Background: Ral GTPases are downstream effectors for Ras oncogenic activity.
Results: RalA ubiquitination is regulated at lipid rafts, and ubiquitinated RalA regulates lipid raft exposure at the plasma membrane.
Conclusion: Ubiquitination represents a novel post-translational modification of Ral GTPases determining RalA subcellular localization and impacting raft trafficking.
Significance: Exploration of the mechanisms of Ral GTPase post-translational modification is crucial to our understanding of Ras-driven signaling in tumorigenesis.
Keywords: Caveolin, GTPase, Lipid Raft, Plasma Membrane, Ubiquitination, Ral GTPases, Anchorage-independent Growth
Abstract
Ras GTPases signal by orchestrating a balance among several effector pathways, of which those driven by the GTPases RalA and RalB are essential to Ras oncogenic functions. RalA and RalB share the same effectors but support different aspects of oncogenesis. One example is the importance of active RalA in anchorage-independent growth and membrane raft trafficking. This study has shown a new post-translational modification of Ral GTPases: nondegradative ubiquitination. RalA (but not RalB) ubiquitination increases in anchorage-independent conditions in a caveolin-dependent manner and when lipid rafts are endocytosed. Forcing RalA mono-ubiquitination (by expressing a protein fusion consisting of ubiquitin fused N-terminally to RalA) leads to RalA enrichment at the plasma membrane and increases raft exposure. This study suggests the existence of an ubiquitination/de-ubiquitination cycle superimposed on the GDP/GTP cycle of RalA, involved in the regulation of RalA activity as well as in membrane raft trafficking.
Introduction
RalA and RalB GTPases are downstream effectors for many Ras-mediated biological functions and are indispensable to Ras oncogenic activity (1–3). Although RalA and RalB are 82% identical and share a set of effectors, they contribute in very different ways to cell (patho)physiology. RalA is required for anchorage-independent proliferation and tumor growth, whereas RalB participates in cancer cell survival and metastasis formation (1, 4), although these roles may be very context-dependent (5, 6). RalA is involved in apicobasal cell polarization (7) and RalB in cell motility (8–10) and autophagy (11). Finally, RalA and B act during different phases of cytokinesis. This temporal difference is based at least in part on their different spatial distribution, together with the action of their activators, the RalGEFs (12). What drives the specific localization of RalA and B and their relationship to specific functions remains elusive.
Phosphorylation of RalA and B by different kinases (13–15) is a mechanism that contributes to driving their specific activation and localization. Here, we have explored whether ubiquitination, another major post-translational modification, takes place on RalA or B, and, if so, under which circumstances and for what purpose. Ubiquitination is important for proteasome-mediated protein degradation in the ubiquitin proteasome system, but also for other processes, including protein activation, localization, and interactions (see examples in Refs. 16–18). It is noteworthy that the proto-oncogenes H-Ras and N-Ras are ubiquitinated (18, 19), and this ubiquitination markedly disrupts the balance between plasma and endomembrane localization of Ras, favoring the latter and disfavoring signal transduction toward ERK1/2 (18, 20, 21). We show here that the endogenous RalA and RalB GTPases are ubiquitinated, which contributes to selective membrane localization. RalA (but not RalB) de-ubiquitination occurs in lipid raft microdomains and promotes raft endocytosis upon loss of cell-matrix interactions.
EXPERIMENTAL PROCEDURES
Cell Culture, DNA, and siRNA
HeLa cells were cultured as described previously (22). Plasmid and siRNA transfections were performed using Jet-PEI (Polyplus Transfection) and Hi-perfect (Qiagen), according to supplier protocols, respectively. siRNA for caveolin was a pool of four siRNAs from Qiagen.
A vector (pRK5-Ubi)8 to express proteins fused by their N terminus to ubiquitin was generated by inserting an oligonucleotide encoding ubiquitin in the polylinker of plasmid pRK5. In the resulting plasmid pRK5-Ubi, the ubiquitin coding region is followed by a polylinker containing unique sites for EcoRI, BamHI, XbaI, SalI, PstI, and HindIII (frame gaa ttc gga tcc tct aga gtc gac ctg cag aag ctt). Coding regions for 3×FLAG-RalA and 3×FLAG-RalB (12) were amplified by polymerase chain reaction (PCR) and cloned between the BamHI and PstI sites of pRK5-Ubi.
Inhibitors
Inhibition of the proteasome was performed using 10 mm MG132 or ALLN for 4 h. Inhibition of lysosomes was performed using a combination of the inhibitors E-64 and pepstatin at 10 mg/ml and 50 mm, respectively, for 4 h. Inhibition of ubiquitination was achieved using the E1 inhibitor PYR41 at 50 mm for 6 h. Inhibition of dynamin-1 and -2 was performed with Dynasore (80 mm for 30 min). Destruction of lipid rafts was performed by treating cells with 15 mm methyl-b-cyclodextrin for 30 min. All inhibitors were purchased from Sigma.
Immunoprecipitation
Cell lysis was performed using a radioimmune precipitation assay buffer (150 mm NaCl, 0.1% Nonidet P-40, 0.1% SDS, protease inhibitors, 50 mm NaF, 1 mm NaVO3, and 50 mm Tris-HCl, pH 8). Lysates were mildly sonicated. Immunoprecipitation of vesicular stomatitis virus- or HA-tagged ubiquitin was carried out with vesicular stomatitis virus (Abcam) or HA antibody (Roche Applied Science) according to standard procedures. Sepharose G and A beads were purchased from GE Healthcare. The bound proteins and whole cell extracts were analyzed by immunoblotting with the antibodies detecting the following proteins: RalA (BD 610222), RalB (Upstate 04-037; Cell Signaling 3523), b-catenin (BD 610154), caveolin-1 (BD 610060), and FLAG epitope (Sigma F1804). Secondary antibodies coupled to peroxidase were obtained from Jackson Laboratories and used at a dilution of 1/25,000.
Cobalt Affinity Chromatography
Purification of ubiquitinated proteins was performed according to published procedures (23). In short, HeLa cells were transfected with expression vectors encoding a His6-tagged-ubiquitin (His6-Ubi) and/or Ral alleles. Then, 10 × 106 cells were lysed for the purification of ubiquitinated overexpressed proteins, and 20 × 106 cells for the purification of ubiquitinated endogenous ones. Cells were harvested in buffer B (PBS, pH 8, 6 m guanidium HCl, 0.1% Nonidet P-40, 10 mm b-mercaptoethanol), lysed by sonication (cycles of 30 s for 15 min at 200 W; Bioruptor, diagenode) and incubated for 4 h at 4 °C with 100 ml of TALON metal affinity beads (Clontech; 1:1 slurry equilibrated in buffer B). Prior to purification, a 100-ml aliquot of the lysate was precipitated with 10% ethanol to eliminate buffer B and resuspended in 50 ml of sample buffer 2×. The resin was washed once with buffer B, twice with buffer 1B/2C (1 volume of buffer B, 2 volumes of buffer C), once in buffer 1B/3C (1 volume of buffer B, 3 volumes of buffer C), and once in buffer C. Buffer C is 0.1% Nonidet P-40, 5% glycerol, and 20 mm imidazole in PBS, pH 8. Proteins were eluted with 100 ml of sample buffer supplemented with 200 mm DTT.
