Background: Notch activity depends notably on the quantity of Notch receptor at the cell surface.
Results: The ubiquitin-specific protease USP12 directly targets Notch and directs it to lysosomal degradation.
Conclusion: USP12 is a novel, conserved negative regulator of Notch signaling.
Significance: Notch signaling regulation by various deubiquitinating enzymes acting at different steps is of crucial importance.
Keywords: Deubiquitination, Notch, Signaling, Trafficking, Ubiquitination, USP12, Deubiquitinating Enzyme
Abstract
Notch signaling is critical for development and adult tissue physiology, controlling cell fate in a context-dependent manner. Upon ligand binding, the transmembrane Notch receptor undergoes two ordered proteolytic cleavages releasing Notch intracellular domain, which regulates the transcription of Notch target genes. The strength of Notch signaling is of crucial importance and depends notably on the quantity of Notch receptor at the cell surface. Using an shRNA library screen monitoring Notch trafficking and degradation in the absence of ligand, we identified mammalian USP12 and its Drosophila melanogaster homolog as novel negative regulators of Notch signaling. USP12 silencing specifically interrupts Notch trafficking to the lysosomes and, as a consequence, leads to an increased amount of receptor at the cell surface and to a higher Notch activity. At the biochemical level, USP12 with its activator UAF1 deubiquitinate the nonactivated form of Notch in cell culture and in vitro. These results characterize a new level of conserved regulation of Notch signaling by the ubiquitin system.
Introduction
The very conserved Notch signaling pathway promotes or suppresses cell differentiation, proliferation, or death, depending on the cellular context. As it is active in a large range of tissues during development and in adulthood, its deregulation is linked to multiple disorders, including developmental syndromes, adult-onset diseases, and cancers. Notch signaling relies on the communication between adjacent cells, mediated by the binding of a transmembrane ligand to its receptor on a neighboring cell. This triggers a series of post-translational modifications and processing events that lead to the release of soluble Notch intracellular domain (Notch-IC),3 which travels to the nucleus, where it participates in a transcription complex that activates downstream target genes (1).
It has been well documented in various systems that Notch signaling is extremely dosage-sensitive. Any perturbation of the efficiency of the overall process of signal transduction may be at the origin of diseases (2) or may result in alternative cell fate decisions. A large number of factors have been described as regulating Notch signaling, acting at various steps: Notch maturation in the endoplasmic reticulum or trans-Golgi network (furin processing, glycosylation events (3, 4)), ligand activity (5), Notch processing during activation (ADAM and γ-secretase proteases (1)), etc. In addition, the strength of Notch signaling is also affected by the quantity of Notch receptor expressed at the cell surface, which depends notably on Notch trafficking under the control of ubiquitination events (6, 7).
Given the importance and the multiplicity of these ubiquitination events, we sought to identify deubiquitinating enzymes (or DUBs) that would specifically target nonactivated Notch. Our search led to the identification of USP12 as a DUB acting as a negative regulator of Notch signaling by regulating the quantity of surface-expressed full-length Notch.
EXPERIMENTAL PROCEDURES
shDUB Library
The shDUB library was described in Refs. 8 and 9 and references therein.
Cell Lines
U2OS-FL, MEF-FL, and OP9-Dll1 cell lines were described in Refs. 9, 10, and 11, respectively.
Constructs
All Myc-tagged Notch constructs (FL, ΔE, and IC) were already described in Refs. 12 and 13 and were gifts from R. Kopan (Washington University, St. Louis, MO). These Notch constructs are all deleted from amino acids 2183 of murine Notch1 and fused to a hexameric Myc tag at the carboxyl terminus. Notch1 retroviral vector (used for U2OS-FL and MEF-FL cell lines) encodes a full-length human Notch1 with an HA-epitope tag inserted between EGF repeats 22 and 23 (gift of J. Aster, Harvard Medical School, Boston, MA).
GFP-fused USP12 construct was a gift from S. Urbé (University of Liverpool, UK). HA-tagged WT and C48S USP12 were generated from pOZ-USP12 constructs described in Ref. 14 by inserting USP12 cDNA (XhoI/NotI) into pCMV-HA backbone vector (Clontech). His6-tagged ubiquitin construct was a gift from M. Treier (European Molecular Biology Laboratory, Heidelberg, Germany). CSL-Luc was a gift from T. Honjo (Kyoto University, Japan) and is referred to as pGa981-6 in Ref. 15. Itch/AIP4 expression vector was described in Ref. 10.
siRNAs and shRNAs
shUSP12 pool belongs to the shDUB library (see above). shUSP12 and shUAF1 were described in Refs. 14 and 16, respectively. siUSP12 pool and si mUSP12 pool correspond to the ON-TARGETplus SMARTpools for human or murine USP12 from Thermo Scientific. siUSP12-2 belongs to the human siUSP12 pool and corresponds to the target sequence 5′-TAGCAGATCTCTTCCATAG-3′. si mUSP8 was designed by Sigma-Aldrich, corresponds to the target sequence 5′-GGACAGGACAGTATAGATA-3′, and was described in Ref. 17.
Antibodies
For Western blot analysis, we used anti-Myc 9E10 antibody. Anti-Notch-IC and anti-Dll1 antibodies were described in Refs. 3 and 11, respectively. Anti-USP12, USP46, and anti-UAF1 were described in Refs. 14 and 16, respectively. Other antibodies were supplied by BD Transduction Laboratories (monoclonal anti-Itch), Covance (monoclonal anti-HA), Invitrogen (polyclonal anti-GFP), Santa Cruz Biotechnology (polyclonal anti-EGFR sc-03), and Sigma (monoclonal anti-α-tubulin and monoclonal anti-FLAG M2). For immunofluorescence, we used anti-Lamp1 developed by J. T. August and obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD, National Institutes of Health, and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA. Secondary antibodies were supplied by Molecular Probes (Alexa Fluor conjugates). For flow cytometry, we used antibodies from Covance (anti-HA) and Santa Cruz Biotechnology (monoclonal anti-EGFR sc-120 recognizing an extracellular epitope).
Fly Strains and Genetics
All the flies were maintained at 25 °C on standard medium. The UAS-CG7023IR was obtained from the Vienna Drosophila Research Center and results from a P-element insertion (P{GD12106}v27799Insertion). The other strains are a kind gift from F. Schweisguth (Institut Pasteur) and are: w1118 strain that was used as a wild-type control, pnr-GAL4 (P{w[+mW.hs] = GawB}pnr[MD237]), Notch−/+ (N[55e11]), and UAS-SpdoIR; pnr-GAL4 (attP[mw, UAS-dsRNA spdo]ID104092 (VDRC); pnr-GAL4) (18).
Quantitative RT-PCR
U2OS-FL or HEK293T cells were first transfected with the siRNAs or shRNAs using jetPRIME transfection reagent (Polyplus Transfection). Cells were lysed, and RNAs were purified using RNeasy mini kit (Qiagen) and submitted to reverse transcription. Quantitative PCR was then performed and analyzed using a CFX96TM real-time PCR detection system and the CFX ManagerTM Software (Bio-Rad). Human USP12 was quantified using 5′-TTCCATACAACAAGGAGGTGAA-3′ (forward) and 5′-AAGAGAAAATGCGTGCCAAT-3′ (reverse) oligonucleotides. USP12 expression was reported to hypoxanthine-guanine phosphoribosyltransferase (HPRT) expression quantified with the oligonucleotides 5′-TAATTGGTGGAGATGATCTCTCAAC-3′ (forward) and 5′-TGCCTGACCAAGGAAAGC-3′ (reverse).
Cell Extracts, Immunoprecipitations, Immunoblots, and Notch-Dll1 Coculture Assay
Each protocol was described in Ref. 9. When necessary, Western blots were quantified using the Quantity One program (Bio-Rad).
Purification of Ubiquitin Conjugates
HEK293T cells were harvested 24 h after transfection and lysed in 8 m urea, 0.1 m NaH2PO4, 10 mm Tris-HCl (pH 8), 1% Triton X-100, and 20 mm imidazole at room temperature. His-Ub-conjugated proteins were purified on chelating Sepharose beads (Amersham Biosciences), previously charged with nickel. Nickel-bound proteins were washed extensively with the same buffer and then with a pH 6.3 buffer and eluted in Laemmli buffer before Western blot analysis.
