Background: The Gram-positive bacterium Bacillus subtilis contains two twin-arginine (Tat) translocases, each specifically secreting one known substrate protein.
Results: The TatAyCy translocase facilitates export of the metallophosphoesterase YkuE to the cell wall.
Conclusion: YkuE is the third identified genuine Tat substrate in Bacillus.
Significance: YkuE is the first protein found to be specifically targeted to a Gram-positive bacterial cell wall via the Tat pathway.
Keywords: Bacillus, Metalloenzymes, Protein Export, Protein Secretion, Protein Translocation, Tat, Twin-arginine Translocation System, Cell Wall
Abstract
The twin-arginine translocation (Tat) pathway is dedicated to the transport of fully folded proteins across the cytoplasmic membranes of many bacteria and the chloroplast thylakoidal membrane. Accordingly, Tat-dependently translocated proteins are known to be delivered to the periplasm of Gram-negative bacteria, the growth medium of Gram-positive bacteria, and the thylakoid lumen. Here, we present the first example of a protein, YkuE of Bacillus subtilis, that is specifically targeted by the Tat pathway to the cell wall of a Gram-positive bacterium. The cell wall binding of YkuE is facilitated by electrostatic interactions. Interestingly, under particular conditions, YkuE can also be targeted to the cell wall in a Tat-independent manner. The biological function of YkuE was so far unknown. Our present studies show that YkuE is a metal-dependent phosphoesterase that preferentially binds manganese and zinc.
Introduction
The twin-arginine protein translocation (Tat)3 pathway is present in the membranes of most bacteria and the thylakoid membranes of plant chloroplasts (1, 2). A key distinguishing feature of the Tat translocase is that it can transport proteins in a folded state (3–7). Moreover, many substrates of the Tat pathway contain a cofactor, which needs to be inserted correctly prior to translocation. If the substrate protein does not fold efficiently or correctly, it is potentially exported in an unfolded state by another transport pathway, such as the general secretion pathway (Sec) (3, 5, 8–10). A second important feature of substrates that are accepted by the Tat translocase is the presence of a specific twin-arginine (RR) signal peptide at the N terminus of the protein. The RR signal peptides are composed of three parts, namely an N-terminal region rich in positively charged residues, a central hydrophobic H-region, and a C-terminal region containing the cleavage site recognized by signal peptidases (11, 12). Importantly, these signal peptides possess a twin-arginine (RR) or a lysine-arginine (KR) recognition motif in the N-region with the consensus sequence (K/R)RX##, where # marks hydrophobic residues and X can be any residue (11–14). This RR motif or its variant with a lysine is specifically recognized by the Tat translocase (15–17). RR signal peptides are generally less effective in protein targeting to the Sec machinery due to the low hydrophobicity of their H-region and the presence of a positively charged residue in the C-region that facilitates “Sec avoidance” (16). Several studies conducted with Gram-negative and Gram-positive bacteria have shown that RR signal peptides are often interchangeable among different proteins and that in some instances proteins can be redirected from the Sec pathway into the Tat pathway by replacing the original Sec-type signal peptide with an RR signal peptide (10, 18, 19).
Interestingly, Tat is a major protein translocation pathway in some bacterial species, whereas other species seem to make only very limited use of their Tat translocase. For example, more than 30 proteins are exported Tat-dependently in streptomycetes (19–23), whereas only two genuine Tat substrates (PhoD and YwbN) have so far been identified in Bacillus subtilis (24, 25) and only one in Staphylococcus aureus (26). In particular, the limited use of Tat in B. subtilis is remarkable in view of this organism's very high capacity for protein export (12, 27). Moreover, B. subtilis contains two Tat translocases named TatAdCd and TatAyCy, which can function independently (25, 28, 29). This suggests that some Tat substrates of B. subtilis may have been overlooked in studies aimed at their identification. Several bioinformatic approaches have been designed with the aim to identify new Tat substrates (20, 30, 31). Indeed such bioinformatics tools predict additional Tat substrates for B. subtilis, the numbers depending on the stringency of the algorithms. To demonstrate the Tat specificity of predicted RR signal peptides, a reporter system was developed based on the agarase of Streptomyces coelicolor, which is secreted via the Tat pathway of this organism (22). Because the agarase is secreted into the growth medium of Streptomyces, its activity can be tested in a simple semi-quantitative way with a colorimetric assay. Using this assay, several potential RR signal peptides have been tested, including the B. subtilis protein YkuE (19). These studies confirmed that the YkuE signal peptide was capable of directing Tat-dependent protein transport, but the actual Tat-dependent secretion of YkuE and other predicted Tat substrates of B. subtilis remained enigmatic.
This study aimed to assess the expression, export, and function of YkuE in B. subtilis. Our results show that YkuE is a Tat-dependently exported metallophosphoesterase. Most noticeably, YkuE is specifically targeted to the cell wall of B. subtilis, making it the first known protein that is targeted via Tat to this particular subcellular compartment. We show that cell wall binding of YkuE is facilitated by electrostatic interactions. Furthermore, we have identified different modes of YkuE export to the cell wall that are specific for particular conditions, such as phosphate starvation or YkuE overexpression.
EXPERIMENTAL PROCEDURES
Plasmids, Bacterial Strains, Media, and Growth Conditions
The plasmids and bacterial strains used in this study are listed in Table 1. Strains were grown with agitation at 37 °C in either Luria Bertani-Miller (LB) medium or Paris minimal (PM) medium. LB medium consisted of 1% tryptone, 0.5% yeast extract, and 1% NaCl, pH 7.4. PM consisted of 10.7 mg ml−1 K2HPO4, 6 mg ml−1 KHPO, 1 mg ml−1 trisodium citrate, 0.02 mg ml−1 MgSO4, 1% glucose, 0.1% casamino acids (Difco), 20 mg ml−1 l-tryptophan, 2.2 mg ml−1 ferric ammonium citrate, and 20 mm potassium glutamate. To trigger a phosphate starvation response, the strains were grown overnight in high phosphate depletion medium, which is rich in phosphate. The next morning, cells were transferred to low phosphate depletion media. Both media were prepared according to Müller et al. (32). Lactococcus lactis was grown at 30 °C in M17 broth supplemented with 0.5% (w/v) glucose and 2 μg ml−1 erythromycin. When required, media for Escherichia coli were supplemented with erythromycin (100 μg ml−1), kanamycin (20 μg ml−1), chloramphenicol (5 μg ml−1), or spectinomycin (100 μg ml−1); media for B. subtilis were supplemented with erythromycin (2 μg ml−1), kanamycin (20 μg ml−1), chloramphenicol (5 μg ml−1), or spectinomycin (100 μg ml−1).
TABLE 1.
