Background: The heart of dry land toads has adapted to sustain circulation in a wide range of body fluid changes.
Results: The toad cardiac muscle expresses exclusively slow skeletal troponin T with cardiac forms of other myofilament proteins and exhibits functional benefit.
Conclusion: This finding reflects a novel adaptation of toad heart.
Significance: The results indicate a molecular mechanism to improve systolic function.
Keywords: Cardiac Muscle, Contractile Protein, Heart, Molecular Evolution, Muscle, Troponin
Abstract
The three isoforms of vertebrate troponin T (TnT) are normally expressed in a muscle type-specific manner. Here we report an exception that the cardiac muscle of toad (Bufo) expresses exclusively slow skeletal muscle TnT (ssTnT) together with cardiac forms of troponin I and myosin as determined using immunoblotting, cDNA cloning, and/or LC-MS/MS. Using RT-PCR and 3′- and 5′-rapid amplification of cDNA ends on toad cardiac mRNA, we cloned full-length cDNAs encoding two alternatively spliced variants of ssTnT. Expression of the cloned cDNAs in Escherichia coli confirmed that the toad cardiac muscle expresses solely ssTnT, predominantly the low molecular weight variant with the exon 5-encoded NH2-terminal segment spliced out. Functional studies were performed in ex vivo working toad hearts and compared with the frog (Rana) hearts. The results showed that toad hearts had higher contractile and relaxation velocities and were able to work against a significantly higher afterload than that of frog hearts. Therefore, the unique evolutionary adaptation of utilizing exclusively ssTnT in toad cardiac muscle corresponded to a fitness value from improving systolic function of the heart. The data demonstrated a physiological importance of the functional diversity of TnT isoforms. The structure-function relationship of TnT may be explored for the development of new treatment of heart failure.
Introduction
Toad and frog are two closely related amphibian species. However, toads are more adapted to living on land than the wet habitats in which frogs reside. In land habitats, the volumes of toad body fluid and blood change with large ranges between rain and dry seasons (1) or with different heat exposures (2). The heart of toad apparently adapts well to working with a big range of blood volume changes (2).
The function of the vertebrate circulatory system is critically dependent on and closely regulated by the volume of circulating blood (3). Decreases of blood volume such as that in humans during hemorrhagic shock would result in decreased cardiac output due to both decreased ventricular filling and increased peripheral arterial resistance from the compensatory increase in sympathetic tone (4). Therefore, to understand the mechanism(s) by which the toad heart is able to cope with the drastic decrease in blood volume such as that occurs during dry season (1) or under experimental conditions (2) and sustain cardiac output is of physiological and medical importance.
A foundation of heart function is the myofilament proteins that form the contractile machinery in cardiac muscle (5). Comparative studies of the myofilament contractile and regulatory proteins in the hearts of toad and frog are valuable in revealing the molecular basis of the functional feature of toad cardiac muscle.
In the present study, we examined the isoform contents of representative myofilament proteins in the toad cardiac muscle. Solely slow skeletal muscle isoform of troponin T (ssTnT)2 was found in toad heart, whereas cardiac muscles of frog and other vertebrate species examined all expressed exclusively cardiac TnT. The unique presence of slow TnT in toad heart was confirmed by cDNA cloning. Normal cardiac troponin I (TnI) and cardiac myosin heavy chain (MHC) were present in toad heart. Functional studies demonstrated that toad hearts had higher contractility and were able to work against higher afterload than that of frog hearts, suggesting a physiological value of the unique evolutionary adaptation of utilizing ssTnT in toad cardiac muscle.
MATERIALS AND METHODS
Animals
American toads (Bufo americanus) and bullfrogs (Rana catesbeiana) with comparable body weights were obtained from commercial sources in non-hibernating season. Toads were kept in moist moss and frogs in a water tank at room temperature for a few days before use. Tadpoles of both species were purchased from a commercial source and euthanized upon arrival to collect tissue samples. The experimental protocols were approved by the Institutional Animal Care and Use Committee.
Muscle Samples
Immediately after euthanasia, atrial and ventricular muscles as well as various skeletal muscles of toad and frog were dissected according to illustrations of frog muscle anatomy. Whole muscle tissue was homogenized in 20 volumes (w/v) of SDS gel sample buffer containing 2% SDS, 10% glycerol, 50 mm Tris base, 2% 2-mercaptoethanol, pH 8.8, using a high speed mechanical homogenizer (PRO Scientific Inc.). The homogenized muscle samples were immediately heated at 80 °C for 5 min, clarified by high speed centrifugation, and stored at −80 °C for SDS-PAGE and Western blot analysis.
Development of Cardiac TnT-specific Monoclonal Antibodies (mAbs)
Using human cardiac TnT expressed from cloned cDNA in Escherichia coli as immunigen, hybridoma cell lines were generated to produce specific mAbs. The expression and purification of human cardiac TnT, immunization of Balb/c mice, hybridoma fusion, screening, and subcloning, mAb production, and immunoglobulin isotyping were carried out as described previously using standard methods (6). Isoform specificity of the anti-TnT mAbs obtained was examined with Western blots on cardiac and skeletal muscle samples from multiple vertebrate species.
SDS-PAGE and Western Blotting
Protein samples were resolved on 14% Laemmli gel with an acrylamide:bisacrylamide ratio of 180:1. The protein bands were visualized by staining the gel with Coomassie Brilliant Blue R-250. Duplicate gels were electrically transferred to blot nitrocellulose membranes. Tris-buffered saline (TBS) containing 1% bovine serum albumin (BSA) was used to block the nitrocellulose membranes at room temperature for 30 min. The membranes were then incubated with anti-TnT mAb 2C8 recognizing all three muscle type-specific isoforms across vertebrate species (7), mAb CT3 recognizing cardiac TnT and slow TnT (8), mAb 4B8 recognizing only cardiac TnT (characterized in the present study), anti-TnI mAb TnI-1 (6), and anti-cardiac MHC mAb FA2 (9) diluted in TBS containing 0.1% BSA at 4 °C overnight. After high stringency washes with TBS plus 0.5% Triton X-100 and 0.05% SDS, the membranes were further incubated with alkaline phosphatase-conjugated goat anti-mouse IgG second antibody (Santa Cruz Biotechnology, Santa Cruz, CA), washed again as above, and developed in 5-bromo-4-chloro-3-indolylphosphate/nitro blue tetrazolium substrate solution. In some experiments, the membrane, after the blot image was documented by scanning, was re-blotted with another mAb at doubled concentration by repeating the above procedure.
Myosin heavy chain isoforms were examined using glycerol/SDS-PAGE as described previously (10). The muscle protein samples were resolved on 8% SDS-polyacrylamide gel containing 30% glycerol run in an icebox at constant voltage of 100 V for 24 h.
Cloning of cDNAs Encoding Toad ssTnT
Total RNA was extracted from toad hearts using the TRIzol reagent as instructed in the product manual (Invitrogen). Integrity of the RNA isolated was verified using agarose gel electrophoresis. Poly(A)+ mRNA was reverse transcribed using an anchored oligo(dT) primer (TV20) at 42 °C for 2 h. Using degenerated primers derived from amino acid sequences in conserved regions of vertebrate ssTnT, partial cDNA encoding toad ssTnT was cloned with polymerase chain reaction (PCR). Full-length toad ssTnT cDNAs were then obtained using 3′- and 5′-rapid amplification of cDNA ends and sequenced.
Cloning of cDNA Encoding Toad Cardiac TnI
From total cardiac cDNA synthesized as above, cDNA encoding toad cardiac TnI was cloned with reverse transcription-coupled PCR using degenerated primers derived from the sequence of Xenopus cardiac TnI (GenBankTM accession number L25721.1) and sequenced.
Expression of Cloned Toad ssTnT cDNA in E. coli
The coding region of toad ssTnT cDNA was inserted into expression plasmid vector pET24. The plasmid DNA was used to transform BL21(ED3)pLysS E. coli for the expression of toad ssTnT. The expressed protein was examined using SDS-PAGE and Western blot as described above.