Anti-HA/Notch Antibody and EGF Uptake
U2OS-FL or MEF-FL cells were grown on glass coverslips. After a 1-h incubation in serum-free medium, cells were incubated at 4 °C for 30 min with an anti-HA antibody coupled to either Alexa Fluor 594 or Alexa Fluor 488 (Invitrogen), and if necessary, with EGF-Alexa Fluor 555 (Invitrogen). Then, cells were washed and incubated in serum-free medium at 37 °C for various periods of time. The cells were finally quickly rinsed in cold PBS and fixed with 4% paraformaldehyde during 20 min. In the case of an additional anti-Lamp1 staining, MEF-FL cells were then permeabilized with PBS containing 0.2% Triton X-100 for 5 min before the incubation with appropriate primary and secondary antibodies. Cells were mounted in Mowiol (Calbiochem, Merck Biosciences), and images were acquired using an AxioImager microscope with ApoTome system with a 63× magnification and AxioVision software (Carl Zeiss MicroImaging Inc.).
In Vitro Deubiquitination Assays
In vitro deubiquitination assays were performed as described previously in Ref. 14. Briefly, the ubiquitin-7-amino-4-methylcoumarin (Ub-AMC) assay was performed with the same complexes in the presence of the fluorogenic substrate Ub-AMC (Boston Biochem), and the reaction was done in a reaction buffer (20 mm HEPES-KOH, pH 7.8, 20 mm NaCl, 0.1 mg/ml ovalbumin, 0.5 mm EDTA, 10 mm dithiothreitol) at 37 °C. The emitted fluorescence (460 nm) from the released AMC was measured after excitation at 380 nm using a FLUOstar Galaxy fluorometer (BMG Labtech).
Notch and EGFR Quantification by Flow Cytometry
MEF-FL cells were harvested and resuspended in cold PBS 48 h after transfection with the siRNAs. Then, cells were fixed in 4% paraformaldehyde in PBS and immediately labeled with anti-HA and anti-EGFR antibodies at 4 °C before the appropriate secondary antibodies and analyzed using a CyAn flow cytometer (Beckman Coulter).
RESULTS
Identification of USP12 as a Candidate DUB Acting on Notch
With the aim of identifying deubiquitinating enzymes regulating trafficking of nonactivated Notch, we first generated a clone of U2OS cells hereafter named U2OS-FL. These cells are devoid of any active Notch ligand and stably express low levels of full-length human Notch, Notch-FL, which remains transcriptionally inactive unless it is activated by its ligands. In this case, it is tagged in the extracellular domain with an HA epitope (9).
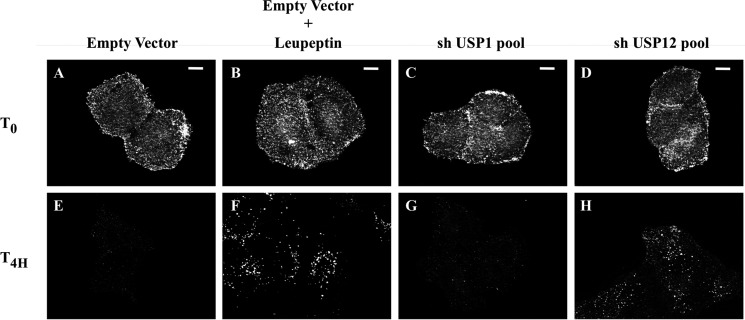
We then designed an immunofluorescence assay where Notch receptor internalization and degradation were assessed in the absence of any ligand, therefore avoiding contamination with deubiquitination events regulating either the ligands or activated Notch trafficking. First of all, the living U2OS-FL cells were shortly labeled at 4 °C with an anti-HA antibody coupled to Alexa Fluor 594 fluorochrome, recognizing the Notch molecules expressed at the cell surface (pulse). Then, after several washes, cells were incubated at 37 °C to allow anti-HA-labeled Notch endocytosis and trafficking (chase) before being fixed. We finally analyzed the anti-HA labeling, which monitors nonactivated Notch localization and quantity in the cells in a time-dependent manner (10) (Fig. 1). As expected, the initial Notch labeling appeared at the plasma membrane in all cases (Fig. 1, T0, panels A–D). During the 4 h of incubation, the Notch signal vanished in untreated cells (Fig. 1E, Empty Vector), whereas it was still persistent and localized in endosomes in cells treated with leupeptin, an inhibitor of the lysosomal proteases (Fig. 1F). This confirmed the published data indicating that nonactivated Notch is degraded through the lysosomal pathway in normal conditions (10). We tested on this screen an shRNA library targeting the 91 known or putative deubiquitinating enzymes encoded by the human genome (called shDUB library, see Refs. 8 and 9). Each pool was transfected in the U2OS-FL cells (expressing Notch) before the uptake experiment to identify the DUBs affecting nonactivated Notch intracellular trafficking. Notch signal disappearance was observed with almost every shDUB library pool (exemplified in Fig. 1, C and G), whereas an endosome-located persistent Notch signal was still observed in cells transfected with a pool targeting USP12 (Fig. 1, D and H). The efficiency of this shUSP12 pool to silence USP12 expression was controlled in supplemental Fig. S1. These observations suggest that USP12 knockdown impairs a step of Notch trafficking taking place between plasma membrane and lysosomes.
FIGURE 1.
Identification of USP12. U2OS-FL cells stably expressing HA-tagged Notch were transfected with empty vector (A, B, E, F) or each one of the 91 pools of shRNAs targeting the human DUBs (USP1 in C and G as a representative example; USP12 in D and H). Living cells were labeled with an anti-HA antibody coupled to Alexa Fluor 594 (shown in white), washed, fixed directly (A–D, T0), or incubated during 4 h at 37 °C before fixation and immunofluorescence analysis (E–H, T4H). When indicated (B and F), cells were treated with 100 μm leupeptin 1 h before adding the antibody and until fixation. Scale bar: 10 μm.
USP12 Is a Negative Regulator of Notch Signaling
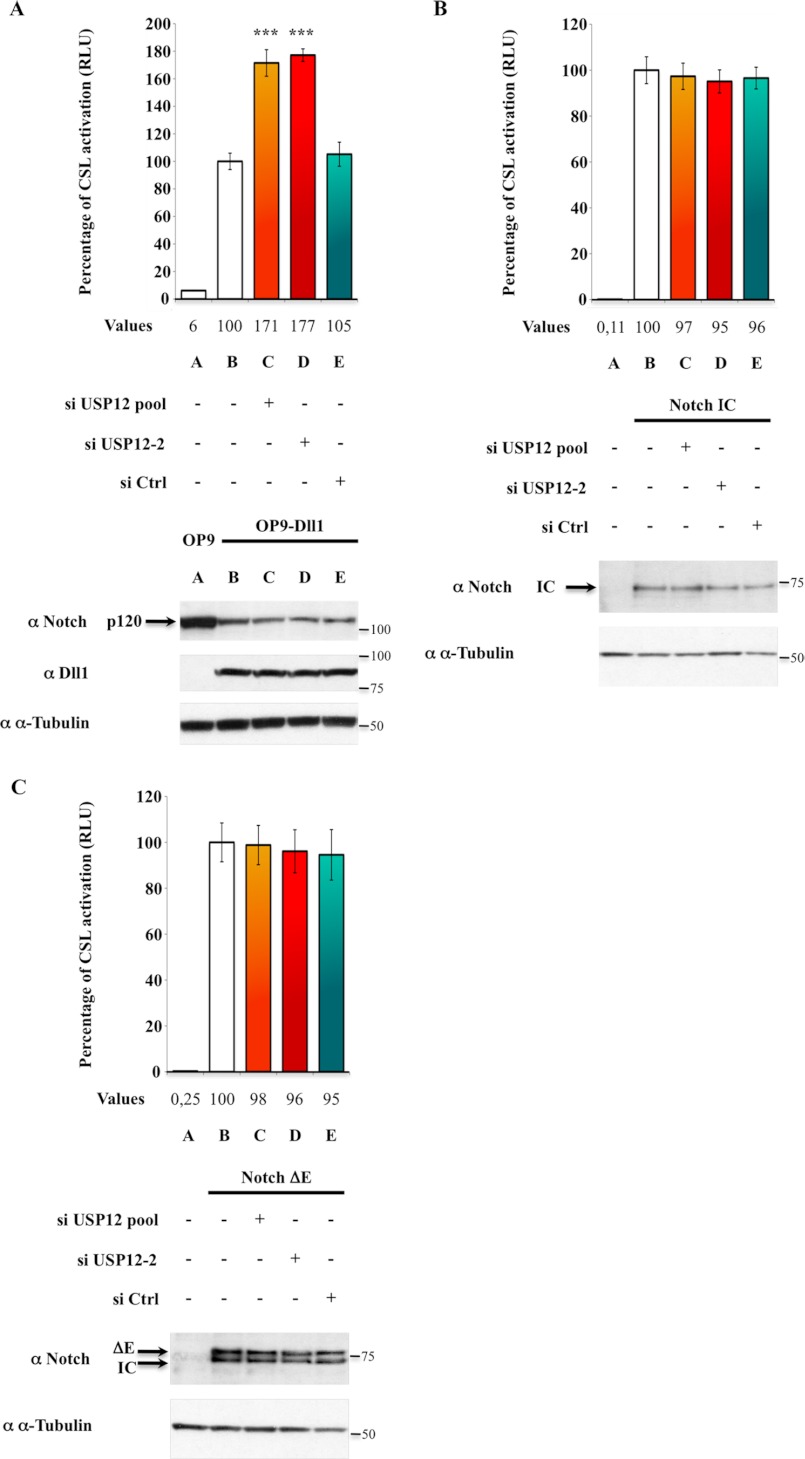
We next sought to assess the role of USP12 in Notch signaling. We monitored physiological activation of Notch signaling by measuring the activation of a Notch reporter gene (CSL-luciferase) in a coculture assay between Notch- and Delta-like1-expressing cells. U2OS-FL cells were first transfected with a control siRNA or with siRNAs targeting USP12 (siUSP12 pool and siUSP12-2), whose target sequence is different from that of the shDUB library pool, thus excluding off-target effect. Their efficiency in knocking down USP12 expression was validated both on ectopic USP12 protein amounts and on USP12 mRNA levels (supplemental Figs. S1A and S2A). Cells were then transfected with a CSL-firefly luciferase Notch reporter gene and with an internal Notch-insensitive control vector encoding Renilla luciferase. U2OS-FL cells were next cocultured with OP9 cells stably expressing or not the Notch ligand Delta-like1 (Dll1) (11). Finally, cells were lysed, and the relative luciferase activity was determined by normalizing firefly luciferase with Renilla luciferase (Fig. 2A). In parallel, the cell extracts were analyzed by Western blot to control levels of Notch and Dll1 (Fig. 2A, bottom).
