enlargement of the ER followed by induction of autophagy and widespread cell death (14). Interestingly, AT-1 is up-regulated during the differentiation of B cells into immunoglobulin-secreting plasma cells, a process that involves expansion of the ER and activation of “nonlethal levels” of ER stress with the purpose of secreting a large quantity of correctly folded immunoglobulins (18). The above should also be viewed together with the fact that a large set of ER-resident proteins involved with synthesis, folding, and disposal of unfolded/misfolded protein intermediates is acetylated in the lumen of the organelle (16). Therefore, it is possible that AT-1 plays an important role in regulating certain ER functions that are essential for cell viability, hence the lethality associated with the lack of AT-1 (14, 17), and that need to be activated under conditions associated with ER stress, hence the up-regulation of AT-1 during the B cell-to-plasma cell transition (18). Perhaps the above considerations can also explain the disease association. In fact, AT-1 is mutated in patients affected by autosomal dominant spastic paraplegia-42 (SPG42) (17). Although the molecular mechanism of the disease association has not been dissected yet, enlarged ER tubules as well as generalized defects of the ER network constitute common morphological features of spastic paraplegia neurons (19).
Here we show that IRE1/XBP1 controls the induction of autophagy/ERAD(II) by activating the expression of the ER membrane transporter AT-1, which ensures continuous supply of acetyl-CoA into the lumen of the ER. Failure to induce AT-1 leads to widespread cell death. Mechanistically, the regulation of the autophagic process involves Nϵ-lysine acetylation of the autophagy protein Atg9A. Finally, neither ATF6 nor PERK signaling appears to be involved in the regulation of the autophagic process.
EXPERIMENTAL PROCEDURES
Cell Cultures
Human neuroglioma (H4) and Chinese hamster ovary (CHO) cell lines were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin/glutamine solution (Mediatech). Stable transfection was performed using Lipofectamine 2000 (Invitrogen), and the culture media were supplemented with 350 μg/ml G418 sulfate (Mediatech). Cells were maintained at 37 °C in a humidified atmosphere with 5% CO2.
For ceramide treatment, cells were plated at a density of 9000 cells/cm2 in DMEM supplemented as above. 24 h later, they were switched to DMEM supplemented with 0.5% FBS and maintained under these conditions for an additional 24 h prior to treatment. C6-ceramide (hereafter referred to as ceramide; Matreya) was added to the culture media at the final concentration of 10 μm.
Antibodies
The following primary antibodies were used: anti-acetylated lysine (1:1000; Abcam); anti-Atg9A (1:2000; Epitomics); anti-c-Myc (1:1000; Santa Cruz Biotechnology); anti-AT1/SLC33A1 (1:500; Abnova); anti-beclin 1 (1:1000; Cell Signaling); and anti-β-actin (1:1000; Cell Signaling).
Immunoprecipitation and Western Blot Analysis
Protein extracts were prepared in GTIP buffer (10 mm Tris, pH 7.6, 2 mm EDTA, 0.15 m NaCl) supplemented with 1% Triton X-100 (Roche Applied Science), 0.25% Nonidet P-40 (Roche Applied Science), Complete protein inhibitor mixture (Roche Applied Science), and phosphatase inhibitors (mixture set I and set II; Calbiochem). Protein concentration was measured by the bicinchoninic acid method (Pierce).
Immunoprecipitation was performed in protein extracts (300 μg) from enriched preparations of internal membranes (cell organelles). Briefly, cells were lysed in homogenization buffer pH 7.4 (10 mm triethanolamine, 10 mm acetic acid, 250 mm sucrose, 1 mm EDTA, 1 mm DTT, and Complete protease inhibitor mixture (Roche Applied Science)) using a Dounce Teflon homogenizer and a 25-gauge 5/8 insulin syringe. Total homogenates were centrifuged at 1500 × g for 5 min at 4 °C, and the supernatant was carefully removed and centrifuged at 20 psi for 15 min using a Beckman-Coulter air-driven ultracentrifuge. The pellet containing the internal membranes was resuspended in GTIP buffer supplemented as described above and incubated for 30 min on ice. After centrifugation at 10,000 rpm for 10 min at 4 °C, the supernatant containing the protein extracts was carefully removed. Immunoprecipitation was performed using either anti-Atg9A (1:50; Epitomics) or anti-acetylated lysine (1:100; Cell Signaling) antibodies and BioMag protein A magnetic particles (Polysciences, Inc.) as described previously (13). Overexpressed Atg9A-Myc fusion protein was purified using the ProFound c-Myc tag IP/Co-IP kit (Pierce) as described (13).
Protein samples prepared in reducing NuPAGE® LDS sample buffer (Invitrogen) were subjected to electrophoresis using precast NuPAGE® Novex 4–12% Bis-Tris gels (Invitrogen) and transferred to nitrocellulose membranes (Invitrogen). Membranes were blocked for 1 h in Tris-buffered saline (TBS) containing 5% bovine serum albumin (BSA; Sigma) followed by an overnight incubation with primary antibody diluted in 5% BSA in TBS, 0.1% Tween® 20 (TBST). After washing with TBST, membranes were incubated with goat anti-rabbit Alexa Fluor 680- or goat anti-mouse Alexa Fluor 800-conjugated secondary antibodies (LI-COR Biosciences). Membranes were imaged and quantified using the LI-COR Odyssey infrared imaging system (LI-COR Biosciences). For AT-1 Western blot, membranes were incubated with peroxidase-conjugated secondary antibody (GE Healthcare). Densitometric analysis was performed using the National Institutes of Health Image program.
Real-time PCR
Real-time PCR was performed as described before (14). The cycling parameters were as follows: 95 °C, 10 s; 55 or 59 °C, 10 s; 72 °C, 15 s. Controls without reverse transcription were included in each assay. Specific primers are shown in supplemental Table S1. Gene expression levels were normalized against GAPDH levels and expressed as the percentage of control.
Plasmid Constructs
Human AT-1 (NM_004733) cDNA was obtained from OriGene (SC117182) and cloned into pcDNA3.1AMyc/His (Invitrogen) and pcDNA3.1V5/His/TOPO (Invitrogen) (14). The cDNA of human Atg9A (NM_024085.2) with a C-terminal fusion of Myc-DDK tag in the expression vector pCMV6-Entry was obtained from OriGene (RC222513).