Preparation of ex Vivo Working Hearts and Functional Measurements
Toad and frog hearts were isolated and cannulated on a perfusion apparatus modified from a working heart system previously used for mouse and rat heart studies (11).
Experiments were performed at 16 °C controlled with circulating water from a refrigerated bath. The left side aorta of toad or frog heart was cannulated and retrograde perfused through a blunted 16-gauge needle. The right side aorta was sutured with a 3-0 silk thread. Preload was established by connecting the anterior vena cava to the preload reservoir through a 16-gauge cannula. All other vena cavae connected to sinus venosus were sutured to prevent leaking. The perfusion solution contains (in mm) 115 NaCl, 2.5 KCl, 1.0 CaCl2, 2.15 Na2HPO4, 0.85 NaH2PO4, and 5.6 d-glucose. pH was adjusted to 7.20 by the addition of Na2HPO4 and equilibrated with air (12).
The heart rate was controlled at 32 beats per minutes (bpm) through pacing with a pair of electrodes attached to the right atrium. Pulse stimulation of 4 ms duration with a constant current 30% above the threshold was given using an isolated stimulator. A pressure transducer (MLT844, Capto, Horten, Norway) was connected to the aortic cannula through a three-way connector at the ventricle level to measure changes in the aortic pressure. A 1.2 French pressure-volume catheter was inserted into the ventricular chamber through the apex to measure ventricular pressure and volume changes. Data were collected continuously using Chart 5 software through an A/D interface (Power Lab).
In functional studies, afterload pressure of 25 cm H2O and preload pressure of 4 cm H2O were set as the baseline conditions with reference to the levels of aortic trunk and left atrium, respectively. Preload and afterload were experimentally altered by moving the reservoirs up or down to test preload and afterload responses.
LC-MS/MS Analysis of MHCs
MHC bands separated in glycerol/SDS-PAGE were stained with GelCode Blue reagent (Thermo Scientific) and manually excised from the gel. For in gel protein digestion, the gel slices were washed with 50 mm ammonium bicarbonate, reduced in 10 mm DTT at 37 °C for 45 min, and then alkylated with 55 mm iodoacetamide at room temperature for 30 min. Subsequently, four proteases, Lys-C, trypsin, Glu-C, and chymotrypsin, were added to four replicate reaction tubes each at 10 ng/μl and the digestion was incubated at 37 °C overnight except for chymotrypsin that was incubated at room temperature.
The resulting peptides were analyzed by nanoLC-MS/MS. The peptides were first separated on a reversed-phase C18 column with a 90-min gradient using a Dionex Ultimate HPLC system. MS and MS/MS spectra were then acquired on an Applied Biosystems QSTAR XL mass analyzer using information dependent acquisition mode. MS scan was performed from m/z 400–1,500 for 1 s followed by product ion scans on the two most intense multiply charged ions. Peak lists were submitted to Mascot server to search against the NCBInr data base for all entries with carbamidomethyl (C) used as a fixed modification and oxidation (M), N-acetylation (protein N terminus) as variable modifications. To confirm the peptide mapping results, an aliquot of the tryptic peptides was also analyzed on a LTQ XL mass spectrometer.
MHC Peptide Mapping and BLAST Analysis
Because the sequence of toad cardiac MHC was unavailable in any public protein sequence databases, our strategy was to obtain a partial sequence of this large protein by mapping the MS/MS data to MHC orthologs from related species. Therefore, we selected all entries in the NCBInr database for the Mascot searches. To maximize the sequence coverage, toad MHC bands were digested with the four different proteases described above.
To determine that the MHC in toad heart was cardiac like or skeletal muscle like, we used the compiled partial protein sequence of toad MHC to perform a BLAST analysis in the NCBInr database.
Data Analysis
DNA and protein sequence analysis was performed using the DNAStar software (Lasergene). Two-dimensional densitometry was done to quantify SDS gel and Western blots on images scanned at 600 dpi. Statistical significance for quantitative data were determined using Student's t test or two-way analysis of variance test.
RESULTS
mAb 4B8 Specifically Recognizes Cardiac TnT in Mammalian, Avian, Amphibian, and Fish Species
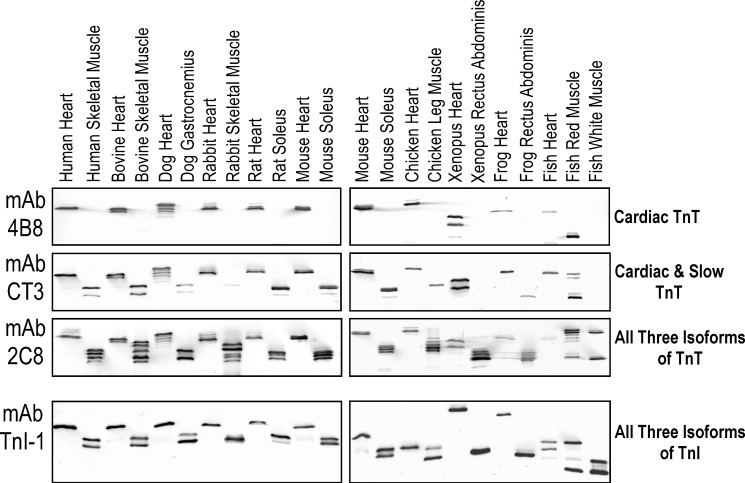
An initial observation of the present study was that the TnT band in toad cardiac muscle is of significantly lower apparent molecular weight as revealed in Western blots (see data below). To characterize the nature of this TnT in toad heart, a cardiac TnT-specific mAb 4B8 was developed after screening of a large number of hybridoma clones obtained from the fusion using spleen cells immunized with human cardiac TnT. The Western blots in Fig. 1 demonstrated that mAb 4B8 specifically recognizes cardiac TnT from human, bovine, dog, rabbit, rat, mouse, chicken, Xenopus, frog, and fish hearts without cross-reaction with fast or slow TnT from mixed fiber skeletal muscles of these species. Immunoglobulin isotyping showed that mAb 4B8 is an IgMκ. The κ light chain reactivity of the alkaline phosphatase-conjugated anti-mouse IgG second antibody was sufficient in producing strong Western blot signals (Fig. 1).
FIGURE 1.
mAb 4B8 specific to cardiac TnT across vertebrate species. Western blots using 14% SDS-PAGE with acrylamide:bisacrylamide ratio of 180:1 characterized the isoform specificity of anti-TnT mAb 4B8 generated against human cardiac TnT immunogen. mAb 4B8 recognized TnT in adult cardiac but not skeletal muscles from all mammalian, avian, amphibian, and fish species examined except for one variant in fish red muscle. The cardiac TnT bands recognized by mAb 4B8 were confirmed in Western blots using mAb CT3 that recognizes both cardiac TnT and ssTnT and blots using mAb 2C8 that recognizes all three muscled type isoforms of TnT across vertebrate species. Western blot using mAb TnI-1 detected cardiac TnI, slow skeletal muscle TnI, and fast skeletal muscle TnI in the muscle samples, verifying the presence of all three fiber types. The results demonstrate that mAb 4B8 is a highly valuable cardiac TnT-specific antibody and can be used for studies in all vertebrate species.