Strains and plasmids used in this study
| Relevant properties | Ref. | |
|---|---|---|
| Plasmids | ||
| pHB201 | B. subtilis-E.coli expression vector; ori-pBR322; ori-pTA1060; cat86::lacZa; CmR; EmR | 43 |
| pHB-YkuE-myc | pHB201 vector carrying the ykuE gene with a Myc tag; CmR; EmR | This study |
| pNZ8910-YkuE-StrepII | pNZ8910 vector containing the ykuE gene with a StrepII tag; EmR | This study |
| pCACy | pGDL48 vector containing the tatAyCy operon; ApR; KmR | 25 |
| pCAy | pGDL48 vector containing the tatAy gene; ApR; KmR | 25 |
| pCCy | pGDL48 vector containing the tatCy gene; ApR; KmR | 25 |
| Strains | ||
| E. coli | ||
| DH5α | supE44; hsdR17; recA1; gyrA96; thi-1; relA1 | 33 |
| L. lactis | ||
| MG1363 | Plasmid-free derivative of NCDO 712 | 67 |
| B. subtilis | ||
| 168 | trpC2 | 68 |
| ykuE | trpC2; pMutin2::ykuE; EmR | 69 |
| ATCC6633 | Subtilin producer | 38 |
| tatAyCy | trpC2; tatAy-tatCy::Sp; SpR | 53 |
| tatAdCd | trpC2; tatAd-tatCd::Km; KmR | 25 |
| tatAdCd | trpC2; tatAd-tatCd::Cm; CmR | 53 |
| total-tat3 | trpC2; tatAd-tatCd::Cm; CmR; tatAy-tatCy::Sp; SpR; tatAc::Tc; TcR | This study |
| AyCy pCACy | trpC2; tatAy-tatCy::Sp; pCACy; KmR; SpR | 70 |
| total-tat3 pCACy | trpC2; tatAd-tatCd::Cm; CmR; tatAy-tatCy::Sp; SpR; tatAc::Tc; TcR pCACy; KmR | This study |
| 168 pHB-YkuE-myc | trpC2; pHB-YkuE-myc EmR; CmR | This study |
| tatAyCy pHB-YkuE-myc | trpC2; tatAy-tatCy::Sp; SpR; pHB-YkuE-myc; EmR; CmR | This study |
| tatAdCd pHB-YkuE-myc | trpC2; tatAd-tatCd::Km; KmR; pHB-YkuE-myc; EmR; CmR | This study |
| total-tat3 pHB-YkuE-myc | trpC2; tatAd-tatCd::Cm; CmR; tatAy-tatCy::Sp; SpR; tatAc::Tc; TcR; pHB-YkuE-myc; EmR; CmR | This study |
| 168 + TatAy | trpC2; pHB-YkuE-myc; pCAy; EmR; KmR | This study |
| 168 + TatCy | trpC2; pHB-YkuE-myc; pCCy; EmR; KmR | This study |
| 168 + TatAyCy | trpC2; pHB-YkuE-myc; pCACy; EmR; KmR | This study |
| tatAyCy + TatAy | trpC2; tatAy-tatCy::Sp; SpR; pHB-YkuE-myc; pCAy; EmR; KmR | This study |
| tatAyCy + TatCy | trpC2; tatAy-tatCy::Sp; SpR; pHB-YkuE-myc; pCCy; EmR; KmR | This study |
| tatAyCy + TatAyCy | trpC2; tatAy-tatCy::Sp; SpR; pHB-YkuE-myc; pCACy; EmR; KmR | This study |
| total-tat3 +TatAy | trpC2; tatAd-tatCd::Cm; tatAy-tatCy::Sp; tatAc::Tc; pHB-YkuE-myc; pCAy; EmR; KmR; TcR; CmR; SpR | This study |
| total-tat3 +TatCy | trpC2; tatAd-tatCd::Cm; tatAy-tatCy::Sp; tatAc::Tc; pHB-YkuE-myc; pCCy; EmR; KmR; TcR; CmR; SpR | This study |
| total-tat3 +TatAyCy | trpC2; tatAd-tatCd::Cm; tatAy-tatCy::Sp; tatAc::Tc; pHB-YkuE-myc; pCACy; EmR; KmR; TcR; CmR; SpR | This study |
| 168 pNZ8910YkuE-StrepII | trpC2; pNZ8910YkuE-StrepII; EmR | This study |
| tatAyCy pNZ8910YkuE-StrepII | trpC2; tatAy-tatCy::Sp; pNZ8910YkuE-StrepII; EmR; SpR | This study |
| tatAdCd pNZ8910-YkuE-StrepII | trpC2; tatAd-tatCd::Km; pNZ8910YkuE-StrepII; EmR; KmR | This study |
| total-tat3 pNZ8910-YkuE-StrepII | trpC2; tatAd-tatCd::Cm; CmR; tatAy-tatCy::Sp; SpR; tatAc::Tc; TcR; pNZ8910YkuE-StrepII; EmR | This study |
DNA Cloning Procedures
Procedures for DNA purification, restriction, ligation, agarose gel electrophoresis, and transformation of competent E. coli cells were carried out as described previously (33). B. subtilis was transformed as described by Kunst and Rapoport (34). PCR was carried out with the Phusion DNA polymerase (Finnzymes). Primers are listed in the supplemental Table 1. PCR products were purified using the PCR purification kit from Roche Applied Science. Restriction enzymes were obtained from New England Biolabs. Plasmid DNA from E. coli was isolated using the alkaline lysis method or by using the Invisorb® plasmid isolation kit (Invitek). All constructs were checked by sequencing. The plasmids pHB-ykuE-myc and pHB-ykuE were constructed by cloning the PCR-amplified ykuE gene into the pHB201 plasmid. The forward primer used to construct pHB-ykuE-myc contained a SalI restriction site and the reverse primer an EcoRI restriction site and a Myc tag. The PCR product was digested with SalI-EcoRI and cloned in the digested SalI-EcoRI pHB201 vector. The forward primer used to construct pHB-ykuE contained an SpeI restriction site, and the reverse primer contained a BamHI restriction site. The PCR product was digested with SpeI-BamHI and cloned into the digested SpeI-BamHI pHB201 vector. To construct pNZ8910-ykuE-StrepII, a synonymous point mutation was introduced into the ykuE gene to delete an internal BspHI restriction site. The mutagenesis was carried out using the F_Mut_ykuE and R_Mut_ykuE primers and the pHB-ykuE plasmid as a template. The resulting plasmid pHB-ykuEmut was verified by sequencing and used as a template for a further PCR with a forward primer containing a BspHI restriction site and a reverse primer with an SpeI restriction site. The fragment was digested with BspHI-SpeI and cloned into NcoI-HindIII digested pNZ8910 plasmid. The ligation mixture was introduced into L. lactis by transformation, resulting in the plasmid pNZ8910-YkuE-StrepII. This plasmid was then introduced into B. subtilis 168 or the tatAyCy, tatAdCd, or total tat3 mutant strains by transformation.
SDS-PAGE and Western Blotting
Cells were separated from the growth medium by centrifugation. Next, the cells were fractionated as described by Zweers et al. (35). Briefly, the cells were incubated with lysozyme in protoplast buffer to liberate cell wall-associated proteins. The resulting protoplasts were collected by centrifugation and disrupted by bead beating. Debris of the disrupted protoplasts was removed through centrifugation. Membranes were then separated from the cytoplasm through ultracentrifugation. Finally, the collected membranes were resuspended in solubilization buffer with 0.1% n-dodecyl β-d-maltoside. To extract cell wall-bound proteins, cells were incubated for 10 min in 25 mm Tris-HCl buffer, pH 8.0, with 1.5 m LiCl, or a solution containing 2 m KSCN as described previously (36, 37). Proteins in the collected fractions were separated by PAGE using pre-cast BisTris NuPAGE gels (Invitrogen). For Western blotting analyses, proteins separated by PAGE were semi-dry blotted (75 min at 1 mA/cm2) onto a nitrocellulose membrane. Subsequently, FeuA, TatAy, TrxA, WprA, YkuE, YwbN, or YfkN were detected with specific polyclonal antibodies raised in rabbits. Visualization of bound antibodies was performed using IRDye 800CW goat anti-rabbit secondary antibodies in combination with the Odyssey infrared imaging system (LiCor Biosciences). Fluorescence was recorded at 800 nm.
Protein Production and Purification
A B. subtilis 168 strain containing a subtilin-inducible ykuE-StrepII-tag construct was grown in 3 liters of LB medium at 37 °C under selective conditions (2 μg ml−1 erythromycin and 10 μg ml−1 kanamycin) until an A600 of 0.6, at which point the cultures were induced by addition of subtilin-containing supernatant from B. subtilis ATCC 6633 prepared as described by Bongers et al. (38). After 3 h of induction, cells were harvested (4 °C, 3500 × g for 20 min). Cell pellets were washed and resuspended in 100 mm Tris, pH 8.0, 150 mm NaCl. Cells were disrupted by using a French pressure cell (Thermo Spectronic) and a French pressure cell press (Aminco), resulting in the liberation of StrepII-tagged YkuE from the cells. The filtrated lysate was subjected to Strep-Tactin chromatography using an FPLC purifier system (Pharmacia) and a column with a 2-ml packed volume of Strep-Tactin Superflow material (IBA). The protein extract was applied to the column at a flow rate of 0.5 ml min−1. The column was washed for 20 min with cell resuspension buffer at 0.8 ml min−1. The bound protein was then eluted with 2.5 mm d-desthiobiotin (IBA) dissolved in resuspension buffer at 0.5 ml min−1. Elution fractions were analyzed by SDS-PAGE and subsequent mass spectrometry for species identification, and those fractions containing pure YkuE protein were concentrated using Amicon ultracentrifugal filter units (Millipore) with a nominal molecular mass limit of 10 kDa at 4 °C and 2000 × g. Protein concentrations were determined with the Bradford method (39) using a BSA calibration curve. Freshly purified protein of different concentrations obtained before and during the concentration process was used for ICP-MS analysis.
MS-based Protein Identification
Protein samples were digested in gel by the addition of sequencing grade modified trypsin (Promega) and incubation at 37 °C overnight. Samples were dissolved in 25 μl of 10% acetonitrile, 0.1% TFA. The mass spectrometric analysis of the samples was performed by using an Orbitrap Velos Pro mass spectrometer (ThermoScientific). An Ultimate nanoRSLC-HPLC system (Dionex), equipped with a nano C18 RP column, was connected on line to the mass spectrometer through a Proxeon nanospray source. A 6-μl aliquot of the tryptic digest was injected onto a C18 pre-concentration column. Automated trapping and desalting of the sample were performed at a flow rate of 6 μl/min using water, 0.05% formic acid as a solvent. Separation of the tryptic peptides was achieved with a gradient of water, 0.045% formic acid (solvent A) and 80% acetonitrile, 0.05% formic acid (solvent B) at a flow rate of 300 nl min−1. The column was connected to a stainless steel nanoemitter (Proxeon), and the eluent was sprayed directly toward the heated capillary of the mass spectrometer using a potential of 2300 V. A survey scan with a resolution of 60,000 was combined with at least three data-dependent MS/MS scans. Data analysis was performed using Proteome Discoverer (ThermoScientific) with SEQUEST and MASCOT (version 2.2; Matrix science) search engines using either the SwissProt or NCBI databases.