FIGURE 2.
USP12 is a negative regulator of Notch signaling in mammals. A, USP12 silencing increases Notch reporter activity in Notch-expressing cells cocultured with Dll1-expressing cells. U2OS-FL cells were first transfected with siRNA control (si Ctrl) or targeting USP12 (siUSP12 pool or siUSP12-2, see supplemental Figs. S1A and S2A) and then with both CSL-firefly luciferase (Notch reporter) and TK-Renilla luciferase (internal control). The CSL-reporter activation corresponds to the ratio between firefly and Renilla luciferase activities. It was defined as 100% in the presence of Dll1-expressing cells (lane B), corresponding to a 17-fold increased as compared with coculture with control OP9 cells (lane A). B and C, USP12 silencing does not affect the activity of Notch-IC and Notch-ΔE. U2OS cells were first transfected with the same doses of siRNA control or targeting USP12 as above and then with Notch and control reporters together with Notch-IC (B) or Notch-ΔE (C). The relative luciferase activity in the absence of any siRNA (lane B) was defined as 100%. In all panels, the CSL-activation values are indicated for each sample under the graph. Error bars represent the S.E. of triplicate experiments, and the Western blot analysis of a representative experiment is shown using α-tubulin as a loading control. In panel A, p values calculated with Student's t test are indicated (***, p < 0.0001 as compared with siRNA control (lane E)). In panel C, Notch-IC originates from the constitutive γ-secretase cleavage of ΔE. The molecular masses (kDa) are also indicated on the right.
In the presence of Dll1, the relative luciferase activity of the Notch reporter gene increased about 20-fold as compared with control cells, showing that Notch signaling is indeed activated by the binding between Dll1 ligand and Notch receptor (Fig. 2A, lanes A and B). We observed a 1.8-fold increase of Notch transcriptional activity in the presence of either the siRNA pool (lane C) or isolated siRNA targeting USP12 (lane D), whereas the control siRNA did not affect Notch activity. This observation indicates that USP12 is a negative regulator of Notch signaling pathway. Notch transcriptional activity in the absence of ligand was not increased when USP12 was silenced, thus indicating that the Notch transcriptional activity increase was not ligand-independent (a phenomenon described in some contexts in Drosophila (7)) (supplemental Fig. S3).
To determine whether silencing USP12 has a direct effect on the Notch activation cascade, we tested the effect of these same siRNAs on ectopic Notch-IC- or Notch-ΔE-induced transcriptional activities. Notch-ΔE, a mutant of Notch deleted of most of the extracellular domain (13), mimics ADAM cleavage product and thus recapitulates the Notch activation pathway between the first ligand-induced Notch cleavage and the nucleus, including monoubiquitination and γ-secretase cleavage (9, 19). Notch-IC is the nuclear form of activated Notch. We did not observe any variation of Notch-IC and Notch-ΔE transcriptional activities, neither with the siUSP12 pool nor with the siUSP12-2 pool (Fig. 2, B and C, lanes B–D). This result means that USP12 is not involved in the Notch activation pathway from ADAM-mediated Notch cleavage to Notch-induced transcriptional activation. Therefore USP12 is a negative regulator of Notch signaling but does not regulate the Notch activation pathway. This strongly suggests that USP12 modulates Notch signaling by acting upstream of Notch activation by its ligands.
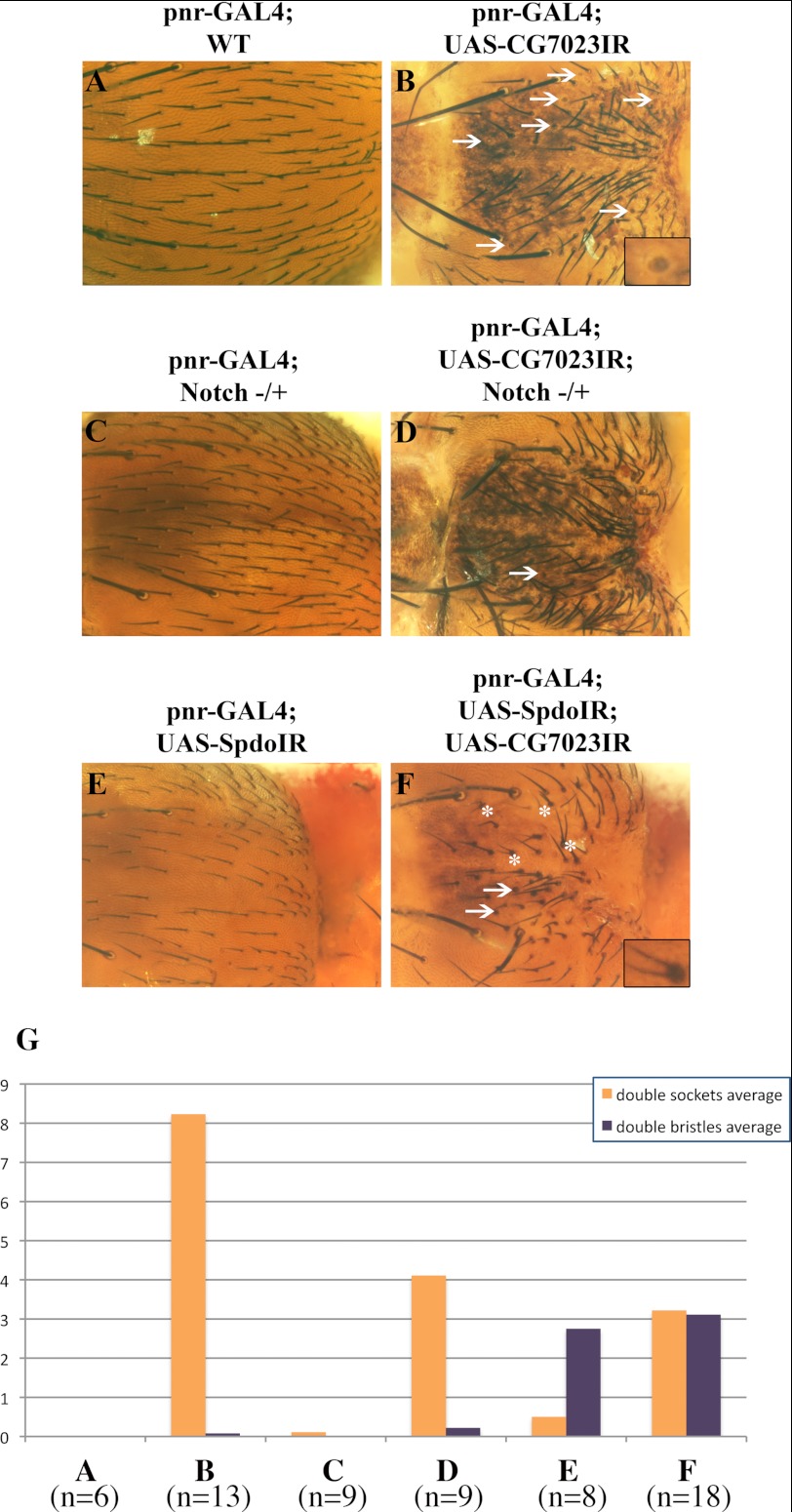
Because Notch signaling pathway is very conserved along evolution, we then asked whether USP12 function could be conserved in Drosophila melanogaster. Among the 21 USPs of the Drosophila genome, CG7023 gene encodes a 424-residue-long protein, which is 69% similar to human USP12 and USP46 (the closest mammalian USP12 homolog). This suggests that CG7023 is the sole bona fide USP12/46 homolog in Drosophila. We used a hairpin interfering RNA (IR) construct under the control of UAS/GAL4 to knockdown CG7023 in a central region of the notum, and we observed the outcome on adult external sensory organ development (summarized in supplemental Fig. S4). In WT flies, the sockets were distributed roughly evenly over the thoracic surface (Fig. 3A). In contrast, CG7023 IR overexpression appeared to disorganize bristle socket pattern (Fig. 3B). It is of note that the overall shape and aspect of the notum was significantly affected, probably because CG7023 also has roles in proliferation, apoptosis, and pigmentation. In addition, loss of bristles and duplication of sockets were frequent with CG7023 IR (Fig. 3, A–F, arrows, and magnification in the inset of Fig. 3B), similarly to what is expected for a Notch gain of function. Therefore the phenotype when CG7023 was knocked down was in accordance with an effect on both processes directed by Notch pathway, lateral inhibition (controlling the number and distribution of the sensory organs), and asymmetric cell division (controlling the differentiation of each precursor to a well shaped organ) (20). To further validate the involvement of CG7023 in the Notch pathway, we examined whether it genetically interacts with Notch and with a positive regulator of Notch signaling, Sanpodo (Spdo) (21). Introducing a mild Notch hypomorphic genotype into the CG7023IR background resulted in a partial restoration of normal phenotype because the cell fate transformation into socket cells was seldom observed, although lateral inhibition problems as well as malformation of the notum were still obvious (Fig. 3, C and D). When expressing an IR targeting Sanpodo, a large number of sensory organ precursor cells adopted a pIIb fate (explained in supplemental Fig. S4), resulting in a partially naked notum (Fig. 3E). When both Spdo and CG7023 were targeted by IR, Notch signaling was partially rescued, resulting in an intermediate phenotype characterized by the production of double-bristles (Fig. 3F, asterisks) or even double-sockets (arrows). Therefore Spdo and CG7023 seem to exert opposing effects on Notch signaling. About 10 flies for each genotype were observed visually, and phenotypic abnormalities (double-bristles or double-sockets) were recorded in Fig. 3G. Together, our results suggest that CG7023 behaves as a negative regulator of Notch signaling, in opposition to Notch and Sanpodo, in perfect accordance with the role of USP12 in mammals.