Toad Cardiac Muscle Expresses Solely ssTnT
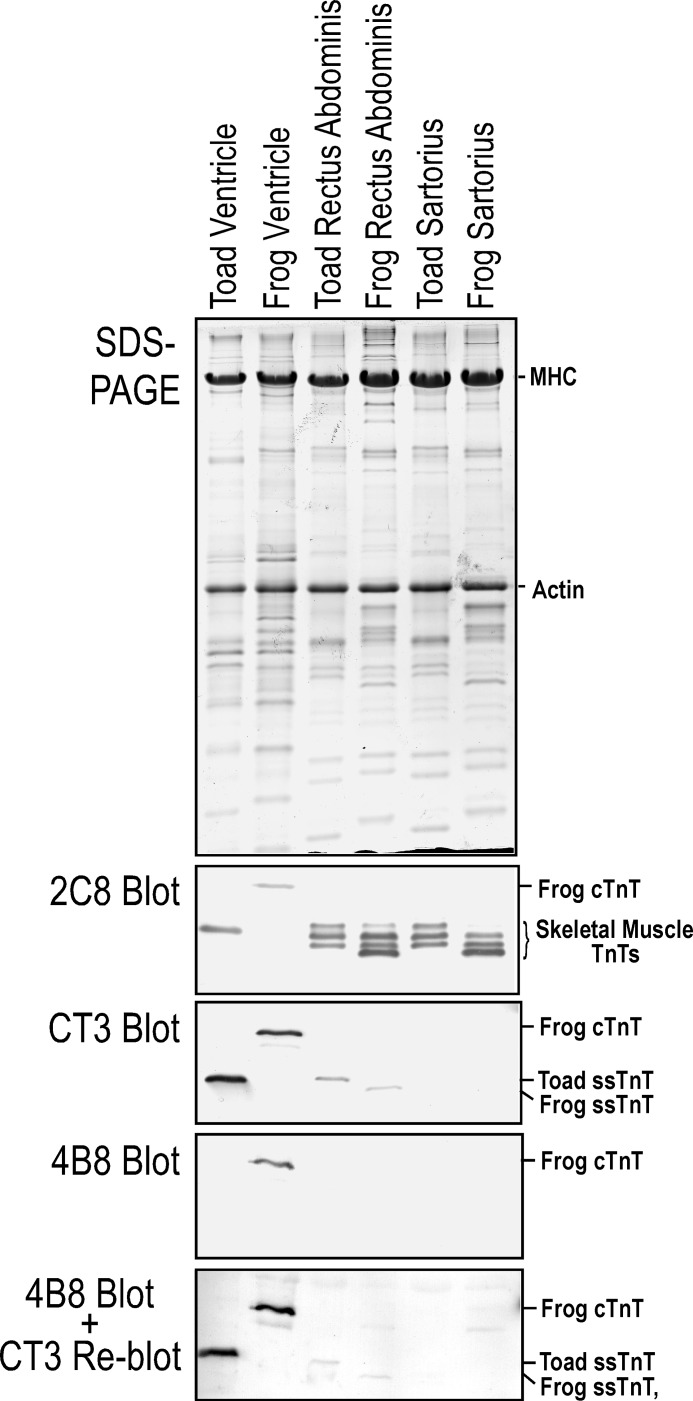
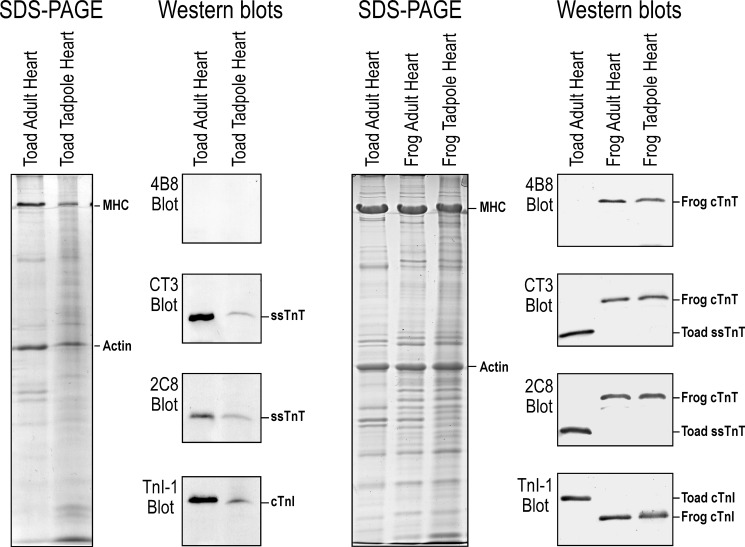
Western blotting analysis of cardiac and skeletal muscles of adult toad and frog using a set of isoform-specific anti-TnT mAbs (Fig. 1) showed cardiac, fast and slow skeletal muscle TnTs are expressed in a muscle type-specific manner in the frog but not toad heart (Fig. 2). Together with that observed in a previous study (7), the other vertebrate species examined also showed muscle type-specific expression of the TnT isoforms (Fig. 1). A single TnT band was detected in the toad heart by the pan-TnT mAb 2C8 with a size in the range of skeletal muscle TnTs (Fig. 2). Western blot using anti-cardiac and ssTnT mAb CT3 identified this toad heart TnT with identical size to ssTnT in toad rectus abdominis muscle (Fig. 2). No band was detected in the Western blot of toad cardiac muscle by the cardiac TnT-specific mAb 4B8 that clearly recognizes cardiac TnT in the frog hearts (Fig. 2). Re-blotting of the 4B8-probed membrane using mAb CT3 confirmed the presence of ssTnT in the toad cardiac muscle blot (Fig. 2). Altogether, the Western blots using TnT isoform-specific mAbs indicated that the toad cardiac muscle expresses a single TnT that is ssTnT and no cardiac TnT was detectable. SDS-PAGE gels and Western blots further showed that toad tadpole heart also expresses exclusively ssTnT (Fig. 3).
FIGURE 2.
Toad cardiac muscle expresses solely ssTnT in the absence of cardiac TnT. A single TnT band was detected in toad heart with a significantly lower molecular weight than that of frog cardiac TnT as shown in the Western blot using mAb 2C8 recognizing all three muscle-type isoforms of TnT (Fig. 1). This TnT band in toad heart was recognized in the Western blot using mAb CT3 that reacts to both cardiac TnT and ssTnT with a molecular weight similar to that of ssTnT in toad and frog skeletal muscles. In contrast, it was not recognized in Western blots using the cardiac TnT-specific mAb 4B8. Reblotting of the 4B8 blot with mAb CT3 confirmed its presence on the membrane. The results demonstrated that the sole TnT expressed in toad heart is ssTnT.
FIGURE 3.
Exclusive expression of ssTnT in toad tadpole heart. The SDS-PAGE gels and Western blots showed expression of ssTnT in the heart of toad tadpoles in the absence of cardiac TnT. Frog tadpole heart and adult toad and frog hearts were used as controls. Same as that in adult toad heart, cardiac TnI was expressed in toad and frog tadpole hearts.
Confirmation of ssTnT in Toad Heart by cDNA Cloning
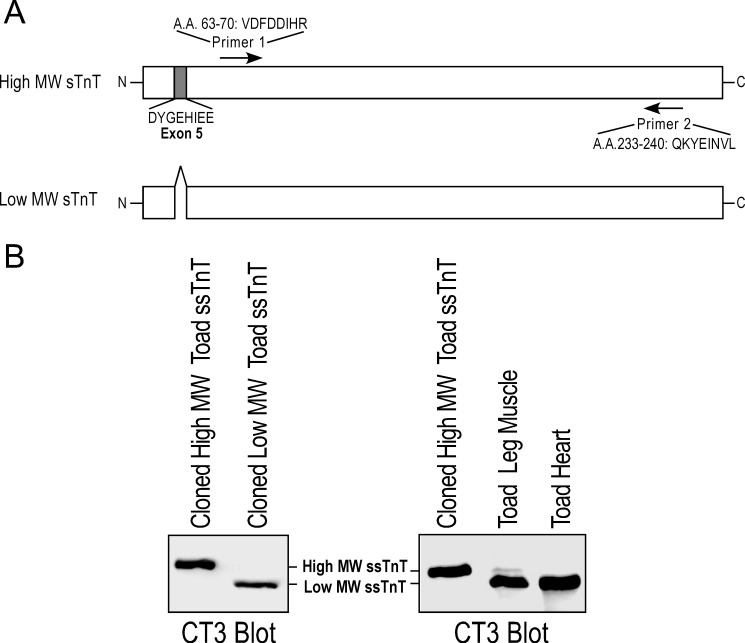
From toad heart poly(A)+ RNA, RT-PCR using two degenerated ssTnT primers followed by 3′- and 5′-rapid amplification of cDNA ends successfully cloned two variants of cDNA encoding toad ssTnT (Fig. 4A). DNA sequencing revealed that the full-length toad ssTnT cDNA cloned included 71-bp of 5′-untranslated region and 251-bp of 3′-untranslated region. The sequences also revealed that the two species of toad ssTnT mRNA in the adult toad heart were generated from alternative splicing with the inclusion or exclusion of the exon 5-encoded segment corresponding to an acidic segment of 8 amino acids in the NH2-terminal variable region (DYGEHIEE) (Fig. 4A). The two splicing variants of toad ssTnT contain 264 and 256 amino acids, with predicted isoelectric points (pI) of 5.45 and 5.81, respectively. The cDNA and amino acid sequences of toad ssTnT have been deposited to GenBank with accession numbers AY773671 and AY773672.