ICP-MS Analysis
Protein samples with defined concentrations were treated with 1:6 suprapure nitric acid (Merck) to dissociate all metals from organic complexors and solutions were further diluted gravimetrically with ultrapure milliQ water. Yttrium was added to all samples as an internal standard, and quantitative analysis of metal contents for magnesium, vanadium, chromium, manganese, iron, cobalt, nickel, copper, zinc, molybdenum, and tungsten was performed using an Agilent 7500ce (Agilent Technologies) together with established calibration and detection protocols (40, 41). External calibration during measurement was carried out using appropriate metal calibration standards (Merck). Data obtained both in the hydrogen and helium collision modes were averaged for all analyzed isotopes. Background metal content, which was detected at substantially low level in buffer control samples, was subtracted from the protein-containing samples after analysis.
Enzymatic Assays
Enzyme activity assays (200-μl reaction mixtures) were carried out in assay buffer containing 50 mm Tris-HCl (pH was varied between 6.5 and 9.5) and 5 mm MnCl2, with varying concentrations of p-nitrophenyl phosphate (0.1–10 mm) and 1 μm purified YkuE-StrepII at 25 °C. The reactions were started by addition of the phosphoester substrate and quenched by addition of 2% SDS (w/v). The release of p-nitrophenol was measured spectrophotometrically at 410 nm. Product quantification was subsequently carried out using a p-nitrophenol standard curve. Control reactions without enzyme were carried out for each pH series to correct for spontaneous p-nitrophenyl phosphate background hydrolysis. Substrate-dependent kinetic data of each pH series were plotted and analyzed by a Michaelis-Menten fitting model (Microcal Origin 5.0 software).
RESULTS
YkuE Is a Tat-dependently Exported Cell Wall Protein
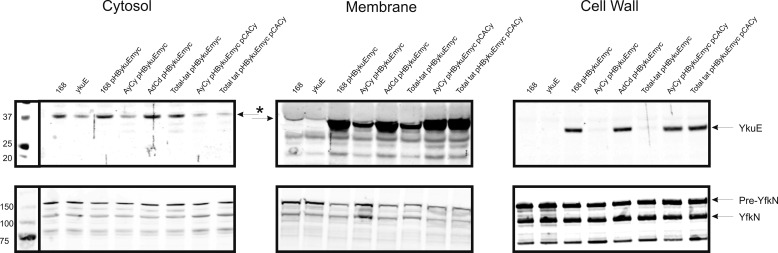
To verify whether the Tat system does indeed translocate the YkuE protein in B. subtilis, a polyclonal antibody was raised against this protein and used to determine the intra- and extracellular levels of YkuE. However, irrespective of the different growth conditions that were tested, we were unable to detect the production of YkuE in B. subtilis by Western blotting (data not shown). In this respect, it should be noted that a recent tiling array analysis of B. subtilis gene expression across 104 conditions revealed that ykuE is expressed at very low levels under all tested conditions (supplemental Fig. S1) (42). For this reason, we expressed a plasmid-borne copy of the ykuE gene from the relatively weak constitutive promoter of the low copy expression vector pHB201, which increases the ykuE expression level to a similar level as that of the ywbN and tatAyCy genes (43, 44). This allowed the detection of the mature-sized YkuE protein in the B. subtilis prototype strain 168 (Fig. 1). Next, the plasmid was introduced in mutant B. subtilis strains lacking either the tatAyCy genes, the tatAdCd genes, or all tat genes (total-tat). To study YkuE export, the transformed strains were grown until stationary phase in LB medium. Cells were separated from the growth medium by centrifugation and incubated with lysozyme to liberate cell wall-bound proteins. Finally, the resulting protoplasts were separated from the liberated cell wall proteins by centrifugation. The collected fractions were analyzed by PAGE and Western blotting using the YkuE-specific antibodies, which revealed that YkuE was only detectable in the protoplast and cell wall fractions. Importantly, the localization of mature YkuE to the cell wall was strictly dependent on the presence of the TatAyCy translocase because this protein was not detectable in the cell wall fractions of the tatAyCy mutant and the total tat mutant, although it was detectable in the cell wall fraction of the tatAdCd mutant. The cell wall localization of YkuE was restored in the tatAyCy mutant and the total tat mutant by ectopic expression of the tatAyCy genes from a plasmid (Fig. 1). In contrast to YkuE, the cell wall localization of the Sec-dependent control protein YfkN was not affected by any of the tat mutations tested. Interestingly, mature YkuE was also detectable in the protoplast fraction irrespective of the presence or absence of functional Tat machinery, although cells lacking TatAyCy contained less YkuE than TatAyCy-proficient cells. Further fractionation studies revealed that the protoplast-associated mature YkuE was localized to the cytoplasmic membrane (Fig. 1). Together, these findings imply that the membrane localization of YkuE is to a large extent Tat-independent, whereas its cell wall localization is strictly TatAyCy-dependent. To investigate whether the Tat-independent membrane localization is specific for YkuE, or a more general feature for Tat substrates in B. subtilis, we also assessed membrane localization of the well studied Tat substrate YwbN in the different tat mutant strains. To this end, we analyzed the localization of YwbN expressed from its authentic promoter using a YwbN-specific polyclonal antibody. As shown in Fig. 2, mature YwbN was localized to the cell wall and growth medium in a strictly TatAyCy-dependent manner. In contrast, the membrane association of mature YwbN was completely Tat-independent, and barely any YwbN was detectable in the cytosolic fraction (Fig. 2). These findings indicate that Tat substrates of B. subtilis can interact with the membrane in a Tat-independent manner. At present, we do not know how this Tat-independent interaction with the membrane is brought about or how the signal peptide is removed in this case. Potentially, signal peptide cleavage is catalyzed by as yet unidentified cytoplasmic or membrane-associated proteases for which there are many candidate proteases in B. subtilis (12, 45).
FIGURE 1.
Subcellular localization of YkuE. To determine the subcellular localization of YkuE in the presence or absence of active Tat translocases, the parental strain B. subtilis 168 and the tat mutant strains tatAyCy, tatAdCd, total-tat3, tatAyCy pCACy, and total-tat3 pCACy were transformed with plasmid pHBykuEmyc for the expression of YkuE. Next, the cells producing YkuE were grown for 7 h in LB medium. The cells were then separated from the growth medium by centrifugation and subjected to subcellular fractionation. Proteins in the obtained fractions were separated by PAGE, and the presence of YkuE and YfkN was monitored by Western blotting with specific polyclonal antibodies. Protein loading on the gels was corrected for A600. Only the results for the cytosol, membrane, and cell wall fractions are shown because no YkuE was detectable in the fractions representing the growth medium. The cell wall-localized protein YfkN was used as a Tat-independent control. The lanes are labeled as follows: 168, B. subtilis 168 Marburg strain; ykuE, B. subtilis 168 with a disrupted ykuE gene; 168 pHBykuEmyc, B. subtilis 168 containing pHBykuEmyc; AyCy pHBykuEmyc, B. subtilis tatAyCy containing pHBykuEmyc; AdCd pHBykuEmyc, B. subtilis tatAdCd pHBykuEmyc; Total-tat pHBykuEmyc, B. subtilis lacking all tat genes but containing pHBykuEmyc; AyCy pHBykuEmyc pCACy, B. subtilis tatAyCy containing pHBykuEmyc and plasmid pCACy for expression of TatAyCy; Total-tat pHBykuEmyc pCACy, B. subtilis lacking all chromosomal tat genes but containing pHBykuEmyc and pCACy. The positions of mature YkuE (YkuE) and the precursor and mature forms of YfkN (pre-Yfkn, YfkN) are marked with arrows. Asterisks mark bands that aspecifically cross-react with anti-YKuE.
FIGURE 2.
Subcellular localization of YwbN. To determine the subcellular localization of YwbN in the presence or absence of active Tat translocases, the parental strain B. subtilis 168 and tat mutant strains were subjected to subcellular fractionation as described in the legend of Fig. 1. The presence of YwbN, the Tat-independently cell wall-localized control protein YfkN, and the Tat-independently secreted control protein FeuA was monitored by Western blotting with specific polyclonal antibodies. The lanes are labeled as follows: ywbN, B. subtilis 168 with a disrupted ywbN gene; 168, B. subtilis 168 Marburg strain; AyCy, B. subtilis tatAyCy; AdCd, B. subtilis tatAdCd; Total-tat, B. subtilis lacking all tat genes; AyCy pCACy, B. subtilis tatAyCy containing plasmid pCACy for expression of TatAyCy; Total-tat pCACy, B. subtilis lacking all chromosomal tat genes but containing pCACy. The positions of mature YwbN (YwbN), the precursor, and mature forms of YfkN (pre-Yfkn and YfkN) and secreted FeuA (FeuA) are marked with arrows.
