Background: PRMT7 symmetrically methylates histones H2A and H4, and modulates cellular response to DNA damage.
Results: PRMT7 interacts with BRG1-hSWI/SNF, targets H2AR3 and H4R3, and represses expression of POLD1.
Conclusion: PRMT7 regulates response to DNA damage and confers resistance to DNA-damaging agents.
Significance: Understanding how PRMT7 regulates response to genotoxic stress will clarify how cancer cells become drug-resistant.
Keywords: Chromatin Remodeling, DNA Damage, DNA Repair, Gene Regulation, Histone Methylation, BRG1-based hSWI/SNF, Genotoxic Stress, Protein Arginine Methyltransferase 7, Polymerase delta Catalytic Subunit POLD1
Abstract
Covalent modification of histones by protein arginine methyltransferases (PRMTs) impacts genome organization and gene expression. In this report, we show that PRMT7 interacts with the BRG1-based hSWI/SNF chromatin remodeling complex and specifically methylates histone H2A Arg-3 (H2AR3) and histone H4 Arg-3 (H4R3). To elucidate the biological function of PRMT7, we knocked down its expression in NIH 3T3 cells and analyzed global gene expression. Our findings show that PRMT7 negatively regulates expression of genes involved in DNA repair, including ALKBH5, APEX2, POLD1, and POLD2. Chromatin immunoprecipitation (ChIP) revealed that PRMT7 and dimethylated H2AR3 and H4R3 are enriched at target DNA repair genes in parental cells, whereas PRMT7 knockdown caused a significant decrease in PRMT7 recruitment and H2AR3/H4R3 methylation. Decreased PRMT7 expression also resulted in derepression of target DNA repair genes and enhanced cell resistance to DNA-damaging agents. Furthermore, we show that BRG1 co-localizes with PRMT7 on target promoters and that expression of a catalytically inactive form of BRG1 results in derepression of PRMT7 target DNA repair genes. Remarkably, reducing expression of individual PRMT7 target DNA repair genes showed that only the catalytic subunit of DNA polymerase, POLD1, was able to resensitize PRMT7 knock-down cells to DNA-damaging agents. These results provide evidence for the important role played by PRMT7 in epigenetic regulation of DNA repair genes and cellular response to DNA damage.
Introduction
Proper and controlled expression of genes involved in various cellular processes, including repair of DNA damage, is essential for normal cell growth and development. Several studies have shown that there is a plethora of enzymes, which are designed to remodel chromatin and maintain genomic fluidity, that can regulate accessibility to regulatory regions and specify transcriptional outcome (1–7). Among these enzymes are members of the PRMT family, which includes enzymes that methylate histone arginine residues either in an asymmetric manner (type I), which generally leads to activation of transcription, or in a symmetric fashion (type II), which often causes transcriptional repression (1, 6). However, as members of each type of PRMT enzyme are being characterized, we are learning that there are exceptions to the rule. For instance, PRMT6 can asymmetrically methylate H3R23 and trigger transcriptional repression (8–10). Studies from three distinct groups have determined that H3R2-induced transcriptional silencing is largely due to the inhibition of H3K4 methylation. Despite progress in understanding how members of the PRMT family function, little is known about their target genes and the pathways they regulate.
The role of histone lysine methylation in transcriptional regulation has been extensively studied; however, the impact of PRMT enzymes on gene regulation remains unexplored. Thus far, there are nine methyltransferases in the PRMT family, and only three members (PRMT5, PRMT7, and PRMT9) have been shown to catalyze symmetric methylation of arginine residues (1, 6). Immunoprecipitation studies using antibodies that can recognize either asymmetrically or symmetrically methylated arginine residues have shown that there are approximately 200 methylated proteins involved in different cellular processes like signal transduction, transcription, and DNA repair, highlighting the wide spectrum of substrates targeted by PRMTs (11). Most PRMTs can methylate arginine residues on a variety of cellular proteins, including histones (1, 6). Currently, there is limited information about PRMT interacting partners, but recent studies have established that both type I and type II PRMTs can interact with chromatin remodeling complexes and modulate target gene expression. For instance, CARM1/PRMT4 has been shown to interact with the BRG1-based hSWI/SNF complex and methylate nucleosomal histones, which can in turn lead to transcriptional activation of target genes (12, 13). In contrast, we have recently shown that PRMT5 targets specific histone H3 and H4 arginine residues and negatively regulates transcription of tumor suppressor genes (14–17).
Although PRMT7 is also a type II PRMT capable of symmetric methylation of histones H2A and H4, little is known about its biological function and the histone residues it methylates (18, 19). Unlike other members of the PRMT family, PRMT7 contains two copies of the S-adenosylmethionine (AdoMet) binding domain, which are both required for activity, because deletion of either AdoMet binding motif leads to loss of PRMT7 methylase activity (20). Previously, it was reported that PRMT7 knockdown in the Chinese hamster lung fibroblast DC-3F/c123 cell line confers resistance to various DNA-damaging agents, suggesting that PRMT7 is involved in modulating cellular response to DNA-damaging agents (21). In a separate study, down-regulation of PRMT7 sensitized HeLa cells to camptothecin, a topoisomerase I inhibitor, reinforcing the idea that modulating the levels of PRMT7 in cancer cells may help potentiate the effects of known cytotoxic drugs (22). Currently, there is no information about PRMT7 target genes, and more work is required to define PRMT7 effects on histone arginine methylation and gene regulation.
To further understand the biochemical and biological functions of PRMT7, we established HeLa S3 stable cell lines to immunopurify and characterize PRMT7 biochemical activity and NIH 3T3 knock-down cell lines to investigate effects of PRMT7 on cell growth and global gene expression. Our results indicate that affinity-purified PRMT7 is able to interact with the BRG1-based hSWI/SNF complex and is capable of specific methylation of histones H2AR3 and H4R3. Global gene profiling using total RNA from control and knock-down PRMT7 cells showed that a wide variety of genes, including genes involved in repair of DNA damage, are derepressed in PRMT7 knockdown cells. Remarkably, many of the identified DNA repair genes are direct targets of PRMT7, as evidenced by decreased PRMT7 recruitment and H2AR3/H4R3 methylation at their promoters in PRMT7 knock-down cells. In addition, we also found that BRG1 co-localizes with PRMT7 on repressed target genes in control parental cells and that expression of catalytically inactive BRG1 results in derepression of PRMT7 target genes. In light of previous studies, which implicated PRMT7 in regulating cellular response to DNA damage, we sought to investigate the role played by PRMT7 and its target genes in drug resistance. Our findings show that PRMT7 knock-down cells, where expression of target DNA repair genes is enhanced, are more resistant to genotoxic drugs compared with control parental cells. Furthermore, analysis of individual PRMT7 target DNA repair genes revealed that reducing expression of POLD1 resensitizes PRMT7 knock-down cells to genotoxic stress. Taken together, these results suggest that PRMT7 is involved in regulating the cellular response to DNA damage through its ability to methylate promoter histones H2AR3 and H4R3 of key target DNA repair genes.
EXPERIMENTAL PROCEDURES
Plasmid DNA Constructs
FLAG-tagged PRMT7 cDNA was generated using the pCMV-SPORT6/PRMT7 plasmid purchased from the American Type Cell Collection (ATCC), which contains PRMT7 cDNA without the 5′-untranslated region (UTR). To ensure that the cloned cDNA expresses full-length PRMT7 protein, we modified the original ATCC PRMT7 cDNA sequence by performing a two-step nested PCR amplification at the 5′-end of the clone using ∼50 base pairs (bp) of the PRMT5 5′-UTR. The first PCR amplification was carried out using a 5′ primer (5′-GTGGACAGCGCGAGGAGAAAGATGATGATGAAGATCTTCTGCAGTCGGGCC-3′), which introduces 21 nucleotides of the PRMT5 5′-UTR region upstream of the PRMT7 start codon, and a 3′ primer (5′-CCGGAATTCTTACTATTTGTCATCGTCGTCCTTGTAGTCGTCTGGGGTATCTGCATGCCTGAA-3′) designed to include a FLAG tag sequence (underlined) followed by two stop codons and an EcoRI restriction site. To further extend the 5′-UTR region of PRMT7, we conducted a second PCR amplification using a second 5′ primer (5′-CCGGAATTCGTGATTGGCTACTAGTATCAAGGAATCCCGGCGTGGACAGCGCGAGGAGAAAG-3′), which incorporates an additional 32 nucleotides of the PRMT5 5′-UTR (underlined) and an EcoRI restriction site, and the same 3′ primer used above. The FLAG-tagged PRMT7 cDNA was first cloned into pBS(KS+) plasmid to generate pBS(KS+)/Fl-PRMT7, and another round of PCR amplification was conducted to subclone FLAG-tagged PRMT7 cDNA into the pBabe-puromycin retroviral vector using a 5′ primer (5′-TCCCCCGGGGGATATCGTGATTGGCTACTAGTATCAAGGA-3′) that harbors an EcoRV restriction site and the same 3′ primer used above. After PCR amplification, the EcoRV-EcoRI-digested DNA fragment was cloned into pBabe-puromycin vector cut with SnaBI and EcoRI. Plasmid pBabe/Fl-Mut/PRMT7 was generated by mutating glycine 74 and threonine 75 to alanines using the QuikChange multisite-directed mutagenesis kit (Agilent Technologies). A single oligonucleotide (5′-GGTTCTCGACATTGGCACTGcCgCGGGACTCTTGTCAATGATGGCGG-3′) was utilized to introduce both mutations.
