Background: Venous ulcers (VUs) are a major health problem, but their molecular pathology remains unknown.
Results: A specific set of miRNAs induced in VUs targets signaling molecules and inhibits healing.
Conclusion: Induction of miRNAs in VUs leads to inhibition of epithelialization and granulation tissue formation.
Significance: This new discovery will enable miRNA use as diagnostic/therapeutic targets in VUs.
Keywords: Epidermis, Keratinocytes, Microarray, MicroRNA, Skin, Wound Healing, Chronic Wounds, Epithelialization, Granulation Tissue, Venous Ulcers
Abstract
Chronic nonhealing wounds, such as venous ulcers (VUs), are a widespread and serious medical problem with high morbidity and mortality. The molecular pathology of VUs remains poorly understood, impeding the development of effective treatment strategies. Using mRNA expression profiling of VUs biopsies and computational analysis, we identified a candidate set of microRNAs with lowered target gene expression. Among these candidates, miR-16, -20a, -21, -106a -130a, and -203 were confirmed to be aberrantly overexpressed in a cohort study of 10 VU patients by quantitative PCR and in situ hybridizations. These microRNAs were predicted to target multiple genes important for wound healing, including early growth response factor 3, vinculin, and leptin receptor (LepR). Overexpression of the top up-regulated miRNAs, miR-21 and miR-130a, in primary human keratinocytes down-regulated expression of the endogenous LepR and early growth response factor 3. The luciferase reporter assay verified LepR as a direct target for miR-21 and miR-130a. Both miR-21 and miR-130a delayed epithelialization in an acute human skin wound model. Furthermore, in vivo overexpression of miR-21 inhibited epithelialization and granulation tissue formation in a rat wound model. Our results identify a novel mechanism in which overexpression of specific set of microRNAs inhibits wound healing, resulting in new potential molecular markers and targets for therapeutic intervention.
Introduction
Chronic wounds, such as venous, diabetic foot, or pressure ulcers, are characterized by physiological impairments manifested by delays in healing, resulting in severe morbidity and mortality (1). It is estimated that each year over 8 million people develop chronic nonhealing wounds in the Unites States (2, 3). Faced with rise of chronic wounds to epidemic proportions, clinical treatments encounter limitations in prevention and therapy deriving from the lack of knowledge and understanding of the cellular and molecular pathology of wound healing inhibition (2, 4). Current Food and Drug Administration-approved biotherapies for VUs3 have a documented failure rate based on the high prevalence and incidence of recalcitrant VUs. Clinical trials of exogenously administered TGFβ, fibroblast growth factor, keratinocyte growth factor, and EGF to human chronic ulcers have achieved very limited efficacy and failed to obtain Food and Drug Administration approval (5, 6), despite early promising animal studies (7, 8).
MicroRNAs (miRNAs) are short (∼22 nucleotides), noncoding RNAs that can suppress the expression of protein-coding genes both through degradation of the transcript and through inhibition of translation (9, 10). miRNAs are generated by sequential processing of long RNA polymerase II transcripts by two key RNase III proteins, Drosha and Dicer (11). Sites that confer mRNA destabilization and translational repression typically pair to the 5′ region of the miRNA, centering on miRNA nucleotides 2–8, known as the miRNA seed (12, 13). These regulators of gene expression are capable of defining and altering cell fate. Importantly, aberrant expression or activity of miRNAs can lead to disease; in particular, miRNAs are often dysregulated in cancer (14–17). Moreover, aberrant miRNA expression may be reflected in statistically detectable changes in mRNA levels of target genes (12, 18, 19).
The importance of miRNAs in epidermal development and adult skin stem cell maintenance has been established (20, 21). When miRNAs are ablated in the skin epithelium by conditionally targeting either Dicer1 or DGCR8, barrier function of the epithelium is compromised (21, 22). Specific miRNAs expressed in developing mouse epidermis and hair follicles have been identified (22). In addition, involvement of miRNAs in the pathology of psoriasis has been shown (23). A recent study has shown that hypoxia-induced miR-210 expression has an inhibiting effect on wound closure and keratinocyte proliferation in an ischemic murine wound model (24). Conversely, antiproliferative effect of miR-483–3p in human keratinocytes was linked to promotion of wound healing in vitro and in vivo (25). Although the role of miRNA in pathogenesis of chronic wounds has been suggested (26, 27), to the best of our knowledge the functional role of miRNAs has not been determined in patients with chronic wounds.
Here we identified induction of specific set of miRNAs (miR-16, -20a, -21, -106a, -130a, and -203) in VUs and show that this induction contributes to a loss of growth factor signaling and delayed epithelialization and granulation tissue formation in the rat and human wound models leading to inhibition of healing, thus identifying new potential diagnostic and therapeutic targets.
EXPERIMENTAL PROCEDURES
Target Prediction
We conducted miRNA target prediction using miRNA sequences grouped into seed families and filtered for conservation based on a 3′-UTR alignment of five species. miRNAs were grouped into families as defined by identical nucleotides in positions 2–8. We searched for target sites for miRNA families in 3′-UTRs using two different types of seed matches: (i) 7-mers (positions 2–8) and (ii) 7-mer positions 2–7 m1A (A at the first position in the mRNA). For target matches, we considered both nonconserved and conserved targets in human 3′-UTRs. 3′-UTR sequences for human (hg18), mouse (mm8), rat (rn4), dog (canFam2), and chicken (galGal2) were derived from RefSeq and the UCSC genome browser. We used multiple genome alignments across the five species as derived by multiZ. The RefSeq annotation with the longest UTR mapped to a single gene was always used. Finally, as a conservation filter, we required that the 7-mer target site in human be present in at least three of the other four species, that is, exact matching in a 7-nucleotide window of the alignment in at least three other species, to be flagged as conserved.
Kolmogorov-Smirnov Statistic
We identified putatively induced miRNA by comparing the expression changes for miRNA target genes versus all (5148) genes. Specifically, we compared their distributions of log(expression change) values using a one-sided Kolmogorov-Smirnov (KS) statistic, which assesses whether the distribution of expression changes for one set (i.e., the miRNA predicted target genes) is significantly shifted downwards (down-regulated) compared with the distribution for the other set. The KS statistic computes the maximum difference in value of the empirical cumulative distribution functions:
where
 |
is the empirical cumulative distribution function for gene set j = 1, 2, based on nj (Z−transformed) log(expression change) values. We used the Matlab function kstest2 to calculate the KS test statistic and asymptotic p value.
Functional Enrichment of miRNA Targets
We annotated the function of each gene using wound ontology information (see Table 1). We used this annotation to perform our functional enrichment analysis. First, given a set of miRNA targets (as defined earlier), we determined whether any functions were (i) significantly over-represented in the miRNA gene targets and (ii) generally down-regulated. We measured significance by performing a Fisher's exact test on the number of genes annotated by the function also targets to some specific miRNA. We set p < 0.05 as our threshold for significance of miRNA gene target function. Next, we assessed down-regulation of function by calculating the median log expression change of all genes annotated by the function. We set −1 as our threshold as the minimum median (log expression change) for genes annotated by some function. Functional annotations passing both thresholds were reported.
TABLE 1.
High confidence candidate miRNAs predicted to be overexpressed in VUs
Candidate miRNAs consist of human miRNAs that passed all statistical, conservation, and functional filters. Members of the seed family experimentally tested with qPCR are underlined. + denotes positively validated; − denotes a negative result.
| MicroRNA seed family | p value | Functional annotation of target genes | miRNA expression evidence | Validated |
|---|---|---|---|---|
| miR-17–5p/20/93/106/519 | p < 2.65E-11 | Transcription factor | Ref. 23 | + |
| miR-181 | p < 3.70E-11 | Receptor growth factor; transcription factor | Ref. 23 | − |
| miR-21/590–5p | p < 3.47E-07 | Transcription factor | Ref. 23 | + |
| miR-15/16/195/424/497 | p < 5.83E-06 | Receptor growth factor | MCF10A cell line | + |
| miR-130/301 | p < 1.51E-05 | Receptor growth factor; transcription factor | MCF10A cell line | + |
| miR-27 | p < 5.26E-05 | Receptor growth factor; transcription factor | MCF10A cell line | − |
| miR-23 | p < 9.81E-05 | Transcription factor | MCF10A cell line | − |
| miR-203 | p < 0.0002 | Receptor growth factor; transcription factor | Ref. 23 | + |
Skin Specimens
Control healthy skin specimens (n = 5) were obtained as discarded tissue from patients 46–72 years of age undergoing elective plastic surgery. Skin biopsies from consented VU patients were collected during surgical debridement procedures, per institutional review board-approved protocol. Patients (n = 10) 43–83 years of age were debrided in the operating room under monitored or general anesthesia (3). The nonhealing edges used in this study were clinically identified by a surgeon as the most proximal skin edge to the ulcer bed. Skin biopsies were then processed as follows: (i) fixation in 4% paraformaldehyde and embedding in OCT compound (Fisher), (ii) stored in formalin for paraffin embedding, and (iii) stored in RNAlater (Ambion; Applied Biosystems, Carlsbad, CA) for subsequent RNA isolation. The samples were standardized as previously described (28). All of the specimens showed characteristic hyperproliferative, hyperkeratotic, and parakeratotic epidermis and the nuclear presence of β-catenin (29).