FIGURE 3.
CG7023 is a negative regulator of Notch signaling in D. melanogaster. A–F, dorsal surface of the thorax of mutant flies, resulting from various crosses. A, pnr-GAL4 x w1118 (WT). B, pnr-GAL4 x UAS-CG7023IR. C and D, Notch−/+ females were crossed with UAS-CG7023IR, the Notch−/+ females of the progeny were then crossed with pnr-GAL4 males, and the resulting females harboring notched wings were observed. Depending on the eye color, they had the UAS-CG7023IR-containing chromosome (D) or not (C). E and F, pnr-GAL4; UAS-SpdoIR flies were crossed with UAS-CG7023IR (F) or w1118 (E) flies as a control. In A and C, all organs have normal shaft and socket, whereas in CG7023IR-expressing flies, there are double-sockets or double-bristles organs, marked by arrows and asterisks, respectively (examples are magnified 3-fold in the insets of B and F). Original magnification is ×150. G, as pnr-GAL4 is expressed in the central region of the notum, this region was observed on 6–18 flies for each cross, and the double-sockets and double-bristles organs were recorded (A–F designate the same fly genotypes as in the representative image of the corresponding panel, n = the number of flies). A schematic view of external sensory organ morphology and lineage is shown in supplemental Fig. S4.
USP12 Regulates the Amount of Notch Receptor Expressed at the Cell Surface
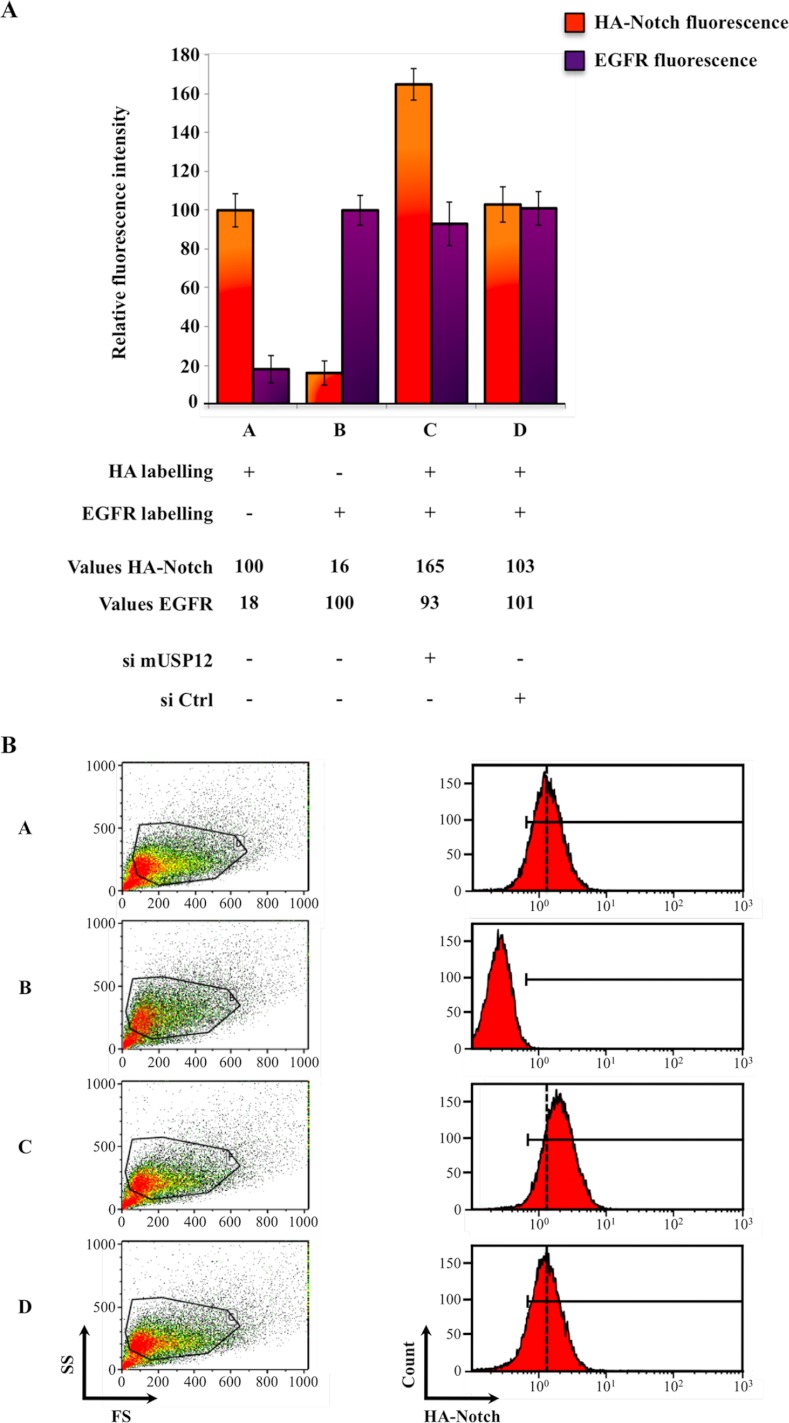
One of the possible consequences of the Notch trafficking defect associated with USP12 silencing, which may result in an increased Notch activation, could be a higher quantity of Notch receptor expressed at the cell surface. To test this possibility, we quantified nonactivated Notch molecules located at the plasma membrane using flow cytometry. For technical reasons, we used a clone of mouse embryonic fibroblast (MEF) cells stably expressing the same HA-tagged Notch-FL as U2OS-FL and where Notch degradation characteristics were first described (called MEF-FL, see Ref. 10). MEF-FL were transfected with an siRNA targeting mUSP12 or with a control siRNA, and we measured the quantity of both membrane Notch and epidermal growth factor receptor (EGFR) receptors using appropriate antibodies. We checked by immunofluorescence that this protocol specifically allowed the detection of cell surface molecules (supplemental Fig. S5). We reported the mean relative fluorescence intensity of Notch and EGFR for each sample (Fig. 4). Interestingly, the siRNA targeting mUSP12 led to a 1.6-fold enhanced Notch fluorescence as compared with the control siRNA (compare lanes C and D with lane A), whereas the EGFR signal was not modified by any of these siRNAs (compare lanes C and D with lane B). These results indicate that the silencing of USP12 does not affect the general sorting of transmembrane proteins toward the plasma membrane, but leads to a specific increase in nonactivated Notch expression at the cell surface.