FIGURE 4.
Cloning of cDNAs encoding ssTnT from toad cardiac mRNA. A, two degenerated primers derived from amino acid sequences conserved in vertebrate ssTnT were used in RT-PCR to clone ssTnT cDNA from total RNA extracted from adult toad cardiac muscle. Extended using 3′- and 5′-rapid amplification of cDNA ends, two full-length ssTnT cDNA variants were cloned. DNA sequencing revealed their difference in the inclusion or exclusion of the exon 5-encoded segment (DYGEHIEE) in the NH2-terminal variable region. B, mAb CT3 Western blots showed that ssTnT proteins expressed from the cloned cDNAs had identical apparent molecular weights (MW) to that of the ssTnT bands in toad cardiac and skeletal muscles. The low Mr ssTnT is the only variant detectable with Western blot in the toad cardiac muscle.
Consistently, two toad ssTnT bands were detected in toad skeletal and cardiac muscles by Western blot using mAb CT3 with the low molecular weight variant being predominant (Fig. 4B). The ssTnT proteins expressed in E. coli from the cloned toad ssTnT cDNA showed sizes identical to the ssTnT proteins in toad heart and skeletal muscle (Fig. 4B). These data further confirm the presence of solely ssTnT in toad cardiac muscle.
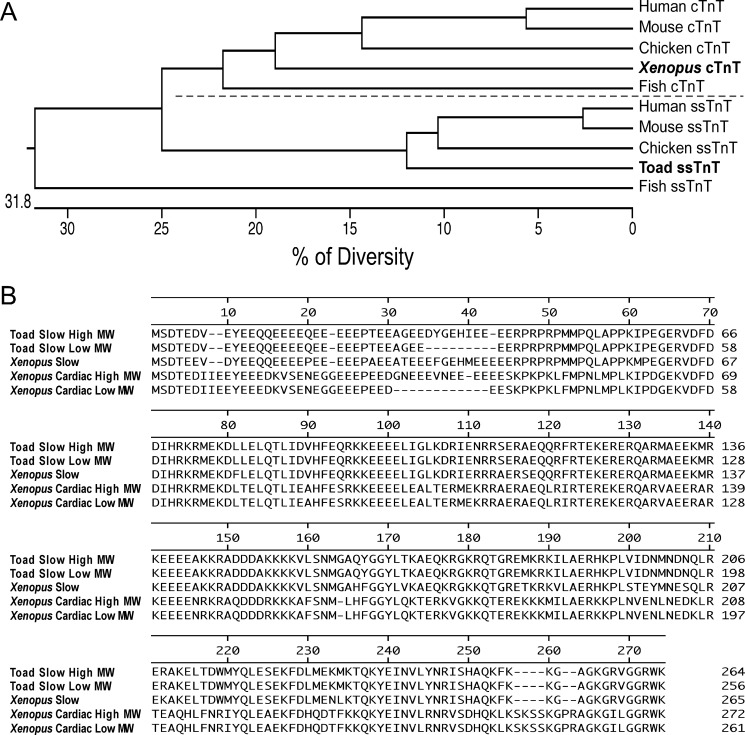
A phylogenetic tree of vertebrate ssTnT was generated from aligning amino acid sequences of cardiac and skeletal muscle TnT isoforms of representative species. The results showed that toad ssTnT is closely related to avian and mammalian ssTnTs (73–80% residue identities) although significantly diverged from cardiac TnT (Fig. 5A).
FIGURE 5.
Phylogenetic and sequence studies of toad ssTnT. A, the phylogenetic tree was constructed from aligning amino acid sequences of vertebrate cardiac TnT and ssTnT isoforms using DNA Star Clustal W software. The result demonstrated a conserved sequence of toad ssTnT, which is distinct from cardiac TnT (cTnT). B, alignment of amino acid sequences of toad ssTnT isoforms, Xenopus ssTnT, and Xenopus cardiac TnT. The results showed that toad ssTnT is similar to Xenopus ssTnT but significantly different from cardiac TnT in the NH2-terminal variable region and a lack of six amino acids near the COOH terminus. GenBank accession numbers: Xenopus ssTnT, NM_001092738.1; Xenopus cardiac TnT high molecular weight (MW) isoform, AF467919.1; Xenopus cardiac TnT low Mr isoform, AF467920.1.
Alignment of amino acid sequences of the toad ssTnT isoforms with the ssTnT and cardiac TnT of Xenopus laevis provided a comparison for their primary structures. In addition to differences in the NH2-terminal hypervariable region, amphibian ssTnT differs from amphibian cardiac TnT by a lack of six amino acids near the COOH terminus (Fig. 5B).
Lower Specific Stroke Volume but Faster Kinetics of Toad Versus Frog Hearts
At similar body weights, toad hearts were found to be larger than frog hearts, resulting in a significantly higher heart weight to body weight ratio (Table 1). This feature may be an evolutionary adaptation with functional significance and further investigation is needed.
TABLE 1.
Heart weight and body weight of toad and frog
| Frog (n = 4) | Toad (n = 6) | |
|---|---|---|
| Body weight (g) | 181.25 ± 16.94 | 187.67 ± 19.92 |
| Heart weight (mg) | 634.48 ± 99.45 | 1016.08 ± 90.75a |
| Heart weight/body weight (mg/g) | 3.46 ± 0.24 | 5.54 ± 0.42b |
a Values are presented as mean ± S.E., p < 0.05 versus frog in Student's t test.
b Values are presented as mean ± S.E., p < 0.01 versus frog in Student's t test.
Ex vivo working heart experiments revealed interesting functional differences between toad and frog hearts. At the baseline condition of 4 cm H2O preload, 25 cm H2O afterload, and heart rate paced at 32 bpm, the toad hearts produced similar absolute stroke volumes as frog hearts (Fig. 6A). However, due to the higher heart weight of toad hearts (Table 1), the stroke volume of toad hearts normalized to heart weight was lower than that of the frog hearts, although no statistical significance was established in two-tail Student's t test (Fig. 6A).
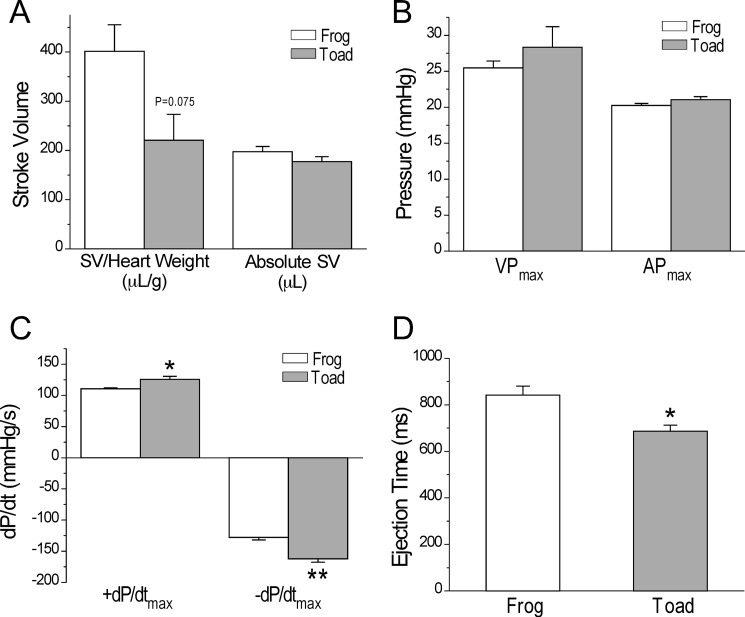
FIGURE 6.
