Background: Sequestosome 1/p62 is a scaffolding protein that plays a critical role in receptor-mediated signal transduction.
Results: p62 interacts with IRS-1 to enhance the Akt phosphorylation, GLUT4 translocation, and glucose uptake upon insulin stimulation.
Conclusion: p62 participates in IRS-1 insulin signaling.
Significance: p62 plays a role in insulin-signaling, which provides a potential mechanism for its connection to type 2 diabetes.
Keywords: Insulin, Protein-Protein Interactions, SH2 Domains, Signaling, Skeletal Muscle, IRS-1, p62, Sequestosome-1
Abstract
Defects in the insulin-signaling pathway may lead to the development of skeletal muscle insulin resistance, which is one of the earliest abnormalities detected in individuals with the metabolic syndrome and predisposes them to develop type 2 diabetes. Previous studies have shown that deletion of the mouse sequestosome 1/p62 gene results in mature-onset obesity that progresses to insulin and leptin resistance and, ultimately, type 2 diabetes. Sequestosome 1/p62 is involved in receptor-mediated signal transduction and functions as an intracellular signal modulator or adaptor protein. Insulin receptor substrate-1 (IRS-1) plays a central role in transducing the insulin signal via phosphorylation, protein-protein interactions, and protein modifications. Mapping studies demonstrated that the SH2 domain at the amino terminus of sequestosome 1/p62 interacts with IRS-1 upon insulin stimulation. Further, IRS-1 interacts with p62 through its YMXM motifs at Tyr-608, Tyr-628, and/or Tyr-658 in a manner similar to its interaction with p85 of phosphoinositol 3-kinase. Overexpression of p62 increased phosphorylation of Akt, GLUT4 translocation, and glucose uptake, providing evidence that p62 participates in the insulin-signaling pathway through its interactions with IRS-1.
Introduction
Insulin is central to regulating glucose uptake in skeletal muscle, and skeletal muscle insulin resistance is a key contributor to the etiology of type 2 diabetes. The intracellular action of insulin is mediated by modification of the activity and subcellular localization of regulatory proteins and enzymes through protein-protein interactions and protein phosphorylation. Abnormalities in insulin signaling, which is crucial for a wide variety of biological processes in skeletal muscle, may contribute to the development of obesity and type 2 diabetes (1). After insulin binds to the insulin receptor (IR)2 on the cell surface, the receptor undergoes autophosphorylation at several tyrosine residues and becomes active (2). The activated IR then phosphorylates tyrosine residues on intracellular substrates such as the insulin receptor substrate (IRS) family proteins, IRS-1–IRS-4. Research has focused on insulin receptor substrate-1 (IRS-1) as a locus for insulin resistance because of its central role in the insulin-signaling cascade. A pleckstrin homology domain located at the amino terminus of IRS-1 is involved in its coupling to the insulin receptor (3, 4). Adjacent to the pleckstrin homology domain, the phosphotyrosine binding domain interacts directly with the phosphorylated NPXY motif of the β-subunit of insulin and insulin-like growth factor receptors (4, 5). The carboxyl-terminal tail of IRS-1 contains several phosphotyrosine (YXXM) motifs that are autophosphorylated by the insulin receptor and serve as docking sites for Src homology-2 (SH2) domain-containing proteins. These include enzymes such as phosphatidylinositol (PI) 3-kinase, Grb-2, SHP-2, Fyn, and Nck, and docking with IRS-1 leads to a cascade of several intracellular signals (6, 7).
Protein kinase B (Akt) is an important component of the signaling cascade and is downstream of PI 3-kinase. Phosphoinositide-dependent kinase 1 (PDK1) phosphorylates and activates several kinases including Akt and atypical protein kinase C ζ (PKCζ). Akt can also be phosphorylated by mammalian target of rapamycin (mTOR) bound to its regulatory protein, rictor (8). The activated Akt enters into the cytoplasm, where it phosphorylates and inactivates glycogen synthase kinase 3 (GSK3). In addition, the activated Akt phosphorylates its substrate proteins, such as AS160, and promotes GLUT4 translocation to the membrane, leading to enhanced glucose uptake.
Sequestosome 1/p62 is an intracellular signal modulator or adaptor protein that plays a major role in receptor-mediated signal transduction. A highly conserved cytosolic 62-kDa protein, p62 functions as a scaffolding protein that interacts with the atypical PKCs (PKCζ and PKCλ/ι) in several signaling pathways (9). p62 was originally identified as a phosphotyrosine-independent ligand of the SH2 domain of p56lck (10). The domain structure of p62 includes an ubiquitin-like domain (UBL) at its amino terminus (11), and within this UBL domain, there are an SH2 binding domain and an acidic interaction domain that bind the atypical PKC (9, 12). Other downstream p62 motifs include a ZZ finger, a binding site for the RING finger protein tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6), two peptide sequences rich in proline, glutamic acid, serine, and threonine (PEST sequences), an LC3-interacting region (13), and a ubiquitin-associated (UBA) domain (11) at the carboxyl terminus. These domains enable p62 to interact with target proteins and to serve as a scaffolding protein (14, 15). p62 is required for shuttling the microtubule-binding protein Tau for proteasomal degradation (16), and disruption of the p62 gene in mice results in an Alzheimer disease phenotype (17). In addition, targeted deletion of the mouse p62 gene results in obesity that progresses to insulin resistance as well as type 2 diabetes (18, 19). Deletion of p62 also leads to leptin resistance, another feature associated with these metabolic disorders (18).