To knock down PRMT7 expression, we generated two lentiviral constructs that express PRMT7-specific shRNAs using the viral RNAi system designed by Campeau et al. (23). Both plasmids pLENTI X2 DEST/sh-PRMT7-1 and pLENTI X2 DEST/sh-PRMT7-2 were constructed by first cloning double-stranded oligonucleotides that encode either PRMT7-specific shRNA1 or shRNA2 into pENTER/pTER+ entry vector. Plasmid pENTER/pTER+/sh-PRMT7-1 was generated using sense-1 (5′-GATCCCAGGAATGGCTTCAGTGATAGTGTGCTGTCCTATCACTGAAGCCATTCCTTTTTTGGAAA-3′) and antisense-1 (5′-AGCTTTTCCAAAAAAGGAATGGCTTCAGTGATAGGACAGCACACTATCACTGAAGCCATTCCTGG-3′) primers, whereas plasmid pENTER/pTER+/sh-PRMT7-2 was constructed using sense-2 (5′-GATCCCGGACAGAACTGATCACTATGTGTGCTGTCCATAGTGATCAGTTCTGTCCTTTTTGGAAA-3′) and antisense-2 (5′-AGCTT TTCCAAAAAGGACAGAACTGATCACTATGGACAGCACACATAGTGATCAGTTCTGTCCGG-3′) primers. Both sh-PRMT7-1 and sh-PRMT7-2 oligonucleotides were designed to include sticky ends compatible with BglII and HindIII and were cloned into BglII and HindIII-linearized pENTER/pTER+ vector. Positive clones were identified by EcoRI digestion and confirmed by DNA sequencing. Next, plasmids pENTER/pTER+/sh-PRMT7-1 and pENTER/pTER+/sh-PRMT7-2 were individually recombined into the pLENTI X2 DEST series of vectors to generate pLENTI X2 DEST/sh-PRMT7-1 and pLENTI X2 DEST/sh-PRMT7-2. Either pENTER/pTER+/sh-PRMT7-1 or pENTER/pTER+/sh-PRMT7-2 plasmid was incubated with pLENTI X2 DEST vector in a reaction containing 2 μl of LR clonase II enzyme mix according to the manufacturer's instructions (Invitrogen). The recombination reaction was then transformed into One Shot Stbl3 chemically competent E. coli cells (Invitrogen), and positive clones were identified by EcoRV digestion followed by DNA sequencing. Plasmids encoding sh-PRMT7 RNA1 and RNA2 were transfected into 293T cells, and 72 h later, lentiviral particles were harvested and used to infect NIH 3T3 cells as described previously (23). Plasmids used to knock down expression of PRMT7 target genes ALKBH5, APEX2, POLD1, and POLD2 were constructed as described above using the oligonucleotides listed in supplemental Table 1.
Plasmid for bacterial expression of glutathione S-transferase (GST)-PRMT7 (amino acids (aa) 2–692) was generated by inserting a 2-kbp PCR-amplified EcoRI fragment into the corresponding site of pGEX-2TK, whereas plasmid pGEX-2TK/PRMT7 (aa 2–199), which expresses the N-terminal region that was used to generate anti-PRMT7 antibody, was constructed by inserting a 597-bp PCR-amplified fragment into EcoRI-linearized pGEX-2TK. The 5′ primer used to generate full-length PRMT7 and the N-terminal region of PRMT7 (5′-CCGGAATTCATAAGATCTTCTGCAGTCGGGCCAAT-3′), the 3′ primer used to PCR-amplify the N-terminal region of PRMT7 (5′-CCGGAATTCTCAGTGGATGGGAAATAGCTTGTTCCACGA-3′), and the 3′ primer used to generate full-length GST-PRMT7 (5′-CCGGAATCCTCAGTCTGGGGTATCTGCATGCCTG-3′) all included an EcoRI restriction site.
Cell Culture, Establishment of Stable Cell Lines, Drug Treatment, and Proliferation Assays
HeLa S3 and NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. NIH 3T3 cells expressing either antisense PRMT5 or FLAG-tagged catalytically inactive BRG1 (Fl-MutBRG1) were grown as described previously (15, 24). To establish HeLa S3 cell lines that stably express epitope-tagged wild type or mutant PRMT7, 70–80% confluent plates were transfected with 2 μg of pBabe/Fl-PRMT7 or pBabe/Fl-Mut/PRMT7 plasmid for 5 h using Lipofectamine 2000 (Invitrogen). To establish NIH 3T3 PRMT7 knock-down stable cell lines, ∼8 × 105 cells were plated 24 h before infection was carried out in 3 ml of DMEM containing 600 μl of lentiviral supernatant, harvested from 293T cells transfected with either pLENTI X2 DEST/sh-GFP or pLENTI X2 DEST/sh-PRMT7, and 6 μg/μl Polybrene. Two days later, drug-resistant HeLa S3 cells expressing FLAG-tagged PRMT7 (Fl-PRMT7), FLAG-tagged mutant PRMT7 (Fl-Mut/PRMT7), and NIH 3T3 cells expressing either sh-GFP or sh-PRMT7 were selected in the presence of puromycin (2.5 μg/ml). Several HeLa S3 and NIH 3T3 puromycin-resistant colonies were expanded, and positive clones were identified by Western blotting using either anti-FLAG antibody in the case of HeLa S3/Fl-PRMT7, HeLa S3/Fl-Mut/PRMT7 cells, or anti-PRMT7 antibody in the case of NIH 3T3/sh-PRMT7-1 and NIH 3T3/sh-PRMT7-2 cells.
To evaluate proliferation of control NIH 3T3, NIH 3T3/antisense PRMT5, NIH 3T3/sh-PRMT7-1, and NIH 3T3/sh-PRMT7-2 cells, an equal number of cells from each cell line was seeded in 6-cm-diameter plates, and the number of viable cells was counted every 2 days for 6 days. To assess the effect of genotoxic drugs on control NIH 3T3, NIH 3T3/antisense PRMT5, NIH 3T3/sh-PRMT7-1, and NIH 3T3/sh-PRMT7-2 cell lines, ∼2 × 105 cells were seeded into 6-cm-diameter plates and treated with either cisplatin (10 μg/ml) or doxorubicin (0.3 μg/ml) for 24 h. Next, control and knock-down cell lines were washed three times with 1× PBS, and fresh medium was added. Proliferation was monitored every 2 days for 6 days by counting the number of viable cells using trypan blue dye exclusion. When cells were treated with chlorambucil (5 μg/ml) or mitomycin C (3 μg/ml), both drugs were not removed from the medium, and proliferation was determined as described above. For rescue experiments, NIH 3T3/sh-PRMT7-1 and NIH 3T3/sh-PRMT7-2 cells were infected with lentiviral particles harvested from 293T cells transfected with pLENTI X2 DEST expression vectors designed to knock down PRMT7 target genes ALKBH5, APEX2, POLD1, or POLD2. Two days later, cells were trypsinized, seeded into 6-cm-diameter plates, and treated with cisplatin as described above. The number of viable cells was determined every 2 days for 6 days.
Fluorescence-activated Cell Sorting (FACS) Analysis
To assess the effects of cisplatin on both control and PRMT7 knock-down cells, 5 × 106 NIH 3T3/sh-GFP or NIH 3T3/sh-PRMT7-1 cells were seeded into 10-cm-diameter plates and treated with cisplatin (10 μg/ml) before harvesting at days 0, 4, and 6. Next, cells were stained with 5 μl of FITC-Annexin V antibody (BD Bioscience) and 5 μl of propidium iodide solution (BD Biosciences) for 15 min at room temperature before they were analyzed on a Beckman Coulter FC500 flow cytometer.
Purification of FLAG-tagged PRMT7, Western Blot Analysis, and Immunoprecipitation Assays
Fl-PRMT7 protein was purified from the HeLa S3 cell line (clone 32) by affinity purification using anti-FLAG M2 agarose beads (Sigma). Approximately 100 mg of either control HeLa S3 or HeLa S3/Fl-PRMT7 nuclear extract was bound to 1.5 ml of anti-FLAG M2 agarose beads for 12 h at 4 °C. Next, beads were washed successively with buffer (20 mm HEPES (pH 7.9), 10% glycerol, 2 mm EDTA, 0.25 mm dithiothreitol (DTT), 0.5 mm phenylmethylsulfonyl fluoride (PMSF)) supplemented with either 150, 300, or 100 mm KCl. Fl-PRMT7 was then eluted with washing buffer containing 100 mm KCl and a 20-fold molar excess of FLAG peptide. Catalytically inactive FLAG-tagged PRMT7 (Fl-Mut/PRMT7) was purified as described above. To analyze affinity-purified PRMT7 fractions (15 μl) or nuclear and radioimmune precipitation assay extracts (30 μg), samples were separated on an SDS-8% PAGE, transferred onto nitrocellulose membrane, and detected by enhanced chemiluminescence reagents using either anti-FLAG (catalog no. sc-807, Santa Cruz Biotechnology, Inc.) or previously described anti-BRG1, anti-BRM, anti-BAF155, anti-BAF57, and anti-BAF45 antibodies (15, 16). Rabbit anti-PRMT7 polyclonal antibody was generated by Covance, Inc. using affinity-purified GST-PRMT7 (aa 2–199) fusion protein. Anti-H2A(Me2)R3 and anti-H4(Me2)R3 antibodies were generated by Covance, Inc. using symmetrically methylated H2AR3- and H4R3-specific peptides. Antibodies specific for CREB-binding protein (catalog no. sc-369) were purchased from Santa Cruz Biotechnology Inc., whereas anti-β-ACTIN (catalog no. A2066) antibody was purchased from Sigma. Antibodies against ALKBH5 (catalog no. ab69325), APEX2 (catalog no. ab13691), POLD1 (catalog no. ab51017), and POLD2 (catalog no. ab38338) were purchased from Abcam.