FIGURE 4.
USP12 silencing specifically increases Notch receptor quantity at the plasma membrane. MEF-FL cells, transfected with siRNA control or targeting USP12 (si mUSP12) as indicated, were analyzed by flow cytometry to quantify HA-Notch and EGFR fluorescence. For each labeling, the mean relative fluorescence was defined as compared with 100% in the absence of any siRNA (A, lanes A and B, respectively, for HA-Notch and EGFR), and their values are indicated. Error bars are standard deviations. The bar graphs are quantifications of the indicated populations shown in the corresponding plots and histograms of B.
USP12 Specifically Regulates Nonactivated Notch Trafficking toward Lysosomal Degradation
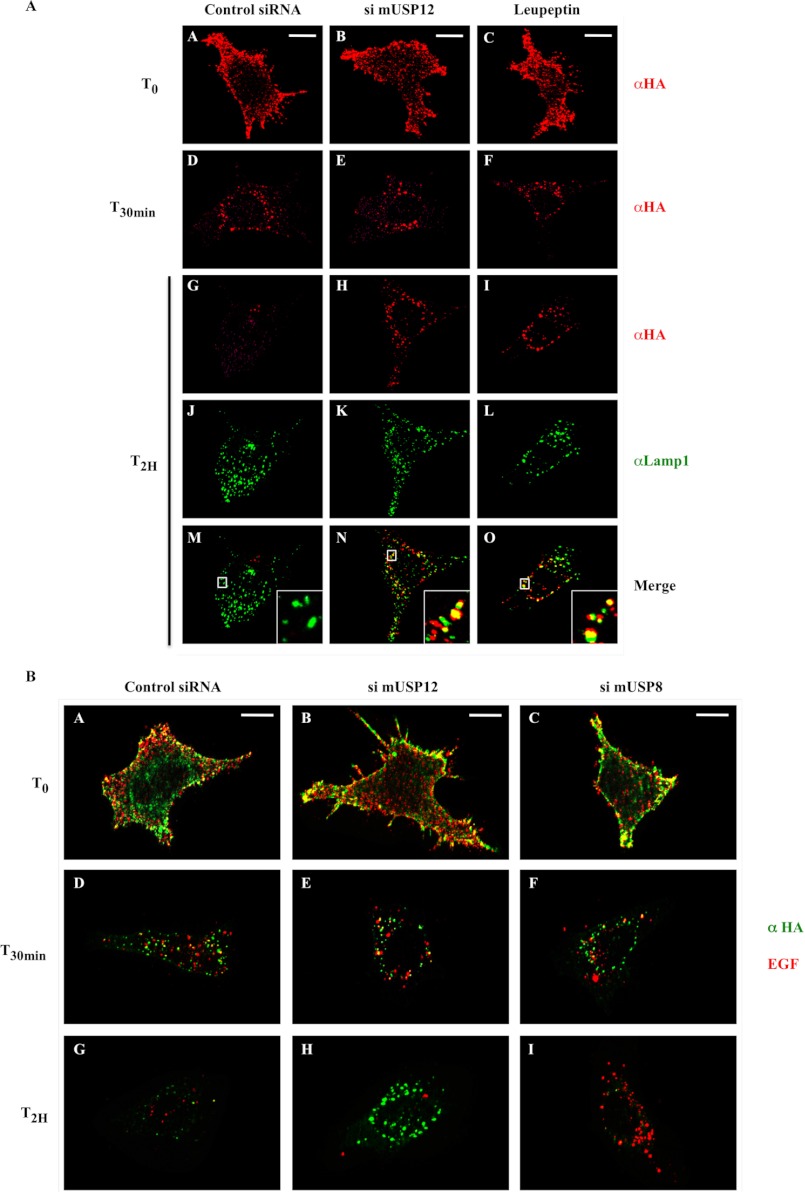
We then tried to characterize the precise step of Notch trafficking regulated by USP12. Anti-HA antibody uptake experiments were performed on siRNA-transfected MEF-FL cells, either nontargeting or targeting the murine USP12 (si mUSP12). At T0 or T30min, Notch molecules were similarly labeled in every condition (Fig. 5A, panels A–F). As observed in Fig. 1 with the human U2OS-FL cells, Notch signal persisted after 2 h in intracellular vesicles in the cells transfected with the siRNA targeting mUSP12 and in the leupeptin-treated cells (panels H and I as compared with panel G). Moreover, this accumulated Notch partially colocalized with Lamp1 (lysosome-associated membrane protein 1, panels N and O), a marker of late endocytic organelles en route to lysosomal degradation (late endosomes, multivesicular bodies (MVBs), and lysosomes). Not only do these last results confirm the involvement of USP12 in nonactivated Notch trafficking, but they also show that USP12 is necessary for Notch degradation in late endosomes/lysosomes. USP12 silencing could impair Notch entry into MVBs and/or fusion of MVBs with late endosomes and lysosomes, thus preventing nonactivated Notch degradation.
FIGURE 5.
Specific regulation of Notch trafficking by USP12. A, USP12 silencing interrupts Notch trafficking in late endosomes/lysosomes. MEF-FL cells stably expressing HA-tagged Notch were transfected with siRNA control (first column) or targeting USP12 (si mUSP12, second column). Living cells were labeled with an anti-HA antibody coupled to Alexa Fluor 594 (shown in red) and incubated or not during 30 min (T30min) or 2 h (T2H) at 37 °C before fixation. For the T2H time point, cells were additionally permeabilized and labeled with an anti-Lamp1 antibody (shown in green). When indicated (third column), cells were treated with 100 μm leupeptin. Insets represent 5-fold enlargements of the boxed regions. Scale bar: 10 μm. T0, time 0. B, USP12 silencing specifically delays Notch degradation. MEF-FL cells were transfected with siRNA control (first column) or targeting USP12 (si mUSP12, second column) or USP8 (si mUSP8, third column). Living cells were labeled with an anti-HA antibody coupled to Alexa Fluor 488 (shown in green) together with EGF coupled to Alexa Fluor 555 (shown in red). Then, cells were incubated during 30 min or 2 h at 37 °C before fixation and immunofluorescence analysis. Scale bar: 10 μm.
We then asked whether USP12 is a general modulator of trafficking and targeting to lysosomes of membrane receptors or whether it specifically acts on nonactivated Notch sorting toward lysosomal degradation. We decided to monitor in parallel the internalization and degradation of Notch and of another receptor. We chose the EGFR as a reference because, upon activation with EGF, it is ubiquitinated by the E3 ubiquitin ligase c-Cbl and consequently endocytosed and degraded in lysosomes (22, 23). Moreover, EGFR degradation in lysosomes requires a deubiquitination step catalyzed by the deubiquitinating enzyme USP8/UBPY that is associated with the endosomal sorting complex required for transport (ESCRT) machinery (17, 24). To assess the role of USP12 in activated EGFR degradation, we set up a dual uptake experiment using MEF-FL cells. The cells were first transfected with a control siRNA or with siRNAs targeting murine USP12 (si mUSP12) or murine USP8 (si mUSP8). Then, living cells were simultaneously incubated with an anti-HA antibody coupled to Alexa Fluor 488 fluorochrome to label nonactivated Notch at the plasma membrane and with EGF coupled to Alexa Fluor 555 to activate and label endogenous EGFR located at the cells surface. We next performed a 2-h uptake experiment at 37 °C to follow both nonactivated Notch and EGFR endocytosis and degradation (Fig. 5B). The initial Notch and EGFR labelings were located at the plasma membrane as expected (panels A–C). In control conditions, both receptors were endocytosed (panel D) and degraded within 2 h (panel G) as expected. With the siRNA targeting USP12, Notch degradation was inhibited as observed previously, whereas EGFR receptor was down-regulated in the normal way with a disappearance of EGFR labeling (panel H). Conversely, the siRNA targeting USP8 prevented EGFR degradation without affecting nonactivated Notch degradation (panel I). Therefore USP12 does not regulate the global trafficking of transmembrane proteins to degradation but instead specifically modulates sorting of nonactivated Notch receptor toward lysosomes.