Baseline function of ex vivo toad and frog working hearts. A, at identical heart rates and normalized to heart weight, toad hearts exhibited a trend of smaller stroke volume than that of frog hearts. This difference was primarily due to the larger size of the toad heart normalized to body weights (Table 1), whereas the actual stoke volumes of toad and frog hearts were similar if normalized to the body weight. B, toad and frog hearts exhibited similar maximum aortic and ventricular pressures (AP and VP, respectively). C, toad hearts had faster systolic and diastolic contractile velocities (±dP/dt) than that of frog hearts. D, toad hearts had shorter ejection time than that of frog hearts. n = 3 in frog and n = 4 in toad groups. Values are presented as mean ± S.E. *, p < 0.05 versus frog heart in two-tail Student's t test.
Under the baseline condition, the ex vivo working toad hearts produced maximum ventricular and aortic pressures similar to that of the frog hearts (Fig. 6B). Compared with the frog hearts, the toad hearts exhibited faster systolic and diastolic velocities as indicated by the faster ±dP/dt (Fig. 6C). The faster contractile kinetics was accompanied by significantly shortened ventricular ejection time (Fig. 6D).
Toad Hearts Were Able to Pump against Higher Afterload Than That of Frog Hearts
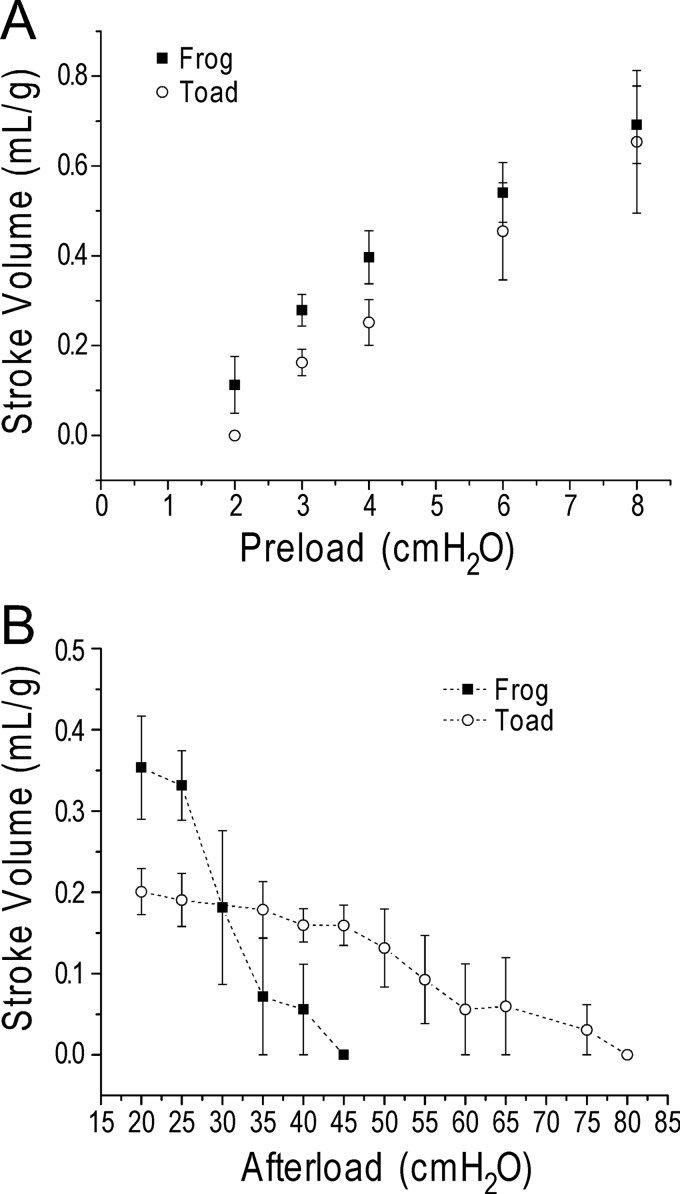
To investigate the functional value of the exclusive utilization of ssTnT in toad cardiac muscle during evolutionary selection, especially in cardiac adaptation to environment-based fluctuation of circulating blood volume, we tested the ex vivo working hearts with a series of preloads ranging from 2 to 8 cm H2O at the afterload of 25 cm H2O. The results showed similar positive preload responses in frog and toad hearts with increases in stroke volume when preload increases (Fig. 7A).
FIGURE 7.
Toad hearts were able to produce output against higher afterload than that of frog hearts. A, at 25 cm H2O afterload, positive stroke volume response to preloads from 2 to 8 cm H2O was detected in both toad and frog hearts. B, at 4 cmH2O preload, toad hearts continued to produce aortic output when afterload was increased from the base line of 25 cm H2O up to 65 cm H2O, whereas frog hearts failed to produce aortic output at afterload above 45 cm H2O (p < 0.05 for the maximum afterload at which aortic output was detectable in Student's t test). n = 3 in frog and n = 3 in toad groups. Values are presented in mean ± S.E.
We further examined the ex vivo working hearts with a series of increasing afterload at a fixed preload of 4 cm H2O. The frog hearts failed to produce aortic output at afterload above 45 cm H2O. In contrast, the toad hearts were able to produce aortic output against afterload up to 65 cm H2O (Fig. 7B). This finding demonstrated an extended pumping ability of toad hearts to overcome increases in afterload, suggesting a possible benefit of the evolutionary adaptation by replacing cardiac TnT with ssTnT in toad cardiac muscle.
Toad Cardiac Muscle Normally Expresses Solely Cardiac TnI
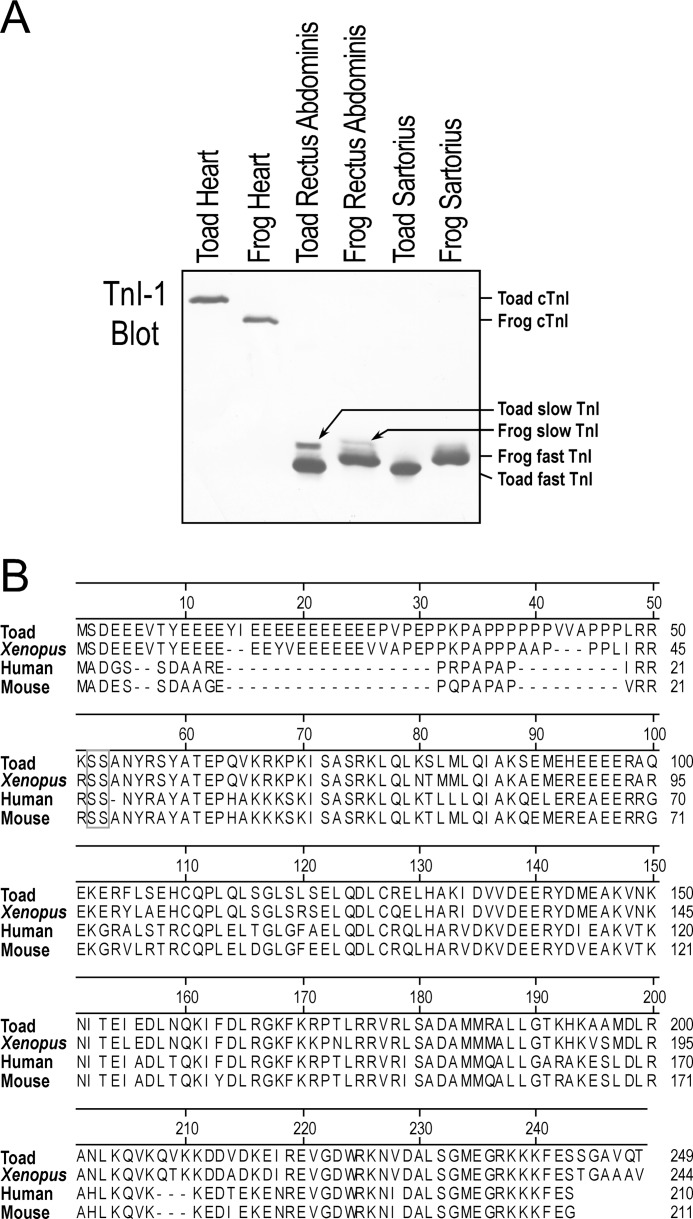
Western blot using mAb TnI-1 showed that similar to that in frog muscles, normal expression patterns of cardiac TnI, slow skeletal muscle TnI, and fast skeletal muscle TnI were found in toad heart and skeletal muscles. Fast and slow TnI were detected in the mixed fiber muscle rectus abdominis and only fast TnI were seen in the fast fiber muscle sartorius. Exclusively cardiac TnI was detected in toad cardiac muscle (Fig. 8A). Same as in the adult heart, only cardiac TnI was detected in the toad tadpole heart (Fig. 3).