Site-directed Mutagenesis
Mutagenesis of Atg9A was performed using the QuikChange Lightning site-directed mutagenesis kit (Stratagene) according to manufacturer's protocol. Residues lysine 359 and lysine 363 were mutated to glutamine using the following primers: forward, 5′-GCCTCAACCGTGGCTACcagCCCGCCTCCcagTACATGAATTGC-3′ and reverse, 5′-GCAATTCATGTActgGGAGGCGGGctgGTAGCCACGGTTGAGGC-3′. The same residues were also mutated to arginine using the following primers: forward, 5′-GCCTCAACCGTGGCTACaggCCCGCCTCCaggTACATGAATTGC-3′ and reverse, 5′-GCAATTCATGTAcctGGAGGCGGGcctGTAGCCACGGTTGAGGC-3′. The presence of the mutations and the full-length gene sequences were confirmed by DNA sequencing (performed at the DNA Sequencing Facility of the University of Wisconsin-Madison Biotechnology Center).
RNA Interference
Cells were plated at a density of 8300 cells/cm2 in DMEM supplemented as described above and transfected with 10 nm siRNA against XBP-1 (Mm_Xbp1_2; Qiagen), ATF6 (Hs_ATF6_5; Qiagen), PERK (Hs_EIF2AK3_5; Qiagen), or AT-1/SLC33A1 (Hs_SLC33A1_5; Qiagen) using the HiPerFect transfection reagent (Qiagen). During the 3- or 4-day experiments, the siRNA was reapplied 48 h after the initial treatment. Nonsilencing siRNA (AllStars negative control; Qiagen) was used as a control.
Electron Microscopy
Transmission electron microscopy was performed at the Electron Microscopy Facility of the William S. Middleton Memorial Veterans Hospital (Madison, WI) and University of Wisconsin-Madison. Briefly, cells were fixed with 2.5% glutaraldehyde in phosphate buffer (pH 7.2) and immediately scraped with a rubber policeman. After cells were fixed for 1 h at 4 °C, they were centrifuged at 5000 × g. Cell pellets were rinsed in Sorenson's phosphate wash buffer, postfixed in Caulfield's osmium tetroxide plus sucrose, and rinsed again in wash buffer. All supplies and reagents were from Electron Microscopy Sciences. Cell pellets were dehydrated in a graded series of ethanol and propylene oxide and embedded in BEEM capsules using EMbed 812 resin. Polymerization was thermally induced by incubating the samples overnight at 60 °C. Thin sections (70–80 nm) were cut and mounted on copper grids, stained with filtered 7.7% uranyl acetate, and counterstained with Reynolds lead citrate. Sections were observed using a transmission electron microscope (H-7650; Hitachi) operated at 80 kV.
Autophagy Assay by LC3B-GFP Imaging
The induction of autophagy was detected with the PremoTM autophagy sensor LC3B-GFP BacMam 2.0 kit (Invitrogen). Briefly, 3 days after siRNA treatment, cells were transduced with BacMam LC3B-GFP at a multiplicity of infection of 30 plaque-forming units per cell. Autophagy-defective LC3B(G120A)-GFP served as negative control, whereas chloroquine diphosphate (100 μm) was used to induce autophagy (positive control). 24 h after transduction, cells were fixed on ice with 4% paraformaldehyde in Dulbecco's phosphate-buffered saline (PBS). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen). Slides were mounted using Gel/Mount aqueous mounting medium (Biomeda) and imaged on a Zeiss AxioVert 200 inverted fluorescent microscope. Autophagy was quantified in a minimum of 500 cells per condition, and cells with more than five LC3B-GFP puncta were considered autophagic.
LC-MS/MS Analysis
MS analysis was performed by the Mass Spectrometry facility at the University of Wisconsin-Madison as described previously (11). Modified peptides were identified with the Mascot search engine (Matrix Science, London, UK) via automated database searching of all tandem mass spectra.
Statistical Analysis
Data analysis was performed using GraphPad InStat 3.06 statistical software (GraphPad Software Inc.). Data are expressed as mean ± S.E. Comparison of the means was performed using Student's t test or one-way analysis of variance followed by Tukey-Kramer multiple comparisons test. Differences were declared statistically significant if p ≤ 0.05.
RESULTS
We initially determined the ability of two commonly used ER stressors, tunicamycin and thapsigargin, to activate the expression of AT-1. In fact, the UPR normally responds to and is activated by conditions that induce ER stress. Therefore, we reasoned that if AT-1 is regulated by UPR signaling, it should respond to the induction of ER stress. The results displayed in Fig. 1, A and B, show that tunicamycin and thapsigargin were able to cause a marked increase of AT-1 at both the mRNA and the protein level, thus supporting our initial hypothesis. These results are consistent with a recent proteomic study where we reported that a large number of ER-resident proteins involved with synthesis, folding, post-translational modification of nascent proteins, as well as degradation of unfolded/misfolded intermediates are acetylated in the lumen of the organelle (16). Interestingly, acetylated ER-resident proteins displayed a high degree of network connectivity (16), suggesting that the Nϵ-lysine acetylation might play an important role in regulating the activity of ER functions that are generally under the control of the UPR.
FIGURE 1.