USP12 and UAF1 Specifically Regulate the Ubiquitination State of Nonactivated Notch
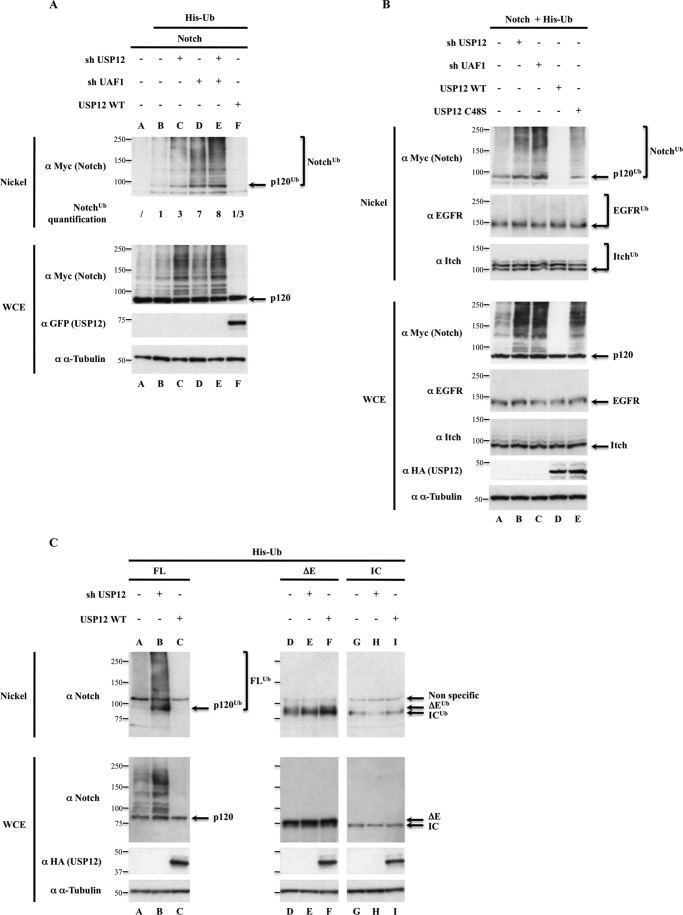
We next sought to determine the target of the deubiquitinating enzyme USP12. Notch has been shown to be polyubiquitinated by the E3 ubiquitin ligase Itch/AIP4 before being targeted to the lysosomes (10); therefore we asked whether USP12 could account for Notch deubiquitination. We made use of another shRNA targeting USP12 (named shUSP12) different from the ones of the shDUB library pool (supplemental Figs. S1B and S2B). HEK293T cells were transfected with vectors encoding for Myc6-tagged Notch and His6-tagged ubiquitin and with shRNAs targeting USP12 and its activator UAF1 (14). The ubiquitinated proteins were purified in denaturing conditions and analyzed with anti-Myc antibody (Fig. 6A). Although no ubiquitinated Notch was detected in the absence of His-Ub (lane A), the targeting of USP12 or UAF1 by shRNAs led to a 3- and 7-fold increase, respectively, of polyubiquitinated Notch p120 (p120Ub and slower migrating forms in Fig. 6A, lanes C and D as compared with lane B, see quantifications under the lane). This increase was even stronger (8-fold) when both shRNAs were mixed (lane E). We obtained the same results when using the siRNAs targeting USP12 (supplemental Fig. S6). Conversely, the overexpression of WT USP12 strongly reduced Notch ubiquitination (Fig. 6A, lane F). In all cases, the levels of nonubiquitinated p120 were comparable in the extracts, reflecting the fact that a small percentage of Notch is undergoing ubiquitination and degradative trafficking.
FIGURE 6.
USP12 and UAF1 regulate deubiquitination of nonactivated Notch. A, effect of shRNAs targeting USP12-UAF1 on Notch ubiquitination. B, the catalytic site of USP12 is required for Notch-specific deubiquitination. C, USP12 acts only on nonactivated Notch. In all panels, HEK293T cells were transfected as indicated above the lanes. Nickel-Sepharose-purified ubiquitinated products and whole cell extracts (WCE, 5% of the total lysates) were resolved on SDS-PAGE and analyzed by Western blot using the antibodies indicated on the left side of each panel. The molecular masses (kDa) are represented on the left of each immunoblot. p120 designates the membrane-anchored form of nonactivated full-length Notch, resulting from furin cleavage, whose apparent molecular mass is lower than 120 kDa due to the deletion of Notch carboxyl terminus end. α-Tubulin was used as a loading control for each Western blot analysis. In A, quantifications of Notch ubiquitination levels are indicated under the lanes. In B, 24 h after transfection, cells were treated with EGF during 30 min before lysis. P120ub, ΔEub, and ICub designate the monoubiquitinated forms of p120, ΔE, and IC, respectively, whereas the polyubiquitinated, slower migrating forms are indicated by brackets (Notchub or FLub).
To assess the contribution of the catalytic activity of USP12 to this function, we compared the overexpression of USP12 and of a catalytically inactive USP12 (USP12 C48S (15)). In contrast to USP12 WT, overexpressed USP12 C48S engendered a slight increase of polyubiquitinated Notch (Fig. 6B, lanes D and E). These effects were also detectable in the whole cell extracts (Fig. 6B, bottom, lanes A, D, and E). Therefore the effect of USP12 and UAF1 relies on USP12 catalytic activity.
To assess the specificity of USP12 enzymatic activity toward Notch, we tested the ubiquitination state of two additional endogenous proteins: the EGFR and the E3 ubiquitin ligase Itch/AIP4. In Fig. 6B, transfected cells were treated with EGF to activate endogenous EGFR and induce its ubiquitination just before protein extraction. In contrast to what was observed for Notch (anti-Myc panel), the ubiquitination status of activated EGFR and Itch/AIP4 was not affected in any condition (lanes A–E). This shows that USP12 specifically targets ubiquitinated Notch, and not any transmembrane protein nor the E3 ubiquitin ligase that polyubiquitinates Notch.
To finally determine whether the effect of USP12 on Notch ubiquitination was specific for the nonactivated receptor, we analyzed in the same way activated Notch in parallel to Notch-FL. Notch-ΔE or Notch-IC ubiquitination levels were neither affected by the shRNA targeting USP12 nor by overexpressed USP12 (Fig. 6C). Thus USP12 affects Notch polyubiquitination by acting specifically on the nonactivated membrane-anchored Notch receptor.
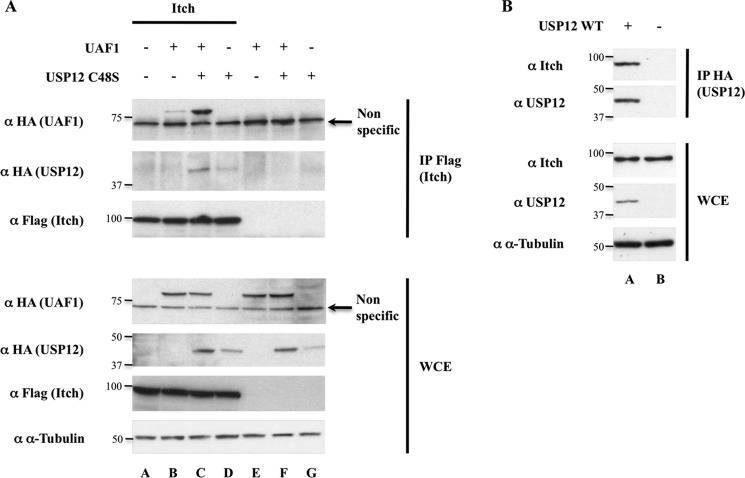
We then asked whether USP12 could interact with its Notch substrate. We were unable to visualize any interaction between overexpressed USP12 WT, C48S, or UAF1 and Notch in HEK293T cells, thus suggesting that substrate recognition could be indirect or that the complexes were not stable or numerous enough to be detected. However, when transfecting these cells with expression vectors encoding for Itch/AIP4, UAF1, and USP12 C48S, we could coimmunoprecipitate UAF1 and USP12 C48S together with Itch/AIP4 (Fig. 7A). The coimmunoprecipitation efficiency was higher when both UAF1 and USP12 C48S were overexpressed, suggesting that they are both required to stabilize the trimeric complex with Itch (compare lane C with lanes B and D). Inversely, overexpressed USP12 could immunoprecipitate endogenous Itch/AIP4, as shown in Fig. 7B. Together, these results show that USP12 can interact with Itch/AIP4 and therefore possibly be recruited to the ubiquitinated Notch en route to lysosomal degradation.
FIGURE 7.
Itch/AIP4 interacts with USP12-UAF1 complex. A, coimmunoprecipitation of UAF1 and USP12C48S with Itch/AIP4. B, coimmunoprecipitation of endogenous Itch/AIP4 with overexpressed USP12. HEK293T cells were transfected as indicated above the lanes. Immunoprecipitations (IP) performed with the FLAG (A) or HA (B) antibodies and whole cell extracts (WCE, 5% of the total lysates) were resolved on SDS-PAGE and analyzed by Western blot using the antibodies indicated on the left side of each panel. The molecular masses (kDa) are represented on the left of each immunoblot.