Immunoprecipitation experiments were performed by incubating 250 μg of HeLa S3 nuclear extract with 10 μl of either preimmune or immune anti-PRMT7 antibody at 4 °C for 4 h. Next, protein A-agarose beads, which were preblocked in 0.5 mg/ml BSA, were added, and samples were incubated at 4 °C overnight. Beads were collected by centrifugation and washed seven times with 1 ml of washing buffer (40 mm Tris-HCl (pH 8.0), 250 mm NaCl, 1% aprotinin). Bound proteins were separated on an SDS-8% polyacrylamide gel and analyzed by Western blotting. For immunoprecipitation experiments using cross-linked nuclear extract, 90% confluent plates of HeLa S3 cells were treated with 2 mm dithiobis(succinimidyl propionate) cross-linking agent (Pierce) for 2 h at 4 °C before stopping the reaction with 20 mm Tris-HCl (pH 8.0) at 25 °C for 20 min (25). Approximately 250 μg of cross-linked HeLa nuclear extract was incubated with either preimmune or immune anti-PRMT7 antibody as described above, and bound proteins were washed seven times with 1 ml of washing buffer (40 mm Tris-HCl (pH 8.0), 350 mm NaCl, 0.75% Nonidet P-40 and 0.05% SDS, 0.25 mm DTT, 0.5 mm PMSF, 1% aprotinin) before they were analyzed by Western blotting.
Histone and Peptide Methylation Assays
Histone methylation was performed using 2 μg of either HeLa S3 core histones or histone N-terminal peptides in the presence or absence of 15 μl of affinity-purified Fl-PRMT7, Fl-Mut/PRMT7, or control fractions purified from HeLa S3 nuclear extracts as described previously (15). Histones were visualized by Coomassie Blue staining before the gel was treated with 1 m salicylic acid (pH 6.0), and methylated products were detected by autoradiography. When either wild type or mutant N-terminal peptides were used as a substrate, reaction mixtures were spotted on Whatman P-81 filter paper, washed five times with 10 ml of 0.1 mm sodium carbonate buffer (pH 9.0) to remove unincorporated [3H]AdoMet, and methylated peptides were detected by scintillation counting.
Reverse Transcription (RT) Real-time PCR and Microarray Analysis
Total RNA was isolated using TRIzol reagent, and RT was performed using 1 μg of total RNA in a 20-μl reaction containing 2.5 μm random hexamer primers and Taqman reverse transcription reagents (Applied Biosystems, Inc). To measure mRNA levels of PRMT7 and its target genes, real-time PCR was carried out using the TaqMan system in a 10-μl reaction as described previously (16). Primer sets and probes used in RT reactions are listed in supplemental Table 2. To normalize mRNA levels, levels of 18S rRNA were measured in both control and PRMT7 knock-down cell lines using 1× pre-mixed 18S primer/probe set (Applied Biosystems, Inc). To identify target genes regulated by PRMT7, 10 μg of total RNA isolated from either puromycin-resistant control NIH 3T3 or PRMT7 knock-down cell lines was used to probe the Affymetrix GeneChip Mouse Genome 430 2.0 Array, which includes more than 39,000 mouse transcripts. Both hybridization and analysis were performed by the Ohio State University Comprehensive Cancer Center microarray shared resource facility. The Affymetrix microarray data were normalized using the GCRMA normalization method with bioconductor (26, 27). After normalization, for each sample, the geometric means of all the genes were computed, and the mean -fold changes for all of the genes between control and test samples were obtained. Genes with mean -fold changes more than 1.5 were selected for further investigation.
ChIP Assay
Cross-linked chromatin was prepared from control NIH 3T3, NIH 3T3/sh-PRMT7, and NIH 3T3/Fl-MutBRG1 cell lines as described previously (15); however, after sonication, cross-linked chromatin was digested with micrococcal nuclease (1 unit/ml) at 37 °C for 20 min before adding 200 μl of stop buffer (100 mm Tris-HCl (pH 8.6), 0.45% SDS, 2.5% Triton X-100, 5 mm EDTA (pH 8.0), protease inhibitors). Chromatin was analyzed by agarose gel electrophoresis to ensure that DNA fragment sizes did not exceed 500 bp, and ChIP assays were carried out essentially as described previously (16, 17). To amplify DNA sequences of PRMT7-regulated genes, preimmune and immune antibodies raised against BRG1, BAF155, BAF57, Fl-MutBRG1, PRMT7, H2A(Me2)R3, and H4(Me2)R3 were used to immunoprecipitate cross-linked chromatin from control NIH 3T3/sh-GFP, NIH 3T3/Fl-MutBRG1, and NIH 3T3/sh-PRMT7 cells. Antibodies that recognize PRMT1 (Millipore, catalog no. 07-404), PRMT4 (Millipore, catalog no. 09-818), asymmetric H4(Me2)R3 (Abcam, catalog no. ab17416), and asymmetric H3(Me2)R17 (Millipore, catalog no. 07-214) were also used to evaluate recruitment of type I PRMTs. After extensive washing, nucleoprotein complexes were eluted in 200 μl of elution buffer (50 mm Tris-HCl (pH 8.0), 10 mm EDTA, 1% SDS), and cross-links were reversed by incubating samples at 65 °C for 12 h. Immunoprecipitated DNA was purified by phenol and chloroform extraction and resuspended in 40 μl of Tris-EDTA (pH 8.0). To amplify promoter sequences of PRMT7 target genes, a 3-μl aliquot of eluted DNA was used in a 10-μl real-time PCR in the presence of primer pairs and specific Roche Applied Science universal probes listed in supplemental Table 2.
Statistical Analysis
Results were expressed as the means ± S.D. unless otherwise specified. Paired t tests and analysis of variance were used to generate p values for comparisons between two groups and when multiple samples within different groups were used, respectively.
RESULTS
PRMT7 Interacts with the BRG1-based hSWI/SNF Chromatin Remodeling Complex
Previous studies have clearly shown that members of the PRMT family interact with ATP-dependent chromatin remodeling complexes in order to efficiently modify histones (13–15, 28). The net outcome of these molecular interactions is either transcriptional activation or repression, and in the case of PRMT5, a type II PRMT that can symmetrically methylate H3R8 and H4R3, interaction with the BRG1 complex triggers transcriptional repression of MYC target genes as well as ST7 and RBL2 tumor suppressors (14–17). Because PRMT7 is also a type II PRMT with little known about its activity and biological function, we wanted to investigate its effects on core histones and further study its impact on global gene expression. Because we have found that bacterially expressed PRMT7 is inactive, we proceeded to express it in a mammalian system. We established several stable HeLa S3 cell lines that express different levels of FLAG-tagged PRMT7 (Fl-PRMT7), and we selected clone 32 for further characterization (Fig. 1A).
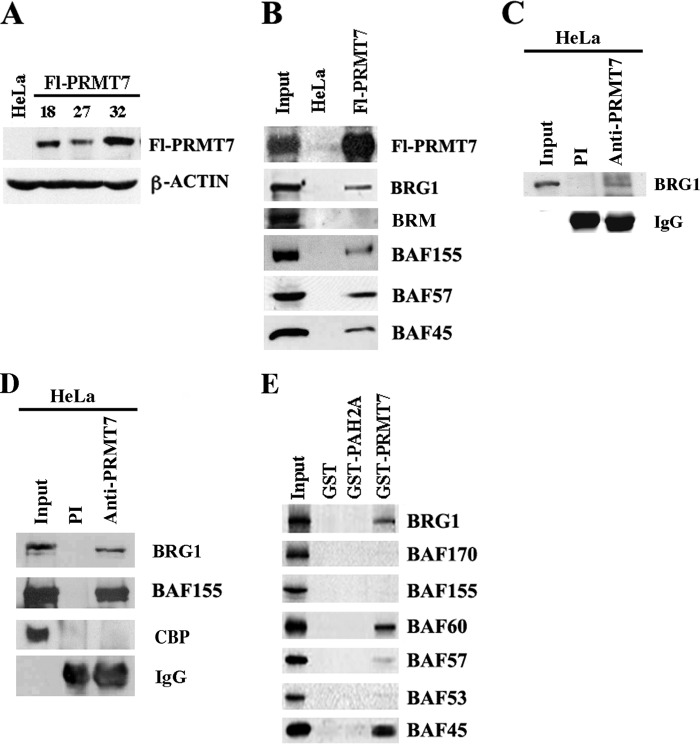
FIGURE 1.
PRMT7 can associate with BRG1-based hSWI/SNF. A, analysis of ectopically expressed FLAG-tagged PRMT7 (Fl-PRMT7) in stable HeLa S3 cell lines. Approximately 30 μg of nuclear extract from either HeLa S3 cells transfected with control vector (HeLa) or HeLa S3 clones 18, 27, and 32 stably transfected with pBabe/Fl-PRMT7 vector were analyzed by Western blotting using anti-FLAG antibody. Anti-β-ACTIN antibody was used as a control. B, components of the BRG1-based hSWI/SNF complex co-purify with Fl-PRMT7. An equal amount (15 μl) of peak fractions purified from control HeLa S3 or Fl-PRMT7 (clone 32) nuclear extract were analyzed by Western blotting using the indicated antibodies. Input represents 20 μg of Fl-PRMT7 crude nuclear extract. C, endogenous PRMT7 can interact with BRG1. Nuclear extract (250 mg) from HeLa S3 cells was immunoprecipitated using either preimmune (PI) (lane 2) or immune anti-PRMT7 antibody (lane 3), and bound proteins were analyzed by Western blot analysis using anti-BRG1 antibody. Input represents 20 mg of HeLa S3 nuclear extract. D, interaction between endogenous PRMT7 and BRG1-based hSWI/SNF is enhanced by the cross-linking agent dithiobis(succinimidyl propionate). Equal amounts of cross-linked HeLa S3 nuclear extract (250 mg) were incubated with PI or immune anti-PRMT7 antibody, and bound proteins were detected by immunoblotting using the indicated antibodies. E, specific BRG1-based hSWI/SNF subunits interact with PRMT7. In vitro-translated hSWI/SNF subunits were synthesized in the presence of [35S]methionine/cysteine using Promega TNT-coupled reticulocyte lysate, as described previously (14) and were incubated with either GST, GST-PAH2A, which includes the second paired amphipathic helix domain of mSIN3A, or GST-PRMT7. After extensive washing, the retained proteins were detected by autoradiography. Input represents 25% of the total amount of 35S-labeled protein used in each reaction. CBP, CREB-binding protein.