In the present study, we observed that sequestosome 1/p62 interacts with IRS-1 upon insulin stimulation. Results showed that the interaction is mediated through the SH2 domain of p62 and the YXXM motif of IRS-1, a motif that is also necessary for its interaction with PI 3-kinase. Herein, we demonstrate that p62 participates in insulin signaling.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Anti-IRS1 was purchased from Millipore, Temecula, CA; anti-p62, anti-V5, anti-HA, anti-Myc, anti-GLUT4, and anti-N-cadherin were obtained from Santa Cruz Biotechnology, Santa Cruz, CA. Phospho-Akt (Thr-308 and Ser-473) and total Akt antibody were purchased from Cell Signaling Technology (Danvers, MA). Anti-rabbit IgG and anti-mouse IgG-HRP linked secondary antibody were from GE Healthcare UK Ltd., and enhanced chemiluminescence (ECL) was from Thermo Scientific. Protein A-Sepharose beads, anti-α-tubulin antibody, and all other reagents were obtained from Sigma-Aldrich.
Cell Culture
Parental L6 cells were maintained in DMEM medium (Invitrogen), and Chinese hamster ovary (CHO) cells overexpressing the human IR (CHO/IR) were grown in Ham's F-12 nutrient mixture (Invitrogen) supplemented with 10% fetal bovine serum and penicillin/streptomycin in a humidified atmosphere containing 5% CO2 and95% air at 37 °C. L6 myoblasts were cultured in DMEM medium (changed daily) containing 2% fetal bovine serum and penicillin/streptomycin for 7 days to promote differentiation into myotubes. CHO/IR cells were transfected using the cationic lipid method with LipofectamineTM 2000 transfection reagent (Invitrogen). The cells were deprived of serum in culture medium for 4 h at 37 °C before cell lysis.
Immunoprecipitation and Western Blotting Analysis
Cells were stimulated with or without insulin (100 nm) for 15 min at 37 °C. The cells were lysed with Triton lysis buffer (50 mm HEPES (pH 7.6), 150 mm NaCl, 20 mm sodium pyrophosphate, 10 mm NaF, 20 mm β-glycerophosphate, 1% Triton, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, and 10 μg/ml leupeptin and aprotinin). Protein was estimated by the Bradford procedure (Bio-Rad) using bovine serum albumin (Sigma-Aldrich) as a standard. Cell lysates (1 mg) were diluted in lysis buffer and incubated with 4 μg of primary antibody. The immunoprecipitates were collected with protein A-Sepharose beads overnight at 4 °C and then washed three times with phosphate-buffered saline (PBS). Samples were boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer; proteins were resolved on 10% SDS-PAGE, transferred to nitrocellulose membranes (Bio-Rad), and analyzed by Western blotting with the appropriate antibodies, and immune complexes were detected by chemiluminescence.
GST Pulldown Assay
GST-p62 in glutathione-agarose beads was prepared from a single colony of Escherichia coli cells as described previously (20). The L6 myotubes were treated with or without 100 nm insulin for 15 min. GST-p62 protein in beads was blocked for 1 h at 4 °C with 0.5% BSA in PBS, and the beads were washed with binding buffer (20 mm Tris, 100 mm NaCl, 2 mm EDTA, 0.1% Triton X-100, 2 mm dithiothreitol, 0.05% BSA, and 5% glycerol). Five μg of GST-p62 was added to the L6 myotube lysates (400 μg), and the samples were mixed by rotation for 90 min at 4 °C and washed with 2.5 mm Tris, 2.5 mm EDTA, 250 mm NaCl, 0.1% Triton X-100, and 10% glycerol. SDS-PAGE sample buffer was added, and the samples were boiled for analysis (14).
Membrane Fractionation
CHO/IR cells were transfected with Myc-p62 or a full-length antisense construct of p62 (ASp62) and serum-starved. The cells were treated with or without 100 nm insulin for 20 min, and cytosolic and membrane fractions were isolated using the ProteoExtract kit (Calbiochem) according to the manufacturer's standard protocol.
Deoxyglucose Transport Assay