For those experiments concerning the impact of adhesion on ubiquitination, 10 × 106 transfected cells were plated on a 10-cm dish coated with 1% agarose (cells in suspension) for 90 min and afterward on fibronectin (25 mg/ml), where cells were allowed to adhere for 20 min prior to lysis and purification.
Immunofluorescence, Cholera Toxin B (CTX), and Transferrin Labeling
For RalA localization, cells were fixed and permeabilized with cold methanol for 10 min followed by 1 min in cold acetone at -20 °C. After saturation in 3% BSA in PBS for 30 min at 37 °C, they were incubated with anti-FLAG antibody (1/1,000 in 0.3% BSA in PBS) for 1 h at 37 °C. Cy3-coupled anti-mouse antibody and DAPI diluted in 0.3% BSA in PBS were incubated for 1 h at 37 °C. Coverslips were mounted using ProLong (Invitrogen).
The “t0” point cells were placed on ice for 20 min to inhibit endocytosis and incubated with CTX coupled to Alexa Fluor 488 (15 mg/ml; Molecular Probes) and transferrin coupled to Alexa Fluor 647 (10 mg/ml; Molecular Probes) for 20 min at 4 °C. Finally, cells were fixed with 3.7% paraformaldehyde (PFA) for 10 min. In t = 40 min, cells were shifted from ice to 37 °C for 40 min prior to fixing with 3.7% PFA for 10 min at 4 °C. All cells were permeabilized using 0.1% Triton X-100 in PBS solution for 10 min on ice and processed for immunofluorescence as above. Cells in suspension were treated as adherent cells for CTX labeling and fixed with 3.7% PFA before being cytospun on slides. Images were acquired on an inverted microscope (model DMIRE2; Leica) equipped with a cool CCD camera (CoolSNAP HQ). The Z-positioning was accomplished using a piezoelectric motor, and stacks were processed and analyzed using the Metamorph software.
RESULTS
Ral GTPases Are Ubiquitinated, but Not for Degradation
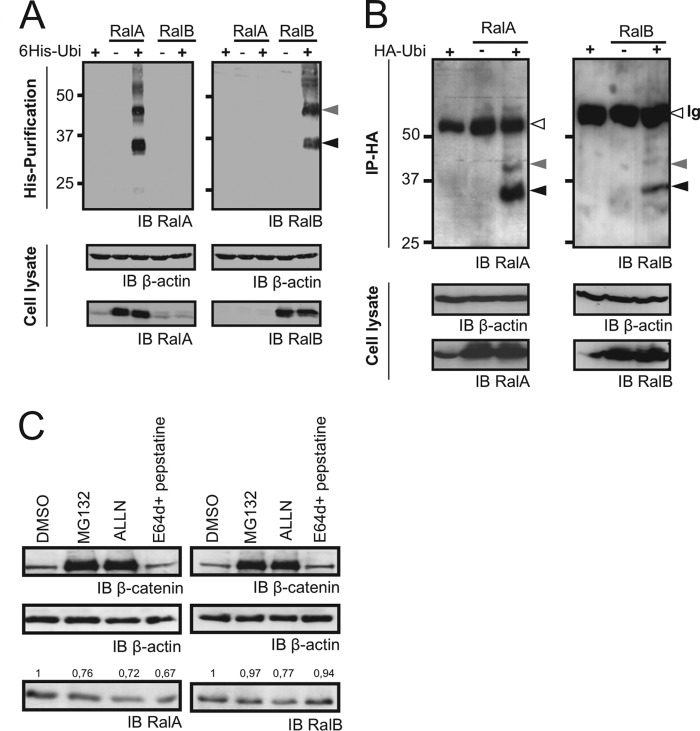
We determined whether RalA and RalB can undergo ubiquitination in vivo (Fig. 1). Plasmids expressing Ral and His-tagged ubiquitin were transfected into HeLa cells together and separately as controls. Cobalt affinity chromatography was used to purify ubiquitin conjugates, followed by Western blotting to detect RalA and RalB, as done previously for Ras GTPases (18). In Fig. 1A, Western blot analysis using specific anti-Ral antibodies revealed two RalA and RalB ubiquitin-conjugated species: a major ubiquitinated band at 42 kDa and a minor ubiquitinated band at 35 kDa (arrowheads). These bands were not detected when His-tagged ubiquitin or Ral GTPases were expressed alone in the cells and disappeared when the experiments were performed in the presence of the E1 ubiquitin-activating enzyme inhibitor PYR41 (24) (supplemental Fig. 1A). Ubiquitination of RalA mutants deleted of the C-terminal domain required for Ral membrane localization (RalA ΔCterm) was strongly decreased (supplemental Fig. 2C). HA-fused ubiquitin was also coexpressed with RalA or RalB, and ubiquitinated proteins were immunoprecipitated with anti-HA antibodies. Both RalA and B were detected in these immunoprecipitates and displayed patterns similar to those obtained by affinity chromatography (Fig. 1B). Ral ubiquitination was neither cell type- nor species-specific. It was observed in murine cells (hamster HET-SR fibroblasts (25) and CHO cells) as well as in human ones (HeLa (Fig. 1) and HEK293 (data not shown)). Considering that a single ubiquitin molecule is ∼8 kDa, Ral (25 kDa) was ubiquitinated mostly by one (minor band at ∼35 kDa) or two ubiquitin moieties (major band at ∼42 kDa), but higher molecular mass species were also observed (Fig. 1A). To distinguish between poly-ubiquitination (a second ubiquitin attached to a lysine of the first ubiquitin, attached directly to Ral) and multi-ubiquitination (a second ubiquitin attached directly to Ral), a plasmid expressing a vesicular stomatitis virus G-tagged ubiquitin mutated on all lysines (K0-ubiquitin) was used. Supplemental Fig. 1B shows that the same pattern of Ral ubiquitination was observed with both wild-type and K0-ubiquitin. The second ubiquitin is therefore not attached to the first one. When bi-ubiquitinated, the two ubiquitins are directly attached to Ral on different lysines. Poly-ubiquitination targets proteins for proteosomal degradation. Cells were treated with proteasome inhibitors MG132 or ALLN and lysosomal degradation inhibitors pepstatin + E64 or (Fig. 1C) leupeptin (data not shown). β-Catenin, which is known to be degraded after ubiquitination, accumulated in cells treated with MG132 or ALLN, whereas Ral never changed. Taken together, these results show that RalA and B undergo multi-ubiquitination in vivo and their ubiquitination does not regulate protein degradation.
FIGURE 1.