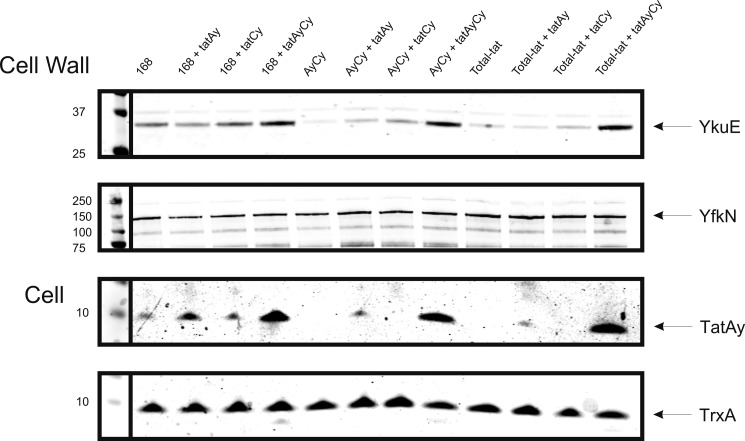
Because YkuE is a predicted metallophosphoesterase, we also investigated the cell wall localization of this protein under phosphate starvation conditions where various secreted phosphatases and phosphodiesterases are produced by B. subtilis (4, 12). However, also under these conditions, no YkuE expression from the authentic ykuE promoter was detectable, which is in line with the low expression level of this gene under phosphate starvation conditions (supplemental Fig. S1, compare the expression profiles for ykuE with those for tatAd, tatCd, and phoD) (42). As shown for cells grown in LB broth, the cell wall localization of YkuE in phosphate-starved cells was strongly dependent on the presence of the TatAyCy translocase (Fig. 3). In fact, the level of cell wall-associated YkuE was increased when the tatAy and tatCy genes were overexpressed from a plasmid, and it was severely reduced in mutant cells lacking the tatAyCy genes or expressing only tatAy or tatCy. The absence of the tatAdCd genes had no effect on YkuE localization to the cell wall showing that the TatAdCd translocase is not involved in YkuE export when this translocase was produced at wild-type levels (supplemental Fig. S2). Surprisingly, very small amounts of YkuE were detectable in the cell wall of the tatAyCy and total tat mutant strains (Fig. 3), suggesting that some Tat-independent YkuE localization to the cell wall can occur under phosphate starvation conditions.
FIGURE 3.
Partially Tat-independent cell wall localization of YkuE under phosphate starvation conditions. To investigate the effects of phosphate starvation on the localization of YkuE, cells containing pHBykuEmyc for the production of YkuE were grown in low phosphate depletion media and subjected to subcellular fractionation as described for Fig. 1. The presence of YkuE, YfkN, TatAy, and TrxA was monitored by Western blotting with specific polyclonal antibodies. The lanes are labeled as follows: 168, B. subtilis 168; 168 + tatAy, B. subtilis 168 containing pCAy for expression of tatAy; 168 + tatCy, B. subtilis 168 containing pCCy for expression of tatCy; 168 + tatAyCy, B. subtilis 168 containing pCACy for expression of tatAyCy; AyCy, B. subtilis tatAyCy; AyCy + tatAy, B. subtilis tatAyCy containing pCAy for expression of tatAy; AyCy + tatCy, B. subtilis tatAyCy containing pCCy for expression of tatCy; AyCy + tatAyCy, B. subtilis tatAyCy containing pCACy for expression of tatAyCy; Total-tat, B. subtilis lacking all tat genes; Total-tat + tatAy, B. subtilis lacking all tat genes containing pCAy for expression of tatAy; Total-tat + tatCy, B. subtilis lacking all tat genes containing pCCy for expression of tatCy; Total-tat + tatAyCy, B. subtilis lacking all tat genes containing pCACy for expression of tatAyCy. The positions of mature YkuE (YkuE), a processed form of YfkN (YfkN), TatAy, and TrxA are marked with arrows.
Tat-independent YkuE Export upon Overexpression
Because some Tat-independent membrane and cell wall localization of YkuE was detectable, we investigated whether this might relate to the production level of this protein. To this end, we made use of the subtilin-regulated gene expression (SURE) system for high level protein production in B. subtilis (38, 46). The ykuE gene was cloned into the SURE plasmid, and its expression was induced with subtilin in different tat mutant strains. As shown in Fig. 4, YkuE was overproduced in high amounts. Most of this overproduced YkuE was localized to the membrane, and relatively small amounts were localized to the cell wall. Furthermore, upon overproduction, YkuE was localized to the cell wall in a mostly Tat-independent manner. Nevertheless, a small decrease in the amount of cell wall-localized YkuE was detectable in the tatAyCy or total tat mutant strains compared with the parental strain 168 and the tatAdCd mutant. Furthermore, high amounts of membrane-associated YkuE were detectable, and also upon overproduction, the membrane association of this protein remained Tat-independent.
FIGURE 4.
Tat-independent export of overproduced YkuE to the cell wall. To overproduce YkuE, a subtilin-inducible copy of the ykuE gene was expressed from the plasmid pNZ8910ykuE-strep in B. subtilis cells expressing the SpaRK two-component regulatory system. YkuE-overproducing cells grown in LB medium were subjected to subcellular fractionation as described for Fig. 1. The presence of YkuE was monitored by Western blotting with specific polyclonal antibodies. The lanes with samples from YkuE (over)producing cells are labeled as follows: 168, B. subtilis 168; AyCy, B. subtilis tatAyCy; AdCd, B. subtilis tatAdCd; total-tat, B. subtilis lacking all tat genes. YkuE-specific protein bands are marked with an arrow.
YkuE Is Retained in the Cell Wall through Electrostatic Interactions
The YkuE protein lacks obvious domains for cell wall binding, such as an LPXTG motif for covalent attachment to peptidoglycan or domains for noncovalent cell wall binding (4, 12). To determine how YkuE can be retained in the cell wall, B. subtilis cells expressing this protein at different levels were subjected to treatment with either 2 m KSCN or 1.5 m LiCl as described previously (36, 37). Treatment with the chaotrope KSCN would disrupt noncovalent cell wall interactions based on hydrogen bonds, van der Waals forces, or hydrophobicity (47), and treatment with LiCl would disrupt electrostatic cell wall interactions. As shown in Fig. 5, treatment of the cells with LiCl was equally effective in extracting YkuE from the cell walls as protoplasting with lysozyme. By contrast, KSCN did not liberate any YkuE from the cells (data not shown). This implies that YkuE is retained in the B. subtilis cell wall through electrostatic interactions, most likely with particular cell wall components.
FIGURE 5.
LiCl extraction of YkuE from the cell wall. Cells of B. subtilis 168, the ykuE mutant BFA1834, B. subtilis 168 pHBykuE, or B. subtilis SURE ykuE were grown and harvested as described in the legend of Fig. 1. Next, the bacterial cells were treated either with 1.5 m LiCl as described by Antelmann et al. (36) or with lysozyme as described under “Experimental Procedures.” The presence of YkuE or the Tat-independent noncovalently cell wall-bound control protein WprA in the supernatant of LiCl-treated cells or in the protoplast supernatant was monitored by Western blotting with specific polyclonal antibodies. The presence of mature YkuE (YkuE), different processed forms of WprA, and an unidentified protein (X) that cross-reacts with the WprA antibodies are marked with arrows.
Purification and Metal Cofactor Analysis of YkuE
Database homology searches suggest that YkuE is a putative metallophosphoesterase. To validate this proposed function, YkuE was produced as a StrepII-tagged variant in its original host B. subtilis to ensure the association with any native metal cofactor(s) that it may carry. The protein was obtained at a yield of about 0.2 mg/liter culture and was analyzed after purification for intrinsic stability (Fig. 6). The protein sample that was analyzed directly after elution (∼0.5 h before SDS-PAGE) was observed to be intact as a full-length variant of the expected size of ∼35 kDa. Mass spectrometric analysis of purified YkuE yielded a total amino acid sequence coverage of ∼75% starting from residue 33 (supplemental Table 2). No peptides relating to the signal peptide were identified, which suggests that YkuE was purified predominantly in the mature form. Furthermore, no evidence for impurities with other B. subtilis proteins was obtained, suggesting that the protein samples contained YkuE only. Interestingly, when purified YkuE samples were incubated for 20 h at 4 °C, some degradation of YkuE was observed. All detectable degradation fragments corresponded to YkuE as shown by mass spectrometry. YkuE degradation was not reduced in the presence of inhibitors for cysteine proteases (N-ethylmaleimide) or serine proteases (phenylmethylsulfonyl fluoride; Fig. 6). In contrast, YkuE degradation was significantly reduced upon addition of EDTA, an effective inhibitor for metallohydrolase activity. Addition of 10 mm EDTA had a slightly higher protective effect than 1 mm EDTA, suggesting a direct relation between competitive metal sequestration and degradation of YkuE. Together these findings suggest a basal level of autohydrolytic activity of the metal-charged enzyme upon storage in aqueous solution. This may indicate a high activation potential of the enzyme for the nucleophilic attack of amide bonds. However, we cannot completely exclude the possibility that small amounts of a metalloprotease (undetectable by mass spectrometry) were co-purified with YkuE causing its subsequent degradation.
FIGURE 6.