Real Time PCR Gene Expression Analysis
MicroRNA real time PCR quantification was performed using TaqMan® MicroRNA Assays (Applied Biosystems) according to the manufacturer's instruction. The IQ5 light cycler system (Bio-Rad) was used for real time PCR. Target miRNA expression was normalized between different samples based on the values of U48 RNA expression. Differences between samples and controls were calculated based on the 2−ΔΔCt method. For quantification of mRNA, 0.5 μg of total RNA from control skin and chronic wounds was reverse transcribed using an Omniscript reverse transcription kit (Qiagen). mRNA real time PCR was performed using iQ SYBR Green Supermix (Bio-Rad). Relative expression was normalized for levels of HPRT1. All of the reactions were performed in triplicate. The primer sequences used were: HPRT1, forward (5′-AAAGGACCCCACGAAGTGTT-3′) and reverse primer (5′-TCAAGGGCATATCCTACAACAA-3′); early growth repose 3 (EGR3), forward (5′-AATGGACATCGGTCTGACCAA-3′) and reverse primer (5′-GGCGAACTTTCCCAAGTAGGT-3′); vinculin (Vcl), forward (5′-ATGGGTCAAGGGGCATCCT-3′) and reverse primer (5′-GGCCCAAGATTCTTTGTGTAAGT-3′); and Lep-R, forward (5′-ACCACACCTCACATTCTCAGA-3′) and reverse primer (5′-GTCAGTCAAAAGCACACCACT-3′). Statistical comparisons of expression levels from chronic wound versus control skin were performed using Student's t test. Statistically significant differences between VUs and control skin controls were defined as p < 0.05.
Western Blot
Protein extraction and Western blot was performed as described previously (30). Antibodies against EGR3 (1:500; Santa Cruz, Santa Cruz, CA), vinculin (1:5000; Abcam, Cambridge, MA), and GAPDH (1:12000; Santa Cruz) were used.
Cell Culture
Human keratinocytes were grown as previously described (30). 24 h prior to transfection, the cells were incubated in basal medium (Invitrogen) (30). Normal human dermal fibroblasts and fibroblasts derived from patients with VUs, AG19641, AG19642, and AG19285 (31) were grown following our previously published protocol (31).
DNA Cloning and Transfection
A 227-bp DNA fragment flanking the pre-miR-21 hairpin was cloned from human genomic DNA with 5′ primer: 5′-CGGGATCCTTATCAAATCCTGCCTGACTG-3′ and 3′ primer: 5′-CCCAAGCTTGACCAGAGTTTCTGATTATAACA-3′, and 328-bp DNA fragment flanking the pre-miR-130a hairpin was cloned from human genomic DNA with 5′ primer: 5′-CGGGATCCGCTGTATTGAAGCAAAGAAGG-3′ and 3′ primer: 5′-CCCAAGCTTGGGTAGCTGACTGGTGCC-3′. The resulting pre-miR-21 and pre-miR-130a fragments were restricted and inserted into the BamHI and HindIII sites of pSilencer 4.1-CMV puro vector (Ambion). LepR 3′-UTR fragment containing miR-130a and miR-21 putative target sites was amplified from human genomic DNA. The primers for the LepR 3′-UTR fragment were 5′ primer: 5′-gctctagaAGTCTAATCATGATCACTACAGATG-3′ and 3′ primer: 5′-gctctagaGAAAAATCCTGCCAAACAACTAC-3′. This fragment was inserted into the XbaI site in the 3′-UTR of pGL3-control plasmid (Promega, Madison, WI) and sequenced. Plasmid containing sense LepR 3′-UTR sequence was used as a reporter (pGL3-LepR 3′S), whereas the plasmid containing the 3′-UTR sequence in the antisense orientation was used as a negative control (pGL3-LepR 3′AS). For dual luciferase assays, pGL3 reporter, pSilencer constructs, and Renilla luciferase control (Promega) were transfected with FuGENE 6 reagent (Roche Applied Science). The relative luciferase activities were determined using the Dual-Glo luciferase assay system (Promega) 48 h after transfection.
Immunohistochemistry
Paraffin sections of human tissue were used for staining with anti-LepR antibody (Abcam). Rat tissue was stained with anti-LepR antibody from Santa Cruz and Vectastain universal kit as previously described (32). Staining with anti-keratin 6 antibody (33) was used for quantification of epithelialization in rat wounds. For visualization, a Nikon Eclipse E800 microscope was used, and digital images were collected using SPOT Camera Advanced program.
Wounding and Treatments with Mimic miRNAs
The experimental animal protocol was approved by the University of Miami Institutional Animal Care and Use Committee. Wounds on 23-day-old Long Evans rats were created using an 8-mm dermal biopsy punch (Acuderm Inc., Fort Lauderdale, FL). Two consecutive days prior to wounding, the animals were injected subcutaneously with 5 μm mimic miR-21 (Dharmacon, Chicago, IL) dissolved in PBS, mimic miRNA negative control (Dharmacon), or PBS. Cy3-labeled miRNA mimic (Ambion) was used to follow the tissue distribution. Six animals/treatment group/time point were used. The size of the closing wound was monitored daily until day 6. Paraffin sections (8 μm) were stained with hematoxylin and eosin to follow the rate of healing. For granulation tissue assessment, sections were stained with Masson's trichrome. Granulation tissue thickness was measured starting from the upper edge of the wound bed descending down to the lower dermis and adipose tissue. Three measurements were taken per three distinct wound areas: left, middle, and right. Granulation tissue formation was quantified by an NIS Elements imaging program (Nikon). Human healthy skin explants (n = 5) were maintained and wounded as described (32, 34). Acute wounds were topically treated at the time of wounding with 5 μm mimic miR-21 and miR-130a (Dharmacon) dissolved in 30% pluronic F-127 gel (Sigma). Frozen sections (7 μm) of ex vivo acute wounds were stained with hematoxylin and eosin to follow the rate of healing. One-way analysis of variance was used to analyze rate of epithelialization and granulation tissue quantification among animal groups; p < 0.05 was considered significant.
In Situ Hybridization (ISH)
In situ transcriptional levels of miRNAs were determined on frozen sections (8 μm) of skin biopsy specimens from six additional patients with chronic VUs and five healthy individuals as described previously (35). The sections were hybridized overnight with digoxygenin-labeled miRCURY LNA probes (Exiqon, Denmark) and incubated with anti-digoxygenin antibody conjugated with alkaline phosphatase. Hybridization with digoxygenin-labeled LNA miRNA-scramble (Exiqon) was used as control.
RESULTS
Computational Analysis of Differential mRNA Expression Predicts Aberrant miRNA Expression in VU
The computational analysis used the rationale that decreased mRNA expression of target genes in VUs may identify miRNAs aberrantly overexpressed in chronic wounds. Previously generated mRNA microarray profiling of VUs (28) was used to screen for human miRNAs whose predicted targets were expressed at significantly lower levels in VUs as compared with normal skin. We took a simple approach of defining the set of putative targets of a miRNA to be all genes containing at least one 7-mer seed match (complementarity to miRNA positions 2–8 or positions 2–7 with nucleotide A across from position 1) in the 3′-UTR, while filtering for site conservation. We assessed whether each the target genes of miRNA were expressed at significantly lower levels, as a distribution, compared with all (∼5000) profiled genes by computing a one-sided Kolmogorov-Smirnov test statistic. We screened 152 human miRNA seed families (miRNA with identical seeds) in this way and took a nominal p value cut-off of p < 0.0002 for our significance threshold (supplemental Table S1).
In addition to looking for significantly lowered miRNA target expression, we applied a set of functional filters for relevance to wound healing. First, for a collection of wound healing-related functional annotations (e.g., receptor growth factor, cell migration and proliferation, and transcriptional regulators), we assessed whether (i) the functional class was over-represented in the miRNA target list and (ii) the median expression change of genes in the functional class was significantly lowered in VUs (see “Experimental Procedures”). Second, we checked whether miRNA profiling evidence from other studies supported the expression of the miRNA in epithelial cells (cell line MFC10A) or in skin (21, 23). Finally, we considered whether the seed class was conserved across vertebrates, because conserved miRNAs may play a more important functional role. The highest confidence predictions from this analysis, i.e., miRNAs that are predicted to be overexpressed in VUs based on the lower target expression and that also pass all additional functional filters, are given in Table 1.