Ral GTPases are ubiquitinated, but not for degradation. A, exogenously expressed RalA and RalB were ubiquitinated. Lysates from HeLa cells overexpressing His6-Ubi and/or RalA or RalB were subjected to affinity chromatography purification on cobalt beads followed by immunodetection using anti-RalA or RalB antibodies after SDS-PAGE and blotting (IB). Whole lysate contents were analyzed by immunoblotting with the indicated antibodies (cell lysate). Black and gray arrowheads indicate the mono- and bi-ubiquitinated forms of Ral, respectively. B, exogenously expressed Ral GTPases immunoprecipitates with HA-ubiquitin. Lysates from HeLa cells overexpressing RalA or RalB and/or HA-ubiquitin were subjected to HA immunoprecipitation (IP) followed by immunoblotting with the indicated antibodies. Whole lysates were analyzed as in A. C, ubiquitination of Ral GTPases was not a signal for degradation. HeLa cells were treated with dimethyl sulfoxide (DMSO), MG132 (10 μm), ALLN (10 μm), or E64 and pepstatin (10 μg/ml and 50 μm, respectively) for 6 h. Whole cell extracts were analyzed by immunoblotting with anti-RalA or anti-RalB antibodies. Ratios between β-actin and RalA or RalB are indicated above the lanes. No increase in the quantity of RalA or B was detectable under these conditions. The regulation of cellular quantities of Ral GTPases did not rely on proteasome or lysosome degradation.
Ral GTPase Ubiquitination Is Dependent on Raft Trafficking
To ascertain whether the ubiquitination of Ral was dependent on its activation, we tested dominant active RalA mutants (G23V) and two dominant negative mutations (S28N and G26A). RalA G23V was more ubiquitinated than wild-type RalA, but the G26A and S28N mutants showed opposite behaviors, with the former more and the latter less ubiquitinated than wild type (supplemental Fig. 2B). Similar results were obtained for RalB (data not shown). Ral GTPase activity therefore did not modulate their ubiquitination.
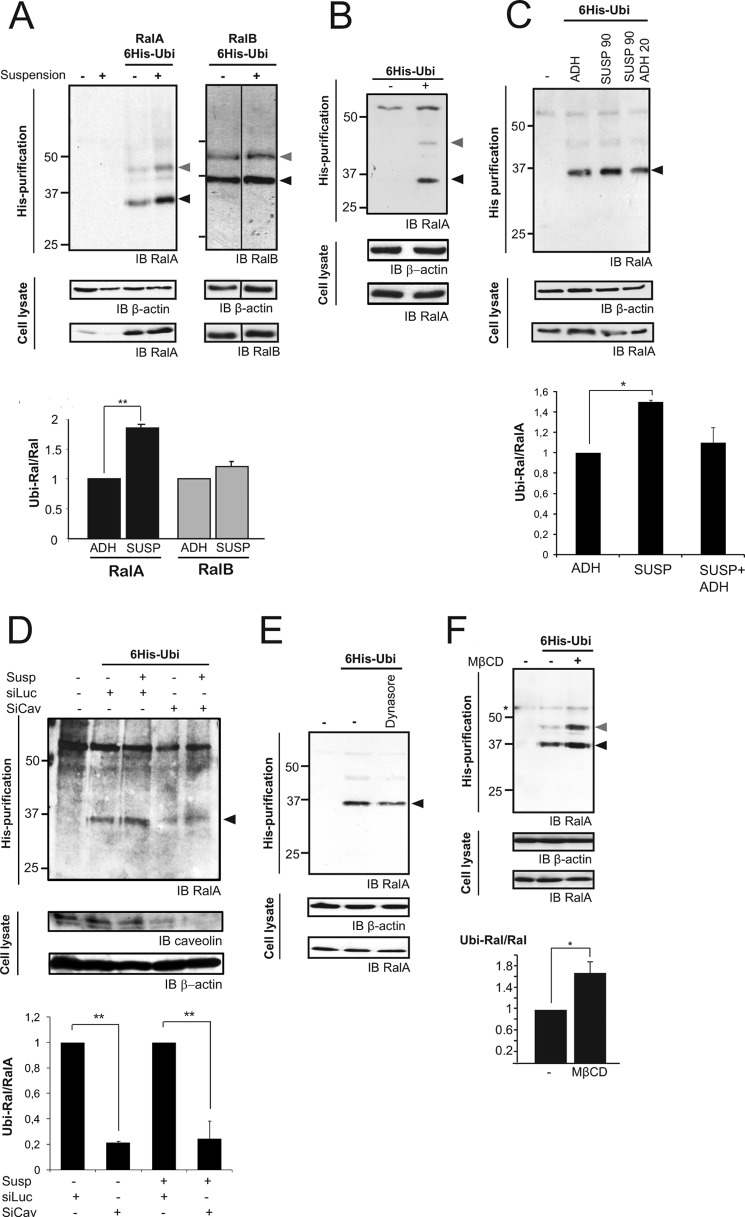
Subsequent investigation concerned the link between Ral GTPase ubiquitination and function. RalA has been reported to support anchorage-independent growth (2). In nonadherent cells, RalA promotes membrane raft exocytosis to sustain proliferative signaling (26). Exogenous Ral GTPase ubiquitination status was therefore explored in nonadherent versus adherent cells. First, the ubiquitination of exogenous Ral GTPases was analyzed. Cells overexpressing RalA or RalB together with His6-Ubi were grown in adherent conditions or in suspension for 48 h (Fig. 2A). Nonadherent cells displayed a substantial and reproducible 2-fold increase in RalA ubiquitination (Fig. 2A). No variation in the ubiquitination level of RalB was detected under these conditions (Fig. 2A).
FIGURE 2.