FIGURE 8.
Toad heart expresses normal cardiac TnI. A, Western blots detected only cardiac TnI (cTnI) in toad and frog hearts, whereas slow and fast skeletal muscle TnI were detectable under the same conditions in skeletal muscles. No skeletal muscle TnI was detected in the toad heart. B, amino acid sequence alignment outlined the similarities and differences between toad cardiac TnI and cardiac TnI of Xenopus, human, and mouse. The main difference between amphibian and mammalian cardiac TnI is in the NH2-terminal extension and a 6-amino acid extension at the COOH terminus. The conserved protein kinase A phosphorylation sites as found in human and mouse cardiac TnI are outlined with a gray box. GenBank accession numbers: Xenopus cardiac TnI, L25721.1; human cardiac TnI, X54163.1; mouse cardiac TnI, NM_009406.
Consistent with the tightly coupled expression of the evolutionarily linked TnI and TnT isoform gene pairs (7, 13), the expressions of TnI and TnT isoforms match well in toad skeletal muscle (Figs. 1 and 2). The results of normally restricted expression of troponin subunits in skeletal muscles as well as the normal expression of cardiac TnI in toad cardiac muscle indicated that the expression of ssTnT instead of cardiac TnT in toad heart is a unique change.
Toad cardiac TnI exhibited significantly slower gel mobility in SDS-PAGE than that of frog and other vertebrate TnI (Figs. 1, 3, and 8A). Sequence of the cloned toad cardiac TnI revealed a correspondingly larger protein size (249 amino acids versus 244 for Xenopus cardiac TnI and ∼210 for mammalian (Fig. 8B) and avian (14) cardiac TnI). The toad cardiac TnI has a long NH2-terminal extension with the two protein kinase A-phosphorylated Ser residues conserved (corresponding to Ser23 and Ser24 in human and mouse cardiac TnI, Fig. 8B). Comparing with mammalian cardiac TnI, amphibian cardiac TnI has an extended COOH terminus (Fig. 8B), mimicking that in the mammalian slow skeletal muscle TnI (14). The cDNA and amino acid sequences of toad cardiac TnI (Tnni3) have been deposited in GenBank with accession number AY773673.
Toad Hearts Express Cardiac Myosin
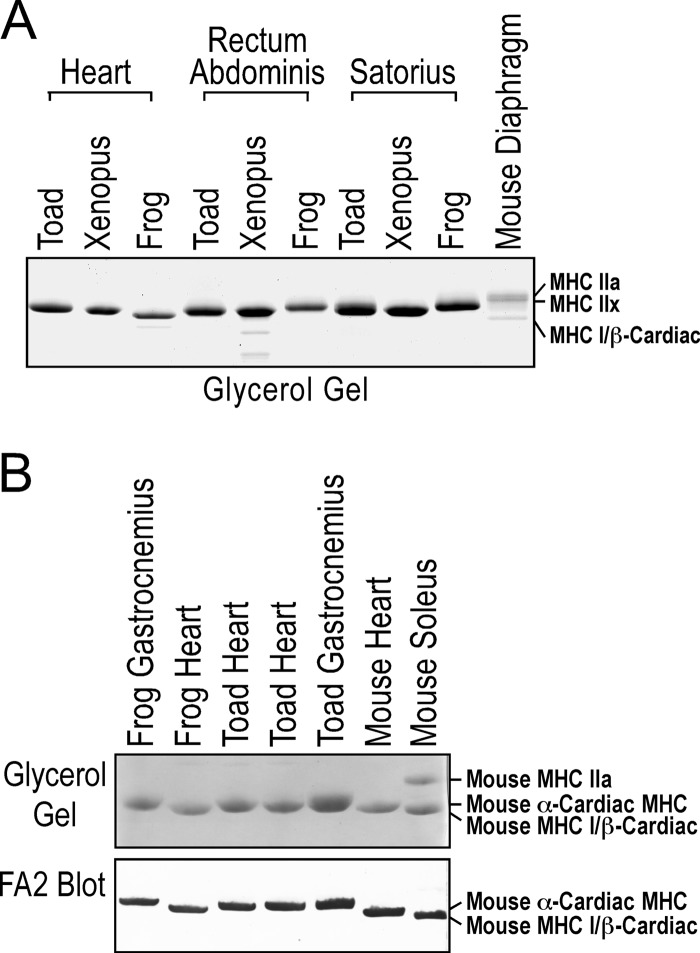
Glycerol/SDS gel detected a single isoform of MHC in toad and frog hearts with a slight difference in gel mobility (Fig. 9A). The MHC in toad cardiac muscle is also of different gel mobility from that of toad skeletal muscle MHC (Fig. 9A). The cardiac nature of toad heart MHC was first identified by Western blot using mAb FA2 recognizing mouse α and β cardiac MHC isoforms (Fig. 9B).
FIGURE 9.
Toad heart expresses cardiac MHC. A, MHC isoforms in the hearts and skeletal muscles of toad, frog, and Xenopus were resolved in glycerol/SDS gel. Mouse diaphragm muscle was analyzed in parallel as control. A single MHC band was detected in toad heart with a slightly different gel mobility from that of frog cardiac MHC or the MHC bands in toad rectum abdominis and sartorius muscles. B, mAb FA2 specific to cardiac α- and β-MHC (cardiac β-MHC is the same as MHC I in slow skeletal muscle) recognized the MHC band in both toad and frog hearts, supporting their nature as cardiac MHC.
To further verify whether the single MHC in toad heart is a cardiac or skeletal muscle isoform, mass spectrometry was carried out. To obtain sufficient sequence information for the toad heart MHC of over 200 kDa in size, we combined the peptide sequences matched to the top ranked proteins from multiple LC-MS/MS experiments using four different proteases (supplemental Table S1). Because there was no sequence information of toad cardiac or skeletal muscle MHC available in public protein sequence databases, we compared the partial amino acid sequence of toad heart MHC with orthologs from closely related species. In the Mascot search of the LC-MS/MS results, the majority of the significant hits were cardiac MHCs from vertebrate species. Among them, the mammalian cardiac β-MHC-like X. laevis myosin heavy polypeptide 15 (gi 148222862) was the top hit in most experiments. Therefore, the MHC 15 sequence was used as a template to demonstrate the partial sequence of toad heart MHC as shown in Fig. 10.
FIGURE 10.
Sequence coverage of toad MHC from LC-MS/MS analyses using four different proteases. A database search showed that cardiac-like myosin heavy polypeptide 15 of X. laevis (gi 148222862) was the top match. Therefore, this sequence was used as a template to demonstrate the peptide mapping results for toad MHC. Amino acids marked with black letters represent sequences detected in LC-MS/MS analyses. The boxed lowercase letters represent amino acids that are different from the Xenopus MHC 15 sequence. X represents amino acid residues in the MHC of the Xenopus MHC 15 sequence, which are not covered in the LC-MS/MS results. Together with a match of the toad heart MHC sequence with other cardiac MHC proteins in the NCBInr database as described in the text, the data confirmed that the MHC in toad heart is a cardiac isoform.