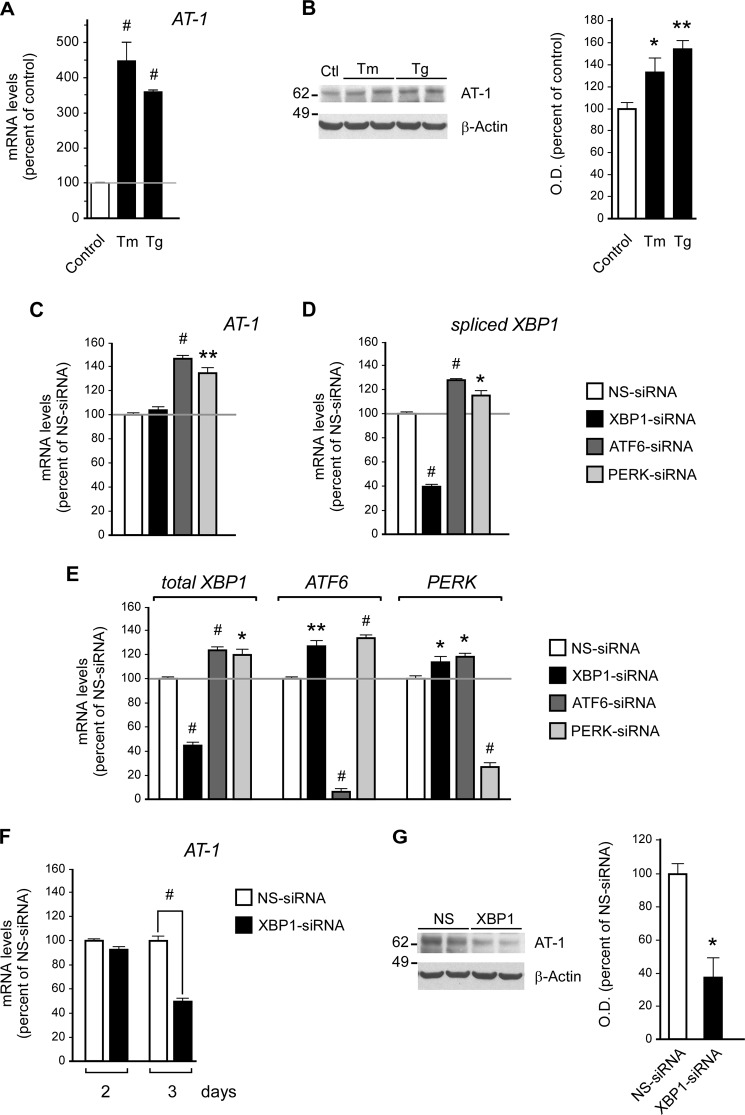
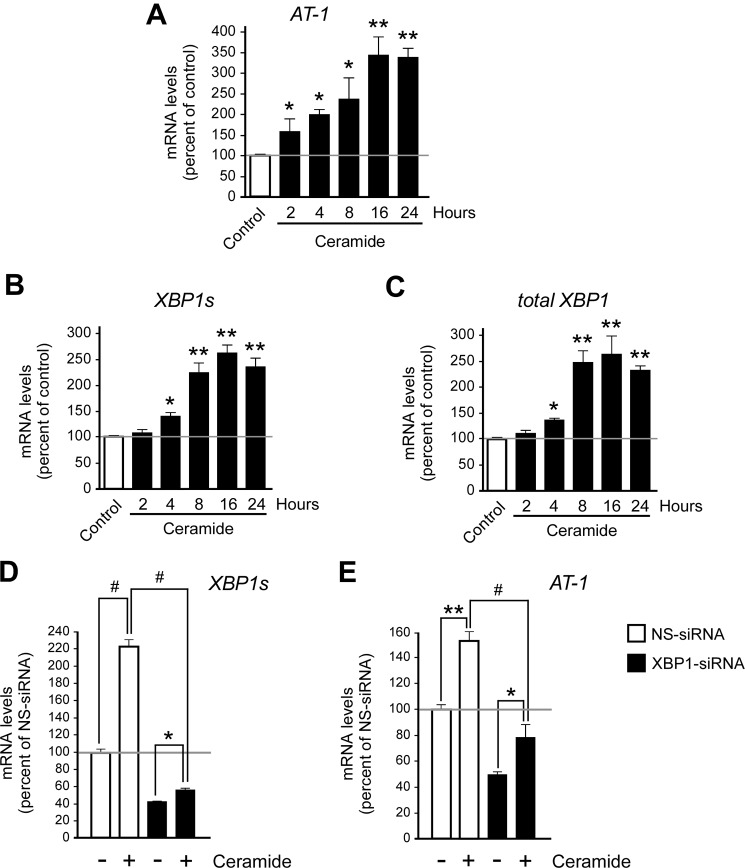
AT-1 acts downstream of XBP1. A and B, H4 cells were treated with either tunicamycin (Tm; 2 μg/ml) or thapsigargin (Tg; 200 nm) prior to quantitative real-time PCR analysis of AT-1 mRNA levels (A, treatment was for 6 h) and Western blot assessment of AT-1 protein levels (B, treatment was for 10 h). Results are expressed as the percentage of control and are the average (n ≥ 3) + S.E. *, p < 0.05; **, p < 0.005; #, p < 0.0005. O. D., optical density. C–E, H4 cells were treated with siRNA for 2 days prior to quantitative real-time PCR. The mRNA levels of AT-1 (C), XBP1s (D), and the other UPR branches (E) are shown. The results in E suggest compensatory cross-talk between the three UPR signaling branches. F, at least 3 days of XBP1 siRNA treatment were necessary to observe significant down-regulation of AT-1 mRNA levels. G, Western blot assessment of AT-1 protein levels following 3 days of XBP1 siRNA treatment. Results in C–G are expressed as the percentage of nonsilencing siRNA (NS-siRNA) and are the average (n = 3) + S.E. *, p < 0.05; **, p < 0.005; #, p < 0.0005.
To assess whether the activation of AT-1 was specifically linked to one of the individual UPR branches, we treated H4 cells with siRNA targeting XBP1, ATF6, or PERK. XBP1 acts immediately downstream of IRE1 and, as such, down-regulation of XBP1 can be used to block IRE1 signaling (8). A 2-day treatment with siRNAs produced changes in AT-1 levels in ATF6- and PERK- but not in XBP1-treated cells (Fig. 1C). However, these changes were the opposite of what we expected. In fact, AT-1 levels increased instead of decreasing. These results might be explained by the finding that the down-regulation of ATF6 or PERK resulted in activation of the spliced version, XBP1s (Fig. 1D), probably as an attempt to compensate for the sudden failure of the UPR signaling machinery. Indeed, down-regulation of each UPR branch resulted in a compensatory up-regulation of the other two branches (Fig. 1E). Therefore, ATF6 and PERK siRNA-treated cells, which were not deficient in XBP1 signaling (Fig. 1D), were able to activate AT-1 (Fig. 1C), whereas XBP1 siRNA-treated cells were not. When taken together, these results might suggest that the activation of AT-1 is under the control of XBP1. To confirm this conclusion, we extended XBP1 siRNA treatment to 3 days and were able to observe a clear down-regulation of both mRNA (Fig. 1F) and protein (Fig. 1G) levels of AT-1.