Endogenous RalA ubiquitination increases in nonadherent cells and is regulated by raft endocytosis. A, exogenously expressed RalA, but not RalB, was more ubiquitinated in nonadherent cells than in adherent ones. HeLa cells overexpressing RalA or RalB with or without (the first two lanes) His6-Ubi were seeded in plates coated (+) or not (−) with agarose to prevent adhesion. After 48 h, cells were lysed, and cell extracts were subjected to affinity chromatography purification on cobalt beads for ubiquitinated proteins. Ubiquitinated forms of RalA and RalB were detected by Western blotting (IB). Arrowheads indicate the mono- (black) and bi-ubiquitinated (gray) forms of Ral. The quantification of mono-ubiquitinated bands was performed, with the ratio between the intensity of mono-ubiquitinated Ral and total Ral shown to be significantly different between adherent cells and cells in suspension (**, p < 0.01; RalA, n = 4; RalB, n = 3). B, ubiquitination of endogenous RalA. HeLa cells expressing His6-Ubi (+) or not (−) were lysed, and cell extracts were subjected to affinity chromatography purification on cobalt beads to analyze endogenous RalA ubiquitination. Arrowheads indicate the mono- (black) and bi-ubiquitinated (gray) forms of RalA. C, endogenous RalA was more ubiquitinated in nonadherent cells. HeLa cells expressing His6-Ubi were seeded for 90 min in plates coated with agarose (SUSP) or in adherent conditions (ADH) or in plates coated with agarose (90 min) followed by 20 min on fibronectin-coated plates (20 μg/ml) (SUSP 90/ADH 20) before being harvested, and cell content was submitted to affinity chromatography for purification of ubiquitinated proteins. Empty vector was used as a specificity control on the first lane. The quantification of mono-ubiquitinated bands was performed, and the ratio between the intensity of mono-ubiquitinated Ral and total Ral was shown to be significantly different between adherent cells and cells in suspension (*, p < 0.05, n = 3). D, caveolin depletion reduced RalA ubiquitination. Adherent or nonadherent HeLa cells expressing His6-Ubi or not (lane 1) were treated with siControl (siLuc) or siCaveolin-1 and analyzed for RalA ubiquitination. The arrowhead indicates the mono-ubiquitinated RalA band. Caveolin depletion is shown. Quantification of mono-ubiquitinated bands was performed, and the ratio between the intensity of mono-ubiquitinated RalA and total Ral was calculated. The ratio was set up as 1 in siLuc-treated cells. In both situations (adherent cells as well as cells in suspension), RalA ubiquitination was substantially and significantly decreased when caveolin-1 was depleted (**, p < 0.01, siCaveolin-1 versus siLuc, n = 3). E, blocking endocytosis reduced RalA ubiquitination. HeLa cells expressing His6-Ubi or not (lane 1) were treated for 30 min with dimethyl sulfoxide (−) or dynasore (100 μm), and the ubiquitination of RalA was analyzed. F, disruption of lipid raft increased Ral ubiquitination. HeLa cells expressing His6-Ubi were treated with methyl-β-cyclodextrin (MβCD) (15 mm, 30 min). Empty vector transfection was used as a control in the first lane. RalA ubiquitination was analyzed as in A, and the increase in ubiquitinated RalA in cells treated with methyl-β-cyclodextrin was found to be statistically significant (*, p < 0.05, n = 3).
When cells are detached from their substrate, lipid rafts are internalized. Upon reattachment to the matrix, rafts are exocytosed for surface display using the RalA/Sec5 pathway (26). To test whether RalA ubiquitination might be regulated by raft trafficking or adhesion, RalA ubiquitination was investigated within the time frame of raft internalization (90 min of suspension) and re-exposure (20 min of re-adhesion). To avoid any effects of overexpressed RalA in cell adhesion, it was decided to follow endogenous RalA ubiquitination (Fig. 2C). Conditions to detect endogenous Ral GTPase ubiquitination were previously established (Fig. 2B and supplemental Fig. 2A). By doubling the cell number (20 × 106) used for purification (compare experiments in Figs. 1 and 2B), it was possible to detect a major mono-ubiquitinated endogenous RalA band at 35 kDa and a weaker bi-ubiquitinated band at 42 kDa, suggesting that endogenous RalA is mostly mono-ubiquitinated. The bi-ubiquitinated band at 42 kDa increased in highly exposed blots (supplemental Fig. 2A), but higher exposure also revealed nonspecific 50-kDa bands. The proportion of mono-ubiquitinated endogenous RalA among RalA proteins increased by a 1.5 factor in cells detached from the substrate for 90 min (Fig. 2C) and returned to steady-state levels after 20 min of cell re-adhesion (Fig. 2C), when rafts are also re-exposed on the plasma membrane. These observations indicated that endogenous RalA ubiquitination was up-regulated upon cell detachment from the substrate and perhaps also upon raft endocytosis. We also confirmed that the activation of endogenous RalA did not regulate its ubiquitination because serum stimulation induced no change in endogenous RalA-ubiquitinated 35-and 42-kDa bands (supplemental Fig. 2A). Lipid raft endocytosis has been reported to be dependent on caveolin-1 and dynamin-2 (27, 28). Caveolin-1 was depleted in His6-Ubi-expressing HeLa cells. The purification of endogenous ubiquitinated RalA was less efficient in RNAi-treated cells, but it was possible to detect the mono-ubiquitinated RalA band increasing 1.5-fold in siControl cells detached from the matrix compared with adherent cells (Fig. 2D). siCaveolin-1-treated cells showed a RalA ubiquitination inhibited by 80% in both adherent and nonadherent cells compared with siControl cells (Fig. 2D). When dynamin-2 was inhibited with the chemical inhibitor dynasore, decrease in endogenous RalA ubiquitination was also observed (Fig. 2E). Because RalA was less ubiquitinated when rafts were at the plasma membrane, one possibility could be that RalA is de-ubiquitinated in rafts at the plasma membrane. This hypothesis is supported by the observation that when raft structures were destroyed (by treating cells for 30 min with methyl-β-cyclodextrin, which extracts cholesterol) (29), endogenous RalA ubiquitination was increased 1.5-fold (Fig. 2F). We conclude that lipid rafts at the plasma membrane are required for RalA de-ubiquitination.
De-ubiquitination of RalA Is Required for Raft Endocytosis
To explore the role of RalA ubiquitination, efforts were devoted to identifying the lysine(s) of Ral that are subjected to ubiquitination. By changing all 21 RalA lysine residues into arginine one by one, it was found that several lysines on RalA could serve as ubiquitination sites (supplemental Table 1). Ubiquitinated RalA was detected in all combinations with the exception of the variant in which all lysines were mutated (data not shown). These results showed that Ral can be ubiquitinated on several lysines, but precluded any genetic approach to unraveling the functional significance of Ral ubiquitination.