CHO/IR cells were transfected with Myc-tagged p62 (OEp62), ASp62, or p62ΔSH2. The transfected cells were treated with or without 100 nm insulin for 20 min. 2-Deoxyglucose uptake was assayed in CHO cells as described previously (21–23).
RESULTS AND DISCUSSION
Insulin Induces the Interaction of Sequestosome 1/p62 with IRS-1
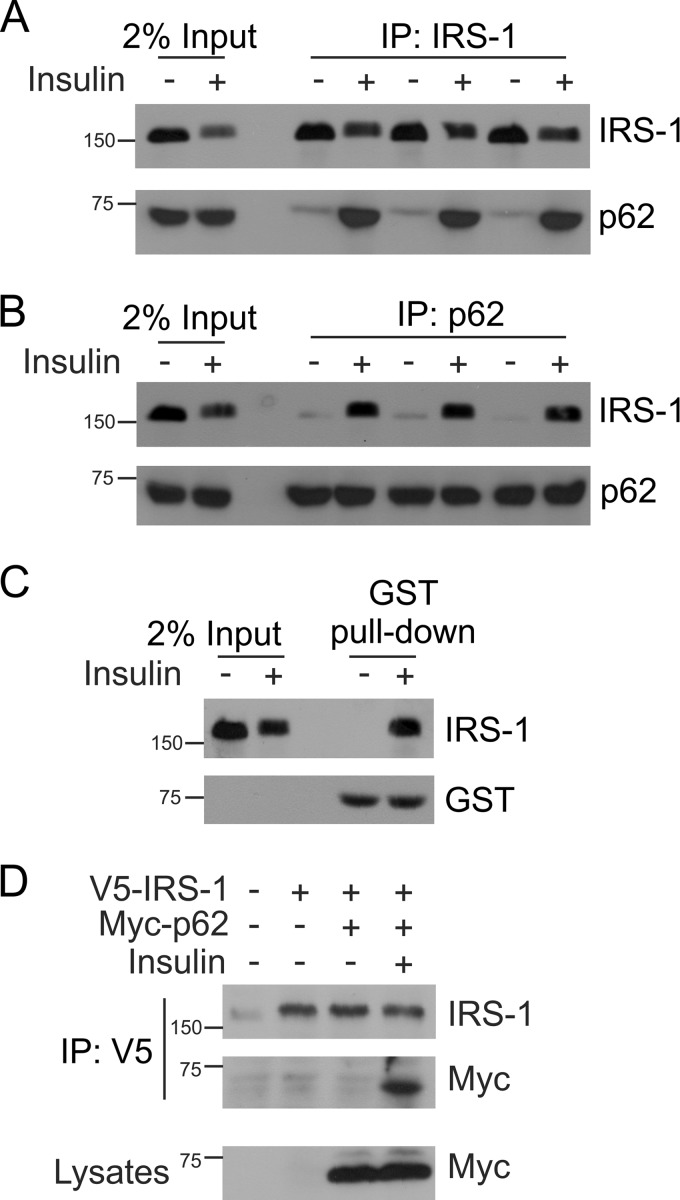
IRS-1 interacts with several SH2 domain-containing proteins such as PI 3-kinase, Grb-2, SHP-2, Fyn, and Nck (24–26). We sought to investigate whether IRS-1 interacts with p62 upon insulin stimulation. L6 myotubes were serum-starved for 4 h and treated with or without insulin (100 nm) for 15 min at 37 °C. The cells were lysed with Triton lysis buffer and immunoprecipitated with anti-IRS-1 or anti-p62 antibody and Western blotted with anti-IRS-1 and anti-p62 antibody. Results showed that p62 interacts with IRS-1 upon insulin stimulation (Fig. 1, A and B). To further support the co-interaction of IRS-1 with p62, GST-p62 pulldown assay was also carried out in L6 myotubes. In the absence of insulin, cells did not exhibit any detectable IRS-1-p62 complex formation. However, stimulation with insulin induced IRS-1 association with p62 (Fig. 1C), which coincided with the coimmunoprecipitation results. In addition, CHO/IR cells were transfected with V5-IRS-1 and Myc-p62. The cells were stimulated with or without insulin for 15 min as shown in Fig. 1D. The cells were lysed, and V5 was immunoprecipitated, which was followed by Western blotting with IRS-1 and Myc antibody. IRS-1 interacted with p62 on insulin stimulation in overexpressed cells (Fig. 1D). The lysates were Western blotted with Myc antibody to confirm the expression of p62. Taken together, these results provide evidence for the formation of an IRS-1-p62 complex upon insulin stimulation.
FIGURE 1.
Insulin-dependent interaction between p62 and IRS-1. A, L6 myotubes were serum-starved for 4 h and either left untreated or stimulated with insulin (100 nm) for 15 min at 37 °C. One mg of lysate protein was immunoprecipitated (IP) with IRS-1 antibody and Western blotted with antibodies against IRS-1 or p62, n = 3. The lanes labeled Input indicate crude lysate. B, the above lysates were also immunoprecipitated with anti-p62 and Western blotted with IRS-1 and p62 antibody. C, L6 myotubes (400 μg) stimulated with or without insulin were interacted with an equivalent amount of GST-p62 in a pulldown assay. The association of IRS-1 was determined by Western blotting with IRS-1 and GST antibody. D, CHO/IR cells were transfected with V5-IRS-1 and Myc-p62 and stimulated with or without insulin for 15 min. The cells were lysed, immunoprecipitated with V5 antibody, and immunoblotted with IRS-1 and Myc antibody. Lysates were Western blotted with anti-Myc to assess the expression of the p62 plasmid. Experiments were replicated three times with similar results.
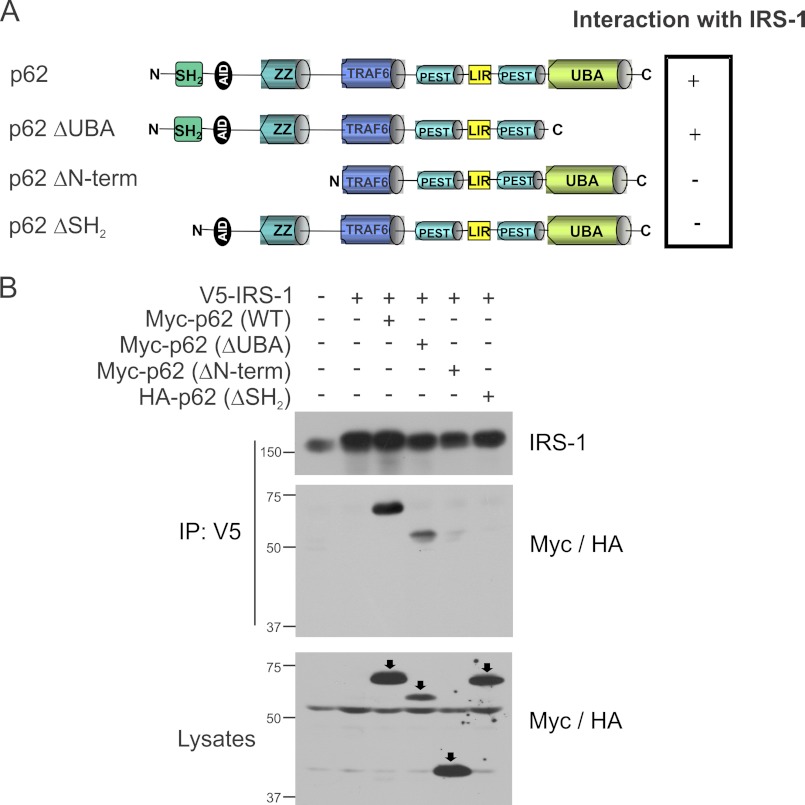
SH2 Domain of p62 Is Essential for Interaction with IRS-1
p62 is a scaffolding protein that interacts with several different proteins (27). We next set out to examine which domain of p62 was involved in the interaction with IRS-1. Several p62 truncated constructs such as Myc-tagged p62ΔN-term, Myc-p62ΔC-term, and HA-p62ΔSH2 domain were used to map the site of the interaction as shown in Fig. 2A. CHO/IR cells were transfected with full-length IRS-1 and p62 truncated constructs. The cells were stimulated with insulin, and the interaction was determined by coimmunoprecipitation of V5 tag of IRS-1 and Western blotted with IRS-1 and Myc/HA antibody. Interestingly, the C-terminal truncated p62 that lacks the UBA domain still interacted with p62. However, the N-terminal truncated p62 (ΔUBL, ΔSH2, ΔZZ) and the p62 with the SH2 domain deleted did not interact with IRS-1 (Fig. 2B). These results suggested that the SH2 domain of p62 is required for the interaction with IRS-1.