miRNA-16, -20a, -21, -106a, -203, and -130a Are Induced in VUs
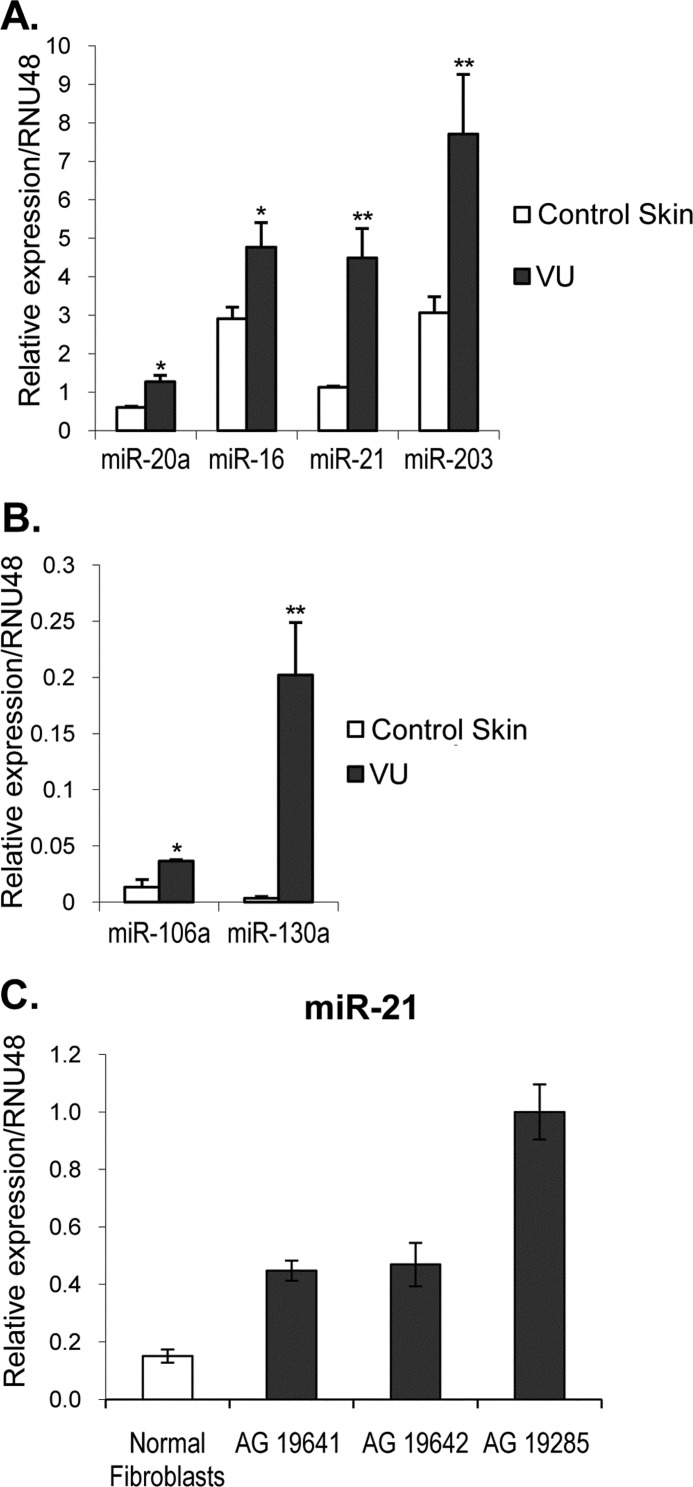
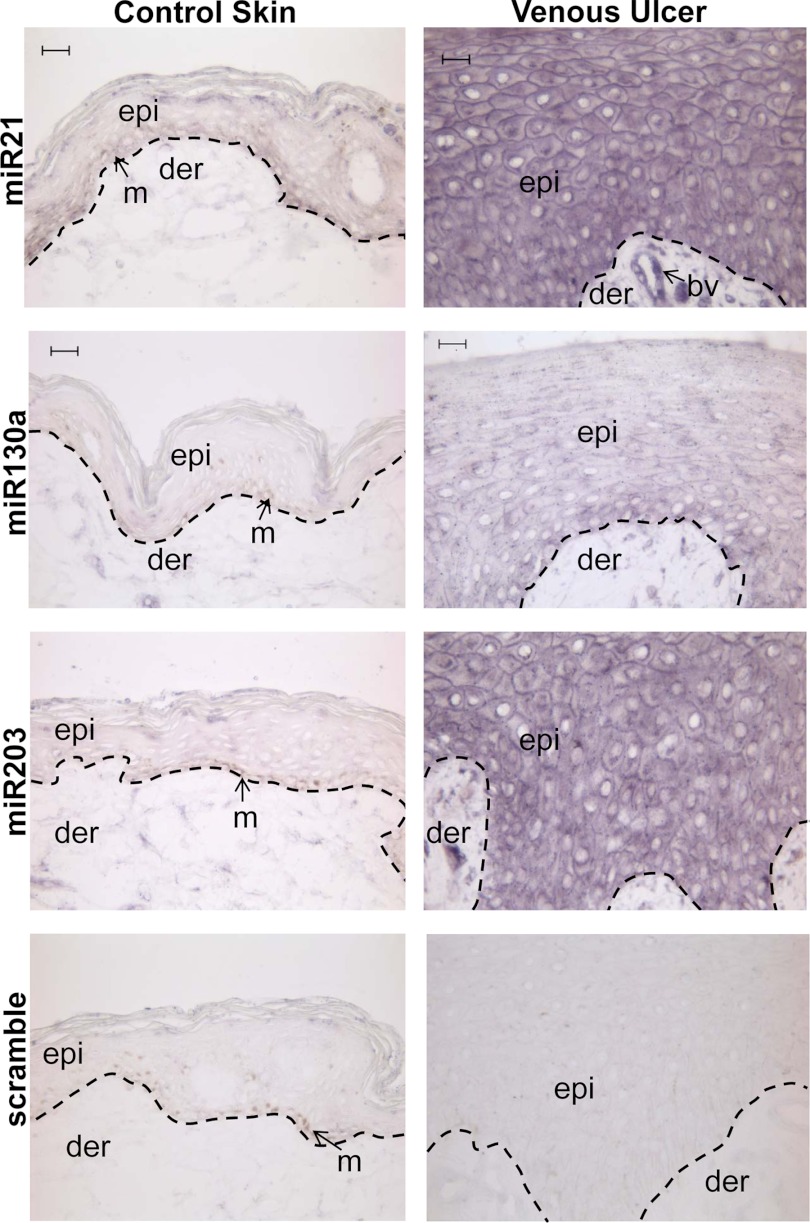
To confirm the results obtained by computational predictions, we performed qPCR quantification of miRNAs selected by statistical analyses. RNA samples were obtained from full thickness skin biopsies after surgical debridement of VUs (n = 10) and age- and gender-matched control skin (n = 5). All of the biopsies were verified for established histological criteria and nuclear presence of pathogenic marker, β-catenin, prior to RNA isolation (29). We used TaqMan primers and probes designed to amplify specifically the mature, active form of 12 miRNAs predicted to be up-regulated in VUs by computational analysis (Table 1). Target miRNA expression was normalized between different samples based on the values of U48 RNA expression. qPCR results showed a significant increase of miR-16, -20a, -21, -203, -106a, and -130a levels in VUs when compared with control skin (Fig. 1, A and B). miR-21 and -203 were previously found to be up-regulated both in psoriasis and atopic eczema (23), suggesting their possible function in hyperproliferative epidermal disorders. The striking overexpression of miR-130a and miR-21 suggests that they may play a leading role in the inhibition of wound healing. We also confirmed the induction of miR-21 in primary human fibroblasts generated from biopsies of VUs (36) (Fig. 1C). Similar to induction of miR-21 in VUs fibroblasts, this miRNA has been found induced in fibroblasts of the failing heart (37).We further showed induction of miR-21, miR-130a, and miR-203 in six additional VUs biopsies using specific LNA probes and ISH (Fig. 2). The specificity of the probes was confirmed by hybridization with an LNA scrambled-miRNA probe (Fig. 2). Increased expression of miR-21 was detected not only in the VUs epidermis but also in the dermis and blood vessels, whereas miR-130a and miR-203 were found be induced mainly in hyperproliferative epidermis of VUs. The intensity of the miR-130a signal detected by ISH was significantly lower than miR-21 (Fig. 2), which correlated with lower relative expression levels of miR-130a obtained by qPCR (Fig. 1B). Brown staining observed in control skin miR hybridizations and hybridizations with a scramble miRNA probe originates from melanin pigment and is not a hybridization signal. We did not observe any melanin in VUs, suggesting a lack of melanocytes in the nonhealing epidermis.
FIGURE 1.
Specific miRNAs are induced in VUs. A and B, miR-20a, -16, -21, -203, -106a, and -130a are up-regulated in VUs in comparison with control healthy skin measured by qPCR. The error bars indicate the means ± S.E. *, p < 0.05; **, p < 0.01. C, miR-21 is up-regulated in primary fibroblasts from VUs. miR-21 is induced in AG19641, AG19642, and AG19285 fibroblasts (31) derived from VUs.
FIGURE 2.
ISH confirmed up-regulation of miR-21, miR-130a, and miR-203 in VUs. ISH with miR-21 LNA probe shows induced levels in epidermis (epi), dermis (der) and blood vessels (bv) of VUs in comparison with control skin. miR-130a and miR-203 are predominantly induced in hyperproliferative epidermis (epi) of VUs. Black dashed lines indicate the dermal-epidermal boundary. Hybridization with digoxygenin-labeled LNA miRNA-scramble was used as control. m, melanin. Brown staining observed in normal skin and control hybridizations with scramble probes corresponds to melanin granules. Scale bars, 20 μm.