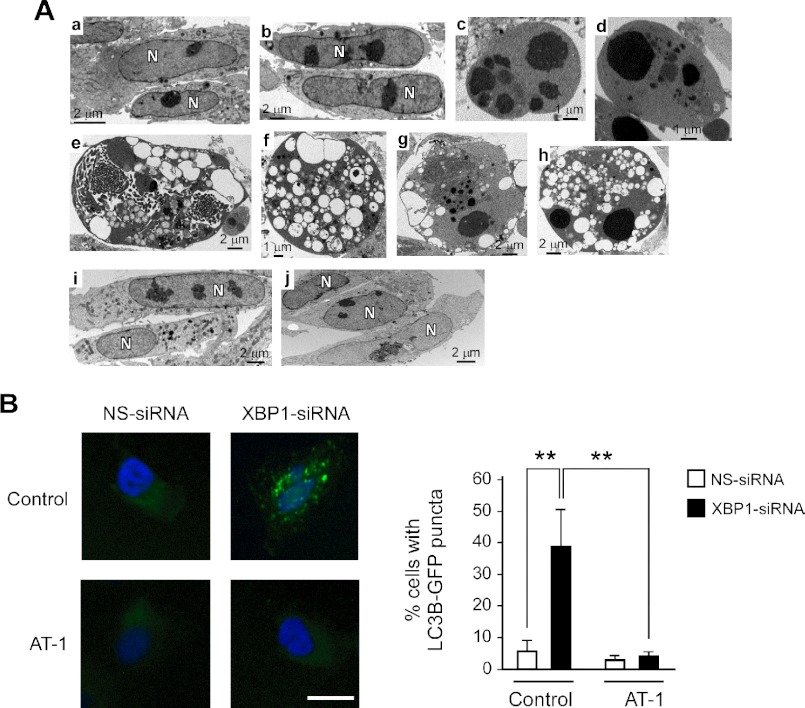
While executing the above experiments, we also noticed a 20–40% cell death, which increased with the duration of the siRNA treatment. However, XBP1 siRNA-treated cells displayed features of autophagic cell death, whereas ATF6 and PERK siRNA-treated cells displayed features of apoptosis (Fig. 2A). In some cases, the down-regulation of XBP1 was accompanied by chromatin aggregation, which could suggest mixed features of autophagic and apoptotic cell death (Fig. 2A, panels g and h). However, a clear apoptotic phenotype was very rare in XBP1 siRNA-treated cells but common in ATF6 and PERK siRNA-treated cells (Fig. 2A; compare panels c and d with panels e–h). Finally, typical autophagosomes were only observed in XBP1 siRNA-treated cells (supplemental Fig. S1A).
FIGURE 2.
AT-1 acts downstream of XBP1 to prevent autophagic cell death. A, H4 cells were treated with XBP1 siRNAs for 4 days prior to transmission EM. Panel a, control (untreated) cells; panel b, NS siRNA-treated cells; panel c, ATF6 siRNA-treated cells; panel d, PERK siRNA-treated cells; panels e–h, XBP1 siRNA-treated cells; panels i–j, XBP1 siRNA treatment of AT-1-overexpressing cells. Panels c and d show chromatin condensation and cytoplasmic changes that are typical of apoptosis. Panels e–h show cells with features that are typical of autophagic cell death. They include cellular blebbing, the presence of several large vacuoles, and no visible organelles. Some of the vacuoles contain dense lysosomal material. Panels i and j show normal features and no evidence of autophagic cell death. B, H4 cells were treated with nonsilencing (NS-siRNA) or XBP1-specific (XBP1-siRNA) siRNAs for 3 days and then transduced with GFP-LC3B BacMam. The induction of autophagy, as assessed by the redistribution of LC3B, was evaluated 24 h later. Total treatment with siRNA was 4 days. The same experiment was performed with both control (empty plasmid) and AT-1-overexpressing H4 cells. Representative images are shown in the left panel, whereas the quantitative data are shown in the right panel. Nuclei were counterstained with DAPI (blue). Bar: 10 μm. Results are the average (n ≥ 6) + S.E. **, p < 0.005. Successful down-regulation of XBP1 in CMV-AT-1-overexpressing cells is shown in supplemental Fig. S4.
To confirm the autophagic nature of the cellular changes observed with electron microscopy (EM), we decided to use cells expressing a GFP-tagged version of the microtubule-associated protein light chain 3B (LC3B), the mammalian homolog of Atg8. In the absence of autophagy, LC3B has a diffuse cytoplasmic distribution. In contrast, upon induction of autophagy, LC3B is recruited on autophagosomal membranes, thus displaying a punctate distribution across the cytoplasm that corresponds to the autophagosomal structures (20). Consistent with the EM assessment, XBP1 siRNA treatment was accompanied by redistribution of GFP-LC3B into clearly identifiable LC3B puncta (supplemental Figs. S1B and S2). Assessment of GFP-LC3B distribution confirmed the lack of autophagy in ATF6 and PERK siRNA-treated cells (supplemental Fig. S2). To further support the above findings, we also assessed cellular levels of beclin, a commonly used protein marker of autophagy, and observed increased levels (supplemental Fig. S1C). In conclusion, three independent approaches confirmed the induction of autophagy following down-regulation of XBP-1. Finally, the above findings are consistent with previous studies, which have also reported abnormal induction of autophagy following down-regulation of XBP1 (9, 10).
We previously reported that down-regulation of AT-1 is accompanied by widespread autophagic cell death (14). Therefore, we concluded that perhaps the presence of autophagic cell death in XBP1 siRNA-treated cells was caused by the inability of these cells to induce AT-1 and secure sufficient influx of acetyl-CoA into the ER lumen. ATF6 and PERK siRNA-treated cells were protected because they were able to induce AT-1 expression through XBP1 signaling (Fig. 1, C and D). To confirm the above conclusion, we down-regulated XBP1 in H4 cells that overexpressed transgenic AT-1 under the control of a cytomegalovirus (CMV) promoter. In fact, being under the control of a CMV promoter, the levels of transgenic AT-1 are independent of XBP1 signaling and should prevent the autophagic cell death. Fig. 2A (panels i and j) clearly shows that AT-1-overexpressing cells were resistant to the induction of autophagy following XBP1 siRNA treatment. Under these conditions, we were not able to detect morphological features corresponding to autophagy or autophagic cell death (Fig. 2A, panels i and j). Similar results were also observed in CHO cells, indicating that these results are not limited to H4 cells (supplemental Fig. S3). The overexpression of CMV-regulated transgenic AT-1 was also able to block the redistribution of GFP-LC3B into the classical autophagy puncta (Fig. 2B), therefore confirming the EM data.
The above results indicate that the influx of acetyl-CoA into the ER lumen regulates the induction of autophagy downstream of IRE1/XBP1. However, it is unclear how the acetylation status of the ER might be “sensed” by the autophagic machinery.