To study the methyltransferase activity of Fl-PRMT7 and to identify its interacting cellular partners, nuclear extracts were prepared from control HeLa S3 cells and Fl-PRMT7 clone 32 and incubated with anti-FLAG M2 affinity gel. After extensive washing, eluates from control and test columns were analyzed by Western blotting (Fig. 1B). As expected, when we used the anti-FLAG antibody to detect Fl-PRMT7, we found that epitope-tagged PRMT7 was enriched in fractions purified from clone 32 but not in control HeLa S3 fractions. We have previously reported that PRMT5 interacts with BRG1 and BRM-based hSWI/SNF chromatin remodeling complexes through both non-conserved N-terminal sequences and highly conserved C-terminal sequences, which include the catalytic domain (14, 15). Knowing that PRMT7 is highly related to PRMT5 through its conserved catalytic domains, we tested its ability to interact with hSWI/SNF chromatin remodelers. Using specific antibodies, we tested affinity-purified Fl-PRMT7 fractions for the presence of hSWI/SNF subunits (Fig. 1B). Our results showed that PRMT7 can interact with the BRG1 complex as determined by the presence of BRG1, BAF155, BAF57, and BAF45 subunits; however, when we tested for the presence of BRM, we were unable to detect it in affinity-purified Fl-PRMT7 fractions, suggesting that PRMT7 preferentially interacts with the BRG1 complex.
To rule out the possibility that interaction with endogenous BRG1-based hSWI/SNF is due to PRMT7 overexpression, we assessed association between PRMT7 and BRG1 using endogenous proteins. Immunoprecipitation experiments using anti-PRMT7 antibody showed a weak interaction between the two proteins under mild washing conditions (250 mm NaCl) (Fig. 1C), which could not be detected using higher salt washing conditions (350 mm NaCl) (data not shown). In contrast, association between endogenous PRMT7 and BRG1-based hSWI/SNF complex was readily detectable using nuclear extracts from HeLa S3 cells treated with the cross-linking agent dithiobis(succinimidyl propionate), which can enhance interaction between neighboring proteins (Fig. 1D) (25). Both BRG1 and BAF155 were detected in immune but not preimmune anti-PRMT7 immunoprecipitates under high salt washing conditions (350 mm NaCl). When we tested for the presence of nuclear CREB-binding protein, we were unable to detect it in anti-PRMT7 immunoprecipitates, indicating that association of PRMT7 with BRG1-based hSWI/SNF is specific.
To further validate our results, we conducted GST pull-down assays using in vitro translated hSWI/SNF subunits (Fig. 1E). Our findings clearly indicated that like PRMT5, PRMT7 associates with specific subunits including BRG1 and BAF45, but unlike PRMT5, which can interact with BAF57 (14), PRMT7 strongly interacts with BAF60, raising the possibility that different contacts with hSWI/SNF subunits might be necessary to specify different epigenetic modifications.
Effects of PRMT7 Knockdown on Cell Growth and Gene Expression
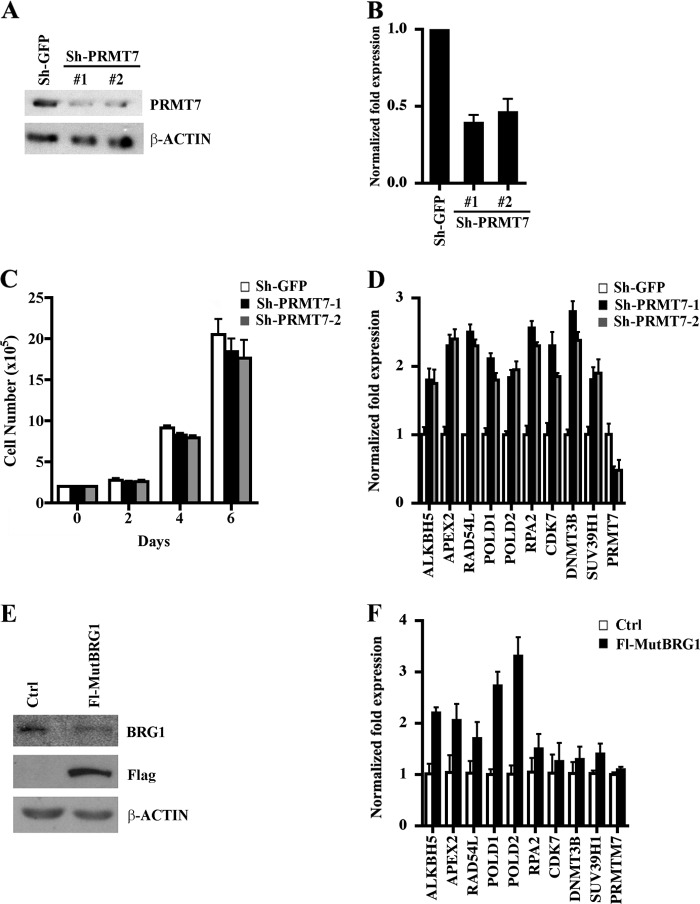
To gain more insight into the biological function of PRMT7, we established NIH 3T3 stable cell lines where PRMT7 expression was knocked down and investigated their growth characteristics as well as global gene expression (Fig. 2). Infection of NIH 3T3 cells with lentiviral particles that expressed either PRMT7-specific shRNA1 or shRNA2 resulted in several puromycin-resistant cell lines, and one representative NIH 3T3/sh-PRMT7 cell line from each shRNA infection, which exhibited 4–5-fold reduction in PRMT7 protein expression (Fig. 2A), was selected for further characterization. Analysis of total RNA by real-time RT-PCR showed that PRMT7 mRNA levels were reduced by 50–60% in both PRMT7 knock-down cell lines (Fig. 2B). Our previous work showed that knocking down expression of PRMT5, another type II PRMT, slows down cell growth and proliferation (15). However, when we measured proliferation of PRMT7 knock-down cells, there was no difference in their ability to grow in comparison with control NIH 3T3/sh-GFP cells, suggesting that PRMT5 and PRMT7 regulate different sets of genes and differ in their ability to control cell growth and proliferation (Fig. 2C).
FIGURE 2.
Effects of PRMT7 on cell growth and target gene expression. A, PRMT7 protein expression was measured by Western blot analysis using 30 μg of whole cell extract from control NIH 3T3/sh-GFP or PRMT7 knock-down cells, NIH 3T3/sh-PRMT7-1, and NIH 3T3/sh-PRMT7-2. Anti-β-ACTIN was used to show equal loading. B, PRMT7 mRNA levels were determined by real-time RT-PCR using 1 μg of total RNA from either control NIH 3T3/sh-GFP, NIH 3T3/sh-PRMT7-1, or NIH 3T3/sh-PRMT7-2. Real time RT-PCR was conducted three times in triplicate, and 18S rRNA was used as an internal control. C, growth rates of control and PRMT7 knock-down cell lines were measured by seeding 2 × 105 cells in each plate, and the number of viable cells was determined by staining with trypan blue every 2 days for 6 days. The experiment was performed three times in triplicate, and the data points represent the average count from nine plates. D, PRMT7 regulates expression of genes involved in DNA repair. Real-time RT-PCR was performed on 1 μg of total RNA from control NIH 3T3/sh-GFP, NIH 3T3/sh-PRMT7-1, or NIH 3T3/sh-PRMT7-2 cells using primers specific for the indicated target genes. 18S rRNA levels were used as internal control. Each reaction was performed at least two times in triplicate. E, nuclear extract from control NIH 3T3 cells (Ctrl) or NIH 3T3 cells that expresses catalytically inactive FLAG-tagged BRG1 (Fl-MutBRG1) were analyzed by Western blotting using the indicated antibodies. F, PRMT7 target genes are transcriptionally derepressed in cells that express Fl-MutBRG1. mRNA levels of PRMT7 target genes were determined by real-time RT-PCR using 1 μg of total RNA from either Ctrl or Fl-MutBRG1 cells. Real-time RT-PCR was conducted three times in triplicate, and 18S rRNA was used as an internal control. Error bars show S.D.
Previously, it has been reported that PRMT7 controls the cell response to cytotoxic agents and that its decreased expression in the DC-3F Chinese hamster cell line is associated with enhanced resistance to certain DNA-damaging agents (21). However, little is known about the mechanism by which PRMT7 exerts its effects, and there is no information about its target genes. To investigate the role played by PRMT7 in gene regulation, we conducted cDNA microarray analysis using total RNA from control and PRMT7 knock-down cell lines. The analysis of target genes that were affected more than 1.5-fold revealed that although 600 genes were down-regulated, another 650 genes were up-regulated, which included a myriad of functional groups ranging from signaling modules to repair of DNA damage (supplemental Tables 3 and 4). To validate the cDNA microarray results and to assess the effect of PRMT7 on target gene repression, we selected nine up-regulated genes, which were primarily involved in repair of DNA damage and regulation of chromatin structure, and verified if they were truly derepressed when PRMT7 protein levels were knocked down (Fig. 2D). Consistent with the cDNA microarray results, we found that all selected genes were derepressed 1.8–2.8-fold (p = 10−3) in both PRMT7 knock-down cell lines compared with control cells, suggesting that PRMT7 is involved in their transcriptional repression.
Western blot analysis of affinity-purified PRMT7 fractions showed that subunits of the BRG1 complex associate with Fl-PRMT7 (Fig. 1B). To determine if BRG1 is involved in PRMT7 target gene regulation, we prepared total RNA from either control NIH 3T3 or NIH 3T3 cells that express catalytically inactive FLAG-tagged BRG1 (Fl-MutBRG1) (Fig. 2E) (24) and measured the mRNA levels of PRMT7 target genes (Fig. 2F). Our results show that although not all PRMT7 target genes were transcriptionally derepressed more than 1.5-fold, ALKBH5, APEX2, POLD1, and POLD2 mRNA levels were increased 2–3.5-fold (p < 10−3) in the presence of mutant BRG1, highlighting the important role played by BRG1 in transcriptional repression of these PRMT7 target genes.