VU-miRNAs Target Multiple Genes/Pathways That Control Wound Healing
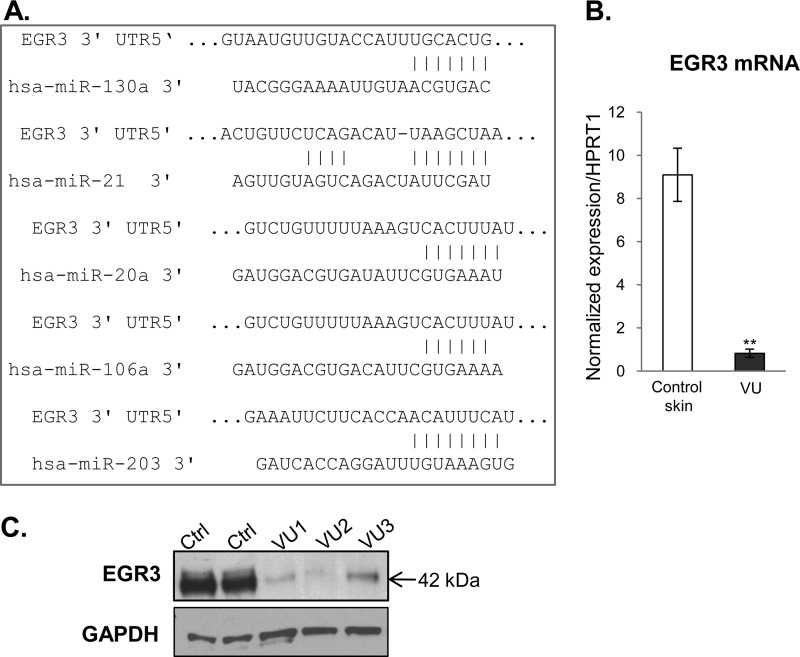
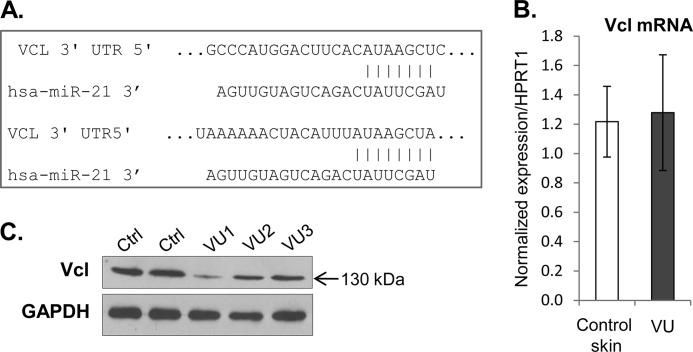
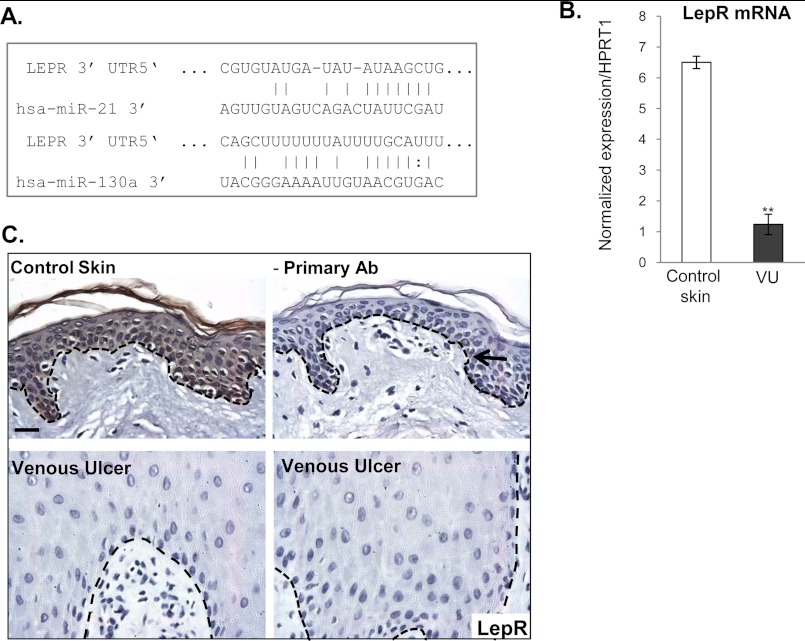
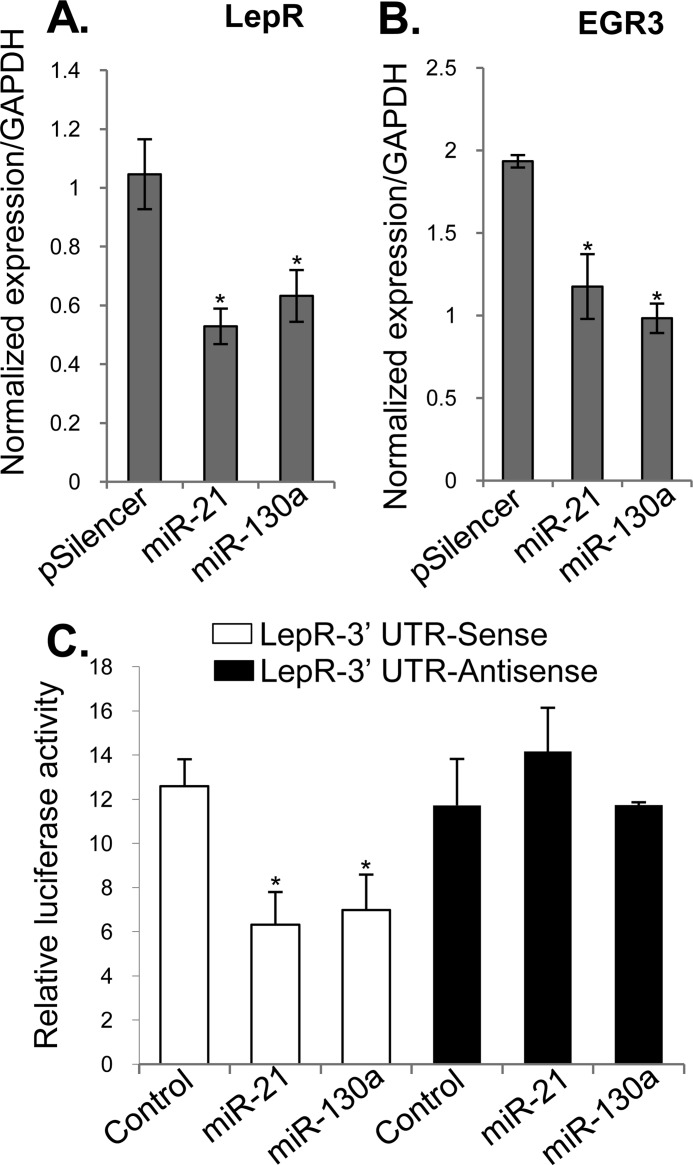
Because our computational analyses approach was to start from downstream targets (genes already found to be deregulated in VUs (28) to identify miRNAs that may target their expression, it was expected that the VU-miRNAs would target multiple cellular processes important for wound healing. Nevertheless, we proceeded to confirm their function in vitro and in vivo. Our computational and qPCR data suggest that miRNAs overexpressed in VUs could be classified on the basis of their predicted targets rather than by their genomic location, e.g., five miRNAs found to be induced in VUs are predicted to target 3′-UTR of EGR3 (Fig. 3A). In support of this, we confirmed down-regulation of EGR3 in VUs at both mRNA and protein levels (Fig. 3, B and C). EGR3 has a regulatory effect on the cellular decision toward keratinocyte migration (38) and acts as one of the first responses upon activation of EGF signaling (39). In addition to inhibition of EGR3, we show suppression of Vcl (Fig. 4), the main component of the focal adhesion complex (40) and a predicted target for miR-21 (Fig. 4A). Vcl mRNA levels were not significantly altered in contrast to the observed down-regulation at the protein level, suggesting the role of miR-21 in translational inhibition of Vcl in VUs. Strong suppression of EGR3 and vinculin by miRNAs coupled with previously shown abnormal, cytoplasmic localization of EGFR (36) may lead to absence of keratinocyte migration, a hallmark of VU (29). In addition, the heptamer and hexamer matches to miR-21 and miR-130a 5′ seed sequences, respectively, were identified within the 3′-UTR of LepR mRNA (Fig. 5A). Expression of LepR was suppressed in VUs (Fig. 5B). Immunohistochemistry confirmed down-regulation of LepR in hyperproliferative epidermis of all tested chronic VUs, in contrast to strong LepR expression throughout the epidermis of control skin (Fig. 5C). The effects of miR-21 and miR-130a, the most induced miRNAs, on the regulation of LepR and EGR3 were further examined by assessing mRNA levels in primary human keratinocytes transfected with pre-miR-21 and pre-miR130a expressed from pSilencer vector. qPCR analysis showed significantly reduced levels of LepR and EGR3 mRNA in miR-21- and miR-130a-expressing cells in comparison with control, pSilencer transfected cells (Fig. 6, A and B).
FIGURE 3.
miRNAs induced in VUs regulate EGR3. A, sequence alignment of miRNAs overexpressed in VUs and 3′-UTR of EGR3. Five miRNAs found to be induced in VUs can target 3′-UTR of EGR3. B, mRNA levels of EGR3 are suppressed in VUs (n = 10) in comparison with control skin (n = 5). mRNA levels of EGR3 measured by qPCR in triplicate; the error bars indicate the means ± S.E. **, p < 0.01. C, representative Western blot showing suppression of EGR3 in VUs in comparison with control skin (Ctrl).