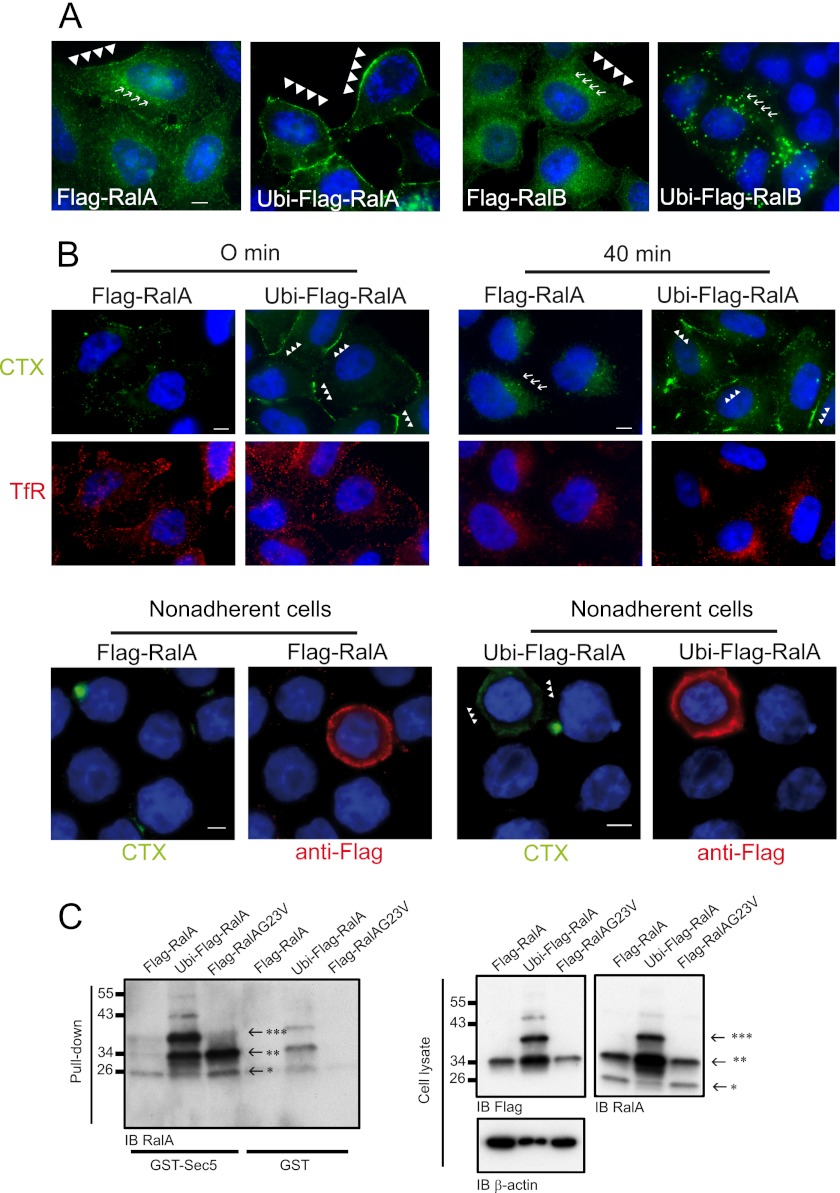
It was therefore decided to use a constitutively ubiquitinated form of Ral in which the ubiquitin moiety was fused via a peptide bound to the N terminus of RalA or RalB. This approach has been successfully employed elsewhere for functional studies of Ras ubiquitination (18). Three FLAG epitopes were inserted between ubiquitin and the Ral protein, leading to a protein with a topology of N-ubiquitin-3×FLAG-Ral-C. The 3×FLAG-RalA and ubiquitin-3×FLAG-RalB fusions served as controls. These Ubi-Ral constructs were consistently highly overexpressed as fusion proteins, suggesting, as expected, that they are not sensitive to de-ubiquitin hydrolases (data not shown). As shown in Fig. 3A and as previously published, RalA was observed at the plasma membrane as well as in endomembranes (30). By contrast, Ubi-RalA was almost totally absent from the cytoplasm and mainly localized at the plasma membrane. However, RalB was observed on endomembranes and at the plasma membrane, whereas Ubi-RalB was absent from the cytoplasm but accumulated in internal punctate structures of 400–900 nm in size (Fig. 3A). The different impacts of ubiquitin fusions on RalA and RalB localization strongly support the notion that ubiquitin fusion itself does not drive nonspecific protein localization and suggest that the ubiquitination of RalA is a signal for plasma membrane targeting or stabilization.
FIGURE 3.
Ubi-RalA fusion protein localized at the plasma membrane and inhibited endocytosis of lipid rafts. A, Ubi-RalA was enriched at the plasma membrane. HeLa cells expressing FLAG-RalA, FLAG-RalB, Ubi-FLAG-RalA, or Ubi-FLAG-RalB were fixed with methanol. FLAG proteins were detected using the anti-FLAG M2 antibody and FITC-conjugated secondary anti-mouse antibodies. Exposure times were identical under all four conditions. Scale bars represent 5 μm. Arrowheads and arrows point to examples of the indicated Ral at the plasma membrane and on endomembranes, respectively. B, Ubi-RalA inhibited endocytosis of lipid rafts. HeLa cells expressing FLAG-RalA or Ubi-FLAG-RalA in adherent (upper panel) or nonadherent conditions (lower panel) were incubated at 4 °C with cholera-toxin coupled to Alexa Fluor 488 (15 μg/ml) and transferrin coupled to Alexa Fluor 647 (10 μg/ml) for 20 min. Cells were fixed with PFA either immediately (t0) (upper and lower panels) or after restoration of endocytosis by incubating cells 40 min at 37 °C (t40). After extensive washing, cells were permeabilized and FLAG-Ral proteins were detected using the anti-FLAG M2 antibody to control cell transfection. To carefully follow CTX signal on the entire Z of cells, we took stack images by using the same exposure parameters within the different conditions. Images in both panels are the results of Z-projection representative of three independent experiments. It should be noted that the detection of RalA at the plasma membrane required cells to be fixed with methanol, which did not allow for detection of bound CTX. In the fields displayed here, all cells expressed the exogenous Ral constructs (data not shown). Arrowheads and arrows indicate examples of CTX enrichment at the plasma membrane and on endomembranes, respectively. C, Ubi-RalA was bound to GTP. HeLa cells were transfected with plasmids expressing the indicated forms of RalA. Twenty-four hours later, cells were lysed. Lysates were split into equal parts, with each part submitted to affinity chromatography using equal amounts of GST fused to the Ral-binding domain of Sec5 (7) and GST as control. Proteins bound to the column were analyzed by Western blotting (IB). For each form, the quantity of protein bound to GST alone was subtracted from the amount bound to GST-Sec5. Evaluation of the relative binding between alleles was obtained by dividing this number by the quantity detected in whole cell lysates. If RalA G23V is considered as 100% bound to GTP, Ubi-RalA was then bound at 96%. *, endogenous RalA; **, FLAG-RalA; ***, Ubi-FLAG-RalA. Note that Ubi-FLAG-RalA can be degraded and lose its ubiquitin moiety. Tfr, transferrin.
To track the fate of rafts as a function of Ral ubiquitination status, the CTX receptor GM1 was used as a marker of lipid rafts, and transferrin receptor as a marker of clathrin-coated vesicles (31). Lipid rafts were found scattered in low quantities at the plasma membrane of control cells either not transfected or transfected with plasmids expressing ubiquitin alone (data not shown) or FLAG-RalA (Fig. 3B; upper panel, t = 0). By contrast, lipid rafts were dramatically enriched at the plasma membrane of cells expressing Ubi-RalA (Fig. 3B; t = 0) but not Ubi-RalB (supplemental Fig. 3). The endocytosis of lipid rafts was then followed upon expression of RalA, RalB, Ubi-RalA, and Ubi-RalB. After binding of CTX-Alexa Fluor 488 to the ganglioside receptor GM1 at 4 °C, cells were washed and then shifted at 37 °C, and GM1 localization was observed 40 min later. In both nontransfected cells and cells transfected to express Ubi alone (data not shown), RalA (Fig. 3B, upper panel, t = 40), or RalB (supplemental Fig. 3), CTX was found to be predominantly internalized as “normal.” In contrast, cells expressing Ubi-RalA did not internalize GM1: almost all of the CTX-Alexa Fluor 488 fluorescence was detected at the plasma membrane (Fig. 3B; t = 40). Transferrin receptor was tracked in the same cells using transferrin-Alexa Fluor 647. The abundance of transferrin receptor at the plasma membrane and its internalization were not affected by the expression of Ubi-RalA (Fig. 3B), demonstrating the specificity of the effect of ubiquitinated RalA on lipid rafts. When Ubi-RalB was expressed, levels of labeled CTX and transferrin, as well as their localization and dynamics, were also equivalent to control (supplemental Fig. 3), indicating that the effect on lipid rafts was specific to ubiquitinated RalA.