YkuE purification and stability in the presence of different protease and hydrolase inhibitors. YkuE was purified from overproducing cells of the native host B. subtilis as described under “Experimental Procedures.” SDS-PAGE analysis of the YkuE-StrepII protein (∼35 kDa) was performed with a freshly purified sample obtained directly after elution from the Strep-Tactin column (lane 1), or with samples obtained after 20 h of incubation at 4 °C in the presence of either 1 mm N-ethylmaleimide (lane 2), 1 mm phenylmethylsulfonyl fluoride (lane 3), 10 mm EDTA (lane 4), or 1 mm EDTA (lane 5). Degradation bands observed upon protein incubation in buffered solution were identified by mass spectrometry to be part of the original full-length protein.
To identify the bound metal cofactor species in a quantitative way, the freshly purified YkuE samples obtained before and after protein concentration by ultrafiltration over a concentration range of 1 order of magnitude were subjected to inductively coupled plasma-mass spectrometry (ICP-MS) analysis. This analysis revealed two dominant metals, zinc and manganese, which were present in a strongly protein concentration-dependent manner (Table 2). The averaged stoichiometries of these metals were for manganese 0.76 ± 0.01 mol and for zinc 1.16 ± 0.02 mol/mol YkuE, which indicates their association with two distinct metal-binding sites. In the higher concentrated samples, small but significant quantities of nickel and copper were also found (Table 2). However, the low stoichiometric proportions of these metals (averaged amounts of 0.08 ± 5 × 10−5 mol of nickel and 0.03 ± 2 × 10−3 mol of copper per mol of YkuE, respectively) point to a fairly low selectivity of these species. Their substoichiometric amounts suggest a nonselective binding site occupation due to a superior metal-ligand affinity within the Irving-Williams series for divalent transition metals, which has to be considered especially if competition with manganese takes place (48, 49). Likewise, the unequal stoichiometric distribution of manganese and zinc with a bound mole ratio of about 0.8 to 1.2 may well be consistent with a partial loss of the low affinity species manganese and second with a partial displacement by the high affinity species zinc during the processes of overproduction and/or purification. This points to the importance and maintenance of proper metal insertion during protein folding and transport between different compartments (50). All further metals identified by ICP-MS were present below a threshold level of 1% compared with the injected protein quantities (Table 2), suggesting that they were not specifically associated with YkuE.
TABLE 2.
Quantitative metal content analysis
Three samples of YkuE-StrepII (containing 1, 5, or 10 nmol of purified protein obtained by Strep-Tactin chromatography) were subjected to ICP-MS analysis, and metal contents were determined by using appropriate internal calibration standards. All values are given in nanomoles.
| YkuE-Strep II | 1.0 | 5.0 | 10.0 |
|---|---|---|---|
| Magnesium | 0.005 | 0.012 | 0.008 |
| Vanadium | 0.002 | 0.004 | 0.007 |
| Chromium | 0.0008 | 0.0007 | 0.001 |
| Manganese | 0.82 | 4.08 | 6.47 |
| Iron | 0.002 | 0.005 | 0.009 |
| Cobalt | 0.003 | 0.002 | 0.005 |
| Nickel | 0.046 | 0.228 | 1.63 |
| Copper | 0.017 | 0.134 | 0.544 |
| Zinc | 1.21 | 5.93 | 10.85 |
| Molybdenum | 0.0001 | 0.00015 | 0.0002 |
| Tungsten | 0.00006 | 0.00008 | 0.0009 |
YkuE Displays Monophosphoesterase Activity in a Strongly pH-dependent Manner
To determine the catalytic activity of YkuE, freshly purified protein was incubated in an enzyme assay buffer containing 5 mm MnCl2 to ensure the quantitative occupation of the proposed manganese-binding site. Enzymatic reactions were carried out with varying concentrations of p-nitrophenyl phosphate as a substrate, which is commonly used for the evaluation of monophosphoesterase activity (51). The reaction series were performed under seven different pH conditions in the range of 6.5 to 9.5 to screen for the pH optimum of enzymatic activity. The product formation rates of p-nitrophenol obtained for each pH were evaluated by the Michaelis-Menten model and revealed relatively constant though slightly decreasing Km values when the pH was changed from acidic to alkaline. In contrast, the kcat value increased over more than 1 order of magnitude within this pH range (Fig. 7A and supplemental Table 3). The obtained catalytic efficiencies at different pH values showed that the pH optimum for YkuE was approximately at pH 9.0, and the pKa value of the activated water species was determined at about 7.8, indicating a nearly 100% active hydroxide complex at pH 9.0 and above (Fig. 7B). Thus, the slight drop of enzyme activity at pH 9.5 pointed to an arising catalytic limitation, which is different from the cofactor-generated activation potential of the water nucleophile. Previous kinetic analyses of zinc-dependent alkaline phosphatase, representing a widely distributed monophosphoesterase, have shown that the rate-limiting process at higher pH is in fact the dissociation rate of the cleaved phosphate product, although a lower pH generally limits the hydrolysis rate of the enzyme-phosphoryl intermediate (51). This substitution of rate-limiting events takes place mainly between pH 7.0 and 8.0, which is also observed for YkuE due to the strong pH dependence of kcat within this range and may relate to the actual pKa value of the proposed metal-bound hydroxide acting as the attacking nucleophile. The presence of this activated water species in bimetallic reaction centers has been confirmed previously from electron densities in the crystal structures of native and phosphoryl-bound alkaline phosphatase (52). Taken together, our present findings show that YkuE apparently acts as a manganese/zinc-dependent metallophosphoesterase that is Tat-dependently localized to the cell wall of B. subtilis.
FIGURE 7.
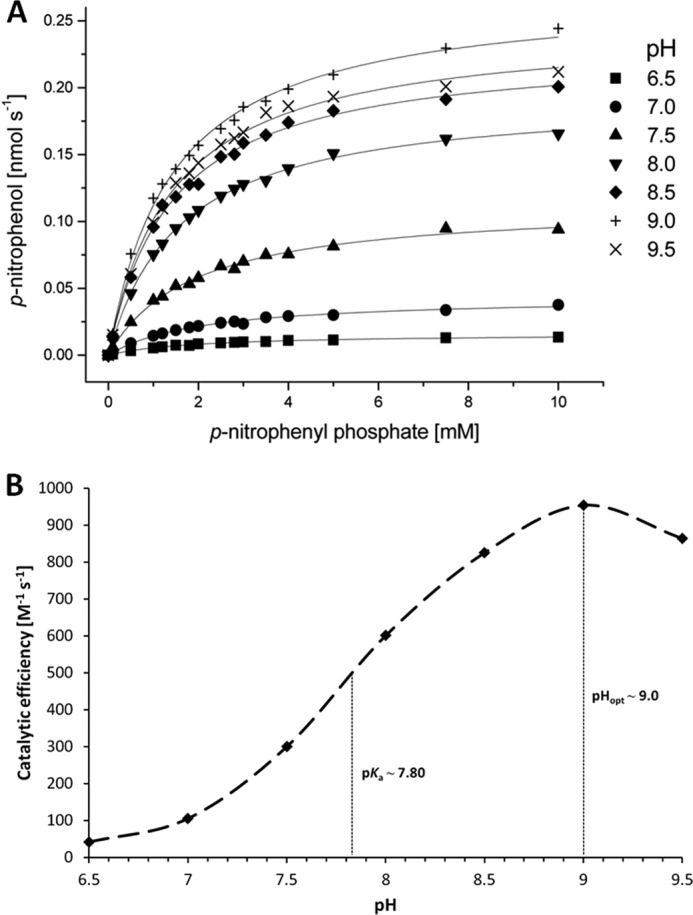
Enzyme kinetics and observed catalytic efficiencies of YkuE under different pH conditions. A, freshly purified YkuE was used under restored conditions of Mn(II) binding for kinetic analysis. The monophosphoester compound p-nitrophenyl phosphate was used as a substrate, and the assays were conducted under varying pH conditions (between pH 6.5 and 9.5) at 25 °C. The formation of p-nitrophenol was monitored spectrophotometrically, and the obtained data were corrected for spontaneous background hydrolysis examined in nonenzymatic control reactions. Final data were plotted and analyzed by Michaelis-Menten-type nonlinear regression. The kinetic parameters for all pH-dependent reaction series are given in supplemental Table 3. B, obtained catalytic efficiencies were plotted versus their cognate pH to determine the optimum catalytic activity and the pKa of the water nucleophile for YkuE-dependent hydrolysis. Because of the strongly pH-dependent kcat values, the catalytic efficiencies vary over 1 order of magnitude within the tested pH range and show an optimum peak at around pH 9.0. The pKa of the hydrated metal center is ∼7.80, revealing the strong activation potential of the binuclear catalytic site of YkuE.