FIGURE 4.
miR-21 targets Vcl. A, sequence alignment of miR-21 and 3′-UTR of Vcl. 3′-UTR of Vcl has two target sites for miR-21. B, mRNA Vcl levels were not down-regulated in VUs (n = 10). mRNA levels of Vcl measured by qPCR in triplicate; the error bars indicate the means ± S.E. C, Vcl protein levels were suppressed in VUs in comparison with control skin (Ctrl) showed by representative immunoblotting.
FIGURE 5.
LepR, predicted target of miR-21 and miR-130a, is down-regulated in VUs. A, sequence alignment of miR-21, miR-130a, and 3′-UTR of LepR. B, mRNA levels of LepR measured by qPCR in VUs (n = 10) and control skin (n = 5); all reactions were performed in triplicate; the error bars indicate the means ± S.E. **, p < 0.01. C, immunohistochemistry shows LepR (brown) in basal and suprabasal layers of control skin. In contrast, no LepR staining was observed in hyperproliferative epidermis of VUs. A black arrow indicates melanin detected as a background signal in the control staining without primary antibody. Hematoxylin (blue) was used as a counterstain for nuclei. The black dashed lines indicate the dermal-epidermal boundary. Scale bar, 50 μm.
FIGURE 6.
miR-21 and 130a target LepR and EGR3 in primary human keratinocytes. A and B, primary human keratinocytes were transfected with miR-21 or miR-130a expressed from pSilencer vector, and total RNA was isolated 24 h after transfection. The qPCR results show that miR-21 and miR-130a overexpression leads to suppression of LepR (A) and EGR3 (B) on mRNA level. C, insertion of miR-21 and miR-130a target sequences in the sense orientation (white bars) within LepR 3′-UTR leads to diminished luciferase reporter activity in the presence of miR-21 or miR-130a expressed from pSilencer vector. Control, pSilencer vector without miRNAs. Antisense orientation (black bars) of LepR 3′-UTR abolishes miR-21- and miR-130a-mediated repression of luciferase activity. Luciferase activity was normalized to Renilla luciferase. The error bars (S.D.) are derived from the three replicates of three independent transfection experiments. *, p < 0.05.
To further determine whether LepR is a direct target of miR-21 and miR-130a, we used a luciferase reporter assay. When introduced into the 3′-UTR of a luciferase reporter gene, a 2259-base pair human LepR 3′-UTR fragment encompassing these two putative target sites (Fig. 5A) caused a reduction in activity in miR-21- and miR-130a-expressing keratinocytes (Fig. 6C). miR-21 and miR-130a did not significantly inhibit luciferase activity in the control with the LepR 3′-UTR in the antisense orientation (Fig. 6C). Together, these results confirmed that miR-21 and miR-130a have a direct effect on LepR, mediated through 3′-UTR target sites. Leptin is a hypoxia-inducible pleiotropic cytokine indispensible for successful wound healing: leptin null (ob/ob) and leptin receptor null (db/db) mouse strains have severe impairment in cutaneous wound healing (41–43). Conversely, systemic and topical application of leptin improves re-epithelialization (44). Our data identified LepR as a novel target for miR-21 and miR-130a, implicating their role in the inhibition of wound healing and pathology of VUs.
miR-21 and miR-130a Inhibit Epithelialization of Human Skin
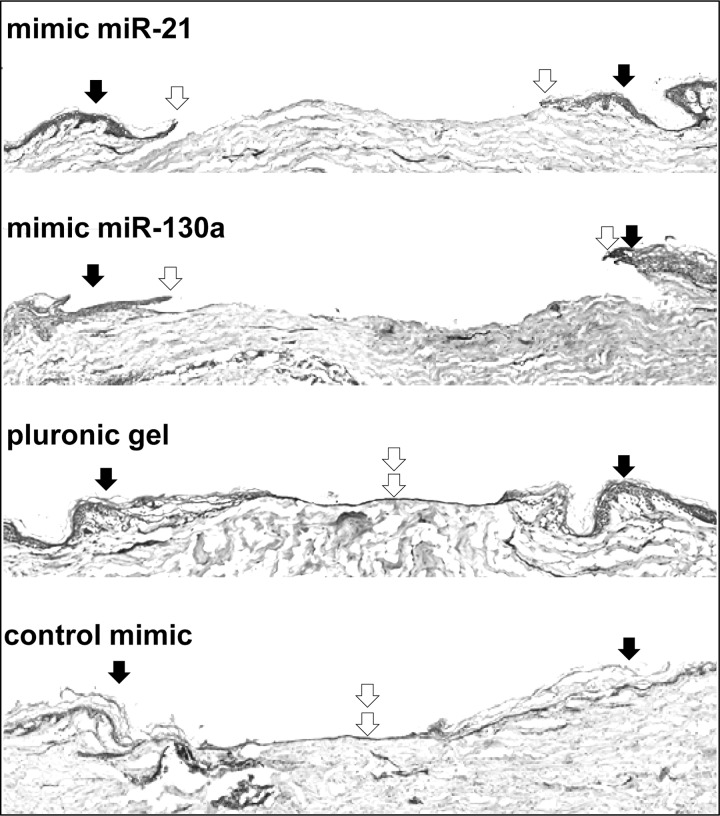
To functionally evaluate miR-21 and miR-130a as two of the most induced miRNAs in VUs, we utilized an established human skin organ culture wound model (32, 34). Healthy human skin was wounded using 3-mm punch biopsy, and the tissue was treated with synthetic mimic miR-21 and miR-130a or control scramble mimic dissolved in pluronic gel (Fig. 7). Pluronic gel has been previously described and used as a vehicle for delivery of synthetic oligonucleotides to wound tissue (45). Four days after the treatment, organ cultures were processed and stained with hematoxilin and eosin for the evaluation of epithelialization. Efficient mimic incorporation into the epidermis and dermis of human skin explants was documented by application of a dye-conjugated mimic (supplemental Fig. S1). A single topical application of mimic miR-21 and mimic miR-130a markedly inhibited epithelialization, i.e., wound edges remained almost at the same initial position 4 days after the treatment, whereas neither negative control mimic nor the vehicle, pluronic gel, showed any effect (Fig. 7). The experiments were repeated in triplicate using skin obtained from three different donors (Fig. 7). We concluded that miR-21 and miR-130a overexpression by synthetic mimics inhibits epithelialization during acute wound healing process ex vivo.
FIGURE 7.
miR-21 and miR-130a inhibit epithelialization in human ex vivo wound model. miR-21 and miR-130a treatment completely inhibited epithelialization when compared with control mimic and pluronic gel (vehicle) treatments in a human skin organ culture model. Black arrows indicate wound edges after initial wounding, whereas open arrows point at the epithelialized edges of the migrating fronts 4 days after wounding. Representative ex vivo wounds stained with hematoxylin and eosin from five independent experiments are shown.