Ubi-RalA was also sufficient to inhibit GM1 internalization in suspension cells (Fig. 3B, lower panel). These results suggest that the de-ubiquitination of RalA is required for the endocytosis of lipid rafts.
Interplay between Ubiquitination and Activation of Ral
Because RalA-GTP is known to promote raft exposure (26), testing was performed to determine whether Ral ubiquitination impacted Ral activity. The quantities of GTP-bound forms of RalA, RalB, Ubi-RalA, and Ubi-RalB were tested using a standard pulldown methodology (32). It was observed that the ubiquitinated form of RalA (Fig. 3C) was extremely enriched in GTP. If RalA G23V is considered 100% bound to GTP, Ubi-RalA was then bound at 96%. Ubiquitination therefore seemed to favor Ral activation. An ubiquitination/de-ubiquitination cycle could be superimposed on the GDP/GTP cycle of RalA for the regulation of membrane raft trafficking.
The Effect of Ubiquitinated RalA in Rafts Is Mediated by Exocyst Complex Components
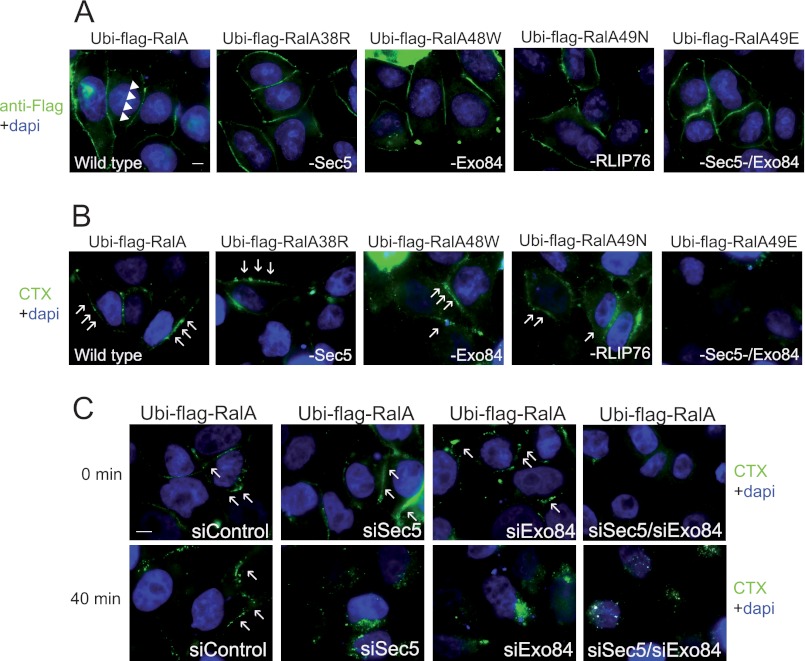
Effector loop mutants of Ubi-RalA have been generated, which selectively impair interactions of Ral with RalBP1, Sec5, Exo84, and Sec5 plus Exo84 (mutations 49N, 38R, 48W and 49E, respectively) (7, 33–35). These mutants were tested for impact on Ubi-RalA enrichment at the plasma membrane and promotion of lipid raft surface localization. As shown in Fig. 4A, all mutants were enriched at the plasma membrane. Furthermore, 38R, 48W, and 49N did not impair Ubi-RalA enrichment of CTX signal at the plasma membrane (Fig. 4B). However, Ubi-RalA 49E, which loses interaction with both Exo84 and Sec5, was inactive in this assay. Interaction with both Exo84 and Sec5 exocyst subunits is therefore likely required to mediate the effects of Ubi-RalA.
FIGURE 4.
Sec5 and Exo84 mediate the inhibition of lipid raft endocytosis induced by Ubi-RalA. A, the enrichment of Ubi-RalA at the plasma membrane was independent of its effectors. HeLa cells expressing Ubi-FLAG-RalA, Ubi-FLAG-RalA38R, Ubi-FLAG-RalA48W, Ubi-FLAG-RalA49N, or Ubi-FLAG-RalA49E were fixed and stained as in Fig. 3A using the anti-FLAG M2 antibody. All of these alleles were detected, as was wild-type Ubi-FLAG-RaA, enriched at the plasma membrane. B, Ubi-FLAG-RalA49E (defective in interaction with both Sec5 and Exo84) could not induce lipid raft enrichment. HeLa cells expressing Ubi-FLAG-RalA, Ubi-FLAG-RalA38R, Ubi-FLAG-RalA48W, Ubi-FLAG-RalA49N, or Ubi-FLAG-RalA49E were incubated at 4 °C with CTX coupled to Alexa Fluor 488 as in Fig. 3B. Images are the results of Z-projection of Z-stacks of three independent experiments as in Fig. 3B. Arrows indicate examples of CTX enrichment at the plasma membrane. All alleles, except RalA49E, which was defective in interaction with both Sec5 and Exo84, induced lipid raft enrichment at the plasma membrane. C, simultaneous depletion of Sec5 and Exo84 abolished the Ubi-RalA-induced enrichment of lipid rafts. HeLa cells expressing Ubi-FLAG-RalA were depleted by siRNA of Sec5, Exo84, or both Sec5 and Exo84, and, 48 h after transfection of siRNAs, were incubated at 4 °C with cholera toxin coupled to Alexa Fluor 488 as in Fig. 3B. Images are the results of Z-projection of Z-stacks of three independent experiments as in Fig. 3B. Arrows indicate examples of CTX enrichment at the plasma membrane.
These results were confirmed using siRNA-mediated depletion of Ral effectors in cells expressing Ubi-RalA. Compared with control, no substantial decrease of lipid raft enrichment was detected at the plasma membrane, as driven by the expression of Ubi-RalA, when RLIP76, Sec5, or Exo84 was depleted. Depletion of Sec5 or Exo84 alone did not impair the Ubi-RalA enrichment of the CTX signal at the plasma membrane (Fig. 4C, upper panel), whereas depletion of both Sec5 and Exo84 suppressed the CTX signal enrichment induced by Ubi-RalA (Fig. 4C, t = 0). The endocytosis of lipid rafts at t = 40 min was then followed, as shown in Fig. 3B (Fig. 4C, t = 40). Ubi-RalA was overexpressed in cells transfected by control siRNA-inhibited lipid raft endocytosis (Fig. 4C, t = 40), as can be seen in Fig. 3 (Fig. 3B, t = 40 min), whereas depletion of Sec5 or Exo84 alone was sufficient to suppress this inhibition. These results suggest a different contribution of exocyst subunits to the UbiRalA-dependent exposure/internalization cycle of lipid rafts.