In addition to Xenopus MHC 15, two other cardiac MHC proteins were also frequently among the top hits: chicken cardiac MHC (gi 45382109) and a bullfrog cardiac MHC isoform (gi 8272640). A few skeletal MHCs were also identified as significant hits in some experiments but with much lower Mascot scores and fewer matched peptides.
To confirm the cardiac nature of toad heart MHC, we used the compiled partial protein sequence of toad MHC (∼50% sequence coverage) to perform a BLAST analysis in the NCBInr data base. The top 30 hits were all cardiac MHCs, whereas several skeletal MHCs were matched with a lower percentage of identity. To validate our conclusion based on BLAST analysis using a partial protein sequence, we also did a BLAST search using peptide mapping from a mouse cardiac MHC (∼57% sequence coverage) obtained in a separate LC-MS/MS study. A similar overall pattern of the BLAST result was obtained (supplemental Fig. S1), which is consistent with the toad heart MHC being a cardiac MHC.
These thorough proteomic analysis concluded that the toad heart MHC is a cardiac MHC. Therefore, the expression solely of ssTnT in toad cardiac muscle is accompanied by normal expressions of other thin and thick filament proteins. Accordingly, the novel functional differences between toad and frog hearts is likely due to the unique replacement of cardiac TnT with ssTnT.
DISCUSSION
More than four decades of extensive studies have established the central position of troponin in the regulation of striated muscle contraction (15–17). TnT is the thin filament anchoring subunit of the troponin complex and plays a central role in the regulation of striated muscle contraction (15). Three homologous genes have evolved encoding the cardiac, slow skeletal and fast skeletal muscle isoforms of TnT in vertebrate cardiac and skeletal muscles (14, 18). Muscle type-specific isoforms of TnT have been identified in vertebrates inclusively from fish to human (7), but their physiological significances are not fully understood.
Multiple studies have documented that genes encoding muscle type-specific TnT and TnI isoforms are expressed in specific fiber types and during development under stringent regulations (14, 18). Cardiac muscle has evolved to have its specific set of myofilament protein isoforms adapted to accomplish the specific function of the heart. With no other myofilament protein isoforms changed, the expression of solely ssTnT to completely replace cardiac TnT in toad cardiac muscle is a unique case.
Two lines of evidence indicated that the exclusive expression of ssTnT to replace cardiac TnT in the toad heart is an evolutionary selection of certain beneficial value to cardiac function. The first is that all other vertebrate species from fish to human including closely related amphibian species such as frog and Xenopus, the immediate ancestor of toads and frogs, all express solely cardiac TnT in the cardiac muscle (7) (Fig. 1). Therefore, the expression of ssTnT in toad hearts is not a residually ancestral trait but an acquired adaptation. The other evidence is that toad heart expresses cardiac forms of other thin and thick filament proteins, such as cardiac TnI (Fig. 8) and cardiac MHC (Fig. 9). Therefore, the exclusive expression of ssTnT in toad cardiac muscle cells is not due to a differentiation of the entire cellular environment but a selective activation of the ssTnT gene and inactivation of the cardiac TnT (Tnnt2) gene in normally differentiated adult cardiomyocytes.
We detected no cardiac TnT expression in either heart or skeletal muscle of the toad. Therefore, the toad cardiac TnT gene is either lost or inactivated during evolution. Because the vertebrate cardiac TnT gene is closely linked to the slow TnI (Tnni1) gene (7) and a chromosomal deletion event to remove the cardiac TnT gene would likely impair the upstream slow TnI gene. However, the slow TnI expression is apparently normal in toad skeletal muscle (Fig. 8A). Therefore, the cardiac TnT gene probably remains in the toad genome but is inactivated.
The molecular mechanisms of this novel gene switch remains to be investigated. Considering the early evolutionary emerging of the three muscle type isoforms of TnT in fish and possibly in the vertebrate ancestor hagfish (7), it would be interesting to further investigate whether the adaptive switch of TnT genes also occurred in the heart of any other amphibians or primitive reptiles.
This intriguing finding provides a novel lead and experimental system to investigate the adaptive values of TnT isoforms in muscle contractility and cardiac function. The ex vivo working heart system used in our study allows analyses of cardiac function excluding neurohumoral and vascular influences. This experimental approach also permits precisely controlling the heart rate and altering preload and/or afterload to explore the functional differences between toad and frog hearts. To cope with the fact that toad heart and frog heart are both typical three-chamber amphibian hearts with two atria and one ventricle, we have successfully modified a system built for mouse and rat heart studies (10, 11) for the study of amphibian hearts.
Two major differences found in the comparison of ex vivo toad and frog working hearts support the proposed functional benefit of utilizing ssTnT in toad cardiac muscle. One is the faster contractile and relaxation velocity of the toad ventricular muscle reflecting higher contractility (Fig. 6C) and the other is the higher afterload that the toad heart could work against to produce aortic output (Fig. 7B). The link between the replacement of cardiac TnT with ssTnT and the enhanced pumping of toad heart provided organ level evidence for a functional benefit.
Previous studies of toads and frogs found that blood pressures (systolic/diastolic) in the pulmocutaneous vessels are the same as those in the systemic and carotid arches, and the average blood pressure is higher in toads (26/19 mm Hg) than in frogs (15/8 mm Hg) (19). The observation that toad hearts are adapted to work under higher afterload supports the notion that the exclusive utilization of ssTnT in toad cardiac muscle was an adaptive change with a functional benefit and, therefore, a significant fitness value during natural selection.
Another previous study reported that maximal blood flow rates in the systemic arches of toads did not decline until blood loss exceeded 5% of body weight, whereas losses of 2% caused a decline in frogs (20). Therefore, the enhanced systolic function of toad hearts may suggest a role of ssTnT in maintaining effective pumping under reduced circulating blood volume and increased afterload due to increased vascular resistance as a part of sympathetic compensation (4).
Although the utilization of ssTnT is a significant difference found with relevance to the functional differences between toad and frog hearts, other differences between the hearts of these two species, for example, other myofilament proteins, may also have contributions and remain to be investigated. Nonetheless, the ability of toad hearts to produce aortic output against higher afterload suggests a likely role of TnT function in determining systolic function. A previous study reported ssTnT gene expression in hypertensive rats, which is an established pressure overload model (21). The exclusive expression of ssTnT in toad heart provides a natural animal model to study the functional significance of TnT modifications in cardiac muscle.
The mechanism for ssTnT to enhance the systolic function of heart remains to be investigated. Knocking down ssTnT induced a slow-to-fast type muscle switch in mice with impaired fatigue tolerance of the diaphragm muscle (22). Although ssTnT may have a unique function in skeletal muscle contractility, its value in cardiac muscle function needs more study. On the other hand, transgenic mice overexpressing fast skeletal muscle TnT in the heart decreased cardiac function (23). Therefore, the apparently benign compatibility of ssTnT in toad cardiac myofilaments merits further investigation.
The present study also raises an interesting question of how the muscle type-specific TnT isoform genes are regulated. Similar to the frog ssTnT gene, the toad ssTnT gene was normally expressed in rectus abdominis muscles containing slow fibers but not in the pure fast fiber sartorius muscle (Fig. 2). Therefore, the specific expression of toad ssTnT gene in slow muscle fibers is preserved while it also became activated in cardiac muscle.
In vertebrate striated muscles, the expression of TnT isoform genes is coupled with TnI isoforms and MHC isoforms (24, 25). Slow skeletal muscle TnI is the sole TnI expressed in embryonic cardiac muscle but replaced by cardiac TnI during perinatal development (26, 27). However, toad heart expresses normal cardiac forms of other myofilament proteins. No skeletal muscle isoforms of other myofilament proteins was seen in the toad heart, indicating no overall change in cellular gene regulations. The selectively activation of the ssTnT gene and inactivation of the cardiac TnT gene in adult toad heart, therefore, suggested a change in specific regulatory factors other than the altered overall cellular environment or a loosened ssTnT gene regulation. To further investigate the transcriptional regulation of the toad ssTnT gene may lead to a better understanding of the regulation of cardiac-specific genes in general. More investigation is also needed to understand why the cardiac TnT gene ceased expression in the toad heart.