miR-21 Delays Wound Healing in Vivo
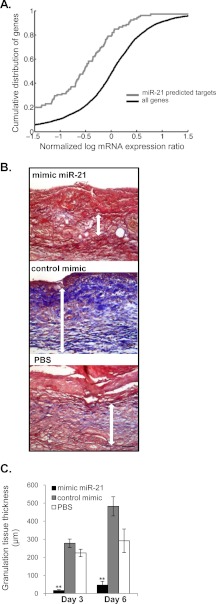
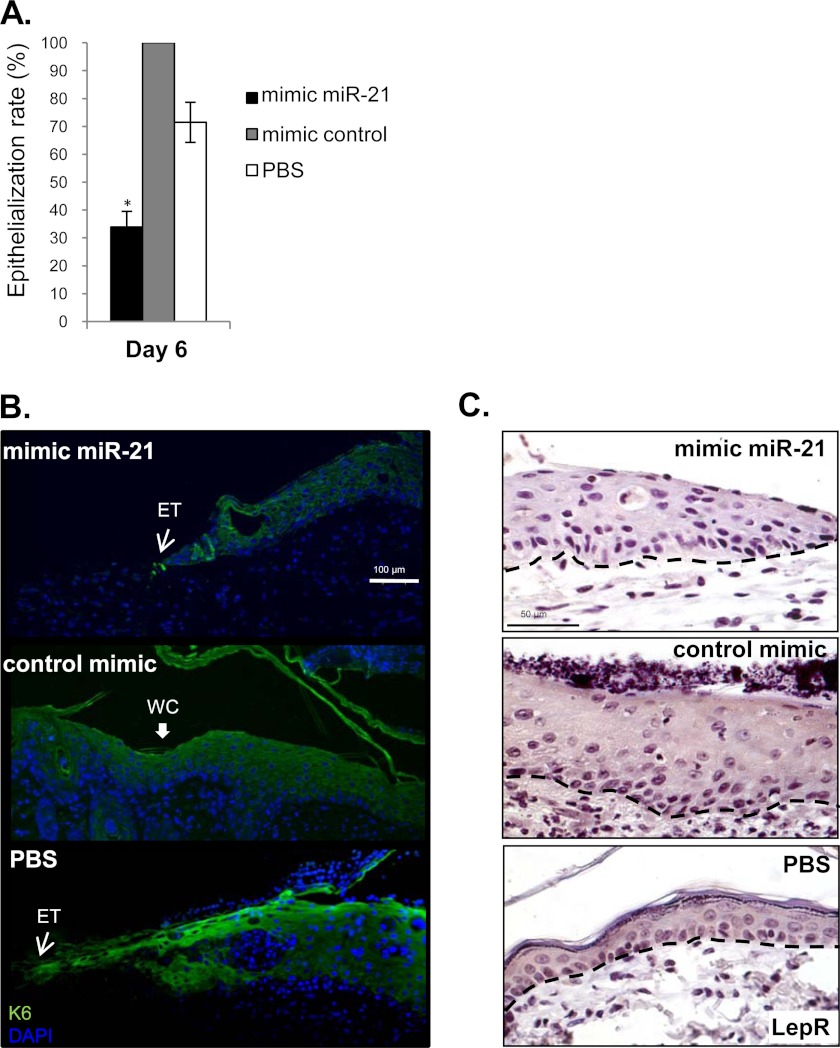
Next we examined biological function of miR-21 in vivo using the rat acute wound healing model. In addition to the highest expression levels of this miRNA (Figs. 1A and 2), the predicted targets of miR-21 were significantly repressed relative to all mRNA expression changes between control skin and VU samples as determined by a KS test (Fig. 8A). To ensure sustained overexpression of miR-21 in vivo, synthetic mimic was injected intradermally 2 consecutive days prior to wounding. Full thickness 8.0-mm excisional wounds were created on each side of the dorsal midline with a punch biopsy. The animals were sacrificed at days 3 and 6 post-wounding, and the tissues were processed for histology and immunohistochemistry. Mimic miR-21-treated wounds show striking reduction in granulation tissue formation at day 3 post-wounding in comparison with mimic control and PBS treatment as evident by almost complete absence of trichrome stain for collagen (Fig. 8, B and C). Significant reduction of granulation tissue formation was also observed at day 6 post-wounding (Fig. 8C). Mimic miR-21 treatment caused increased infiltration of immune cells, whereas inflammation has already ceased in control wounds at day 3 post-wounding (Fig. 8B). Furthermore mimic miR-21 had an inhibitory effect on epithelialization, whereas control (scramble mimic) treated wounds were fully closed at day 6 post-wounding (Fig. 9, A and B). Staining with keratin 6 (K6)-specific antibody revealed reduced epithelial thickness and confirmed delayed epithelialization in mimic miR-21-treated wounds (Fig. 9B). Lastly, LepR was suppressed in epidermal edges of miR-21-treated wounds, confirming LepR as a target for miR-21 in vivo (Fig. 9C). In summary, overexpression of miR-21 not only inhibited granulation tissue formation but also affected epithelialization and prolonged the inflammation, turning an acute wound into a nonhealing wound.
FIGURE 8.
miR-21 inhibits granulation tissue formation wound healing in rat wound model. A, empirical cumulative distribution function for miR-21 target genes. Predicted targets of miR-21 (105 genes) are significantly repressed relative to all mRNA expression changes between control skin and VU samples (p < 3.47E-07; KS test). B, mimic miR-21 inhibited granulation tissue formation and wound healing in rat acute wounds in comparison with control mimic and PBS treatments. Histological sections of rat wounds 3 days after wounding were investigated using trichrome stain (blue) to detect collagen; two-sided white arrows illustrate representative granulation tissue thickness used for measurements and quantification. Abundant granulation tissue was observed in the control mimic and PBS-treated wounds. Only minor granulation tissue was observed in mimic miR-21-treated wounds. Scale bar, 50 μm. C, quantification of wound granulation tissue thickness at days 3 and 6 post-wounding is shown. Granulation tissue thickness was determined based on positive trichrome stain (blue); three measurements were taken per three distinct areas (left, middle, and right) for each wound. The data are the means and S.E. **, p < 0.01, n = 6 animals/experiment/group.
FIGURE 9.
miR-21 treatment delays epithelialization in vivo. A, graph represents quantification of epithelialization using planimetry 6 days after wounding. Mimic miR-21 treatment delayed wound closure in comparison with control mimic and PBS treatments in full thickness rat acute wounds. *, p < 0.05. B, immunolocalization of keratin 6 (K6, green) marker of wound healing at the wound edge on day 6 post-wounding. Mimic miR-21 inhibits epithelialization and decreases thickness of the epithelial tongue (ET). WC, wound closure. Scale bar, 100 μm. C, miR-21 overexpression by mimics in rat wounds suppresses LepR; immunolocalization of LepR at the wound edge at day 6 post-wounding. The black dashed lines indicate the dermal-epidermal boundary. Scale bar, 50 μm.
DISCUSSION
We herein provide, to the best of our knowledge, the first evidence that aberrantly expressed miRNAs play a critical role in the pathology of nonhealing VUs. Computational analyses of mRNA expression profiles identified miRNAs overexpressed in VUs, which was further confirmed in a cohort study of 10 patients. Induction of six specific miRNAs in VUs suggests that these regulators may also play different and cell type-specific roles in pathogenesis of nonhealing VUs.
Contiguous and perfect base pairing of the miRNA nucleotides 2–8 is the most stringent requirement for efficient target recognition (46), and our computational analysis confirmed that decreased mRNA expression of target genes in VUs can identify miRNAs that are aberrantly overexpressed in chronic wounds. Using TaqMan qPCR, we showed that a small subset of miRNAs identified by a bioinformatics approach is significantly induced in 10 nonhealing VUs (miR-16, -20a, -21, -203, -106a, and -130a). In addition, miR-21, -130a, and -203 up-regulation was confirmed by ISH in biopsies originated from six additional patients, therefore underscoring the importance of these miRNAs in VU pathogenesis. Because VU is a complex condition involving changes in both epidermis and dermis coupled with prolonged inflammation, aberrant regulation of this subset of miRNAs may contribute to multiple aspects of chronic wound disorders. In addition, this method provides an alternative, cost effective approach to identification of miRNAs by using mRNA microarray data.
Our in vitro and in vivo studies were focused on deciphering the impact of miRNAs up-regulated in VUs on growth factor signaling, because the growth factory therapy approach to chronic wounds did not achieve the expected outcomes in clinic (47). Using primary human keratinocytes overexpressing miR-21 and miR-130a, we provide evidence that aberrant expression of these miRNAs leads to inhibition of EGF pathway through EGR3, implicating their role in pathology of VUs. Overexpression of miR-21 and miR-130a in VUs may be the underlying factor for failure of exogenous EGF to accelerate healing in chronic wounds. Suppression of EGR3 and vinculin by VU-specific miRNAs can also contribute to inhibition of keratinocytes migration at the nonhealing wound edge. We recently reported attenuation of another wound healing relevant pathway, TGFβ signaling, which is markedly suppressed in VUs epidermis (32). Interestingly, mRNA levels of TGFβRII were not significantly altered in contrast to the marked down-regulation at the protein level, suggesting the potential role of miRNAs in translational inhibition. Indeed, TGFβRII has been recently confirmed as target for miR-20a in human keratinocytes (48), suggesting that induction of this miRNA in VUs can be responsible for suppression of TGFβ signaling. Further studies are needed to confirm the role of miR-20a in wound healing; however, induction of miR-20a can potentially provide the explanation for limited success of TGFβ in clinical trials, in addition to promising animal studies (7).
Leptin signaling is also well recognized for its stimulative pleiotropic effects on wound healing (44, 49, 50). Although expressed in keratinocytes, fibroblasts, and endothelial cells, LepR expression in epidermis is of crucial importance for successful wound healing (51). Our study identified and confirmed LepR as a novel target for the most induced miR-21 and miR-130a in epidermis, suggesting that exogenous leptin would not improve healing of chronic VUs. We further deciphered the role of miR-21 and miR-130a in primary keratinocytes, human skin, and rat wound models. Overexpression of both miR-21 and miR-130a in human skin organ culture model had an inhibitory effect on epithelialization. Furthermore treatment with synthetic miR-21 inhibited epithelialization and granulation tissue formation in vivo, while inducing prolonged inflammation, therefore turning acute into nonhealing wounds (Fig. 10). miR-21 may represent a common marker of chronic inflammation because it has been reported to be induced in many inflammatory states, including osteoarthritis (52), psoriasis (53), allergic airway inflammation (54), active ulcerative colitis tissue (55), inflammatory response to LPS (56), and cardiac muscle injury (57). Overexpression of miR-21 has also been reported in many types of cancer and linked to a loss of cell cycle control and enhanced proliferation (26, 58–61). We have previously reported induction of c-Myc in hyperproliferative nonmigratory epidermis of VUs (29). miR-21 inhibition was shown to reduce expression of c-Myc in breast cancer cells (61), suggesting that suppression of miR-21 in VUs could lead to reduced proliferation and restored migration of chronic wound keratinocytes. A recent study using the HaCaT cell line has indicated the opposite role of miR-21, suggesting a beneficial role for this miRNA in wound healing (62). Suppression of miR-21 by antisense oligonucleotides led to inhibition of migration and did not affect keratinocytes proliferation in vitro and in vivo (62). However, contradictory results obtained from studies using different approaches to inhibit miR-21 in other tissues such as cardiac suggest that caution is needed when interpreting results using antisense oligonucleotides or antagomirs (63). Overall our results implicate the role of miR-21 in the epidermal hyperproliferation and fibroblast dysfunction of the chronic VUs.