DISCUSSION
Previous studies have shown that the Tat translocases of B. subtilis are highly restrictive with respect to the acceptance of potential substrate proteins (53, 54). Thus, despite extensive investigations, merely two strictly Tat-dependent proteins of this organism, PhoD and YwbN, have so far been identified. In this study, we show that the YkuE protein is also a genuine substrate of the B. subtilis Tat pathway and that the TatAyCy translocase specifically directs this protein to the cell wall. No evidence for an involvement of the TatAdCd translocase in YkuE export was obtained under any of the tested conditions. Furthermore, our data show that the translocated YkuE protein is retained in the cell wall through electrostatic interactions. Notably, this is the first time that a cell wall protein has been identified as a specific substrate of the Tat system. In contrast, the two other known Tat substrates of B. subtilis, PhoD and YwbN, are secreted into the growth medium. Interestingly, the Tat dependence of YkuE is not as strict as that of the two other substrates because we have observed at least two conditions where YkuE is exported Tat-independently, albeit to different extents. Although the cell wall localization of YkuE in cells growing in LB medium seems strictly Tat-dependent, a very low but detectable level of Tat-independent YkuE export takes place when cells are grown under phosphate starvation conditions. Under the latter conditions, the amount of cell wall-associated YkuE was found to be increased upon overexpression of the TatAyCy translocase, which underscores the view that YkuE is most effectively exported via this translocase. This finding is also fully consistent with the previously observed increase in the Tat-dependent secretion of the YwbN protein into the growth medium upon overexpression of TatAyCy (25). We therefore conclude that TatAyCy is present in limiting amounts when substrates like YkuE and YwbN are overproduced. This is actually very clearly observed upon high level overexpression of YkuE (Fig. 3). Under these high level production conditions, most YkuE is Tat-independently localized to the cell wall, showing that the saturation of TatAyCy leads to YkuE export via a Tat-independent route, which is probably the Sec pathway (4, 12).
Several known Tat substrates are proteins that contain a metal cofactor that may need to be inserted in the folded protein prior to translocation (55). For example, the MncA protein in Synechocystis is secreted via the Tat pathway. MncA is believed to bind its manganese cofactor during folding in the cytoplasm where metal species such as copper and zinc are tightly bound and thus are less competitive than in the extracytosolic compartment. This study suggests that a similar mechanism for distinct incorporation of transition metal species with different binding affinities could have evolved in the case of YkuE. The determined metal-protein stoichiometries indicate that one manganese and one zinc atom are incorporated at the predicted binuclear metal center of YkuE. This would place YkuE in the group of binuclear metallohydrolases that selectively bind magnesium, manganese, zinc, iron, nickel, or cobalt at their distinct metal recognition sites (56). However, it remains to be shown whether metal species with different ligand affinities are generally bound in a successive way to the binuclear site before and after transport of the folded protein through a secretion pathway, such as the Tat system. Indeed, many metallohydrolases such as the E. coli alkaline phosphatase PhoA (57) and the aminopeptidase Iap (58, 59) as well as several virulence-associated metalloproteases in Vibrio (60), Streptococcus (61, 62), or Burkholderia (63, 64) are secreted in an unfolded state via the Sec pathway. However, these proteins usually contain only one transition metal species, such as zinc, that does not face strong binding site competition in the extracytosolic environment due to its high binding site affinity.
Comparisons of the sequences of various binuclear metallophosphoesterases with the sequence of YkuE and its conserved binding site motifs suggest mixed nitrogen/oxygen ligand donor sites for both metals in the binuclear center, primarily formed by histidine and aspartate residues (Fig. 8). Binding site 1, which contains a higher proportion of oxygen donors, may be suspected to favor the binding of manganese, although site 2 with higher nitrogen proportion may in contrast favor interaction with zinc, due to the corresponding metal-ligand affinity series (65). However, metal selectivities are also dependent on binding site-specific geometric features, and thus, predictions of selectivity for one or other of the species can only be tentatively based on the currently available sequence data. Nonetheless, the binding site compositions suggest that zinc will bind at least 2 orders of magnitude stronger to both of these sites than manganese according to the affinity ranges of the Irving-Williams series for divalent cations. The indicated partial displacement of manganese by zinc may further suggest that the selectivity of the lower affinity site 1 may be relaxed. At present, it cannot be excluded that catalytic activity may be provided upon occupation with either one or the other of these two species or even further divalent species of the transition metal series displaying considerable binding affinity.
FIGURE 8.

YkuE active site model. The proposed binuclear metal reaction center of YkuE is shown, and amino acid residues are indicated that are predicted to act as ligand donors at the metal cofactor site. Predictions were made upon comparison with various metallophosphoesterase consensus sequences, and their metal-binding motifs were based on protein database information (UniProt, NCBI). Putative metal selectivities at the distinct binding sites are further suggested according to the experimentally found dominant metal species associated with natively purified YkuE and based on known metal-ligand complex stabilities for varied N/O donor sites. The core processes of the proposed catalytic mechanism of YkuE are indicated by interaction of a substrate phosphoryl moiety with the bi-metal cofactor site, which also activates water for nucleophilic attack and subsequent hydrolysis of the currently not further specified phosphoester substrate compound(s).
The simplest catalytic mechanism of the YkuE metallophosphoesterase may compose four core processes, each of which can be further divided into particular equilibrium steps. First, the association of the preferably deprotonated oxygen donor groups of the phosphoryl substrate molecule takes place at the free metal coordination sites of the binuclear metal center. As the investigation of native enzyme crystal structures of this superfamily showed (56), water coordination at the bi-metal center is expected to precede this event, leading to a readily activated hydroxyl group, which nucleophilically attacks the substrate phosphorus atom within the second process. This then leads to hydrolysis by electron pair shift toward the alcohol function of the phosphoester. Third, dissociation of the alcohol and phosphate hydrolysis products takes place, followed by a fourth process of repeated water binding and activation at the metal center. Further detailed investigations of the catalytic properties of YkuE may include a closer inspection of its phosphoryl substrate specificity as well as the detailed analysis of the metal-binding site by metal replacement analyses and binding site mutagenesis.
In conclusion, our novel finding that the B. subtilis YkuE protein is a mixed site metallophosphoesterase, which is specifically targeted to the cell wall, sheds new light on the versatility of Tat translocases in the targeting of proteins to different subcellular compartments. To date, no specific Tat-dependent protein targeting to a bacterial cell wall has been documented. Although previous studies have shown that several Tat-dependently exported proteins can be extracted from the cell wall of streptomycetes, there is no published evidence that these proteins are exclusively localized in this subcellular compartment (22, 23). Furthermore, our present data show that particular Tat-dependently secreted proteins of B. subtilis can bypass the Tat pathway when the required Tat translocase is present in limiting amounts. This is also a novel finding, because secretion of the two other known Tat substrates of this organism, PhoD and YwbN, remains strictly Tat-dependent when TatAdCd or TatAyCy becomes limiting. This finding is important, because it may explain, at least in part, why only a very limited number of Tat-dependently secreted proteins were identified so far in B. subtilis. Another reason for the detection of only few Tat-dependently exported proteins in B. subtilis could be that some of these proteins have been overlooked due to specific targeting to the cell wall, as is the case for YkuE that is retained in the cell wall through electrostatic interactions. Finally, our findings support the view that, during the evolution of B. subtilis as a colonizer of ecological niches in the soil and plant rhizosphere, there must have been a particularly high selective pressure on phosphate acquisition because this organism produces at least five exported proteins with phosphoesterase activity, namely PhoA, PhoB, PhoD, YfkN, and as shown in the present studies YkuE. All these enzymes contribute to the acquisition of phosphate. A similar functional redundancy was previously shown for other proteins that fulfill critical physiological roles in B. subtilis as exemplified by eight exported proteases that cooperate in nitrogen acquisition (12) or essential protein secretion machinery components such as the five type I signal peptidases that catalyze the maturation of secretory precursor proteins (66). In this light, we believe that the demonstration of the metallophosphoesterase activity of YkuE is of physiological relevance.
Acknowledgment
We thank Emma Denham for critical reading of the manuscript.
This work was supported in part by Commission of the European Union Projects LSHG-CT-2004-005257, LSHM-CT-2006-019064, LSHG-CT-2006-037469, PITN-GA-2008-215524, and -244093 and Transnational SysMO Initiative Projects BACELL SysMO1 and -2 through the Research Council for Earth and Life Sciences of the Netherlands Organization for Scientific Research (to C. G. M., R. v. d. P., C. G., and J. M. v. D.).

This article contains supplemental Figs. S1 and S2 and Tables 1–3.
- Tat
- twin-arginine translocation
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Sec
- secretion
- ICP
- inductively coupled plasma.