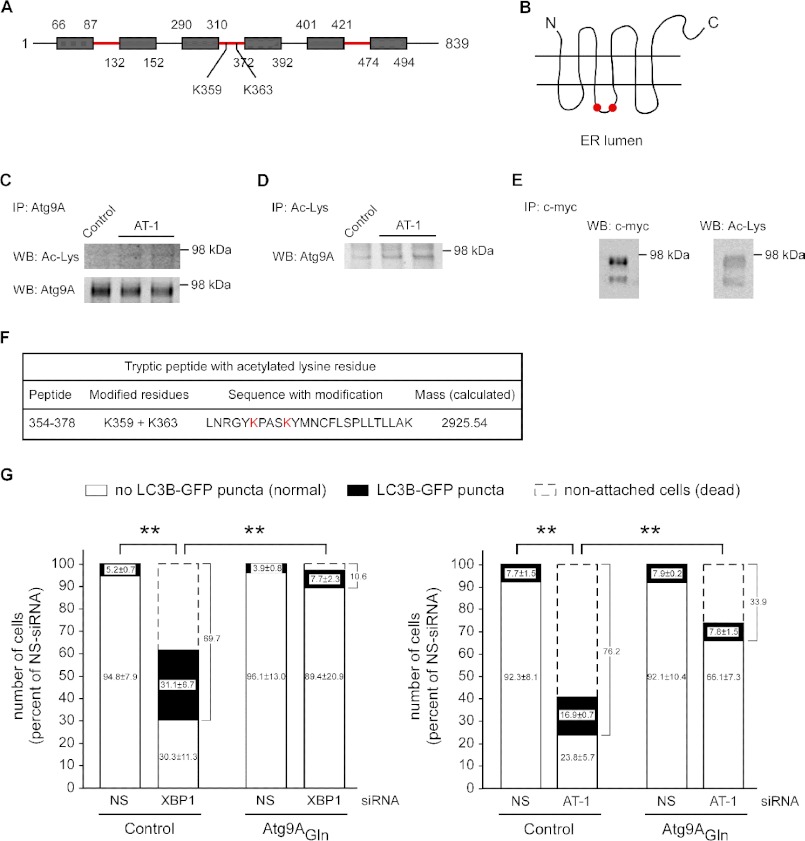
The Nϵ-lysine acetylation status of several molecular components of the autophagic machinery, including Atg5, Atg7, Atg8, and Atg12, regulates the induction of autophagy. Specifically, increased acetylation inhibits autophagy, whereas reduced acetylation stimulates autophagy (21, 22). However, the above proteins are all located in the cytosol and, therefore, not directly affected by the influx of acetyl-CoA into the ER lumen. The only known integral membrane autophagy protein is Atg9A, an ER-based protein that appears to translocate out of the ER during the induction of autophagy (23–25). The topology of Atg9A predicts six transmembrane domains with both the C termini and the N termini facing the cytosol (Fig. 3, A and B). Three loops face the lumen of the ER with a total of 6 lysine residues (Lys-96, Lys-109, Lys-321, Lys-359, Lys-363, and Lys-472) available for possible acetylation. To assess whether Atg9A is acetylated, we first immunoprecipitated the endogenous protein. Under these conditions, we only detected one band that migrated with the expected mass of the N-glycosylated form of the protein (Fig. 3C, lower panel). The same band was also identified with an antibody against acetylated lysine residues (Fig. 3C, upper panel). Importantly, overexpression of AT-1 appeared to increase the acetylation of Atg9A (Fig. 3C, upper panel). Atg9A was also observed following immunoprecipitation with an antibody against acetylated lysine residues (Fig. 3D). Again, overexpression of AT-1 appeared to increase the acetylation of Atg9A (Fig. 3D). To confirm these results, we generated H4 cells overexpressing transgenic Atg9A with a Myc tag at the C terminus. Overexpression of Atg9A improved detection with an anti-acetylated lysine antibody and confirmed the acetylation status of the protein (Fig. 3E). As reported previously (23), under these conditions, we could detect both the immature and the N-glycosylated forms of the protein; both appeared to be acetylated (Fig. 3E). Finally, transgenic Atg9A was purified with an anti-Myc immobilized column and used to map the modified lysine residues by LC-MS/MS. Only 2 residues, Lys-359 and Lys-363, were identified as acetylated (Fig. 3F). Both residues are located in the third loop and are exposed to the luminal site of the ER (Fig. 3, A and B), thus confirming the acetylation status of Atg9A and implicating the ER-based acetylation machinery, which depends on AT-1. Mass spectrometry also identified a nonacetylated digest (data not shown), suggesting that under steady-state conditions, acetylated and nonacetylated species of Atg9A coexist.
FIGURE 3.
Atg9A is acetylated in the lumen of the ER and acts downstream of AT-1 to prevent the induction of autophagy in XBP1 siRNA-treated cells. A, topology of Atg9A. Transmembrane domains are indicated as gray boxes, whereas regions that face the ER lumen are in red. The location of modified lysines Lys-359 and Lys-363 is also indicated. B, schematic view of Atg9A arrangement across the ER membrane. Modified lysines Lys-359 and Lys-363 are indicated by two red dots. C, endogenous Atg9A was immunoprecipitated (IP) from a crude preparation of intracellular membranes and then analyzed with both anti-acetylated lysine (upper panel) and anti-Atg9A (lower panel) antibodies. WB, Western blot. D, the same membrane preparation was also immunoprecipitated with an antibody against acetylated lysine residues and then analyzed with an anti-Atg9A antibody. E, a Myc-tagged version of Atg9A was immunoprecipitated from stable transfected cells and analyzed with the indicated antibodies. F, tryptic peptide with the acetylated lysine residues identified by mass spectrometry. For LC-MS/MS, Atg9A was purified from cells that did not overexpress AT-1. G, control (empty plasmid) and Atg9AGln-overexpressing H4 cells were treated with nonsilencing (NS-siRNA), XBP1-specific (XBP1-siRNA), or AT-1-specific (AT-1-siRNA) siRNAs for 3 days and then transduced with GFP-LC3B BacMam. The induction of autophagy, as assessed by the redistribution of LC3B, was evaluated 24 h later. Total treatment with siRNA was 4 days. The number of dead cells was also calculated. Results are the average (n ≥ 6) + S.E. **, p < 0.005.
The fact that overexpression of AT-1 increases the acetylation of Atg9A supports the hypothesis that the acetylation status of Atg9A might act as a “sensor” for the activation of autophagy in the ER. Increased acetyl-CoA influx into the ER lumen would increase the acetylation of Atg9A and prevent the induction of autophagy. Conversely, decreased AT-1 activity would achieve the opposite effect. This scenario would be consistent with previous results with Atg5, Atg7, Atg8, and Atg12 (21, 22). To test this possibility, we mutated both Lys-359 and Lys-363 to glutamine (Atg9AGln). In fact, the Lys-to-Gln substitution mimics the effect of the lysine acetylation and is predicted to generate a “gain-of-acetylation” mutant form of Atg9A (11, 26). As shown in Fig. 3G, expression of the K359Q/K363Q mutant prevented the formation of autophagosomal structures as well as the overall mortality of the cells. This protective effect was observed after down-regulation of both XBP1 (Fig. 3G, left panel) and AT-1 (Fig. 3G, right panel), indicating that Atg9A is the last output of the molecular pathway that controls the induction of autophagy as part of the UPR.