FIGURE 2.
p62 interacts with IRS-1 through its SH2 domain. A, domain structures of full-length p62 and p62 mutants (ΔUBA, ΔN-term, and ΔSH2) were used to map the interaction with IRS-1. N, N terminus; AID, acidic interaction domain; LIR, LC3-interacting region; C, C terminus. B, CHO/IR cells were transfected with IRS-1 and wild-type Myc-tagged p62, ΔUBA, ΔN-term, or HA-p62ΔSH2 domain. The cells were stimulated with insulin (100 nm) for 15 min and lysed in Triton lysis buffer. The cell lysates were immunoprecipitated (IP) with anti-V5 followed by Western blotting with anti-IRS-1 and anti-Myc/HA. The lysates were also blotted with anti-Myc/HA to verify the expression of the p62 constructs. Experiments were replicated three times with similar results.
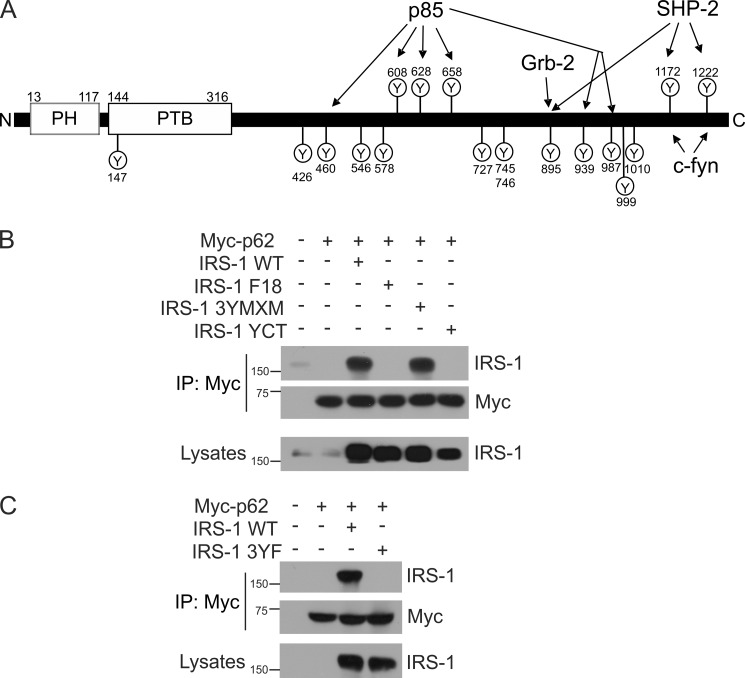
Mapping the Interaction of IRS-1 with p62
IRS-1 contains several phosphotyrosine (YXXM) motifs at its carboxyl terminus that serve as docking sites for several proteins (Fig. 3A) (15). Therefore, we hypothesized that IRS-1 might also interact with the p62 through its YXXM motif. To determine specifically whether YXXM motifs may be involved in the interaction with p62, IRS-1 mutants in which 18 tyrosine residues (Tyr-147, -426, -460, -526, -578, -608, -628, -658, -727, -745, -746, -895, -939, -987, -999, -1010, -1172, and -1222) were all substituted with phenylalanine (F18), as well as 3YMXM (with only three phosphorylation sites: Tyr-608, Tyr-628, and Tyr-658) and YCT (with only two phosphorylation sites: Tyr-1172 and Tyr-1222) were examined. The three tyrosine sites in IRS-1 3YMXM are located in the middle of the IRS-1 protein and share a common sequence (YMXMSP). When phosphorylated, Tyr-608 and Tyr-628 (and, perhaps, Tyr-658) bind the p85 regulatory subunit (28, 29). Tyr-1172 and Tyr-1222 sites are located in the C terminus of IRS-1 and are known to be activated by phosphorylation to form the binding sites for SHP-2 and Fyn (30–32). To examine the site of the interaction between IRS-1 and p62, wild-type IRS-1 or its mutants, F18, 3YMXM, or YCT, were each transfected together with Myc-tagged p62 in CHO cells that overexpressed the insulin receptor. The transfected cells were stimulated with insulin, Myc-tagged p62 was immunoprecipitated, and Western blotting was performed to detect IRS-1 and p62. The wild-type IRS-1 and 3YXXM mutant protein interacted with p62, whereas the IRS-1 F18 and YCT mutants did not interact with p62 (Fig. 3B). Thus, IRS-1 YCT, with Tyr-1172 and Tyr-1222 that were previously shown to be the binding sites for SHP-2 and Fyn, does not participate in the interaction with p62. However, IRS-1 3YXXM, which retains only the three YMXMSP motifs at Tyr-608, Tyr-628, and Tyr-658, is involved in the interaction with p62. This interaction of IRS-1 with p62 may be similar to its interaction with p85. However, this does not rule out the possibility that other tyrosine residues of the YXXM motif may be involved in the IRS-1/p62 interaction. To confirm the involvement of YXXM motifs only at positions Tyr-608, Tyr-628, and Tyr-658, we mutated these three tyrosine amino acids to phenylalanine in the full-length IRS-1 plasmid (IRS-1 3YF) (GenScript, Piscataway, NJ). CHO/IR cells were transfected with wild-type IRS-1 or the mutants IRS-1 3YF along with Myc-tagged p62 and were stimulated with insulin. The Myc-tagged p62 was immunoprecipitated, and Western blotting was performed to detect IRS-1 and p62. The wild-type IRS-1 was found associated with p62, whereas the IRS-1 3YF mutant did not interact with p62 (Fig. 3C). The lysates were also Western blotted with anti-IRS-1 to check the expression of IRS-1 plasmids. These data suggested that YXXM motifs at positions Tyr-608, Tyr-628, and Tyr-658 are required for IRS-1 to interact with p62.