REFERENCES
- 1. Robinson C., Matos C. F., Beck D., Ren C., Lawrence J., Vasisht N., Mendel S. (2011) Transport and proofreading of proteins by the twin-arginine translocation (Tat) system in bacteria. Biochim. Biophys. Acta 1808, 876–884 [DOI] [PubMed] [Google Scholar]
- 2. Fröbel J., Rose P., Müller M. (2012) Twin-arginine-dependent translocation of folded proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1029–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Robinson C, Bolhuis A. (2004) Tat-dependent protein targeting in prokaryotes and chloroplasts. Biochim. Biophys. Acta 1694, 135–147 [DOI] [PubMed] [Google Scholar]
- 4. Tjalsma H., Antelmann H., Jongbloed J. D., Braun P. G., Darmon E., Dorenbos R., Dubois J. Y., Westers H., Zanen G., Quax W. J., Kuipers O. P., Bron S., Hecker M., van Dijl J. M. (2004) Proteomics of protein secretion by Bacillus subtilis. Separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68, 207–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Palmer T., Sargent F., Berks B. C. (2005) Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol. 13, 175–180 [DOI] [PubMed] [Google Scholar]
- 6. Natale P., Brüser T., Driessen A. J. (2008) Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane. Distinct translocases and mechanisms. Biochim. Biophys. Acta 1778, 1735–1756 [DOI] [PubMed] [Google Scholar]
- 7. Driessen A. J., Nouwen N. (2008) Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77, 643–667 [DOI] [PubMed] [Google Scholar]
- 8. Pommier J., Méjean V., Giordano G., Iobbi-Nivol C. (1998) TorD, a cytoplasmic chaperone that interacts with the unfolded trimethylamine N-oxide reductase enzyme (TorA) in Escherichia coli. J. Biol. Chem. 273, 16615–16620 [DOI] [PubMed] [Google Scholar]
- 9. Sargent F. (2007) The twin-arginine transport system. Moving folded proteins across membranes. Biochem. Soc. Trans. 35, 835–847 [DOI] [PubMed] [Google Scholar]
- 10. Kolkman M. A., van der Ploeg R., Bertels M., van Dijk M., van der Laan J., van Dijl J. M., Ferrari E. (2008) The twin-arginine signal peptide of Bacillus subtilis YwbN can direct either Tat- or Sec-dependent secretion of different cargo proteins. Secretion of active subtilisin via the B. subtilis Tat pathway. Appl. Environ. Microbiol. 74, 7507–7513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cristóbal S., de Gier J. W., Nielsen H., von Heijne G. (1999) Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J. 18, 2982–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tjalsma H., Bolhuis A., Jongbloed J. D., Bron S., van Dijl J. M. (2000) Signal peptide-dependent protein transport in Bacillus subtilis. A genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64, 515–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Berks B. C. (1996) A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22, 393–404 [DOI] [PubMed] [Google Scholar]
- 14. Stanley N. R., Palmer T., Berks B. C. (2000) The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J. Biol. Chem. 275, 11591–11596 [DOI] [PubMed] [Google Scholar]
- 15. Bolhuis A., Mathers J. E., Thomas J. D., Barrett C. M., Robinson C. (2001) TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J. Biol. Chem. 276, 20213–20219 [DOI] [PubMed] [Google Scholar]
- 16. Chaddock A. M., Mant A., Karnauchov I., Brink S., Herrmann R. G., Klösgen R. B., Robinson C. (1995) A new type of signal peptide. Central role of a twin-arginine motif in transfer signals for the delta pH-dependent thylakoidal protein translocase. EMBO J. 14, 2715–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alami M., Lüke I., Deitermann S., Eisner G., Koch H. G., Brunner J., Müller M. (2003) Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell 12, 937–946 [DOI] [PubMed] [Google Scholar]
- 18. Tullman-Ercek D., DeLisa M. P., Kawarasaki Y., Iranpour P., Ribnicky B., Palmer T., Georgiou G. (2007) Export pathway selectivity of Escherichia coli twin arginine translocation signal peptides. J. Biol. Chem. 282, 8309–8316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Widdick D. A., Eijlander R. T., van Dijl J. M., Kuipers O. P., Palmer T. (2008) A facile reporter system for the experimental identification of twin-arginine translocation (Tat) signal peptides from all kingdoms of life. J. Mol. Biol. 375, 595–603 [DOI] [PubMed] [Google Scholar]
- 20. Bendtsen J. D., Nielsen H., Widdick D., Palmer T., Brunak S. (2005) Prediction of twin-arginine signal peptides. BMC Bioinformatics 6, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li H., Jacques P. E., Ghinet M. G., Brzezinski R., Morosoli R. (2005) Determining the functionality of putative Tat-dependent signal peptides in Streptomyces coelicolor A3(2) by using two different reporter proteins. Microbiology 151, 2189–2198 [DOI] [PubMed] [Google Scholar]
- 22. Widdick D. A., Dilks K., Chandra G., Bottrill A., Naldrett M., Pohlschröder M., Palmer T. (2006) The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. Proc. Natl. Acad. Sci. U.S.A. 103, 17927–17932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Joshi M. V., Mann S. G., Antelmann H., Widdick D. A., Fyans J. K., Chandra G., Hutchings M. I., Toth I., Hecker M., Loria R., Palmer T. (2010) The twin arginine protein transport pathway exports multiple virulence proteins in the plant pathogen Streptomyces scabies. Mol. Microbiol. 77, 252–271 [DOI] [PubMed] [Google Scholar]
- 24. Pop O., Martin U., Abel C., Müller J. P. (2002) The twin-arginine signal peptide of PhoD and the TatAd/Cd proteins of Bacillus subtilis form an autonomous Tat translocation system. J. Biol. Chem. 277, 3268–3273 [DOI] [PubMed] [Google Scholar]
- 25. Jongbloed J. D., Grieger U., Antelmann H., Hecker M., Nijland R., Bron S., van Dijl J. M. (2004) Two minimal Tat translocases in Bacillus. Mol. Microbiol. 54, 1319–1325 [DOI] [PubMed] [Google Scholar]
- 26. Biswas L., Biswas R., Nerz C., Ohlsen K., Schlag M., Schäfer T., Lamkemeyer T., Ziebandt A. K., Hantke K., Rosenstein R., Götz F. (2009) Role of the twin-arginine translocation pathway in Staphylococcus. J. Bacteriol. 191, 5921–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harwood C. R., Cranenburgh R. (2008) Bacillus protein secretion. An unfolding story. Trends Microbiol. 16, 73–79 [DOI] [PubMed] [Google Scholar]
- 28. Jongbloed J. D., Martin U., Antelmann H., Hecker M., Tjalsma H., Venema G., Bron S., van Dijl J. M., Müller J. (2000) TatC is a specificity determinant for protein secretion via the twin-arginine translocation pathway. J. Biol. Chem. 275, 41350–41357 [DOI] [PubMed] [Google Scholar]
- 29. Jongbloed J. D., van der Ploeg R., van Dijl J. M. (2006) Bifunctional TatA subunits in minimal Tat protein translocases. Trends Microbiol. 14, 2–4 [DOI] [PubMed] [Google Scholar]
- 30. Dilks K., Rose R. W., Hartmann E., Pohlschröder M. (2003) Prokaryotic utilization of the twin-arginine translocation pathway. A genomic survey. J. Bacteriol. 185, 1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taylor P. D., Toseland C. P., Attwood T. K., Flower D. R. (2006) TATPred. A Bayesian method for the identification of twin arginine translocation pathway signal sequences. Bioinformation 1, 184–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Müller J. P., An Z., Merad T., Hancock I. C., Harwood C. R. (1997) Influence of Bacillus subtilis phoR on cell wall anionic polymers. Microbiology 143, 947–956 [DOI] [PubMed] [Google Scholar]
- 33. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, New York [Google Scholar]
- 34. Kunst F., Rapoport G. (1995) Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177, 2403–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zweers J. C., Wiegert T., van Dijl J. M. (2009) Stress-responsive systems set specific limits to the overproduction of membrane proteins in Bacillus subtilis. Appl. Environ. Microbiol. 75, 7356–7364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Antelmann H., Yamamoto H., Sekiguchi J., Hecker M. (2002) Stabilization of cell wall proteins in Bacillus subtilis. A proteomic approach. Proteomics 2, 591–602 [DOI] [PubMed] [Google Scholar]