USP12 and UAF1 Catalyze Nonactivated Notch Deubiquitination
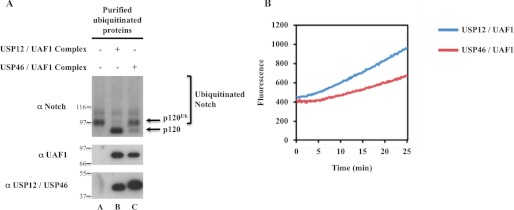
From the previous data, we could not discriminate from USP12 acting as a DUB on ubiquitinated Notch or impairing polyubiquitination of Notch by targeting another factor, different from Notch and Itch. To discriminate between both possibilities, we performed an in vitro deubiquitination assay using ubiquitinated Notch as a substrate. Ubiquitinated products were purified in denaturing conditions from Notch and His-Ub-transfected HEK293T, thus clearing out any interacting protein, and then renatured before incubation with the recombinant USP12-UAF1 or USP46-UAF1 complexes (14). Notch in vitro deubiquitination was efficient when using the USP12-UAF1 complex (Fig. 8A, lanes A and B), but barely detectable with the USP46-UAF1 complex (lane C), despite the strong homology between USP12 and USP46 and despite the fact that both are similarly active in a Ub-AMC experiment performed in parallel (Fig. 8B).
FIGURE 8.
USP12-UAF1 complex deubiquitinates Notch in vitro. A, in vitro Notch deubiquitination assay. HEK293T cells were transfected with Notch full-length and His-ubiquitin. Ubiquitinated proteins were purified in denaturing conditions on nickel-charged beads before being renatured and finally eluted with imidazole. The sample was divided in three, incubated or not with purified recombinant USP12-UAF1 or USP46-UAF1 complexes for 1 h, and analyzed by Western blot using the indicated antibodies. The molecular masses are also indicated. p120 designates the membrane-anchored form of nonactivated full-length Notch, resulting from furin cleavage, whose apparent molecular mass is lower than 120 kDa due to the deletion of Notch carboxyl terminus end. B, USP12-UAF1 and USP46-UAF1 complexes are catalytically active. Purified recombinant protein complexes were incubated with the fluorogenic substrate Ub-AMC, and the relative emitted fluorescence was measured during 25 min.
These results indicate that the USP12-UAF1 complex can recognize and directly deubiquitinate ubiquitinated Notch in vitro. Together with the fact that shRNA pools targeting USP46 were not able to affect Notch trafficking in the initial screen, these results confirm the specificity of USP12 versus USP46 in this deubiquitination process.
DISCUSSION
USP12 Is a Conserved Negative Regulator of Notch Signaling
Our results demonstrate that USP12 deubiquitinates nonactivated Notch and is required for its lysosomal degradation. USP12 down-regulation leads to an increased level of Notch molecules at the cell surface. This increase in Notch receptor availability at the surface probably accounts for the increase in Notch activation, although it remains possible that it indirectly influences it, for example, by titrating a negative factor of signal transduction. However, the consequence of USP12 silencing on Notch expression at the cell surface can be explained by two (nonmutually exclusive) mechanisms; the interruption of Notch trafficking could either decrease the Notch endocytosis rate or induce its recycling by default to the plasma membrane. Therefore USP12 probably regulates the quantity of Notch receptor at the cell surface and behaves as a negative regulator of Notch signaling.
In mammals, there are nearly 100 deubiquitinating enzymes, divided into five subfamilies. USP12 belongs to the largest, the ubiquitin-specific protease (USP) family (58 enzymes (25)). USP12, USP46, and USP1 are members of a subclass of USPs because each one is present in the cells in complexes containing UAF1 (for USP1-associated factor 1), which activates these enzymes (14). In a global proteomic analysis of DUBs and their associated protein complexes, Sowa et al. (26) found USP12 and USP46 as sharing five interacting proteins: WDR48 (UAF1), WDR20, DMWD, PHLPP, and PHLPPL. Whether these proteins are all involved in Notch deubiquitination and regulation remains to be determined. Despite the strong homology between USP12 and USP46 (92.2% similarity in humans), our results suggest a specific role for USP12 in nonactivated Notch trafficking. First, we did not identify USP46 and USP1 in the shDUB screen. Additionally, the in vitro deubiquitination assay showed a much stronger deubiquitination of nonactivated Notch by USP12 as compared with USP46.
Our results support genetic interactions between D. melanogaster CG7023, the closest homolog of USP12 and USP46, and the Notch pathway during adult sensory organ development. In perfect accordance with the mammalian data, CG7023 acts as a negative regulator of Notch pathway, although the observed phenotypes in a whole organism are probably complicated by the fact that CG7023 targets several proteins beside Notch. These broader and more severe phenotypes can explain why this gene was not specifically identified in previous RNAi screens for Notch regulators in Drosophila (20, 27). Nevertheless CG7023 encodes the first DUB identified in Drosophila acting on Notch. It is tempting to speculate that its mechanism of action is also conserved.
USP12 Deubiquitinates Nonactivated Notch
When we tested the effect of USP12 on Notch ubiquitination, we observed that silencing USP12 and/or UAF1 or expressing a catalytically inactive form of USP12 in cells stabilized polyubiquitinated Notch, whereas overexpression of USP12 WT drastically decreased it, thus demonstrating the involvement of USP12 in the regulation of nonactivated Notch ubiquitination. USP12 activity is specific for nonactivated Notch as compared with activated forms of Notch (Notch-ΔE and Notch-IC) and to other cargo proteins during their trafficking. Notch degradation in lysosomes is directed by Itch/AIP4-dependent polyubiquitination (10). Because Itch activity has been shown to depend on its ubiquitination status (28), the effect of USP12 on Notch ubiquitination could have been due to an action of USP12 on Itch/AIP4. We excluded this possibility by showing that endogenous Itch/AIP4 ubiquitination is not modified by USP12, although both proteins do interact. Together with the in vitro reconstituted deubiquitination assay, this allowed us to conclude that the USP12 substrate in Notch signaling is nonactivated Notch.
When and How Is USP12 Recruited to Notch?
When reconstituting physiological activation of Notch signaling by the ligand Dll1 using a coculture assay, we observed that Notch-dependent transcriptional activation of a reporter gene was specifically increased in the presence of siRNAs targeting USP12. Moreover, we showed that the activation steps, from Notch receptor activation at the plasma membrane to the transcriptional activation in the nucleus (i.e. endocytosis, monoubiquitination, deubiquitination, γ-secretase cleavage, and nuclear targeting) were not stimulated in the case of USP12 silencing by siRNAs, meaning that USP12 regulates Notch signaling upstream of Notch activation by its ligand. This conclusion is in agreement with the fact that USP12 knockdown interrupts the trafficking of nonactivated Notch receptor and also with the fact that we did not identify USP12 as a regulator of Notch activation in our first screen of the shDUB library (described in Ref. 9). USP12 silencing engenders an accumulation of nonactivated Notch in endosomes that we have characterized as Lamp1-positive late endosomes/MVBs/lysosomes, thus indicating that the action of USP12 is required for a proper addressing of nonactivated Notch to lysosomal degradation. Although USP12 is not associated with a specific compartment, its localization may depend on its recruitment to its substrates, as it is the case when it deubiquitinates the histones H2A and H2B (29). We have not been able to observe any interaction between USP12 and Notch in coimmunoprecipitation experiments. One possible explanation could be that a direct association between the two proteins exists, but either it is too labile to be detected or it requires polyubiquitinated Notch, which is a poorly represented species. The in vitro assay showing a deubiquitination of Notch by USP12-UAF1 strongly suggests that this direct interaction may exist, at least transiently. An alternative, nonexclusive possibility could be that a third protein is required as an intermediate or a recruiting factor for the formation of the Notch-USP12-containing complexes. A candidate for the recruitment of USP12 to Notch would be Itch/AIP4 because it is able to interact with both proteins. This E3 ubiquitin ligase is associated with endosomes and accounts for Notch polyubiquitination (10). A very recent study of enzymatic properties of the USP family (30) suggests that, in contrast to most USPs, the USP12-UAF1 complex is able to cleave all types of chains (except linear) and could theoretically hydrolyze the Lys-29-linked chains formed on Notch by virtue of Itch/AIP4 E3 ubiquitin ligase activity (10). All this favors the model whereby USP12-UAF1 complex is recruited to Notch-Itch and allows the delivery of Notch receptor to intraluminal vesicles of MVBs and its final degradation.