FIGURE 3.
IRS-1 interacts with p62 through its YXXM motif. A, schematic representation of IRS-1 showing the various tyrosine residues. N, N terminus; PH, pleckstrin homology domain; PTB, phosphotyrosine binding domain; C, C terminus. B, CHO/IR cells were expressed with wild-type IRS-1 and the mutants F18, 3YMXM, or YCT along with Myc-tagged p62. The cells were serum-starved and stimulated with insulin (100 nm) for 15 min. p62 was immunoprecipitated (IP) using Myc antibody and Western blotted for IRS-1 and p62. C, CHO/IR cells were transfected with Myc-p62 along with wild-type IRS-1 or 3YF mutant and treated with insulin for 15 min. The co-interaction of p62 and IRS-1 was determined by immunoprecipitation with anti-Myc and Western blotting for anti-IRS-1 and anti-Myc. The lysates were Western blotted with anti-IRS-1 to verify the expression of the constructs. Experiments were replicated three times with similar results.
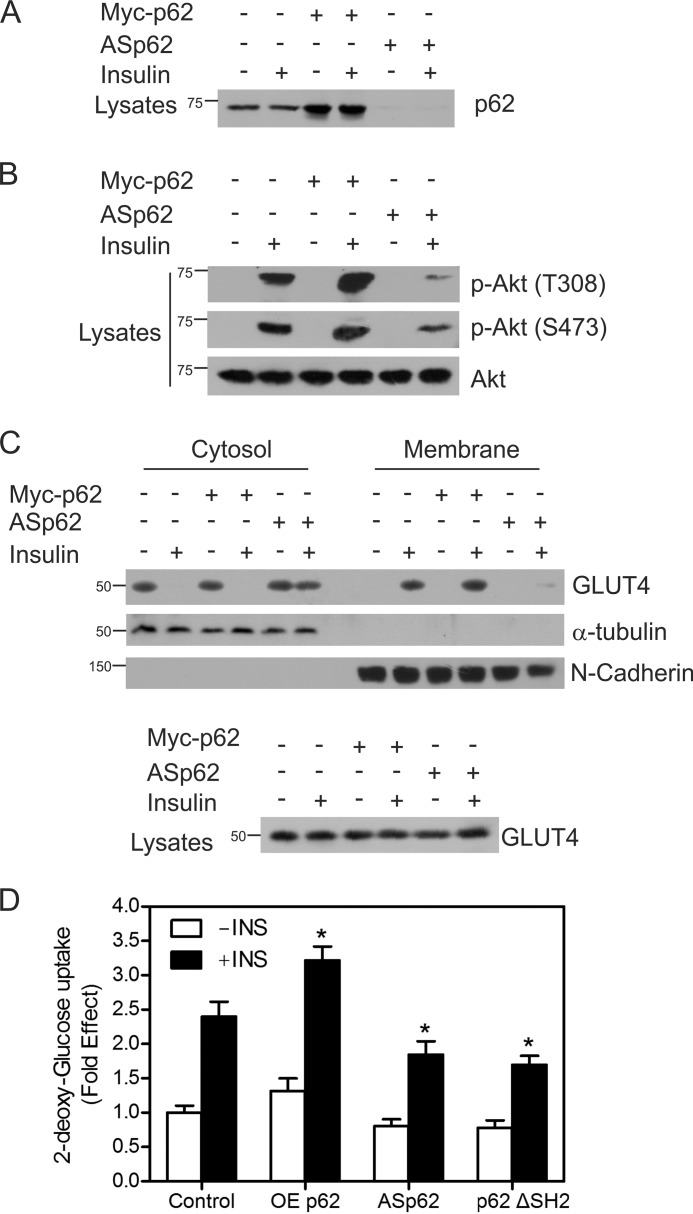
p62 Influences the Insulin Receptor Signaling
Because p62 interacts with IRS-1 on insulin stimulation, we sought to investigate whether p62 may affect insulin receptor signaling. CHO/IR cells were transfected with OEp62 or ASp62, which has been shown to deplete the levels of p62 upon transfection (33, 34). As control, the expression level of p62 was examined employing equal protein concentrations of whole cell lysates (Fig. 4A). The level of p62 expression increased in cells transfected with Myc-tagged p62 (third and fourth lanes) and was notably decreased in cells transfected with ASp62 (fifth and sixth lanes).
FIGURE 4.
p62 influences insulin receptor signaling. A, CHO/IR cells were transfected with either Myc-tagged p62 or ASp62. The transfected cells were treated with or without insulin (100 nm) for 15 min at 37 °C. Lysates from control and transfected cells were Western blotted with p62. B, the above lysates (A) were immunoblotted for phospho-Akt Thr-308 (p-Akt (T308)) and phospho-Akt Ser-473 (p-Akt (S473)) and total Akt (Akt). C, the transfected cells from above (A) were incubated in the presence or absence of insulin for 20 min, fractionated into membrane and cytosolic fractions, and Western blotted with GLUT4, α-tubulin, and N-cadherin. The lysates were immunoblotted for total GLUT4. D, OEp62, ASp62, or p62ΔSH2 mutants were expressed in CHO/IR cells, cells were stimulated in the presence or absence of 100 nm insulin for 20 min, and the rates of 2-deoxy-d-[3H]glucose uptake were determined. Each bar in the graph indicates the -fold change relative to the control cells unstimulated with insulin, which was taken to be 1. Differences from the control value treated with insulin and other groups are statistically significant (*, p < 0.001). Error bars indicate S.D.