FIGURE 10.
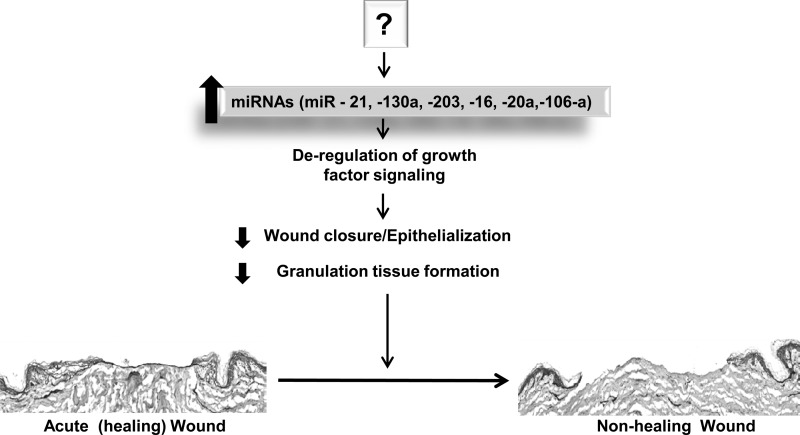
Induction of specific miRNAs inhibits wound healing. Summary figure showing the role of miRNAs in pathology of nonhealing wounds. Induction of miR-21, -130a, -16, -203, -20a, and -106a by yet unidentified mechanism results in suppression of epithelialization and granulation tissue formation, turning an acute wound into chronic wound.
Taken together, our data identify a set of miRNAs that play a role in pathology of VUs by targeting multiple signaling pathways, including leptin and EGF signaling, thus contributing to a lack of keratinocyte migration and deregulation of granulation tissue formation. Furthermore, treatment with miR-21 and miR-130a mimic ex vivo, and miR-21 in vivo replicates the phenotype found in VU patients, providing functional confirmation that overexpression of these miRNAs inhibits wound healing. Therapeutic targeting of VU-specific miRNAs may affect multiple pathways associated with VUs pathology to stimulate wound healing. However, tissue and cell specificity of the action of miRNA along with simultaneous targeting of large number of mRNAs must be studied in detail to confirm beneficial effects. Compared with currently used therapies, the possibility to regulate multiple targets by miRNAs may have a potential to more efficiently achieve wound closure. Our results support the notion that VU-specific miRNAs may serve as a new class of diagnostic and potentially therapeutic targets.
Acknowledgments
We are grateful to S. V. Jackson, L. M. Mauro, and K. A. Gordon for the technical support and Dr. Pierre Coulombe for keratin 6 antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants NR013881 (to M. T.-C.) and ULIRR024996 Clinical and Translational Science Center pilot grant (to M. T.-C.) and Grant 1-U24-CA143840 (to C. L.). This work was also supported by Starr Cancer Consortium Grant I4-A411 (to C. L.).

This article contains supplemental Table S1 and Fig. S1.
- VU
- venous ulcer
- qPCR
- quantitative PCR
- EGR
- early growth response factor
- LepR
- leptin receptor
- miRNA
- microRNA
- KS
- Kolmogorov-Smirnov
- Vcl
- vinculin
- ISH
- in situ hybridization
- LNA
- locked nucleic acid.
REFERENCES
- 1. Herschthal J., Kirsner R. S. (2008) Current management of venous ulcers. An evidence-based review. Surg. Technol. Int. 17, 77–83 [PubMed] [Google Scholar]
- 2. Sen C. K., Gordillo G. M., Roy S., Kirsner R., Lambert L., Hunt T. K., Gottrup F., Gurtner G. C., Longaker M. T. (2009) Human skin wounds. A major and snowballing threat to public health and the economy. Wound Repair Regen. 17, 763–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harsha A., Stojadinovic O., Brem H., Sehara-Fujisawa A., Wewer U., Loomis C. A., Blobel C. P., Tomic-Canic M. (2008) ADAM12. A potential target for the treatment of chronic wounds. J. Mol. Med. 86, 961–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brem H., Tomic-Canic M. (2007) Cellular and molecular basis of wound healing in diabetes. J. Clin. Invest. 117, 1219–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Robson M. C., Phillip L. G., Cooper D. M., Lyle W. G., Robson L. E., Odom L., Hill D. P., Hanham A. F., Ksander G. A. (1995) Safety and effect of transforming growth factor-β2 for treatment of venous stasis ulcers. Wound Repair Regen. 3, 157–167 [DOI] [PubMed] [Google Scholar]
- 6. Bennett S. P., Griffiths G. D., Schor A. M., Leese G. P., Schor S. L. (2003) Growth factors in the treatment of diabetic foot ulcers. Br. J. Surg. 90, 133–146 [DOI] [PubMed] [Google Scholar]
- 7. Mustoe T. A., Pierce G. F., Thomason A., Gramates P., Sporn M. B., Deuel T. F. (1987) Accelerated healing of incisional wounds in rats induced by transforming growth factor-β. Science 237, 1333–1336 [DOI] [PubMed] [Google Scholar]
- 8. Sporn M. B., Roberts A. B. (1993) A major advance in the use of growth factors to enhance wound healing. J. Clin. Invest. 92, 2565–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ambros V. (2004) The functions of animal microRNAs. Nature 431, 350–355 [DOI] [PubMed] [Google Scholar]
- 10. Bartel D. P. (2004) MicroRNAs. Genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 11. Kim V. N. (2005) MicroRNA biogenesis. Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 6, 376–385 [DOI] [PubMed] [Google Scholar]
- 12. Lewis B. P., Burge C. B., Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 13. Bartel D. P. (2009) MicroRNAs. Target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Croce C. M., Calin G. A. (2005) miRNAs, cancer, and stem cell division. Cell 122, 6–7 [DOI] [PubMed] [Google Scholar]
- 15. Kloosterman W. P., Plasterk R. H. (2006) The diverse functions of microRNAs in animal development and disease. Dev. Cell 11, 441–450 [DOI] [PubMed] [Google Scholar]
- 16. Pillai R. S., Bhattacharyya S. N., Filipowicz W. (2007) Repression of protein synthesis by miRNAs. How many mechanisms? Trends Cell Biol. 17, 118–126 [DOI] [PubMed] [Google Scholar]
- 17. Voorhoeve P. M., le Sage C., Schrier M., Gillis A. J., Stoop H., Nagel R., Liu Y. P., van Duijse J., Drost J., Griekspoor A., Zlotorynski E., Yabuta N., De Vita G., Nojima H., Looijenga L. H., Agami R. (2006) A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124, 1169–1181 [DOI] [PubMed] [Google Scholar]
- 18. Baek D., Villén J., Shin C., Camargo F. D., Gygi S. P., Bartel D. P. (2008) The impact of microRNAs on protein output. Nature 455, 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 [DOI] [PubMed] [Google Scholar]
- 20. Andl T., Murchison E. P., Liu F., Zhang Y., Yunta-Gonzalez M., Tobias J. W., Andl C. D., Seykora J. T., Hannon G. J., Millar S. E. (2006) The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr. Biol. 16, 1041–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yi R., Pasolli H. A., Landthaler M., Hafner M., Ojo T., Sheridan R., Sander C., O'Carroll D., Stoffel M., Tuschl T., Fuchs E. (2009) DGCR8-dependent microRNA biogenesis is essential for skin development. Proc. Natl. Acad. Sci. U.S.A. 106, 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yi R., O'Carroll D., Pasolli H. A., Zhang Z., Dietrich F. S., Tarakhovsky A., Fuchs E. (2006) Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 38, 356–362 [DOI] [PubMed] [Google Scholar]
- 23. Sonkoly E., Wei T., Janson P. C., Sääf A., Lundeberg L., Tengvall-Linder M., Norstedt G., Alenius H., Homey B., Scheynius A., Ståhle M., Pivarcsi A. (2007) MicroRNAs. Novel regulators involved in the pathogenesis of psoriasis? PLoS One 2, e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Biswas S., Roy S., Banerjee J., Hussain S. R., Khanna S., Meenakshisundaram G., Kuppusamy P., Friedman A., Sen C. K. (2010) Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc. Natl. Acad. Sci. U.S.A. 107, 6976–6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bertero T., Gastaldi C., Bourget-Ponzio I., Imbert V., Loubat A., Selva E., Busca R., Mari B., Hofman P., Barbry P., Meneguzzi G., Ponzio G., Rezzonico R. (2011) miR-483–3p controls proliferation in wounded epithelial cells. FASEB J. 25, 3092–3105 [DOI] [PubMed] [Google Scholar]
- 26. Pastar I., Ramirez H., Stojadinovic O., Brem H., Kirsner R. S., Tomic-Canic M. (2012) Micro-RNAs. New regulators of wound healing. Surg. Technol. Int. XXI, 51–60 [PubMed] [Google Scholar]
- 27. Shilo S., Roy S., Khanna S., Sen C. K. (2007) MicroRNA in cutaneous wound healing. A new paradigm. DNA Cell Biol. 26, 227–237 [DOI] [PubMed] [Google Scholar]