In summary, the significance of our findings is 2-fold. The first is that cardiac muscle cells have the potential, at least in lower vertebrates, to switch TnT isoform gene expression in functional adaptations. The other is that by changing the function of a single thin filament regulatory protein, TnT, it is possible to increase the systolic function of the heart. The finding that the function of TnT may improve systolic function of the heart suggests a potential target for the treatment of heart failure, in which pharmacologically altering the function of normal cardiac TnT, rather than switching to ssTnT, may provide similar therapeutic benefits.
Supplementary Material
Acknowledgments
We thank Hui Wang for technical assistance. Dr. Jimin Gao participated in the cloning of cDNAs encoding toad ssTnT and cardiac TnI when this research group was at the ENH Research Institute and Northwestern University Feinberg School of Medicine.
This work was supported, in whole or in part, by National Institutes of Health Grants AR048816 and HL098945 (to J. P. J.).

This article contains supplemental Fig. S1 and Table S1.
- ssTnT
- slow skeletal muscle troponin T
- bpm
- beat per minute
- MHC
- myosin heavy chain
- TnI
- troponin I
- TnT
- troponin T.
REFERENCES
- 1. Boral M. C., Deb C. (1970) Seasonal changes in body fluids and hematology in toad (Bufo melanostictus)—a poikilothermic cold torpor. Proc. Indian Natl. Sci. Acad. 36, 369–373 [Google Scholar]
- 2. Deb C., Chatterjee S., Boral M. C. (1974) Body fluid and hematological changes in toads following heat exposure. Am. J. Physiol. 226, 408–410 [DOI] [PubMed] [Google Scholar]
- 3. Schadt J. C., Ludbrook J. (1991) Hemodynamic and neurohumoral responses to acute hypovolemia in conscious mammals. Am. J. Physiol. 260, H305-H318 [DOI] [PubMed] [Google Scholar]
- 4. Peitzman A. B., Billiar T. R., Harbrecht B. G., Kelly E., Udekwu A. O., Simmons R. L. (1995) Hemorrhagic shock. Curr. Probl. Surg. 32, 925–1002 [DOI] [PubMed] [Google Scholar]
- 5. Gordon A. M., Homsher E., Regnier M. (2000) Regulation of contraction in striated muscle. Physiol. Rev. 80, 853–924 [DOI] [PubMed] [Google Scholar]
- 6. Jin J. P., Yang F. W., Yu Z. B., Ruse C. I., Bond M., Chen A. (2001) The highly conserved COOH terminus of troponin I forms a Ca2+-modulated allosteric domain in the troponin complex. Biochemistry 40, 2623–2631 [DOI] [PubMed] [Google Scholar]
- 7. Chong S. M., Jin J. P. (2009) To investigate protein evolution by detecting suppressed epitope structures. J. Mol. Evol. 68, 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jin J. P., Chen A., Ogut O., Huang Q. Q. (2000) Conformational modulation of slow skeletal muscle troponin T by an NH2-terminal metal-binding extension. Am. J. Physiol. Cell Physiol. 279, C1067-C1077 [DOI] [PubMed] [Google Scholar]
- 9. Jin J. P., Malik M. L., Lin J. J. (1990) Monoclonal antibodies against cardiac myosin heavy chain. Hybridoma 9, 597–608 [DOI] [PubMed] [Google Scholar]
- 10. Feng H. Z., Chen M., Weinstein L. S., Jin J. P. (2008) Removal of the N-terminal extension of cardiac troponin I as a functional compensation for impaired myocardial β-adrenergic signaling. J. Biol. Chem. 283, 33384–33393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feng H. Z., Biesiadecki B. J., Yu Z. B., Hossain M. M., Jin J. P. (2008) Restricted N-terminal truncation of cardiac troponin T. A novel mechanism for functional adaptation to energetic crisis. J. Physiol 586, 3537–3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mazza R., Gattuso A., Mannarino C., Brar B. K., Barbieri S. F., Tota B., Mahata S. K. (2008) Catestatin (chromogranin A344–364) is a novel cardiosuppressive agent. Inhibition of isoproterenol and endothelin signaling in the frog heart. Am. J. Physiol. Heart Circ. Physiol 295, H113-H122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brotto M. A., Biesiadecki B. J., Brotto L. S., Nosek T. M., Jin J. P. (2006) Coupled expression of troponin T and troponin I isoforms in single skeletal muscle fibers correlates with contractility. Am. J. Physiol. Cell Physiol. 290, C567-C576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jin J. P., Zhang Z., Bautista J. A. (2008) Isoform diversity, regulation, and functional adaptation of troponin and calponin. Crit. Rev. Eukaryot. Gene Expr. 18, 93–124 [DOI] [PubMed] [Google Scholar]
- 15. Perry S. V. (1998) Troponin T. Genetics, properties and function. J. Muscle Res. Cell Motil. 19, 575–602 [DOI] [PubMed] [Google Scholar]
- 16. Perry S. V. (1999) Troponin I. Inhibitor or facilitator. Mol. Cell. Biochem. 190, 9–32 [PubMed] [Google Scholar]
- 17. Perry S. V. (2008) Background to the discovery of troponin and Setsuro Ebashi's contribution to our knowledge of the mechanism of relaxation in striated muscle. Biochem. Biophys. Res. Commun. 369, 43–48 [DOI] [PubMed] [Google Scholar]
- 18. Wei B., Jin J. P. (2011) Troponin T isoforms and posttranscriptional modifications. Evolution, regulation, and function. Arch. Biochem. Biophys. 505, 144–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Simons J. R. (1957) J. Physiol. 137, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hillman S. S., Withers P. C. (1988) The hemodynamic consequences of hemorrhage and hypernatremia in two amphibians. J. Comp. Physiol. B 157, 807–812 [DOI] [PubMed] [Google Scholar]
- 21. Barton P. J., Felkin L. E., Koban M. U., Cullen M. E., Brand N. J., Dhoot G. K. (2004) The slow skeletal muscle troponin T gene is expressed in developing and diseased human heart. Mol. Cell Biochem. 263, 91–97 [DOI] [PubMed] [Google Scholar]
- 22. Feng H. Z., Wei B., Jin J. P. (2009) Deletion of a genomic segment containing the cardiac troponin I gene knocks down expression of the slow troponin T gene and impairs fatigue tolerance of diaphragm muscle. J. Biol. Chem. 284, 31798–31806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang Q. Q., Feng H. Z., Liu J., Du J., Stull L. B., Moravec C. S., Huang X., Jin J. P. (2008) Co-expression of skeletal and cardiac troponin T decreases mouse cardiac function. Am. J. Physiol. Cell Physiol 294, C213-C222 [DOI] [PubMed] [Google Scholar]
- 24. Feng H. Z., Chen M., Weinstein L. S., Jin J. P. (2011) Improved fatigue resistance in Gαs-deficient and aging mouse skeletal muscles due to adaptive increases in slow fibers. J. Appl. Physiol. 111, 834–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu Z. B., Gao F., Feng H. Z., Jin J. P. (2007) Differential regulation of myofilament protein isoforms underlying the contractility changes in skeletal muscle unloading. Am. J. Physiol. Cell Physiol. 292, C1192–C1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jin J. P. (1996) Alternative RNA splicing-generated cardiac troponin T isoform switching. A non-heart-restricted genetic programming synchronized in developing cardiac and skeletal muscles. Biochem. Biophys. Res. Commun. 225, 883–889 [DOI] [PubMed] [Google Scholar]
- 27. Saggin L., Gorza L., Ausoni S., Schiaffino S. (1989) Troponin I switching in the developing heart. J. Biol. Chem. 264, 16299–16302 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.