DISCUSSION
The nondegradative ubiquitination of proteins introduces a degree of diversity to their biological functions by increasing their repertoire in terms of activity, localization, and/or interaction. In this study, we show that the RalA and RalB GTPases, two critical participants in Ras-driven oncogenesis (36, 37), are ubiquitinated proteins binding mainly one and occasionally two ubiquitin moieties. In particular, it was found that this modification could selectively modulate RalA and RalB localization and played a key role in the RalA regulation of lipid raft dynamics at the plasma membrane.
Ubiquitination Is a Signal Localizing RalA to the Plasma Membrane
Despite high sequence similarity and common effectors, RalA and RalB support different aspects of oncogenesis and cell homeostasis. In cytokinesis, we proposed that the different functions of RalA and RalB are correlated to a regulated subcellular localization (12). RalA and RalB subcellular localization have also been suggested to exhibit different impacts on oncogenesis (14, 38). However, the manner in which RalA and RalB achieve and maintain their different localizations is still poorly understood. Assuming that Ubi-Ral fusions mimic the physiological effect of ubiquitination, data from this investigation indicated that one function of ubiquitination was to direct RalA, but not RalB, to its plasma membrane destination. Ubi-RalA was not sensitive to de-ubiquitin hydrolases and was enriched at the plasma membrane. If ubiquitination targets RalA to the plasma membrane, de-ubiquitination might be responsible for delocalizing RalA from it.
RalA De-ubiquitination Occurs in Raft Microdomains and Regulates Raft Endocytosis
RalA ubiquitination increased in nonadherent cells, where raft microdomains were endocytosed, and decreased when cells re-adhered and rafts were re-exposed to the plasma membrane. Loss of raft microdomains at the plasma membrane therefore caused an increase in RalA ubiquitination, whereas blocking raft endocytosis caused a decrease. This suggests that RalA is de-ubiquitinated in raft microdomains.
The expression of Ubi-RalA fusion protein induced an accumulation of raft microdomains at the plasma membrane. Ral ubiquitination and raft trafficking seemed to regulate mutually. Mutants unable to interact with RalBP1, Exo84, or Sec5 were not impaired in the Ubi-Ral capacity to force lipid raft exposure at the plasma membrane, as opposed to mutants unable to interact with either Sec5 or Exo84. Exocyst complex mediated the Ubi-RalA effect on lipid rafts, and the two Ral effectors Exo84 and Sec5 appeared to compensate for one another. This was supported by the impact of the depletion of Sec5, Exo84, or both. The silencing of both subunits was required to abolish Ubi-RalA-induced raft exposure. Interestingly, membrane targeting of Ubi-RalA did not require interaction with its effectors involved in endocytosis and exocytosis. Mutants unable to interact with the exocyst complex or RalBP1 successfully reached the plasma membrane. We propose the existence of a molecular choreography in which ubiquitination targets RalA to the plasma membrane, where it is de-ubiquitinated in raft microdomains. In this scenario, RalA de-ubiquitination would be necessary for raft dynamics via Sec5 and Exo84.
Ubi-RalA induced lipid raft plasma membrane accumulation in nonadherent cells as well. RalA ubiquitination may play a role in promoting raft exocytosis of tumoral nonadherent cells, as does active RalA (26). However, because Ral ubiquitination was not influenced by Ral activation, it could be argued that the cycle of ubiquitination/de-ubiquitination is superimposed on the GDP/GTP RalA cycle. The ubiquitination/de-ubiquitination cycle represents a novel post-translational modification of Ral GTPases determining RalA subcellular localization and raft trafficking, and which could have a major impact on tumor growth. One might speculate that the E3 ligase(s) responsible for RalA ubiquitination should support RalA function in raft exocytosis and contribute positively to anchorage-independent growth and tumorigenesis. In return, the de-ubiquitin hydrolases with ubiquitinated RalA as a target would then be tumor suppressors.
Supplementary Material
Acknowledgments
We thank Drs. Gacon, Lemichez, Lamaze, and Hagenauer for the indispensable reagents, stimulating discussions, and insightful suggestions; Nathalie Brandon for skillful technical assistance; and Michael White and Maria Carla Parrini for critical reading of the manuscript.
This research was supported by grants from ANR and ARC (to J. C.).

This article contains supplemental Figs. 1–3 and Table 1.
- Ubi
- ubiquitin
- CTX
- cholera toxin B
- GM1
- monosialotetrahexosylganglioside
- PFA
- paraformaldehyde
- GEF
- guanosine exchange factor.
REFERENCES
- 1. Chien Y., Kim S., Bumeister R., Loo Y. M., Kwon S. W., Johnson C. L., Balakireva M. G., Romeo Y., Kopelovich L., Gale M., Jr., Yeaman C., Camonis J. H., Zhao Y., White M. A. (2006) RalB GTPase-mediated activation of the IκB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 127, 157–170 [DOI] [PubMed] [Google Scholar]
- 2. Chien Y., White M. A. (2003) RAL GTPases are linchpin modulators of human tumour cell proliferation and survival. EMBO Rep. 4, 800–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Drosten M., Dhawahir A., Sum E. Y., Urosevic J, Lechuga C. G., Esteban L. M., Castellano E., Guerra C., Santos E., Barbacid M. (2010) Genetic analysis of Ras signalling pathways in cell proliferation, migration and survival. EMBO J. 29, 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lim K. H., O'Hayer K., Adam S. J., Kendall S. D., Campbell P. M., Der C. J., Counter C. M. (2006) Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr. Biol. 16, 2385–2394 [DOI] [PubMed] [Google Scholar]
- 5. Martin T. D., Samuel J. C., Routh E. D., Der C. J., Yeh J. J. (2011) Activation and involvement of Ral GTPases in colorectal cancer. Cancer Res. 71, 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sowalsky A. G., Alt-Holland A., Shamis Y., Garlick J. A., Feig L. A. (2010) RalA suppresses early stages of Ras-induced squamous cell carcinoma progression. Oncogene 29, 45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moskalenko S., Henry D. O., Rosse C., Mirey G., Camonis J. H., White M. A. (2002) The exocyst is a Ral effector complex. Nat. Cell Biol. 4, 66–72 [DOI] [PubMed] [Google Scholar]
- 8. Oxford G., Theodorescu D. (2003) Ras superfamily monomeric G proteins in carcinoma cell motility. Cancer Lett. 189, 117–128 [DOI] [PubMed] [Google Scholar]
- 9. Rossé C., Hatzoglou A., Parrini M. C., White M. A., Chavrier P., Camonis J. (2006) RalB mobilizes the exocyst to drive cell migration. Mol. Cell. Biol. 26, 727–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oxford G., Owens C. R., Titus B. J., Foreman T. L., Herlevsen M. C., Smith S. C., Theodorescu D. (2005) RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 65, 7111–7120 [DOI] [PubMed] [Google Scholar]