- 28. Stojadinovic O., Pastar I., Vukelic S., Mahoney M. G., Brennan D., Krzyzanowska A., Golinko M., Brem H., Tomic-Canic M. (2008) Deregulation of keratinocyte differentiation and activation. A hallmark of venous ulcers. J. Cell Mol. Med. 12, 2675–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stojadinovic O., Brem H., Vouthounis C., Lee B., Fallon J., Stallcup M., Merchant A., Galiano R. D., Tomic-Canic M. (2005) Molecular pathogenesis of chronic wounds. The role of β-catenin and c-myc in the inhibition of epithelialization and wound healing. Am. J. Pathol. 167, 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vukelic S., Stojadinovic O., Pastar I., Vouthounis C., Krzyzanowska A., Das S., Samuels H. H., Tomic-Canic M. (2010) Farnesyl pyrophosphate inhibits epithelialization and wound healing through the glucocorticoid receptor. J. Biol. Chem. 285, 1980–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brem H., Golinko M. S., Stojadinovic O., Kodra A., Diegelmann R. F., Vukelic S., Entero H., Coppock D. L., Tomic-Canic M. (2008) Primary cultured fibroblasts derived from patients with chronic wounds. A methodology to produce human cell lines and test putative growth factor therapy such as GMCSF. J. Transl. Med. 6, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pastar I., Stojadinovic O., Krzyzanowska A., Barrientos S., Stuelten C., Zimmerman K., Blumenberg M., Brem H., Tomic-Canic M. (2010) Attenuation of the transforming growth factor β-signaling pathway in chronic venous ulcers. Mol. Med. 16, 92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Takahashi K., Paladini R. D., Coulombe P. A. (1995) Cloning and characterization of multiple human genes and cDNAs encoding highly related type II keratin 6 isoforms. J. Biol. Chem. 270, 18581–18592 [DOI] [PubMed] [Google Scholar]
- 34. Tomic-Canic M., Mamber S. W., Stojadinovic O., Lee B., Radoja N., McMichael J. (2007) Streptolysin O enhances keratinocyte migration and proliferation and promotes skin organ culture wound healing in vitro. Wound Repair Regen. 15, 71–79 [DOI] [PubMed] [Google Scholar]
- 35. Obernosterer G., Martinez J., Alenius M. (2007) Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat. Protoc. 2, 1508–1514 [DOI] [PubMed] [Google Scholar]
- 36. Brem H., Stojadinovic O., Diegelmann R. F., Entero H., Lee B., Pastar I., Golinko M., Rosenberg H., Tomic-Canic M. (2007) Molecular markers in patients with chronic wounds to guide surgical debridement. Mol. Med. 13, 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thum T., Gross C., Fiedler J., Fischer T., Kissler S., Bussen M., Galuppo P., Just S., Rottbauer W., Frantz S., Castoldi M., Soutschek J., Koteliansky V., Rosenwald A., Basson M. A., Licht J. D., Pena J. T., Rouhanifard S. H., Muckenthaler M. U., Tuschl T., Martin G. R., Bauersachs J., Engelhardt S. (2008) MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456, 980–984 [DOI] [PubMed] [Google Scholar]
- 38. Busch H., Camacho-Trullio D., Rogon Z., Breuhahn K., Angel P., Eils R., Szabowski A. (2008) Gene network dynamics controlling keratinocyte migration. Mol. Syst. Biol. 4, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kasper M., Schnidar H., Neill G. W., Hanneder M., Klingler S., Blaas L., Schmid C., Hauser-Kronberger C., Regl G., Philpott M. P., Aberger F. (2006) Selective modulation of Hedgehog/GLI target gene expression by epidermal growth factor signaling in human keratinocytes. Mol. Cell Biol. 26, 6283–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Burridge K., Fath K. (1989) Focal contacts. Transmembrane links between the extracellular matrix and the cytoskeleton. Bioessays 10, 104–108 [DOI] [PubMed] [Google Scholar]
- 41. Greenhalgh D. G., Sprugel K. H., Murray M. J., Ross R. (1990) PDGF and FGF stimulate wound healing in the genetically diabetic mouse. Am. J. Pathol. 136, 1235–1246 [PMC free article] [PubMed] [Google Scholar]
- 42. Tsuboi R., Shi C. M., Rifkin D. B., Ogawa H. (1992) A wound healing model using healing-impaired diabetic mice. J. Dermatol. 19, 673–675 [DOI] [PubMed] [Google Scholar]
- 43. Brem H., Tomic-Canic M., Entero H., Hanflik A. M., Wang V. M., Fallon J. T., Ehrlich H. P. (2007) The synergism of age and db/db genotype impairs wound healing. Exp. Gerontol. 42, 523–531 [DOI] [PubMed] [Google Scholar]
- 44. Frank S., Stallmeyer B., Kämpfer H., Kolb N., Pfeilschifter J. (2000) Leptin enhances wound re-epithelialization and constitutes a direct function of leptin in skin repair. J. Clin. Invest. 106, 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mori R., Shaw T. J., Martin P. (2008) Molecular mechanisms linking wound inflammation and fibrosis. Knockdown of osteopontin leads to rapid repair and reduced scarring. J. Exp. Med. 205, 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Khan A. A., Betel D., Miller M. L., Sander C., Leslie C. S., Marks D. S. (2009) Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 27, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Barrientos S., Stojadinovic O., Golinko M. S., Brem H., Tomic-Canic M. (2008) Growth factors and cytokines in wound healing. Wound Repair Regen. 16, 585–601 [DOI] [PubMed] [Google Scholar]
- 48. Schultz N., Marenstein D. R., De Angelis D. A., Wang W. Q., Nelander S., Jacobsen A., Marks D. S., Massagué J., Sander C. (2011) Off-target effects dominate a large-scale RNAi screen for modulators of the TGF-β pathway and reveal microRNA regulation of TGFBR2. Silence 2, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stallmeyer B., Kämpfer H., Podda M., Kaufmann R., Pfeilschifter J., Frank S. (2001) A novel keratinocyte mitogen. Regulation of leptin and its functional receptor in skin repair. J. Invest. Dermatol. 117, 98–105 [DOI] [PubMed] [Google Scholar]
- 50. Murad A., Nath A. K., Cha S. T., Demir E., Flores-Riveros J., Sierra-Honigmann M. R. (2003) Leptin is an autocrine/paracrine regulator of wound healing. FASEB J. 17, 1895–1897 [DOI] [PubMed] [Google Scholar]
- 51. Larcher F., Del Rio M., Serrano F., Segovia J. C., Ramírez A., Meana A., Page A., Abad J. L., González M. A., Bueren J., Bernad A., Jorcano J. L. (2001) A cutaneous gene therapy approach to human leptin deficiencies. Correction of the murine ob/ob phenotype using leptin-targeted keratinocyte grafts. FASEB J. 15, 1529–1538 [DOI] [PubMed] [Google Scholar]
- 52. Iliopoulos D., Malizos K. N., Oikonomou P., Tsezou A. (2008) Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PLoS One 3, e3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Meisgen F., Xu N., Wei T., Janson P. C., Obad S., Broom O., Nagy N., Kauppinen S., Kemény L., Ståhle M., Pivarcsi A., Sonkoly E. (2012) MiR-21 is up-regulated in psoriasis and suppresses T cell apoptosis. Exp. Dermatol. 21, 312–314 [DOI] [PubMed] [Google Scholar]
- 54. Lu T. X., Munitz A., Rothenberg M. E. (2009) MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J. Immunol. 182, 4994–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu F., Zikusoka M., Trindade A., Dassopoulos T., Harris M. L., Bayless T. M., Brant S. R., Chakravarti S., Kwon J. H. (2008) MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2α. Gastroenterology 135, 1624–1635.e24 [DOI] [PubMed] [Google Scholar]
- 56. Sheedy F. J., Palsson-McDermott E., Hennessy E. J., Martin C., O'Leary J. J., Ruan Q., Johnson D. S., Chen Y., O'Neill L. A. (2010) Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 11, 141–147 [DOI] [PubMed] [Google Scholar]
- 57. Ji R., Cheng Y., Yue J., Yang J., Liu X., Chen H., Dean D. B., Zhang C. (2007) MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ. Res. 100, 1579–1588 [DOI] [PubMed] [Google Scholar]
- 58. Calin G. A., Croce C. M. (2006) MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866 [DOI] [PubMed] [Google Scholar]
- 59. Cho W. C. (2007) OncomiRs. The discovery and progress of microRNAs in cancers. Mol. Cancer 6, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Krichevsky A. M., Gabriely G. (2009) miR-21. A small multi-faceted RNA. J. Cell Mol. Med. 13, 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Frankel L. B., Christoffersen N. R., Jacobsen A., Lindow M., Krogh A., Lund A. H. (2008) Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 283, 1026–1033 [DOI] [PubMed] [Google Scholar]
- 62. Yang X., Wang J., Guo S. L., Fan K. J., Li J., Wang Y. L., Teng Y. (2011) miR-21 promotes keratinocyte migration and re-epithelialization during wound healing. Int. J. Biol. Sci. 7, 685–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Patrick D. M., Montgomery R. L., Qi X., Obad S., Kauppinen S., Hill J. A., van Rooij E., Olson E. N. (2010) Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J. Clin. Invest. 120, 3912–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]