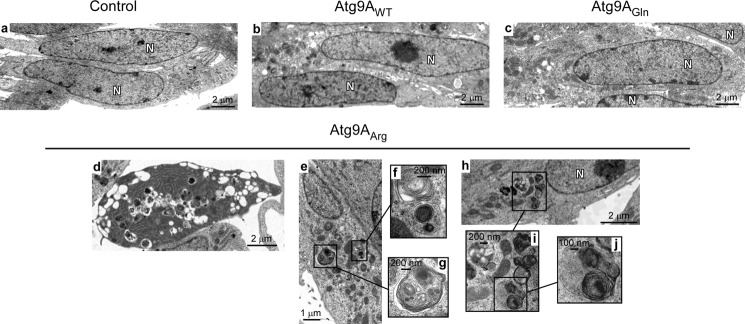
Finally, we assessed whether the acetylation status of Atg9A itself could induce autophagy and affect cell morphology. For this purpose, we mutated both Lys-359 and Lys-363 to arginine and generated a “loss-of-acetylation” mutant form of Atg9A. The overexpression of either the wild-type (Atg9AWT) or the gain-of-acetylation mutant (Atg9AGln) versions of Atg9A did not affect cell morphology or induce autophagic features (Fig. 4). In contrast, overexpression of the loss-of-acetylation mutant form (Atg9AArg) caused autophagic cell death (Fig. 4, panel d) and the appearance of typical intracellular autophagosomes (Fig. 4, panels d–j), thus impeding functional assessment. Importantly, these features were observed in the presence of endogenous Atg9A, suggesting that Atg9AArg can act as a dominant mutant.
FIGURE 4.
The loss-of-acetylation mutant form of Atg9A induces cell death as well as features of autophagy. Panels a–j, H4 cells overexpressing WT (Atg9AWT), K359Q/K363Q (Atg9AGln; gain-of-acetylation) or K359R/K363R (Atg9AArg; loss-of-acetylation) versions of Atg9A were analyzed with transmission electron microscopy in the absence of any treatment. Different cellular features that are typical of the early stages of autophagy were observed in cells overexpressing Atg9AArg (panels d–j). Nuclei are labeled with N. Mitochondria appear normal. Control indicates cells transfected with an empty plasmid.
When taken together, the above results indicate that AT-1 acts downstream of IRE1/XBP1 to ensure the acetylation status of the ER. To confirm the relevance of these results under a more physiologic context, we decided to analyze the effect of the lipid second messenger ceramide. In fact, we previously published that the expression levels of AT-1 are tightly regulated by both exogenous and endogenous ceramide (14). Because ceramide has already been linked to the induction of cellular stress (27), we decided to assess whether ceramide acts through IRE1/XBP1.
As an initial strategy, we monitored the activation status of XBP1 in the presence of physiologically relevant concentrations of ceramide. In fact, we reasoned that if ceramide activates AT-1 through IRE1/XBP1, the up-regulation of AT-1 levels should be concomitant with the activation of IRE1/XBP1 signaling. The kinetics of AT-1 expression activation, as induced by ceramide treatment, is shown in Fig. 5A. Next, we analyzed changes in the mRNA levels of XBP1s. It is worth remembering that IRE1 has endoribonuclease activity, and when activated, as a result of ER stress, it removes a small fragment from the unspliced mRNA of XBP1 (XBP1u) to generate the spliced version, XBP1s (8). As a result of ATF6 activation (28) and perhaps XBP1s activation (29), the total levels of XBP1 also increase. Therefore, increased mRNA levels of XBP1s are an indication of IRE1 activation, whereas increased levels of total XBP1 may be an indication of both ATF6 and XBP1s activation. As shown here, ceramide treatment resulted in a marked increase of both XBP1s (Fig. 5B) and total XBP1 (Fig. 5C) mRNA levels, supporting the conclusion that ceramide treatment results in activation of IRE1/XBP1 signaling. Importantly, the up-regulation of AT-1 induced by ceramide was prevented by the concomitant down-regulation of XBP1 (Fig. 5, D and E), indicating that the induction of IRE1/XBP1 and AT-1 is functionally linked. The fact that the block of AT-1 up-regulation in XBP1 siRNA-treated cells was never absolute is probably due to the remaining, and still active, XBP1 (compare Fig. 5, D and E; see also Fig. 1, D and F). It is also worth mentioning that the ability of ceramide to induce the UPR was not limited to IRE1/XBP1 signaling (data not shown).
FIGURE 5.
The induction of AT-1 as a result of ceramide treatment is concomitant with the activation of IRE1/XBP1 signaling. A–C, human neuroglioma (H4) cells were treated with 10 μm ceramide for the indicated periods of time prior to the analysis of the kinetics of activation of AT-1 and XBP1. A, quantitative real-time PCR of AT-1 mRNA levels. B and C, quantitative real-time PCR of XBP1s (B) and total XBP1 (C) mRNA levels. Results are expressed as the percentage of control and are the average (n ≥ 3) + S.E. *, p < 0.05; **, p < 0.005. D and E, H4 cells were treated with nonsilencing (NS-siRNA) or XBP1-specific (XBP1-siRNA) siRNA in the presence or absence of ceramide. Total treatment with siRNA was 3 days, whereas ceramide exposure was limited to the last 8 h. Changes in XBP1s (D) and AT-1 (E) mRNA levels are shown. Results are expressed as the percentage of nonsilencing siRNA and are the average (n ≥ 5) + S.E. *, p < 0.05; **, p < 0.005; #, p < 0.0005.
DISCUSSION
Collectively, our studies suggest that the acetylation status of the ER is regulated by IRE1/XBP1, which acts by controlling the influx of acetyl-CoA through the membrane transporter AT-1. Failure to maintain active transport of acetyl-CoA into the ER lumen is sensed by Atg9A, which regulates the induction of autophagy/ERAD(II).