PRMT7 and BRG1 Co-localize and Are Enriched at Target DNA Repair Genes
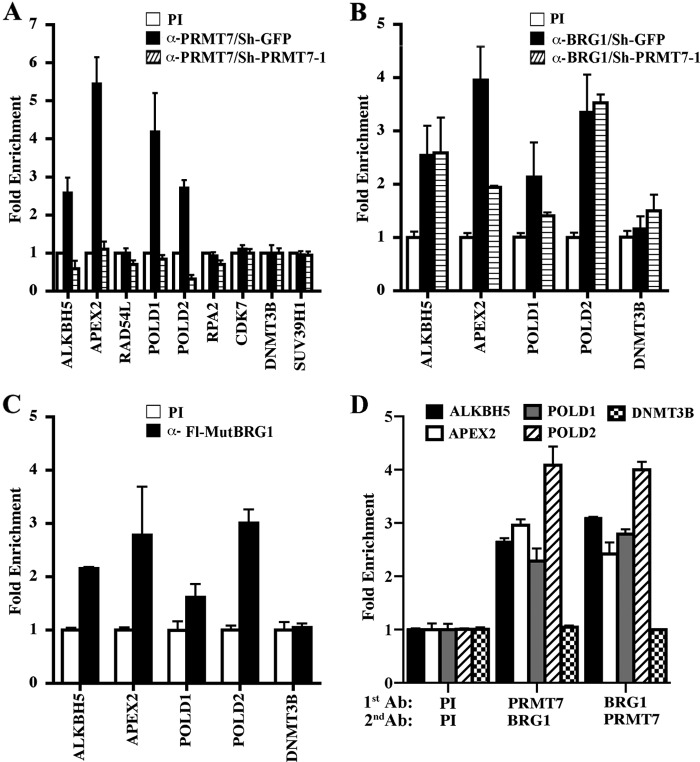
Our real-time RT-PCR results clearly showed that the PRMT7 target genes examined become derepressed when PRMT7 levels are knocked down; however, it is not clear if PRMT7 is directly involved in their transcriptional regulation. To address this question, we performed ChIP experiments using cross-linked and micrococcal nuclease-treated chromatin from control and PRMT7 knock-down cells (Fig. 3A). We discovered that PRMT7 binding was enriched 2.5–5.5-fold (p < 10−3) in the promoter region of four target genes involved in repair of DNA damage, including ALKBH5, APEX2, POLD1, and POLD2; however, PRMT7 recruitment was not enhanced at RAD54L, RPA2, CDK7, DNMT3B, and SUV39H1 promoters. When anti-PRMT7 antibody was used to immunoprecipitate chromatin from PRMT7 knock-down cells, there was a significant decrease in PRMT7 recruitment to all four direct target genes, suggesting that lack of PRMT7 binding contributes to their transcriptional derepression.
FIGURE 3.
PRMT7 is recruited to a select set of DNA repair genes. A, ChIP assays were carried out using cross-linked chromatin from control NIH 3T3/sh-GFP and NIH 3T3/sh-PRMT7-1 cells. Nucleoprotein complexes were immunoprecipitated with either PI or immune anti-PRMT7 antibodies, and bound DNA was analyzed by real-time PCR using gene-specific primers. -Fold enrichment was determined relative to the PI sample, and ChIP experiments were repeated three times in triplicate. B, BRG1 is recruited to the promoter region of PRMT7 target DNA repair genes. Cross-linked chromatin from control NIH 3T3/sh-GFP or NIH 3T3/sh-PRMT7-1 cells was immunoprecipitated with either PI or anti-BRG1 antibody, and the purified DNA was checked for enrichment of PRMT7 target promoters using gene-specific primers. C, catalytically inactive Fl-MutBRG1 is recruited to PRMT7 target genes. Chromatin from control NIH 3T3 or NIH 3T3 cells that express Fl-MutBRG1 was immunoprecipitated with either PI or anti-FLAG antibody, and DNA sequences of PRMT7 target promoters were detected by real-time PCR using gene-specific primers. D, PRMT7 and BRG1 co-localize on target DNA repair promoters. ChIP-re-ChIP assays were performed on cross-linked chromatin from NIH 3T3 cells using the indicated antibodies (1st Ab). Next, bound nucleoprotein complexes were released in the presence of 20 mm DTT, and subjected to a second immunoprecipitation (2nd Ab) using either PI, anti-PRMT7, or anti-BRG1. Real-time PCR was conducted as described above. Error bars show S.D.
Having found that PRMT7 can interact with the BRG1 catalytic subunit of hSWI/SNF and that catalytically inactive BRG1 interferes with transcriptional repression of ALKBH5, APEX2, POLD1, and POLD2, we wanted to investigate recruitment of both wild type and mutant BRG1 to these PRMT7 target genes. ChIP analysis using cross-linked chromatin from control and PRMT7 knock-down cells showed that BRG1 binding to PRMT7 target promoters was enhanced 2.2–4-fold (p < 10−3) in control NIH 3T3/sh-GFP cells (Fig. 3B). Interestingly, PRMT7 knockdown did not affect recruitment of BRG1 to ALKBH5 and POLD2 promoters; however, both APEX2 and POLD1 promoters showed partial reduction in BRG1 binding, suggesting that PRMT7 may positively contribute to BRG1 recruitment in a promoter-specific manner. We also analyzed recruitment of two other hSWI/SNF subunits, BAF57 and BAF155, to PRMT7 target promoters (supplemental Fig. 1). Our findings show that both BAF57 and BAF155 are enriched 2–3-fold in control NIH 3T3/sh-GFP and NIH 3T3/sh-PRMT7 cells.
To evaluate recruitment of catalytically inactive BRG1 (Fl-MutBRG1) to PRMT7 target genes, we performed ChIP assays on chromatin form NIH 3T3 cells that express mutant BRG1 using anti-FLAG antibody (Fig. 3C). Real-time PCR analysis revealed that recruitment of Fl-MutBRG1 to target promoters is enriched 1.8–3-fold (p < 10−3). Taken together, these results show that both PRMT7 and BRG1 are directly involved in and are both required for transcriptional repression of ALKBH5, APEX2, POLD1, and POLD2.
Although ChIP assays indicated that PRMT7 may impact BRG1 recruitment in a promoter-specific fashion and that both enzymes bind to the same target genes, we wished to determine if both proteins co-localize on the same nucleosomes of target promoters (Fig. 3D). ChIP-re-ChIP experiments were performed on cross-linked chromatin from NIH 3T3 cells using anti-PRMT7 and anti-BRG1 antibodies. When anti-PRMT7 antibody was first used to immunoprecipitate chromatin of target DNA repair genes, BRG1 recruitment was enhanced 2.5–4.1-fold. Similarly, when anti-BRG1 antibody was used first, PRMT7 binding to target DNA repair promoters was enriched 2.5–4-fold. Binding of BRG1 and PRMT7 to the DNMT3B promoter showed no enrichment. These findings indicate that PRMT7 and BRG1 co-localize on target DNA repair promoters.
PRMT7 target DNA repair genes become derepressed in knock-down cells. To determine whether type I PRMTs are involved in transcriptional reactivation of PRMT7 target genes, we examined recruitment of PRMT1 and PRMT4 as well as their induced epigenetic marks (supplemental Fig. 2). ChIP assays revealed that although PRMT1 and PRMT4 were enriched at known target promoters (29, 30), their binding to PRMT7 target promoters was unaffected in both control and PRMT7 knockdown cells. These results indicate that both of these co-activator enzymes are not involved during transcriptional derepression of PRMT7 target DNA repair genes.
PRMT7 Preferentially Methylates Histones H2A and H4
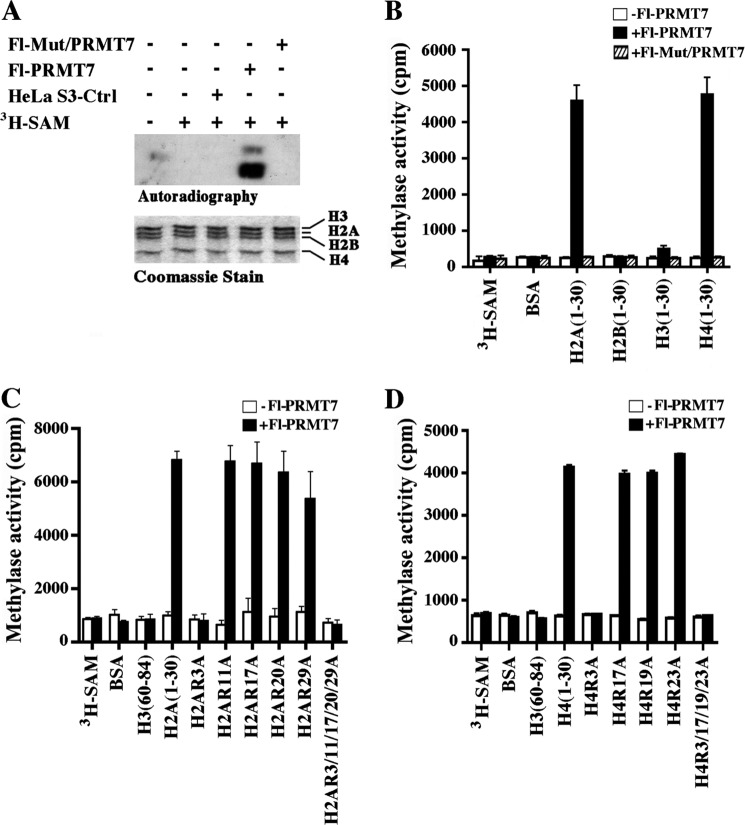
Having established that PRMT7 interacts with the BRG1 complex and regulates expression of DNA repair genes, we sought to evaluate its methyltransferase activity (Fig. 4). Previous work had shown that PRMT7 modifies histones H2A and H4 (18, 19); however, it is unclear which arginine residues it methylates. When we performed in vitro histone methyltransferase assays using H1-depleted HeLa core histones, we found that affinity-purified Fl-PRMT7 specifically methylates histones H2A and H4 (Fig. 4A), which is consistent with previously published results (18, 19). In stark contrast, immunopurified fractions from control HeLa S3 or HeLa S3 cells that express catalytically inactive Fl-PRMT7 (Fl-Mut/PRMT7) did not exhibit any methyltransferase activity. To confirm our findings and to determine if Fl-PRMT7 can methylate arginine residues in the H2A and H4 N-terminal tails, we incubated H2A, H2B, H3, and H4 N-terminal peptides with [3H]AdoMet) in the presence and absence of affinity-purified Fl-PRMT7 or Fl-Mut/PRMT7 (Fig. 4B). As expected, Fl-Mut/PRMT7 was inactive, whereas Fl-PRMT7 was able to specifically methylate arginine residues in the H2A and H4 N-terminal tails. When other substrates that contain arginine residues, such as BSA, H2B (aa 1–30), and H3 (aa 1–30), were incubated with either wild type or mutant Fl-PRMT7, no methylated products were detected, further confirming the specificity of PRMT7-mediated H2A and H4 methylation.