- 37. Sibbald M. J., Winter T., van der Kooi-Pol M. M., Buist G., Tsompanidou E., Bosma T., Schäfer T., Ohlsen K., Hecker M., Antelmann H., Engelmann S., van Dijl J. M. (2010) Synthetic effects of secG and secY2 mutations on exoproteome biogenesis in Staphylococcus aureus. J. Bacteriol. 192, 3788–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bongers R. S., Veening J. W., Van Wieringen M., Kuipers O. P., Kleerebezem M. (2005) Development and characterization of a subtilin-regulated expression system in Bacillus subtilis. Strict control of gene expression by addition of subtilin. Appl. Environ. Microbiol. 71, 8818–8824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 40. Eickhorst T., Seubert A. (2004) Germanium dioxide as internal standard for simplified trace determination of bromate, bromide, iodate, and iodide by on-line coupling ion chromatography inductively coupled plasma mass spectrometry. J. Chromatogr. A. 1050, 103–109 [PubMed] [Google Scholar]
- 41. Albrecht A. G., Landmann H., Nette D., Burghaus O., Peuckert F., Seubert A., Miethke M., Marahiel M. A. (2011) The frataxin homologue Fra plays a key role in intracellular iron channeling in Bacillus subtilis. ChemBioChem. 12, 2052–2061 [DOI] [PubMed] [Google Scholar]
- 42. Nicolas P., Mäder U., Dervyn E., Rochat T., Leduc A., Pigeonneau N., Bidnenko E., Marchadier E., Hoebeke M., Aymerich S., Becher D., Bisicchia P., Botella E., Delumeau O., Doherty G., Denham E. L., Fogg M. J., Fromion V., Goelzer A., Hansen A., Härtig E., Harwood C. R., Homuth G., Jarmer H., Jules M., Klipp E., Le Chat L., Lecointe F., Lewis P., Liebermeister W., March A., Mars R. A., Nannapaneni P., Noone D., Pohl S., Rinn B., Rügheimer F., Sappa P. K., Samson F., Schaffer M., Schwikowski B., Steil L., Stülke J., Wiegert T., Devine K. M., Wilkinson A. J., van Dijl J. M., Hecker M., Völker U., Bessières P., Noirot P. (2012) Condition-dependent transcriptome reveals high level regulatory architecture in Bacillus subtilis. Science 335, 1103–1106 [DOI] [PubMed] [Google Scholar]
- 43. Bron S., Bolhuis A., Tjalsma H., Holsappel S., Venema G., van Dijl J. M. (1998) Protein secretion and possible roles for multiple signal peptidases for precursor processing in bacilli. J. Biotechnol. 64, 3–13 [DOI] [PubMed] [Google Scholar]
- 44. van der Ploeg R., Barnett J. P., Vasisht N., Goosens V. J., Pöther D. C., Robinson C., van Dijl J. M. (2011) Salt sensitivity of minimal twin arginine translocases. J. Biol. Chem. 286, 43759–43770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dalbey R. E., Wang P., van Dijl J. M. (2012) Membrane proteases in the bacterial protein secretion and quality control pathway. Microbiol. Mol. Biol. Rev. 76, 311–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vavrová L., Muchová K., Barák I. (2010) Comparison of different Bacillus subtilis expression systems. Res. Microbiol. 161, 791–797 [DOI] [PubMed] [Google Scholar]
- 47. Hatefi Y., Hanstein W. G. (1969) Solubilization of particulate proteins and nonelectrolytes by chaotropic agents. Proc. Natl. Acad. Sci. U.S.A. 62, 1129–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Irving H., Williams R. J. (1948) Order of stability of metal complexes. Nature 162, 746–747 [Google Scholar]
- 49. Waldron K. J., Rutherford J. C., Ford D., Robinson N. J. (2009) Metalloproteins and metal sensing. Nature 460, 823–830 [DOI] [PubMed] [Google Scholar]
- 50. Waldron K. J., Robinson N. J. (2009) How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 7, 25–35 [DOI] [PubMed] [Google Scholar]
- 51. Hull W. E., Halford S. E., Gutfreund H., Sykes B. D. (1976) 31P nuclear magnetic resonance study of alkaline phosphatase. The role of inorganic phosphate in limiting the enzyme turnover rate at alkaline pH. Biochemistry 15, 1547–1561 [DOI] [PubMed] [Google Scholar]
- 52. Kim E. E., Wyckoff H. W. (1991) Reaction mechanism of alkaline phosphatase based on crystal structures. Two-metal ion catalysis. J. Mol. Biol. 218, 449–464 [DOI] [PubMed] [Google Scholar]
- 53. Jongbloed J. D., Antelmann H., Hecker M., Nijland R., Bron S., Airaksinen U., Pries F., Quax W. J., van Dijl J. M., Braun P. G. (2002) Selective contribution of the twin-arginine translocation pathway to protein secretion in Bacillus subtilis. J. Biol. Chem. 277, 44068–44078 [DOI] [PubMed] [Google Scholar]
- 54. van Dijl J. M., Braun P. G., Robinson C., Quax W. J., Antelmann H., Hecker M., Müller J., Tjalsma H., Bron S., Jongbloed J. D. (2002) Functional genomic analysis of the Bacillus subtilis Tat pathway for protein secretion. J. Biotechnol. 98, 243–254 [DOI] [PubMed] [Google Scholar]
- 55. Tottey S., Waldron K. J., Firbank S. J., Reale B., Bessant C., Sato K., Cheek T. R., Gray J., Banfield M. J., Dennison C., Robinson N. J. (2008) Protein-folding location can regulate manganese-binding versus copper or zinc binding. Nature 455, 1138–1142 [DOI] [PubMed] [Google Scholar]
- 56. Wilcox D. E. (1996) Binuclear metallohydrolases. Chem. Rev. 96, 2435–2458 [DOI] [PubMed] [Google Scholar]
- 57. Pugsley A. P. (1993) The complete general secretory pathway in Gram-negative bacteria. Microbiol. Rev. 57, 50–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A. (1987) Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169, 5429–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Håkansson K., Miller C. G. (2002) Structure of peptidase T from Salmonella typhimurium. Eur. J. Biochem. 269, 443–450 [DOI] [PubMed] [Google Scholar]
- 60. Norqvist A., Norrman B., Wolf-Watz H. (1990) Identification and characterization of a zinc metalloprotease associated with invasion by the fish pathogen Vibrio anguillarum. Infect. Immun. 58, 3731–3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Novak R., Tuomanen E., Charpentier E. (2000) The mystery of psaA and penicillin tolerance in Streptococcus pneumoniae. Mol. Microbiol. 36, 1504–1505 [DOI] [PubMed] [Google Scholar]
- 62. Chiavolini D., Memmi G., Maggi T., Iannelli F., Pozzi G., Oggioni M. R. (2003) The three extracellular zinc metalloproteinases of Streptococcus pneumoniae have a different impact on virulence in mice. BMC Microbiol. 3, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Corbett C. R., Burtnick M. N., Kooi C., Woods D. E., Sokol P. A. (2003) An extracellular zinc metalloprotease gene of Burkholderia cepacia. Microbiology 149, 2263–2271 [DOI] [PubMed] [Google Scholar]
- 64. Kooi C., Subsin B., Chen R., Pohorelic B., Sokol P. A. (2006) Burkholderia cenocepacia ZmpB is a broad specificity zinc metalloprotease involved in virulence. Infect. Immun. 74, 4083–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fraústo da Silva J. J., Williams R. J. (2001) The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. Oxford University Press, Oxford, UK [Google Scholar]
- 66. van Dijl J. M. (2012) Biofilm research uncovers a novel nonenzymatic signal peptidase function in Bacillus. J. Bacteriol. 194, 2779–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gasson M. J. (1983) Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Antelmann H., Darmon E., Noone D., Veening J. W., Westers H., Bron S., Kuipers O. P., Devine K. M., Hecker M., van Dijl J. M. (2003) The extracellular proteome of Bacillus subtilis under secretion stress conditions. Mol. Microbiol. 49, 143–156 [DOI] [PubMed] [Google Scholar]
- 69. Kobayashi K., Ehrlich S. D., Albertini A., Amati G., Andersen K. K., Arnaud M., Asai K., Ashikaga S., Aymerich S., Bessieres P., Boland F., Brignell S. C., Bron S., Bunai K., Chapuis J., Christiansen L. C., Danchin A., Débarbouille M., Dervyn E., Deuerling E., Devine K., Devine S. K., Dreesen O., Errington J., Fillinger S., Foster S. J., Fujita Y., Galizzi A., Gardan R., Eschevins C., Fukushima T., Haga K., Harwood C. R., Hecker M., Hosoya D., Hullo M. F., Kakeshita H., Karamata D., Kasahara Y., Kawamura F., Koga K., Koski P., Kuwana R., Imamura D., Ishimaru M., Ishikawa S., Ishio I., Le Coq D., Masson A., Mauël C., Meima R., Mellado R. P., Moir A., Moriya S., Nagakawa E., Nanamiya H., Nakai S., Nygaard P., Ogura M., Ohanan T., O'Reilly M., O'Rourke M., Pragai Z., Pooley H. M., Rapoport G., Rawlins J. P., Rivas L. A., Rivolta C., Sadaie A., Sadaie Y., Sarvas M., Sato T., Saxild H. H., Scanlan E., Schumann W., Seegers J. F., Sekiguchi J., Sekowska A., Séror S. J., Simon M., Stragier P., Studer R., Takamatsu H., Tanaka T., Takeuchi M., Thomaides H. B., Vagner V., van Dijl J. M., Watabe K., Wipat A., Yamamoto H., Yamamoto M., Yamamoto Y., Yamane K., Yata K., Yoshida K., Yoshikawa H., Zuber U., Ogasawara N. (2003) Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A. 100, 4678–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. van der Ploeg R., Mäder U., Homuth G., Schaffer M., Denham E. L., Monteferrante C. G., Miethke M., Marahiel M. A., Harwood C. R., Winter T., Hecker M., Antelmann H., van Dijl J. M. (2011) Environmental salinity determines the specificity and need for Tat-dependent secretion of the YwbN protein in Bacillus subtilis. PLoS One 6, e18140. [DOI] [PMC free article] [PubMed] [Google Scholar]