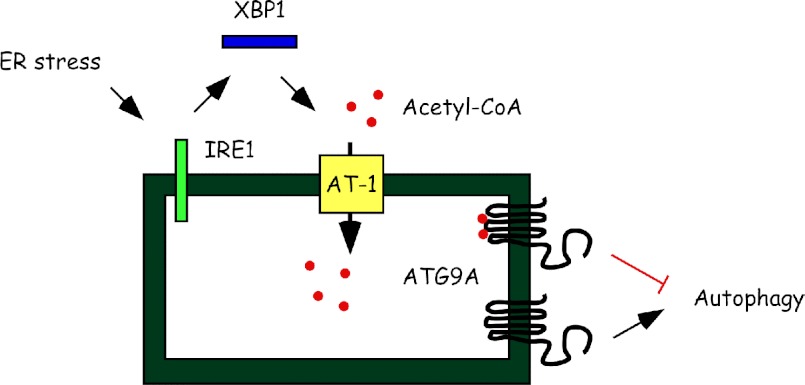
The ultimate function of the UPR is to correct possible imbalances that result in ER stress, thus ensuring folding of nascent membrane/secreted proteins and disposal of unfolded/misfolded or aggregated proteins (1, 6, 7). This is achieved by activating signaling pathways that, in mammalian cells, act downstream of three main UPR initiators: IRE1/XBP1, ATF6, and PERK. Although there is extensive cross-talk and functional overlap, individual UPR branches can preferentially activate or repress specific subsets of UPR genes to provide the most appropriate response. Here we propose that the import of acetyl-CoA into the ER lumen by the ER membrane transporter AT-1 is part of the UPR. Specifically, AT-1 acts downstream of IRE1/XBP1 to ensure the acetylation status of the ER and control the induction of autophagy/ERAD(II) (Fig. 6).
FIGURE 6.
Schematic summary and proposed model. ER stress activates IRE1/XBP1 signaling to regulate the influx of acetyl-CoA into the ER lumen through the membrane transporter AT-1. Acetyl-CoA levels in the lumen of the ER determine the acetylation status of Atg9A, which acts as the last output for the induction of autophagy/ERAD(II) during the UPR. Acetylated (with red dots) Atg9A prevents autophagy, whereas nonacetylated (without red dots) Atg9A activates autophagy.
Autophagy is a general process that allows the cell to dispose of damaged or toxic components generated as a result of a variety of stress stimuli, which include ER stress and nutrient/energy deprivation (3, 20). The elimination of toxic protein aggregates in the ER is an essential feature of this organelle. Large ER protein aggregates are mostly dealt with by expanding the ER and activating autophagy/ERAD(II) (Refs. 30–33; also reviewed in Ref. 3). Failure to clear these protein aggregates from the ER lumen has been linked to a variety of diseases (3, 34). Therefore, when tightly controlled, autophagy can allow the cell to recover from the transient accumulation of protein aggregates; however, when unchecked, it can be detrimental and cause autophagic/Type 2 cell death (4, 5). Because one of the main outcomes of the UPR is to eliminate toxic protein aggregates, it is likely that under nonlethal levels of ER stress, the UPR maintains the autophagic process under strict control (20). Recent work in the mouse has revealed that IRE1/XBP1 signaling protects from excessive autophagy (9). Our results suggest that AT-1 might be the downstream effector of the above pathway. In fact, increased dosage of AT-1 was able to prevent the induction of autophagy and the cell death caused by the down-regulation of XBP1. Therefore, we propose that the ability of the cell to up-regulate AT-1 levels and secure sufficient influx of acetyl-CoA into the ER lumen is crucial to prevent cell death during ER stress. It is worth stressing that AT-1 has been identified as one of the downstream targets of XBP1 in different cell types and under different forms of nonlethal ER stress (18, 35). Our results also suggest that ATF6 and PERK signaling controls the induction of apoptosis but not autophagy, indicating functional separation between the three UPR branches.
Recent work has shown that the acetyl-CoA:lysine acetyltransferase p300 controls the Nϵ-lysine acetylation of Atg5, Atg7, Atg8, and Atg12, all known components of the autophagic machinery (22). Knockdown of p300 resulted in reduced acetylation and stimulated autophagy. Conversely, overexpression of p300 achieved the opposite effects (22). Sirt1, a known deacetylase, is also a regulator of autophagy (21). Overexpression of Sirt1 activates the basal levels of autophagy. Conversely, down-regulation of Sirt1 achieves the opposite effect (21). Once again, this effect was linked to the lysine acetylation status of known components of the autophagic machinery, specifically Atg5, Atg7, and Atg8 (21). Therefore, different lines of experiments indicate that autophagy is tightly regulated by the Nϵ-lysine acetylation of the molecular components of the autophagic machinery; increased acetylation inhibits autophagy, whereas reduced acetylation stimulates autophagy. Because the above autophagic components are cytosolic, it is not surprising that the biochemical components of the acetylation machinery that modifies them (acetyltransferases and deacetylases) are cytosolic as well.
Here we show that Atg9A, the only integral membrane autophagy protein, is also acetylated. The acetylation occurs on lysine residues that face the lumen of the ER, thus implicating the ER-based acetylation machinery, which depends on AT-1 activity (14). Importantly, a gain-of-acetylation mutant form of Atg9A was able to rescue the phenotype caused by the down-regulation of XBP1 or AT-1, whereas a loss-of-acetylation mutant form was able to act in a dominant fashion and induce cell death as well as autophagy in the absence of any treatment. Therefore, based on the results reported here as well as the previous studies with other Atg proteins (21, 22), we can conclude that the acetylation status of Atg9A regulates its ability to participate in the induction of autophagy (Fig. 6).
Based on our results, we can predict that deregulated activity of AT-1 would have deleterious effects on the ability of the cell to control autophagy and dispose of large protein aggregates. In fact, increased activity (or gain-of-function) would prevent autophagy but also cause abnormal accumulation of protein aggregates. Conversely, decreased activity (or loss-of-function) would lead to very efficient removal of protein aggregates but also to autophagic cell death. Therefore, a tight balance must be kept to ensure ER homeostasis. It is also possible that the intrinsic need for autophagy has to be constantly adjusted depending on the status of the ER. As such, levels of autophagy that are toxic for normal cells (under nonlethal forms of ER stress) might be beneficial for cells that accumulate abnormal levels of protein aggregates, such as neurons expressing ALS-linked mutant Cu,Zn-superoxide dismutase (SOD1) (9, 36).
Supplementary Material
Acknowledgments
We thank Dr. Nansi Jo Colley and Dr. Barry Ganetzki for critical reading of an early version of this manuscript. We are grateful to Joan Sempf at the Veterans Administration Hospital/University of Wisconsin-Madison (VAH/UW) Electron Microscopy Facility and Grzegorz Sabat at the UW Mass Spectrometry Facility for their help. We are also grateful for use of the resources and facilities of the William S. Middleton Memorial Veterans Hospital, Madison, WI.
This work was supported, in whole or in part, by National Institutes of Health Grants AG028569 and AG033514 from the NIA (to L. P.).

This article contains supplemental Table S1 and Figs. S1–S4.