To examine the effect of p62 on Akt phosphorylation, the transfected cells were treated with or without insulin and immunoblotted for phospho and total Akt. Overexpression of p62 enhanced the phosphorylation of Akt at Thr-308 and Ser-473 on insulin treatment. However, the reduction of p62 with the antisense construct decreased the phosphorylation of Akt when compared with either the control cells or those overexpressing p62 (Fig. 4B). TRAF6 was found to be a direct ubiquitin ligase (E3) for Akt and essential for Akt ubiquitination and phosphorylation (35). The activity of TRAF6 is regulated by its polyubiquitination and oligomerization (36, 37). p62 is found to interact with TRAF6 (33) and is known to induce polyubiquitination, and oligomerization of TRAF6 thereby enhances the E3 ubiquitin ligase activity of TRAF6 (38). Depletion of p62 blocks TRAF6 polyubiquitination and activation, which would inhibit the Akt ubiquitination and phosphorylation. This might explain the reduction in the phosphorylation of Akt on depletion of p62 shown in Fig. 4B. Efforts are underway to examine the effect of p62 on Akt ubiquitination.
As Akt phosphorylation regulates GLUT4 translocation (39, 40), we tested whether p62 influenced membrane recruitment of GLUT4. We isolated the cytoplasmic and membrane fractions from control, cells transfected with antisense p62, and cells overexpressing p62. The membrane-to-cytoplasmic distribution of the GLUT4 was examined by Western blotting analysis. In control and p62-overexpressing cells, insulin stimulation translocated the GLUT4 from the cytosol to the membrane fractions, whereas in ASp62-transfected cells, the GLUT4 translocation to the membrane was impaired (Fig. 4C). The purity of the fractions was verified by blotting with marker antibodies such as α-tubulin and N-cadherin. We hypothesized that the overexpression of p62 would increase the glucose uptake. To test this, we used CHO/IR cells transfected with OEp62 or ASp62 mutant stimulated in the presence or absence of insulin for 20 min. We also overexpressed the ΔSH2 domain of p62, which lacks the SH2 domain that is necessary to interact with IRS-1. The cells were incubated with 2-deoxy-d-[3H]glucose, and the uptake of glucose was measured. We found that the overexpression of p62 displayed a 0.8-fold increase in glucose uptake when compared with the control cells on insulin treatment. Reduction of p62 decreased the glucose uptake 0.6-fold, and p62ΔSH2 mutant decreased the glucose uptake by 0.7-fold when compared with control upon insulin stimulation (Fig. 4D). These results indicate that the ability of p62 to regulate glucose uptake is dependent upon its interaction with IRS-1. Together, these findings reveal that p62 may play an important role in insulin receptor signaling.
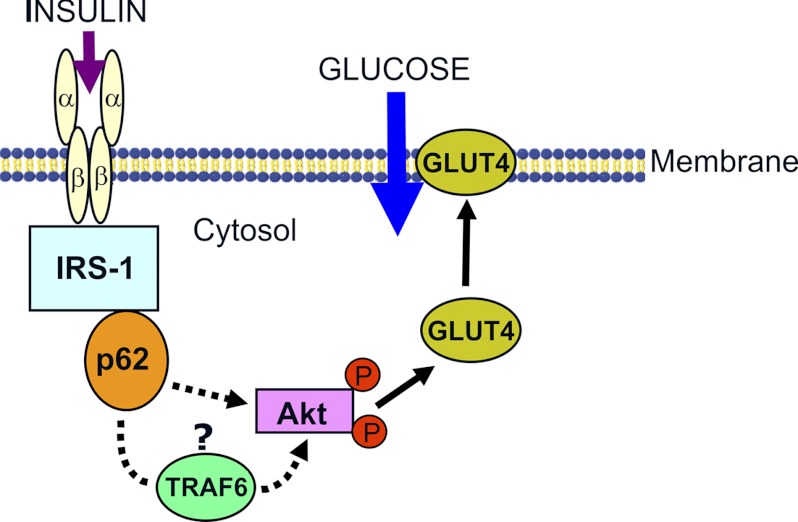
Our findings support a model where the interaction of p62 with IRS-1 regulates Akt phosphorylation, GLUT4 translocation to the membrane, and glucose uptake (Fig. 5). Results of coimmunoprecipitation and GST pulldown experiments confirm the interaction of p62 with IRS-1 upon insulin stimulation. The interaction between p62 and IRS-1 is mediated through the SH2 domain of p62 and YXXM motifs at Tyr-608, Tyr-628, and Tyr-658 of IRS-1. The interaction site is similar to that of the p85 subunit of PI 3-kinase and IRS-1 (28, 29).
FIGURE 5.
Model depicting the role of p62 in insulin signaling. p62 interacts with IRS-1 on insulin stimulation and leads to Akt activation, GLUT4 translocation, and glucose uptake. Effect of p62 on Akt phosphorylation (indicated by circled P) may involve other proteins such as TRAF6 (35, 38).
Insulin plays a major role in the disposal of glucose from the plasma following a meal. Insulin stimulation leads to the activation of PI 3-kinase, which is essential for Akt phosphorylation and downstream signaling. Insulin drives the translocation of GLUT4 into the plasma membrane (39, 40), thereby increasing glucose uptake into tissues and restoring glucose levels. Previous studies (18) have shown that p62 knock-out mice develop mature-onset obesity and insulin resistance. It has been demonstrated that p62 influences insulin signaling and may play a role in insulin resistance. Indeed, insulin injection in p62-deleted young nonobese mice did not impair the activation of Akt in muscle and fat tissue (18). However, in obese p62 knock-out mice, the phosphorylation of Akt was impaired in muscle and fat but not in liver (18). Our results reveal that overexpression of p62 increased the phosphorylation of Akt, GLUT4 translocation and glucose uptake on insulin stimulation. Depletion of p62 by use of the antisense p62 construct has been shown to abrogate the insulin signaling. Together, these findings reveal that interaction of p62 with IRS-1 links p62 and the insulin-signaling pathway.