FIGURE 4.
Affinity-purified FLAG-tagged PRMT7 preferentially methylates histones H2A and H4. A, approximately 10 μl of affinity-purified peak fractions from either control HeLa S3, Fl-PRMT7, or Fl-Mut/PRMT7 cells were incubated with 2 μg of HeLa S3 core histones in the presence of [3H]AdoMet. Methylated histones were visualized by autoradiography, and Coomassie Blue staining was used to show equal amounts of core histones in each reaction. B, PRMT7 methylates H2A and H4 N-terminal tails. H2A, H2B, H3, and H4 peptides (2 μg) were incubated with or without affinity-purified Fl-PRMT7 and Fl-Mut/PRMT7 in the presence of [3H]AdoMet, and samples were spotted onto Whatman P-81 filter paper before methylation of histone peptides was quantified by liquid scintillation counting. To show specificity, methylation of BSA was also measured. C, wild type and mutant H2A peptides (2 μg) containing either single or multiple arginine to alanine substitutions at positions 3, 11, 17, 20, and 29 were incubated with or without affinity-purified Fl-PRMT7 in the presence of [3H]AdoMet, and methylation was measured as described in B. D, approximately 2 μg of either wild type or mutant H4 N-terminal peptides with single point mutations (R3A, R17A, R19A, or R23A) or mutations at all four positions were incubated with or without affinity-purified Fl-PRMT7 in the presence of [3H]AdoMet as described in C. Peptide methylation assays were performed in triplicate and repeated three times. Error bars show S.D.
To identify the histone arginine residues methylated by Fl-PRMT7, we used wild type and mutant H2A and H4 N-terminal peptides with arginine to alanine substitutions in in vitro methylation assays. Our results showed that Fl-PRMT7 specifically methylates all H2A peptides, except mutant H2A peptides bearing arginine to alanine substitutions at position 3 (Fig. 4C). Similarly, when immunopurified Fl-PRMT7 was incubated with wild type and mutant H4 N-terminal peptides, there was complete loss of methylation when H4 peptides containing arginine to alanine at position 3 were used (Fig. 4D). These experiments indicate that PRMT7 can specifically methylate H2AR3 and H4R3.
PRMT7 Epigenetic Marks Are Enriched at Target DNA Repair Genes
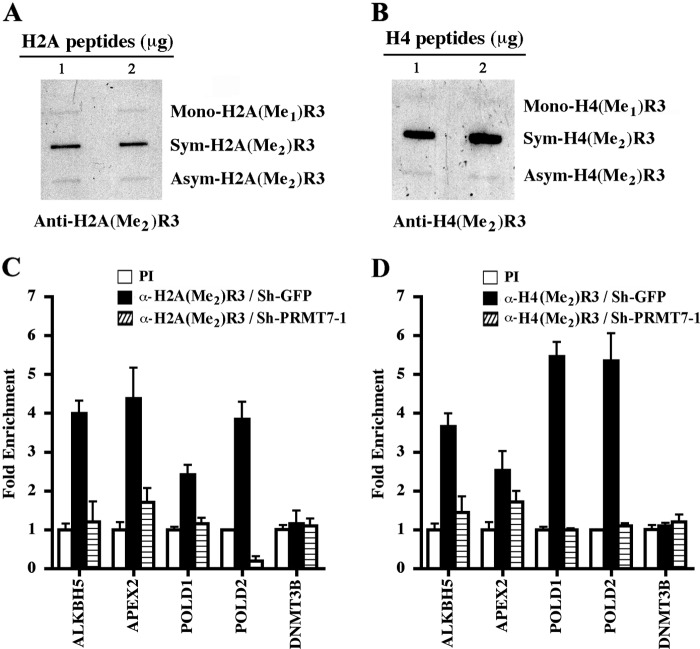
Biochemical characterization demonstrated that both H2AR3 and H4R3 are specific PRMT7 methylation marks (Fig. 4). Because we had determined that PRMT7 is directly involved in transcriptional regulation of target DNA repair genes, we wanted to assess if both PRMT7-induced epigenetic marks were enriched in the promoter region of affected genes. Using rabbit polyclonal antibodies, which can specifically recognize symmetrically methylated H2AR3 and H4R3 (Fig. 5, A and B, and supplemental Fig. 3), we conducted ChIP assays on cross-linked and micrococcal nuclease-treated chromatin from control NIH 3T3/sh-GFP and NIH 3T3/sh-PRMT7 knock-down cells (Fig. 5, C and D). We found that symmetric methylation of promoter H2AR3 was enhanced 2.5–4.2-fold (p < 10−4) in control cells, whereas epigenetic modification of this residue was significantly reduced in PRMT7 knock-down cells (Fig. 5C). Similarly, symmetric methylation of H4R3 was enriched 2.8–5.6-fold (p < 10−3) at target promoters in control cells and significantly reduced in PRMT7 knock-down cells (Fig. 5D). When H2AR3 and H4R3 methylation was evaluated at the DNMT3B promoter, which is not directly targeted by PRMT7, there was no noticeable enrichment. Collectively, these results suggest that PRMT7 is involved in transcriptional regulation of DNA repair genes through its ability to methylate promoter histones H2AR3 and H4R3.
FIGURE 5.
PRMT7-induced epigenetic marks are enriched at target DNA repair genes. Anti-H2A(Me2)R3 (A) and anti-H4(Me2)R3 (B) antibodies are highly specific and do not cross-react with other methylation marks. Approximately 1 and 2 μg of either monomethylated, symmetrically methylated, or asymmetrically methylated H2AR3 and H4R3 peptides were slot-blotted on nitrocellulose, and anti-H2A(Me2)R3 and anti-H4(Me2)R3 antibodies were used to detect methylation marks. ChIP assays were conducted essentially as described in Fig. 3A using chromatin from control NIH 3T3/sh-GFP and NIH 3T3/sh-PRMT7-1 cells using either PI or immune anti-H2A(Me2)R3 (C) and anti-H4(Me2)R3 (D). DNA sequences subjected to ChIP were analyzed as described in Fig. 3A. Each ChIP experiment was repeated twice in triplicate. Error bars show S.D.
Down-regulation of PRMT7 Expression Results in Enhanced Resistance to Genotoxic Stress
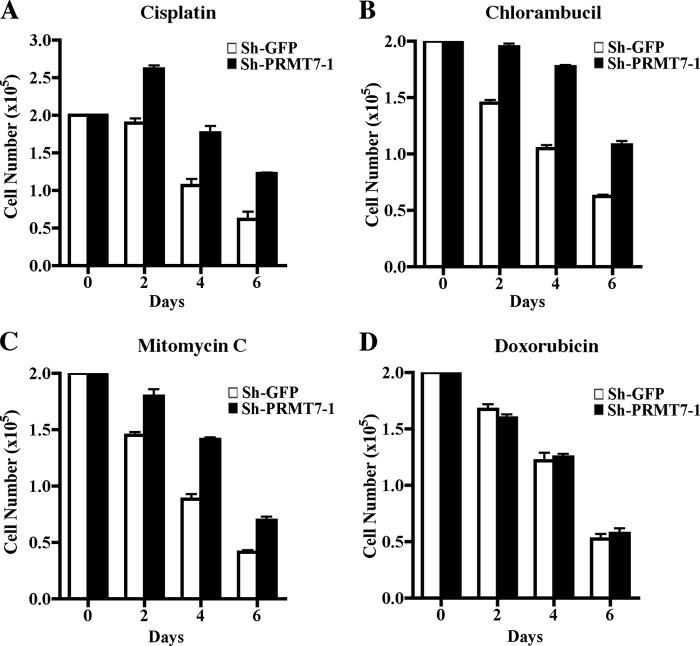
Previous studies have implicated PRMT7 in the regulation of DNA damage repair (21, 22); however, details about the molecular mechanisms by which PRMT7 impacts response to DNA-damaging agents remain unclear. To verify if down-regulation of PRMT7 can alter cell growth in the presence of DNA-damaging agents, we treated both control and PRMT7 knock-down cells with either cisplatin (which induces 1,2 intrastrand d(GpG) cross-links), chlorambucil (which alkylates DNA at the guanine base), or mitomycin C (which cross-links DNA by alkylating guanine nucleoside in d(CpG) sequences) and measured the number of viable cells. All three DNA-damaging agents inhibited growth of control and PRMT7 knock-down cell lines; however, PRMT7 knock-down cells exhibited 1.5–2-fold more resistance in the presence of cisplatin, chlorambucil, and mitomycin C (Fig. 6, A–C). When we treated cells with doxorubicin, which intercalates DNA and inhibits progression of topoisomerase II, there was no difference in growth between control and PRMT7 knock-down cells (Fig. 6D), suggesting that not all genotoxic drugs have the same effect on pathways regulated by PRMT7. Similar results were observed with a second PRMT7 knock-down cell line generated using PRMT7-specific shRNA2 (supplemental Fig. 4). Furthermore, when PRMT5 knock-down NIH 3T3 cells were treated with cisplatin, chlorambucil, or mitomycin C, they were equally sensitive to DNA damage as control NIH 3T3 cells (supplemental Fig. 5), indicating that resistance to DNA-damaging agents is a unique trait acquired by PRMT7 knock-down cells. We also performed FACS analysis to measure the number of cells undergoing cell death upon treatment with cisplatin (supplemental Fig. 6). Annexin V/propidium iodide staining revealed that at day 4, more control NIH 3T3/sh-GFP cells were apoptotic (34%) compared with PRMT7 knock-down cells (17%). By day 6, the trend continued with more control cells staining positive for Annexin V (55.5%) in comparison with sh-PRMT7 cells (35%). These results confirm that PRMT7 knock-down cells are more resistant to DNA damage.