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- IRE1
- inositol-requiring protein-1
- XBP1
- X-box-binding-1 protein
- ATF6
- activating transcription factor-6
- LC3B
- light chain 3B
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
- UPR
- unfolded protein response
- PERK
- PKR-like ER kinase
- PKR
- protein kinase RNA
- NS
- nonsilencing.
REFERENCES
- 1. Trombetta E. S., Parodi A. J. (2003) Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 19, 649–676 [DOI] [PubMed] [Google Scholar]
- 2. Kleizen B., Braakman I. (2004) Protein folding and quality control in the endoplasmic reticulum. Curr. Opin. Cell Biol. 16, 343–349 [DOI] [PubMed] [Google Scholar]
- 3. Buchberger A., Bukau B., Sommer T. (2010) Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol. Cell 40, 238–252 [DOI] [PubMed] [Google Scholar]
- 4. Klionsky D. J. (2006) Neurodegeneration: good riddance to bad rubbish. Nature 441, 819–820 [DOI] [PubMed] [Google Scholar]
- 5. Bergmann A. (2007) Autophagy and cell death: no longer at odds. Cell 131, 1032–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mori K. (2000) Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell 101, 451–454 [DOI] [PubMed] [Google Scholar]
- 7. Patil C., Walter P. (2001) Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13, 349–355 [DOI] [PubMed] [Google Scholar]
- 8. Ron D., Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 9. Hetz C., Thielen P., Matus S., Nassif M., Court F., Kiffin R., Martinez G., Cuervo A. M., Brown R. H., Glimcher L. H. (2009) XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 23, 2294–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matus S., Nassif M., Glimcher L. H., Hetz C. (2009) XBP-1 deficiency in the nervous system reveals a homeostatic switch to activate autophagy. Autophagy 5, 1226–1228 [DOI] [PubMed] [Google Scholar]
- 11. Costantini C., Ko M. H., Jonas M. C., Puglielli L. (2007) A reversible form of lysine acetylation in the ER and Golgi lumen controls the molecular stabilization of BACE1. Biochem. J. 407, 383–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jonas M. C., Costantini C., Puglielli L. (2008) PCSK9 is required for the disposal of nonacetylated intermediates of the nascent membrane protein BACE1. EMBO Rep. 9, 916–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ko M. H., Puglielli L. (2009) Two endoplasmic reticulum (ER)/ER Golgi intermediate compartment-based lysine acetyltransferases post-translationally regulate BACE1 levels. J. Biol. Chem. 284, 2482–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jonas M. C., Pehar M., Puglielli L. (2010) AT-1 is the ER membrane acetyl-CoA transporter and is essential for cell viability. J. Cell Sci. 123, 3378–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 16. Pehar M., Lehnus M., Karst A., Puglielli L. (2012) Proteomic assessment shows that many endoplasmic reticulum (ER)-resident proteins are targeted by Nϵ-lysine acetylation in the lumen of the organelle and predicts broad biological impact. J. Biol. Chem. 287, 22436–22440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin P., Li J., Liu Q., Mao F., Li J., Qiu R., Hu H., Song Y., Yang Y., Gao G., Yan C., Yang W., Shao C., Gong Y. (2008) A missense mutation in SLC33A1, which encodes the acetyl-CoA transporter, causes autosomal-dominant spastic paraplegia (SPG42). Am. J. Hum. Genet. 83, 752–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shaffer A. L., Shapiro-Shelef M., Iwakoshi N. N., Lee A. H., Qian S. B., Zhao H., Yu X., Yang L., Tan B. K., Rosenwald A., Hurt E. M., Petroulakis E., Sonenberg N., Yewdell J. W., Calame K., Glimcher L. H., Staudt L. M. (2004) XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21, 81–93 [DOI] [PubMed] [Google Scholar]
- 19. Park S. H., Blackstone C. (2010) Further assembly required: construction and dynamics of the endoplasmic reticulum network. EMBO Rep. 11, 515–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kroemer G., Mariño G., Levine B. (2010) Autophagy and the integrated stress response. Mol. Cell 40, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee I. H., Cao L., Mostoslavsky R., Lombard D. B., Liu J., Bruns N. E., Tsokos M., Alt F. W., Finkel T. (2008) A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. U.S.A. 105, 3374–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee I. H., Finkel T. (2009) Regulation of autophagy by the p300 acetyltransferase. J. Biol. Chem. 284, 6322–6328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Young A. R., Chan E. Y., Hu X. W., Köchl R., Crawshaw S. G., High S., Hailey D. W., Lippincott-Schwartz J., Tooze S. A. (2006) Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119, 3888–3900 [DOI] [PubMed] [Google Scholar]
- 24. Tamura H., Shibata M., Koike M., Sasaki M., Uchiyama Y. (2010) Atg9A protein, an autophagy-related membrane protein, is localized in the neurons of mouse brains. J. Histochem. Cytochem. 58, 443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohashi Y., Munro S. (2010) Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol. Biol. Cell 21, 3998–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Masumoto H., Hawke D., Kobayashi R., Verreault A. (2005) A role for cell cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436, 294–298 [DOI] [PubMed] [Google Scholar]
- 27. Hannun Y. A. (1996) Functions of ceramide in coordinating cellular responses to stress. Science 274, 1855–1859 [DOI] [PubMed] [Google Scholar]
- 28. Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 [DOI] [PubMed] [Google Scholar]
- 29. Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., Mori K. (2007) Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev. Cell 13, 365–376 [DOI] [PubMed] [Google Scholar]
- 30. Bernales S., McDonald K. L., Walter P. (2006) Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 4, e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S., Murakami T., Taniguchi M., Tanii I., Yoshinaga K., Shiosaka S., Hammarback J. A., Urano F., Imaizumi K. (2006) Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 26, 9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ding W. X., Ni H. M., Gao W., Hou Y. F., Melan M. A., Chen X., Stolz D. B., Shao Z. M., Yin X. M. (2007) Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J. Biol. Chem. 282, 4702–4710 [DOI] [PubMed] [Google Scholar]
- 33. Axe E. L., Walker S. A., Manifava M., Chandra P., Roderick H. L., Habermann A., Griffiths G., Ktistakis N. T. (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182, 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Acosta-Alvear D., Zhou Y., Blais A., Tsikitis M., Lents N. H., Arias C., Lennon C. J., Kluger Y., Dynlacht B. D. (2007) XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol. Cell 27, 53–66 [DOI] [PubMed] [Google Scholar]
- 36. Madeo F., Eisenberg T., Kroemer G. (2009) Autophagy for the avoidance of neurodegeneration. Genes Dev. 23, 2253–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.