Acknowledgments
We thank Dr. Marie Wooten, Auburn University, for providing the p62 plasmids, which were generated by Dr. Jorge Moscat. Also, we thank Dr. Martin Myers, University of Michigan, for IRS-1 plasmids. Many thanks go to Dr. Xiao Jian Sun, University of Maryland, for providing the CHO/IR cells and Dr. Catherine Wernette, Auburn University, for editing this manuscript. We thank Drs. Suresh Mathews, Kevin Huggins, Doug White, and Jianzhong Shen, Auburn University, for valuable suggestions.
This work was supported by the New Faculty Start-up Fund from Auburn University and Alabama Agricultural Experiment Station, Hatch/Multistate Funding Program (to J. R. B.).
- IR
- insulin receptor
- IRS-1
- insulin receptor substrate-1
- PI 3-kinase
- phosphoinositol 3-kinase
- SH2
- Src homology-2
- CHO/IR
- Chinese hamster ovary cells overexpressing the human insulin receptor
- GLUT4
- glucose transporter type 4
- UBL
- ubiquitin-like domain
- TRAF6
- TNF receptor-associated factor 6
- UBA
- ubiquitin-associated
- term
- terminus.
REFERENCES
- 1. Bouzakri K., Koistinen H. A., Zierath J. R. (2005) Molecular mechanisms of skeletal muscle insulin resistance in type 2 diabetes. Curr. Diabetes Rev. 1, 167–174 [DOI] [PubMed] [Google Scholar]
- 2. Lee J., Pilch P. F., Shoelson S. E., Scarlata S. F. (1997) Conformational changes of the insulin receptor upon insulin binding and activation as monitored by fluorescence spectroscopy. Biochemistry 36, 2701–2708 [DOI] [PubMed] [Google Scholar]
- 3. Myers M. G., Jr., Grammer T. C., Brooks J., Glasheen E. M., Wang L. M., Sun X. J., Blenis J., Pierce J. H., White M. F. (1995) The pleckstrin homology domain in insulin receptor substrate-1 sensitizes insulin signaling. J. Biol. Chem. 270, 11715–11718 [DOI] [PubMed] [Google Scholar]
- 4. Myers M. G., Jr., White M. F. (1996) Insulin signal transduction and the IRS proteins. Annu. Rev. Pharmacol. Toxicol. 36, 615–658 [DOI] [PubMed] [Google Scholar]
- 5. Craparo A., O'Neill T. J., Gustafson T. A. (1995) Non-SH2 domains within insulin receptor substrate-1 and SHC mediate their phosphotyrosine-dependent interaction with the NPEY motif of the insulin-like growth factor I receptor. J. Biol. Chem. 270, 15639–15643 [DOI] [PubMed] [Google Scholar]
- 6. Quon M. J., Butte A. J., Taylor S. I. (1994) Insulin signal transduction pathways. Trends Endocrinol. Metab. 5, 369–376 [DOI] [PubMed] [Google Scholar]
- 7. Cheatham B., Kahn C. R. (1995) Insulin action and the insulin-signaling network. Endocr. Rev. 16, 117–142 [DOI] [PubMed] [Google Scholar]
- 8. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 9. Moscat J., Diaz-Meco M. T. (2000) The atypical protein kinase Cs: functional specificity mediated by specific protein adapters. EMBO Rep. 1, 399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Joung I., Strominger J. L., Shin J. (1996) Molecular cloning of a phosphotyrosine-independent ligand of the p56lck SH2 domain. Proc. Natl. Acad. Sci. U.S.A. 93, 5991–5995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Seibenhener M. L., Babu J. R., Geetha T., Wong H. C., Krishna N. R., Wooten M. W. (2004) Sequestosome 1/p62 is a polyubiquitin chain-binding protein involved in ubiquitin proteasome degradation. Mol. Cell. Biol. 24, 8055–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Geetha T., Wooten M. W. (2002) Structure and functional properties of the ubiquitin-binding protein p62. FEBS Lett. 512, 19–24 [DOI] [PubMed] [Google Scholar]
- 13. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 14. Geetha T., Wooten M. W. (2003) Association of the atypical protein kinase C-interacting protein p62/ZIP with nerve growth factor receptor TrkA regulates receptor trafficking and Erk5 signaling. J. Biol. Chem. 278, 4730–4739 [DOI] [PubMed] [Google Scholar]
- 15. Geetha T., Jiang J., Wooten M. W. (2005) Lysine 63 polyubiquitination of the nerve growth factor receptor TrkA directs internalization and signaling. Mol. Cell 20, 301–312 [DOI] [PubMed] [Google Scholar]
- 16. Babu J. R., Geetha T., Wooten M. W. (2005) Sequestosome 1/p62 shuttles polyubiquitinated Tau for proteasomal degradation. J. Neurochem. 94, 192–203 [DOI] [PubMed] [Google Scholar]
- 17. Ramesh Babu J., Lamar Seibenhener M., Peng J., Strom A. L., Kemppainen R., Cox N., Zhu H., Wooten M. C., Diaz-Meco M. T., Moscat J., Wooten M. W. (2008) Genetic inactivation of p62 leads to accumulation of hyperphosphorylated Tau and neurodegeneration. J. Neurochem. 106, 107–120 [DOI] [PubMed] [Google Scholar]
- 18. Rodriguez A., Durán A., Selloum M., Champy M. F., Diez-Guerra F. J., Flores J. M., Serrano M., Auwerx J., Diaz-Meco M. T., Moscat J. (2006) Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 3, 211–222 [DOI] [PubMed] [Google Scholar]
- 19. Okada K., Yanagawa T., Warabi E., Yamastu K., Uwayama J., Takeda K., Utsunomiya H., Yoshida H., Shoda J., Ishii T. (2009) The α-glucosidase inhibitor acarbose prevents obesity and simple steatosis in sequestosome 1/A170/p62-deficient mice. Hepatol. Res. 39, 490–500 [DOI] [PubMed] [Google Scholar]