FIGURE 6.
PRMT7 knock-down cells a more resistant to genotoxic stress. A–D, drug treatments were carried out as described under “Experimental Procedures” using an equal number (2 × 105) of control NIH 3T3/sh-GFP and NIH 3T3/sh-PRMT7-1, and proliferation was monitored every 2 days for 6 days. The number of viable cells was determined by trypan blue staining. Each drug treatment was conducted twice in triplicate. Error bars show S.D.
Knockdown of the DNA Polymerase δ Catalytic Subunit POLD1 Resensitizes PRMT7 Knock-down Cells to DNA Damage
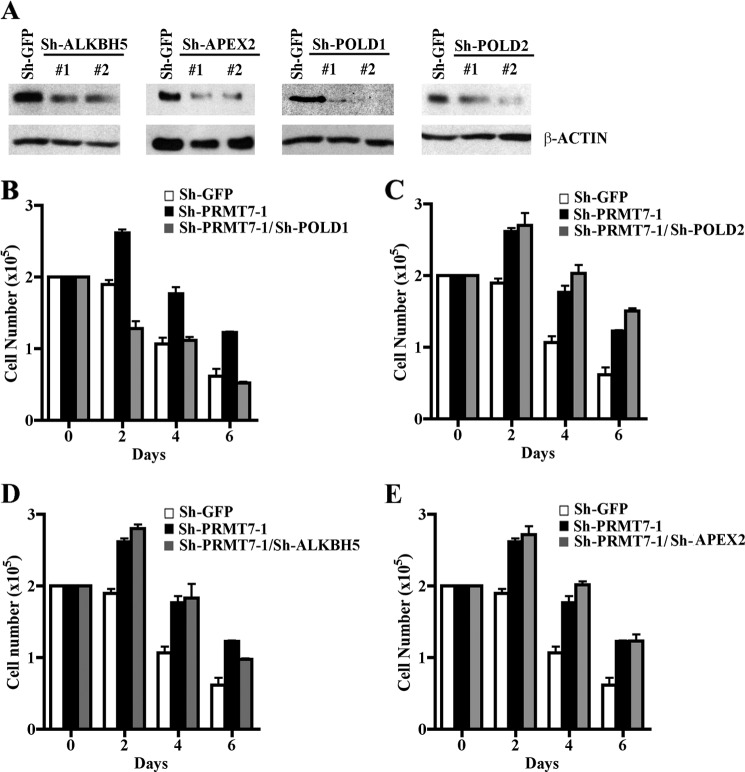
Because both PRMT7 knock-down cell lines, sh-PRMT7-1 and sh-PRMT7-2, showed derepression of four distinct target DNA repair genes and were resistant to DNA-damaging agents, we reasoned that the identified target genes might be involved in rendering cells more tolerant of DNA damage. Therefore, we set out to assess their contribution to the resistant phenotype of PRMT7 knock-down cells. First, NIH 3T3/sh-PRMT7-1 knock-down cells were infected with two distinct lentiviruses that expressed either shRNA1 or shRNA2, which are designed to reduce expression of PRMT7 target genes, and levels of ALKBH5, APEX2, POLD1, and POLD2 proteins were measured by Western blot analysis (Fig. 7A). Expression of all four PRMT7 target genes was decreased by more than 50% (Fig. 7A). Next, we treated control NIH 3T3/sh-GFP, NIH 3T3/sh-PRMT7-1, and each of the four NIH 3T3/sh-PRMT7-1 knock-down cells, where expression of PRMT7 target genes was reduced, with cisplatin and measured cell viability (Fig. 7, B–E). Our findings showed that of the four PRMT7 target genes, only POLD1 knockdown resulted in loss of resistance and rendered cells sensitive to DNA damage (Fig. 7B). To further confirm these results, we knocked down expression of PRMT7 target DNA repair genes in NIH 3T3/sh-PRMT7-2 cells and measured the number of viable cells (supplemental Fig. 7). Consistent with our results using NIH 3T3/sh-PRMT7-1 cells, reduced expression of POLD1 resensitized NIH 3T3/sh-PRMT7-2 knock-down cells to DNA damage. These results suggest that PRMT7 is able to modulate cellular response to DNA damage by regulating expression of the polymerase δ catalytic subunit POLD1.
FIGURE 7.
Knockdown of the polymerase δ catalytic subunit POLD1 resensitizes PRMT7 knock-down cells to DNA damage. A, expression of PRMT7 target genes was knocked down using lentiviruses that express two distinct shRNAs (#1 and #2) against either ALKBH5, APEX2, POLD1, or POLD2. Protein expression of each PRMT7 target gene was evaluated by Western blot analysis using equal amounts of whole cell extracts (30 μg) from control NIH 3T3/sh-PRMT7-1 infected with lentivirus that expresses either sh-GFP or the indicated shRNAs against ALKBH5, APEX2, POLD1, and POLD2. PRMT7 target gene expression was determined using either anti-ALKBH5, anti-APEX2, anti-POLD1, or anti-POLD2 antibody. Anti-β-ACTIN was used a control. B–E, growth rates of control NIH 3T3/sh-GFP, NIH 3T3/sh-PRMT7-1, and NIH 3T3/sh-PRMT7-1 cells where expression of individual PRMT7 target DNA repair genes has been knocked down was measured by seeding 2 × 105 cells in each plate and treating cells with cisplatin as described under “Experimental Procedures.” The number of viable cells was determined by trypan blue staining every 2 days for 6 days. Each experiment was conducted in triplicate. Error bars show S.D.
DISCUSSION
Understanding how PRMT7-mediated epigenetic modification of specific histone arginine residues contributes to the control of gene expression and elucidating mechanisms by which PRMT7 regulates the cell response to genotoxic agents are of paramount importance, because most aggressive and recurring cancers develop the ability to become resistant to DNA-damaging drugs. In this study, we have shown that PRMT7 can be found in association with the BRG1-based hSWI/SNF complex and interacts with specific components of the complex, including the catalytic subunit BRG1. We have also shown that both PRMT7 and BRG1 are involved in transcriptional repression of a select set of DNA repair genes, including ALKBH5, APEX2, POLD1, and POLD2. ChIP experiments clearly showed that PRMT7 and BRG1 co-localize on target DNA repair genes and that, whereas BRG1 recruitment to ALKBH5 and POLD2 is PRMT7-independent, BRG1 binding to APEX2 and POLD1 is negatively impacted by reduced PRMT7 expression. We also characterized the histone-modifying activity of PRMT7, identified H2AR3 and H4R3 as its targets in vitro, and demonstrated using highly specific antibodies that both epigenetic marks are highly enriched in the promoter region of PRMT7 target genes in drug-sensitive parental NIH 3T3 cells.
Global gene profiling studies showed that several genes involved in repair of DNA damage were derepressed in PRMT7 knock-down cells, suggesting that reduced expression of PRMT7 might render cells more resistant to DNA-damaging agents. In fact, previous work has shown that PRMT7 knockdown enhances the ability of the Chinese hamster lung fibroblast cell line DC-3F to survive in the presence of cytotoxic amounts of DNA-damaging agents; however, the mechanism by which PRMT7 controls cell resistance to DNA damage was unclear (21). Consistent with these results, we showed that PRMT7 knock-down cells are more resistant to cisplatin, chlorambucil, and mitomycin C, and more importantly, we identified POLD1 as the only PRMT7 target DNA repair gene capable of resensitizing PRMT7 knock-down cells to DNA damage.
PRMT7 Is Involved in Transcriptional Regulation of Target DNA Repair Genes
Cells possess a battery of DNA repair enzymes and factors specialized in detecting and correcting DNA damage, and there are several pathways whose primary function is to ensure genome integrity. In most cases, whether DNA damage is caused by endogenous or exogenous factors, altered DNA is first detected, and specific enzymes are dispatched in an ordered fashion to incise the DNA strand containing the damage to repair and restore the genetic information (31). Recent studies have shown that reduced expression of PRMT7 contributes to cisplatin resistance (21). We have confirmed these results, and we have also found that PRMT7 knock-down cells are more resistant to chlorambucil and mitomycin C (Fig. 6). A detailed examination of the list of genes affected by PRMT7 revealed that although key target genes involved in methylation of histones and DNA are not directly regulated by PRMT7, genes specialized in repair of DNA damage turned out to be direct targets of PRMT7, including ALKBH5, APEX2, POLD1, and POLD2 (Figs. 2D and 3A).
Initial studies in E. coli identified ALKB as a gene involved in protecting bacterial DNA from cytotoxic effects of alkylation. More recently, ALKB has been found to be highly conserved in various species, and currently there are eight homologs in human cells, including ALKBH5 (hABH5) (32–34). Although all hABH proteins share sequence similarities with bacterial ALKB, only hABH2 and hABH3 have DNA repair functions and catalyze oxidative demethylation in the presence of Fe(II) and α-ketoglutarate. The function of the other human ALKB homologs, including ALKBH5, remains unknown due to their lack of activity in in vitro demethylation assays (34). Another important enzyme in repair of DNA damage is APEX2, which functions in base excision repair by cleaving 5′ to the abasic site after a DNA glycosylase has removed a damaged nucleoside, thereby allowing POLβ or, in the case of a long patch of base excision repair, POLβ, POLδ, and POLϵ to fill in the missing sequence of nucleotides (35).