USP12 appears to be the second DUB identified as a key regulator of Notch signaling. eIF3f deubiquitinates activated Notch and positively regulates Notch activation (9). When comparing the two Notch substrates targeted by DUBs, namely nonactivated versus activated Notch, targeted by USP12 and eIF3f (9), respectively, they share the same intracellular domain sequence and are both sent to endocytic vesicles before deubiquitination. Interestingly, they differ in the type of ubiquitination they undergo, polyubiquitination in the case of nonactivated Notch and monoubiquitination in the case of activated Notch. Notch recognition by USP12 versus eIF3f depends on the substrate sequence (Notch), on its specific ubiquitination state (poly- versus monoubiquitination), but also on its localization in the cell (endocytic vesicles) and on cofactor recruitment (UAF1, Itch/AIP4 versus Deltex). In both cases, DUB recruitment is required for pursuing the process, showing that these enzymes are key negative and positive regulators of Notch signaling.
Supplementary Material
Acknowledgments
We thank J. Aster (Boston, MA), M. O. Fauvarque (Commissariat à l'Energie Atomique Grenoble, France), and F. Schweisguth (Institut Pasteur, Paris, France) for generous gifts of materials; P. H. Commere from the Flow Cytometry platform of the Institut Pasteur; and R. Bernards (The Netherlands Cancer Institute, Amsterdam, The Netherlands) for providing the shDUB library. We also thank L. Puca, D. Kachaner, A. Plessis, A. Dirac, R. Haguenauer-Tsapis, and F. Logeat for discussions or critical reading of the manuscript and L. Couturier for technical advice for experiments with flies.
This work was supported in part by the Institut Pasteur, CNRS, Association pour la Recherche sur le Cancer (Grant SFI20101201664, to C. B.), Ligue Nationale Contre le Cancer (Grant LNCC RS11/75-21, to C. B.), and the European Community (Grant 018683, Network of Excellence RUBICON, to A. I.).

This article contains supplemental Figs. S1–S6.
- Notch-IC
- Notch intracellular domain
- Notch-FL
- Notch full-length
- DUB
- deubiquitinating enzyme
- USP
- ubiquitin-specific protease
- mUSP
- mouse USP
- Ub
- ubiquitin
- Ub-AMC
- ubiquitin-7-amino-4-methylcoumarin
- ADAM
- a disintegrin and metalloproteinase
- MVBs
- multivesicular bodies
- MEF
- mouse embryonic fibroblast(s)
- EGFR
- epidermal growth factor receptor
- Lamp1
- lysosome-associated membrane protein 1
- UAF1
- USP1-associated factor 1
- IR
- interfering RNA
- si
- small interfering
- sh
- short hairpin.
REFERENCES
- 1. Kopan R., Ilagan M. X. (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. High F. A., Epstein J. A. (2008) The multifaceted role of Notch in cardiac development and disease. Nat. Rev. Genet. 9, 49–61 [DOI] [PubMed] [Google Scholar]
- 3. Logeat F., Bessia C., Brou C., LeBail O., Jarriault S., Seidah N. G., Israël A. (1998) The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. U.S.A. 95, 8108–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vodovar N., Schweisguth F. (2008) Functions of O-fucosyltransferase in Notch trafficking and signaling: toward the end of a controversy? J. Biol. 7, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heuss S. F., Ndiaye-Lobry D., Six E. M., Israël A., Logeat F. (2008) The intracellular region of Notch ligands Dll1 and Dll3 regulates their trafficking and signaling activity. Proc. Natl. Acad. Sci. U.S.A. 105, 11212–11217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brou C. (2009) Intracellular trafficking of Notch receptors and ligands. Exp. Cell Res. 315, 1549–1555 [DOI] [PubMed] [Google Scholar]
- 7. Vaccari T., Lu H., Kanwar R., Fortini M. E., Bilder D. (2008) Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J. Cell Biol. 180, 755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dirac A. M., Nijman S. M., Brummelkamp T. R., Bernards R. (2005) Functional annotation of deubiquitinating enzymes using RNA interference. Methods Enzymol. 398, 554–567 [DOI] [PubMed] [Google Scholar]
- 9. Moretti J., Chastagner P., Gastaldello S., Heuss S. F., Dirac A. M., Bernards R., Masucci M. G., Israël A., Brou C. (2010) The translation initiation factor 3f (eIF3f) exhibits a deubiquitinase activity regulating Notch activation. PLoS Biol. 8, e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chastagner P., Israël A., Brou C. (2008) AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS ONE 3, e2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Six E. M., Ndiaye D., Sauer G., Laâbi Y., Athman R., Cumano A., Brou C., Israël A., Logeat F. (2004) The Notch ligand Delta1 recruits Dlg1 at cell-cell contacts and regulates cell migration. J. Biol. Chem. 279, 55818–55826 [DOI] [PubMed] [Google Scholar]
- 12. Brou C., Logeat F., Gupta N., Bessia C., LeBail O., Doedens J. R., Cumano A., Roux P., Black R. A., Israël A. (2000) A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol. Cell 5, 207–216 [DOI] [PubMed] [Google Scholar]
- 13. Schroeter E. H., Kisslinger J. A., Kopan R. (1998) Notch-1 signaling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 [DOI] [PubMed] [Google Scholar]
- 14. Cohn M. A., Kee Y., Haas W., Gygi S. P., D'Andrea A. D. (2009) UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J. Biol. Chem. 284, 5343–5351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Minoguchi S., Taniguchi Y., Kato H., Okazaki T., Strobl L. J., Zimber-Strobl U., Bornkamm G. W., Honjo T. (1997) RBP-L, a transcription factor related to RBP-Jκ. Mol. Cell Biol. 17, 2679–2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cohn M. A., Kowal P., Yang K., Haas W., Huang T. T., Gygi S. P., D'Andrea A. D. (2007) A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell 28, 786–797 [DOI] [PubMed] [Google Scholar]
- 17. Row P. E., Prior I. A., McCullough J., Clague M. J., Urbé S. (2006) The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J. Biol. Chem. 281, 12618–12624 [DOI] [PubMed] [Google Scholar]
- 18. Couturier L., Vodovar N., Schweisguth F. (2012) Endocytosis by Numb breaks Notch symmetry at cytokinesis. Nat. Cell Biol. 14, 131–139 [DOI] [PubMed] [Google Scholar]
- 19. Gupta-Rossi N., Six E., LeBail O., Logeat F., Chastagner P., Olry A., Israël A., Brou C. (2004) Monoubiquitination and endocytosis direct γ-secretase cleavage of activated Notch receptor. J. Cell Biol. 166, 73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mummery-Widmer J. L., Yamazaki M., Stoeger T., Novatchkova M., Bhalerao S., Chen D., Dietzl G., Dickson B. J., Knoblich J. A. (2009) Genome-wide analysis of Notch signaling in Drosophila by transgenic RNAi. Nature 458, 987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Babaoglan A. B., O'Connor-Giles K. M., Mistry H., Schickedanz A., Wilson B. A., Skeath J. (2009) Sanpodo: a context-dependent activator and inhibitor of Notch signaling during asymmetric divisions. Development 136, 4089–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ravid T., Heidinger J. M., Gee P., Khan E. M., Goldkorn T. (2004) c-Cbl-mediated ubiquitinylation is required for epidermal growth factor receptor exit from the early endosomes. J. Biol. Chem. 279, 37153–37162 [DOI] [PubMed] [Google Scholar]
- 23. Stang E., Blystad F. D., Kazazic M., Bertelsen V., Brodahl T., Raiborg C., Stenmark H., Madshus I. H. (2004) Cbl-dependent ubiquitination is required for progression of EGF receptors into clathrin-coated pits. Mol. Biol. Cell 15, 3591–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alwan H. A., van Leeuwen J. E. (2007) UBPY-mediated epidermal growth factor receptor (EGFR) deubiquitination promotes EGFR degradation. J. Biol. Chem. 282, 1658–1669 [DOI] [PubMed] [Google Scholar]
- 25. Komander D., Clague M. J., Urbé S. (2009) Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 26. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saj A., Arziman Z., Stempfle D., van Belle W., Sauder U., Horn T., Dürrenberger M., Paro R., Boutros M., Merdes G. (2010) A combined ex vivo and in vivo RNAi screen for Notch regulators in Drosophila reveals an extensive Notch interaction network. Dev. Cell 18, 862–876 [DOI] [PubMed] [Google Scholar]
- 28. Mund T., Pelham H. (2009) Control of the activity of WW-HECT domain E3 ubiquitin ligases by NDFIP proteins. EMBO Rep. 10, 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Joo H. Y., Jones A., Yang C., Zhai L., Smith A. D., 4th, Zhang Z., Chandrasekharan M. B., Sun Z. W., Renfrow M. B., Wang Y., Chang C., Wang H. (2011) Regulation of histone H2A and H2B deubiquitination and Xenopus development by USP12 and USP46. J. Biol. Chem. 286, 7190–7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Faesen A. C., Luna-Vargas M. P., Geurink P. P., Clerici M., Merkx R., van Dijk W. J., Hameed D. S., El Oualid F., Ovaa H., Sixma T. K. (2011) The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem. Biol. 18, 1550–1561 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.