- 20. Samuels I. S., Seibenhener M. L., Neidigh K. B., Wooten M. W. (2001) Nerve growth factor stimulates the interaction of ZIP/p62 with atypical protein kinase C and targets endosomal localization: evidence for regulation of nerve growth factor-induced differentiation. J. Cell. Biochem. 82, 452–466 [DOI] [PubMed] [Google Scholar]
- 21. Sale E. M., Atkinson P. P., Arnott C. H., Chad J. E., Sale G. J. (1999) Role of ERK1/ERK2 and p70S6K pathway in insulin signaling of protein synthesis. FEBS Lett. 446, 122–126 [DOI] [PubMed] [Google Scholar]
- 22. Hodgkinson C. P., Mander A., Sale G. J. (2005) Protein kinase-ζ interacts with munc18c: role in GLUT4 trafficking. Diabetologia 48, 1627–1636 [DOI] [PubMed] [Google Scholar]
- 23. Olianas M. C., Dedoni S., Onali P. (2011) δ-Opioid receptors stimulate GLUT1-mediated glucose uptake through Src- and IGF-1 receptor-dependent activation of PI3-kinase signaling in CHO cells. Br. J. Pharmacol. 163, 624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Giovannone B., Scaldaferri M. L., Federici M., Porzio O., Lauro D., Fusco A., Sbraccia P., Borboni P., Lauro R., Sesti G. (2000) Insulin receptor substrate (IRS) transduction system: distinct and overlapping signaling potential. Diabetes Metab. Res. Rev. 16, 434–441 [DOI] [PubMed] [Google Scholar]
- 25. Sesti G., Federici M., Hribal M. L., Lauro D., Sbraccia P., Lauro R. (2001) Defects of the insulin receptor substrate (IRS) system in human metabolic disorders. FASEB J. 15, 2099–2111 [DOI] [PubMed] [Google Scholar]
- 26. Virkamäki A., Ueki K., Kahn C. R. (1999) Protein-protein interaction in insulin signaling and the molecular mechanisms of insulin resistance. J. Clin. Invest. 103, 931–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moscat J., Diaz-Meco M. T., Wooten M. W. (2007) Signal integration and diversification through the p62 scaffold protein. Trends Biochem. Sci. 32, 95–100 [DOI] [PubMed] [Google Scholar]
- 28. Esposito D. L., Li Y., Cama A., Quon M. J. (2001) Tyr-612 and Tyr-632 in human insulin receptor substrate-1 are important for full activation of insulin-stimulated phosphatidylinositol 3-kinase activity and translocation of GLUT4 in adipose cells. Endocrinology 142, 2833–2840 [DOI] [PubMed] [Google Scholar]
- 29. Rordorf-Nikolic T., Van Horn D. J., Chen D., White M. F., Backer J. M. (1995) Regulation of phosphatidylinositol 3′-kinase by tyrosyl phosphoproteins: full activation requires occupancy of both SH2 domains in the 85-kDa regulatory subunit. J. Biol. Chem. 270, 3662–3666 [DOI] [PubMed] [Google Scholar]
- 30. Sun X. J., Crimmins D. L., Myers M. G., Jr., Miralpeix M., White M. F. (1993) Pleiotropic insulin signals are engaged by multisite phosphorylation of IRS-1. Mol. Cell. Biol. 13, 7418–7428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuhné M. R., Pawson T., Lienhard G. E., Feng G. S. (1993) The insulin receptor substrate 1 associates with the SH2-containing phosphotyrosine phosphatase Syp. J. Biol. Chem. 268, 11479–11481 [PubMed] [Google Scholar]
- 32. Sun X. J., Pons S., Asano T., Myers M. G., Jr., Glasheen E., White M. F. (1996) The Fyn tyrosine kinase binds Irs-1 and forms a distinct signaling complex during insulin stimulation. J. Biol. Chem. 271, 10583–10587 [DOI] [PubMed] [Google Scholar]
- 33. Sanz L., Diaz-Meco M. T., Nakano H., Moscat J. (2000) The atypical PKC-interacting protein p62 channels NF-κB activation by the IL-1-TRAF6 pathway. EMBO J. 19, 1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wooten M. W., Seibenhener M. L., Mamidipudi V., Diaz-Meco M. T., Barker P. A., Moscat J. (2001) The atypical protein kinase C-interacting protein p62 is a scaffold for NF-κB activation by nerve growth factor. J. Biol. Chem. 276, 7709–7712 [DOI] [PubMed] [Google Scholar]
- 35. Yang W. L., Wang J., Chan C. H., Lee S. W., Campos A. D., Lamothe B., Hur L., Grabiner B. C., Lin X., Darnay B. G., Lin H. K. (2009) The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 325, 1134–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang C., Deng L., Hong M., Akkaraju G. R., Inoue J., Chen Z. J. (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351 [DOI] [PubMed] [Google Scholar]
- 37. Ea C. K., Sun L., Inoue J., Chen Z. J. (2004) TIFA activates IκB kinase (IKK) by promoting oligomerization and ubiquitination of TRAF6. Proc. Natl. Acad. Sci. U.S.A. 101, 15318–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wooten M. W., Geetha T., Seibenhener M. L., Babu J. R., Diaz-Meco M. T., Moscat J. (2005) The p62 scaffold regulates nerve growth factor-induced NF-κB activation by influencing TRAF6 polyubiquitination. J. Biol. Chem. 280, 35625–35629 [DOI] [PubMed] [Google Scholar]
- 39. Leney S. E., Tavaré J. M. (2009) The molecular basis of insulin-stimulated glucose uptake: signaling, trafficking, and potential drug targets. J. Endocrinol. 203, 1–18 [DOI] [PubMed] [Google Scholar]
- 40. Saltiel A. R., Kahn C. R. (2001) Insulin signaling and the regulation of glucose and lipid metabolism. Nature 414, 799–806 [DOI] [PubMed] [Google Scholar]