We have found that PRMT7 and its epigenetic marks are enriched in the promoter region of target DNA repair genes in drug-sensitive parental NIH 3T3 cells and that PRMT7 recruitment and methylation of its target arginine residues were significantly reduced in the promoter region of derepressed target genes in the more resistant PRMT7 knock-down cells, highlighting the importance of PRMT7-mediated epigenetic regulation of DNA repair. In addition, we have discovered that mRNA levels of ALKBH5, APEX2, POLD1, and POLD2 become derepressed in PRMT7 knock-down cells and that cells with elevated levels of these PRMT7 target genes are more resistant to DNA damage induced by either cisplatin, chlorambucil, or mitomycin C. By knocking down expression of individual PRMT7 target DNA repair genes, we have determined that the DNA polymerase catalytic subunit POLD1 is directly involved in rendering PRMT7 knock-down cells resistant to DNA damage. Reduced expression of other PRMT7 target DNA repair genes, including POLD2, which is also a subunit of POLδ, did not affect the resistant phenotype of PRMT7 knock-down cells, indicating that these PRMT7 target genes do not contribute to drug resistance. Taken together, these results suggest that PRMT7 modulates cellular response to DNA damage through its ability to methylate promoter histones H2AR3 and H4R3 of key target DNA repair genes.
Supplementary Material
Acknowledgments
We thank members of the laboratory for help and critical reading of the manuscript. We especially thank P. Shamulailatpam, J. Sundaram, and S. Wei for technical help.
This work was supported, in whole or in part, by National Institutes of Health, NCI, Grants CA116093 and CA101956 (to S. S.), GM56244 (to A. N. I.), and DK084278 (to A. N. I. and S. S.).

This article contains supplemental Tables 1–4 and Figs. 1–7.
- H3R2
- H3K4, and H3R8, histone H3 Arg-2, Lys-4, and Arg-8, respectively
- H4R3
- histone H4 Arg-3
- H2AR3
- histone H2A Arg-3
- H2A(Me2)R3
- H4(Me2)R3, and H2A(Me2)R3, dimethylated H2AR3, H4R3, and H2AR17, respectively
- AdoMet
- S-adenosylmethionine
- CREB
- cAMP-response element-binding protein
- aa
- amino acids
- PI
- preimmune.
REFERENCES
- 1. Bedford M. T., Clarke S. G. (2009) Protein arginine methylation in mammals. Who, what, and why. Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cloos P. A., Christensen J., Agger K., Helin K. (2008) Erasing the methyl mark. Histone demethylases at the center of cellular differentiation and disease. Genes Dev. 22, 1115–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Henikoff S. (2008) Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 9, 15–26 [DOI] [PubMed] [Google Scholar]
- 4. Klose R. J., Zhang Y. (2007) Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 8, 307–318 [DOI] [PubMed] [Google Scholar]
- 5. Lee K. K., Workman J. L. (2007) Histone acetyltransferase complexes. One size doesn't fit all. Nat. Rev. Mol. Cell Biol. 8, 284–295 [DOI] [PubMed] [Google Scholar]
- 6. Pal S., Sif S. (2007) Interplay between chromatin remodelers and protein arginine methyltransferases. J. Cell Physiol. 213, 306–315 [DOI] [PubMed] [Google Scholar]
- 7. Shilatifard A. (2006) Chromatin modifications by methylation and ubiquitination. Implications in the regulation of gene expression. Annu. Rev. Biochem. 75, 243–269 [DOI] [PubMed] [Google Scholar]
- 8. Guccione E., Bassi C., Casadio F., Martinato F., Cesaroni M., Schuchlautz H., Lüscher B., Amati B. (2007) Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449, 933–937 [DOI] [PubMed] [Google Scholar]
- 9. Hyllus D., Stein C., Schnabel K., Schiltz E., Imhof A., Dou Y., Hsieh J., Bauer U. M. (2007) PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 21, 3369–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kirmizis A., Santos-Rosa H., Penkett C. J., Singer M. A., Vermeulen M., Mann M., Bähler J., Green R. D., Kouzarides T. (2007) Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449, 928–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boisvert F. M., Côté J., Boulanger M. C., Richard S. (2003) A proteomic analysis of arginine-methylated protein complexes. Mol. Cell Proteomics 2, 1319–1330 [DOI] [PubMed] [Google Scholar]
- 12. Chen D., Ma H., Hong H., Koh S. S., Huang S. M., Schurter B. T., Aswad D. W., Stallcup M. R. (1999) Regulation of transcription by a protein methyltransferase. Science 284, 2174–2177 [DOI] [PubMed] [Google Scholar]
- 13. Xu W., Cho H., Kadam S., Banayo E. M., Anderson S., Yates J. R., 3rd, Emerson B. M., Evans R. M. (2004) A methylation-mediator complex in hormone signaling. Genes Dev. 18, 144–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pal S., Yun R., Datta A., Lacomis L., Erdjument-Bromage H., Kumar J., Tempst P., Sif S. (2003) mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell Biol. 23, 7475–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pal S., Vishwanath S. N., Erdjument-Bromage H., Tempst P., Sif S. (2004) Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell Biol. 24, 9630–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pal S., Baiocchi R. A., Byrd J. C., Grever M. R., Jacob S. T., Sif S. (2007) Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 26, 3558–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang L., Pal S., Sif S. (2008) Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol. Cell Biol. 28, 6262–6277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee J. H., Cook J. R., Yang Z. H., Mirochnitchenko O., Gunderson S. I., Felix A. M., Herth N., Hoffmann R., Pestka S. (2005) PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine. J. Biol. Chem. 280, 3656–3664 [DOI] [PubMed] [Google Scholar]
- 19. Jelinic P., Stehle J. C., Shaw P. (2006) The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol. 4, e355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miranda T. B., Miranda M., Frankel A., Clarke S. (2004) PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity. J. Biol. Chem. 279, 22902–22907 [DOI] [PubMed] [Google Scholar]
- 21. Gros L., Delaporte C., Frey S., Decesse J., de Saint-Vincent B. R., Cavarec L., Dubart A., Gudkov A. V., Jacquemin-Sablon A. (2003) Identification of new drug sensitivity genes using genetic suppressor elements. Protein arginine N-methyltransferase mediates cell sensitivity to DNA-damaging agents. Cancer Res. 63, 164–171 [PubMed] [Google Scholar]
- 22. Verbiest V., Montaudon D., Tautu M. T., Moukarzel J., Portail J. P., Markovits J., Robert J., Ichas F., Pourquier P. (2008) Protein arginine (N)-methyl transferase 7 (PRMT7) as a potential target for the sensitization of tumor cells to camptothecins. FEBS Lett. 582, 1483–1489 [DOI] [PubMed] [Google Scholar]
- 23. Campeau E., Ruhl V. E., Rodier F., Smith C. L., Rahmberg B. L., Fuss J. O., Campisi J., Yaswen P., Cooper P. K., Kaufman P. D. (2009) A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS ONE 4, e6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de La Serna I. L., Carlson K. A., Hill D. A., Guidi C. J., Stephenson R. O., Sif S., Kingston R. E., Imbalzano A. N. (2000) Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol. Cell Biol. 20, 2839–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang L., Rayner S., Katoku-Kikyo N., Romanova L., Kikyo N. (2007) Successful co-immunoprecipitation of Oct4 and Nanog using cross-linking. Biochem. Biophys. Res. Commun. 361, 611–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu Z., Irizarry R. A., Gentleman R., Murillo F. M., Spencer F. (2004) A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99, 909–917 [Google Scholar]
- 27. Wu Z., Irizarry R. A. (2005) Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J. Comput. Biol. 12, 882–893 [DOI] [PubMed] [Google Scholar]
- 28. Le Guezennec X., Vermeulen M., Brinkman A. B., Hoeijmakers W. A., Cohen A., Lasonder E., Stunnenberg H. G. (2006) MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell Biol. 26, 843–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Du K., Arai S., Kawamura T., Matsushita A., Kurokawa R. (2011) TLS and PRMT1 synergistically coactivate transcription at the survivin promoter through TLS arginine methylation. Biochem. Biophys. Res. Commun. 404, 991–996 [DOI] [PubMed] [Google Scholar]
- 30. Ou C. Y., LaBonte M. J., Manegold P. C., So A. Y., Ianculescu I., Gerke D. S., Yamamoto K. R., Ladner R. D., Kahn M., Kim J. H., Stallcup M. R. (2011) A coactivator role of CARM1 in the dysregulation of β-catenin activity in colorectal cancer cell growth and gene expression. Mol. Cancer Res. 9, 660–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lindahl T., Wood R. D. (1999) Quality control by DNA repair. Science 286, 1897–1905 [DOI] [PubMed] [Google Scholar]
- 32. Drabløs F., Feyzi E., Aas P. A., Vaagbø C. B., Kavli B., Bratlie M. S., Peña-Diaz J., Otterlei M., Slupphaug G., Krokan H. E. (2004) Alkylation damage in DNA and RNA. Repair mechanisms and medical significance. DNA Repair 3, 1389–1407 [DOI] [PubMed] [Google Scholar]
- 33. Mishina Y., He C. (2006) Oxidative dealkylation DNA repair mediated by the mononuclear non-heme iron AlkB proteins. J. Inorg. Biochem. 100, 670–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sedgwick B., Robins P., Lindahl T. (2006) Direct removal of alkylation damage from DNA by AlkB and related DNA dioxygenases. Methods Enzymol. 408, 108–120 [DOI] [PubMed] [Google Scholar]
- 35. Wood R. D., Shivji M. K. (1997) Which DNA polymerases are used for DNA repair in eukaryotes? Carcinogenesis 18, 605–610 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.