- 11. Bodemann B. O., Orvedahl A., Cheng T., Ram R. R., Ou Y. H., Formstecher E., Maiti M., Hazelett C. C., Wauson E. M., Balakireva M., Camonis J. H., Yeaman C., Levine B., White M. A. (2011) RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell 144, 253–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cascone I., Selimoglu R., Ozdemir C., Del Nery E., Yeaman C., White M., Camonis J. (2008) Distinct roles of RalA and RalB in the progression of cytokinesis are supported by distinct RalGEFs. EMBO J. 27, 2375–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu J. C., Chen T. Y., Yu C. T., Tsai S. J., Hsu J. M., Tang M. J., Chou C. K., Lin W. J., Yuan C. J., Huang C. Y. (2005) Identification of V23RalA-Ser194 as a critical mediator for Aurora-A-induced cellular motility and transformation by small pool expression screening. J. Biol. Chem. 280, 9013–9022 [DOI] [PubMed] [Google Scholar]
- 14. Lim K. H., Brady D. C., Kashatus D. F., Ancrile B. B., Der C. J., Cox A. D., Counter C. M. (2010) Aurora-A phosphorylates, activates, and relocalizes the small GTPase RalA. Mol. Cell. Biol. 30, 508–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang H., Owens C., Chandra N., Conaway M. R., Brautigan D. L., Theodorescu D. (2010) Phosphorylation of RalB is important for bladder cancer cell growth and metastasis. Cancer Res. 70, 8760–8769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoeller D., Crosetto N., Blagoev B., Raiborg C., Tikkanen R., Wagner S., Kowanetz K., Breitling R., Mann M., Stenmark H., Dikic I. (2006) Regulation of ubiquitin-binding proteins by mono-ubiquitination. Nat. Cell Biol. 8, 163–169 [DOI] [PubMed] [Google Scholar]
- 17. Dikic I. (2009) Journal club: a new ubiquitin chain, a new signal. Nat. Rev. Mol. Cell Biol. 10, 306. [DOI] [PubMed] [Google Scholar]
- 18. Jura N., Scotto-Lavino E., Sobczyk A., Bar-Sagi D. (2006) Differential modification of Ras proteins by ubiquitination. Mol. Cell 21, 679–687 [DOI] [PubMed] [Google Scholar]
- 19. Kim S. E., Yoon J. Y., Jeong W. J., Jeon S. H., Park Y., Yoon J. B., Park Y. N., Kim H., Choi K. Y. (2009) H-Ras is degraded by Wnt/β-catenin signaling via β-TrCP-mediated poly-ubiquitylation. J. Cell Sci. 122, 842–848 [DOI] [PubMed] [Google Scholar]
- 20. Xu L., Lubkov V., Taylor L. J., Bar-Sagi D. (2010) Feedback regulation of Ras signaling by Rabex-5-mediated ubiquitination. Curr. Biol. 20, 1372–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yan H., Jahanshahi M., Horvath E. A., Liu H. Y., Pfleger C. M. (2010) Rabex-5 ubiquitin ligase activity restricts Ras signaling to establish pathway homeostasis in Drosophila. Curr. Biol. 20, 1378–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Balakireva M., Rossé C., Langevin J., Chien Y. C., Gho M., Gonzy-Treboul G., Voegeling-Lemaire S., Aresta S., Lepesant J. A., Bellaiche Y., White M., Camonis J. (2006) The Ral/exocyst effector complex counters c-Jun N-terminal kinase-dependent apoptosis in Drosophila melanogaster. Mol. Cell. Biol. 26, 8953–8963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laney J. D., Hochstrasser M. (2002) Analysis of protein ubiquitination. Curr. Protoc. Protein Sci. Chapter 14:Unit 14 5 [DOI] [PubMed] [Google Scholar]
- 24. Yang Y., Kitagaki J., Dai R. M., Tsai Y. C., Lorick K. L., Ludwig R. L., Pierre S. A., Jensen J. P., Davydov I. V., Oberoi P., Li C. C., Kenten J. H., Beutler J. A., Vousden K. H., Weissman A. M. (2007) Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 67, 9472–9481 [DOI] [PubMed] [Google Scholar]
- 25. Deichman G. I., Kashleva H. A., Kluchareva T. E., Matveeva V. A. (1989) Clustering of discrete cell properties essential for tumorigenicity and metastasis. II. Studies of Syrian hamster embryo fibroblasts transformed by Rous sarcoma virus. Int. J. Cancer 44, 908–910 [DOI] [PubMed] [Google Scholar]
- 26. Balasubramanian N., Meier J. A., Scott D. W., Norambuena A., White M. A., Schwartz M. A. (2010) RalA-exocyst complex regulates integrin-dependent membrane raft exocytosis and growth signaling. Curr. Biol. 20, 75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Del Pozo M. A. (2004) Integrin signaling and lipid rafts. Cell Cycle 3, 725–728 [PubMed] [Google Scholar]
- 28. del Pozo M. A., Balasubramanian N., Alderson N. B., Kiosses W. B., Grande-García A., Anderson R. G., Schwartz M. A. (2005) Phospho-caveolin-1 mediates integrin-regulated membrane domain internalization. Nat. Cell Biol. 7, 901–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kilsdonk E. P., Yancey P. G., Stoudt G. W., Bangerter F. W., Johnson W. J., Phillips M. C., Rothblat G. H. (1995) Cellular cholesterol efflux mediated by cyclodextrins. J. Biol. Chem. 270, 17250–17256 [DOI] [PubMed] [Google Scholar]
- 30. Shipitsin M., Feig L. A. (2004) RalA but not RalB enhances polarized delivery of membrane proteins to the basolateral surface of epithelial cells. Mol. Cell. Biol. 24, 5746–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harder T., Scheiffele P., Verkade P., Simons K. (1998) Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 141, 929–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wolthuis R. M., Franke B., van Triest M., Bauer B., Cool R. H., Camonis J. H., Akkerman J. W., Bos J. L. (1998) Activation of the small GTPase Ral in platelets. Mol. Cell. Biol. 18, 2486–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bauer B., Mirey G., Vetter I. R., García-Ranea J. A., Valencia A., Wittinghofer A., Camonis J. H., Cool R. H. (1999) Effector recognition by the small GTP-binding proteins Ras and Ral. J. Biol. Chem. 274, 17763–17770 [DOI] [PubMed] [Google Scholar]
- 34. Jin R., Junutula J. R., Matern H. T., Ervin K. E., Scheller R. H., Brunger A. T. (2005) Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase. EMBO J. 24, 2064–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moskalenko S., Tong C., Rosse C., Mirey G., Formstecher E., Daviet L., Camonis J., White M. A. (2003) Ral GTPases regulate exocyst assembly through dual subunit interactions. J. Biol. Chem. 278, 51743–51748 [DOI] [PubMed] [Google Scholar]
- 36. Camonis J. H., White M. A. (2005) Ral GTPases: corrupting the exocyst in cancer cells. Trends Cell Biol. 15, 327–332 [DOI] [PubMed] [Google Scholar]
- 37. Bodemann B. O., White M. A. (2008) Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat. Rev. Cancer 8, 133–140 [DOI] [PubMed] [Google Scholar]
- 38. Lim K. H., Baines A. T., Fiordalisi J. J., Shipitsin M., Feig L. A., Cox A. D., Der C. J., Counter C. M. (2005) Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell 7, 533–545 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.