Abstract
Background
The identification of the JAK2V617F mutation is mandatory in the diagnostic work-up of Philadelphia chromosome-negative myeloproliferative neoplasms. Several molecular techniques to detect this mutation are currently available, but each of them has some limits.
Design and Methods
We set up a novel molecular method for the identification of the JAK2V617F mutation based on an allele-specific loop-mediated amplification, not polymerase chain reaction analysis. This innovative technique amplifies DNA targets under isothermal conditions with high specificity, efficiency and rapidity. The method does not require either a thermal cycler or gel separation and the DNA amplification reaction is visible to the naked eye and can be monitored by turbidimetry. This method was validated on DNA from cell lines as well as from patients with myeloproliferative neoplasms. The results were compared with those obtained by conventional polymerase chain reaction methods.
Results
This assay detects, within 1 hour, the JAK2V617F mutation down to an allele burden of 0.1–0.01%. All samples positive by polymerase chain reaction (n=146) proved positive when tested by allele-specific loop-mediated amplification and none of the 80 negative controls gave false positive results. In addition, six patients with essential thrombocythemia previously diagnosed as being JAK2V617F negative by polymerase chain reaction analysis were found to be positive (at a low level) by allele-specific loop-mediated amplification. Furthermore, this assay discriminated the amount of JAK2V617F tumor allele within intervals of positivity, above 50%, between 50% and 10% and below 10%.
Conclusions
Allele-specific loop-mediated amplification is a simple, robust and easily applicable method for the molecular diagnosis and monitoring of JAK2V617F mutation in patients with chronic myeloproliferative neoplasms.
Keywords: myeloproliferative neoplasms, JAK2V617F mutation, LAMP
Introduction
Polycythemia vera, essential thrombocythemia and myelofibrosis are heterogeneous clonal stem cell disorders classified as Philadelphia chromosome-negative chronic myeloproliferative neoplasms, which are characterized by increased proliferation of erythroid, megakaryocytic and myeloid lineages.1 Despite the recent discoveries of various different genetic abnormalities involved in the pathogenesis of myeloproliferative neoplasms, the JAK2V617F point mutation continues to occupy a unique position, since it is the most common lesion found in these diseases, occurring in the majority of cases of polycythemia vera (>95%) and in more than half of patients with essential thrombocythemia and myelofibrosis. This mutation leads to constitutive activation of the JAK-STAT signaling pathway2,3 and there is strong evidence that hematopoietic precursors bearing this mutation acquire proliferative and survival advantages.4–6 The presence of the JAK2V617F mutation is now included as a major criterion in the revised World Health Organization (WHO) diagnostic criteria for polycythemia vera, whereas it is considered as a clonal marker for essential thrombocythemia and myelofibrosis.7 Molecular analysis of this mutation is, therefore, mandatory in the diagnostic work-up of myeloproliferative neoplasms and it is no surprise that a wide variety of methodologies have been described for its detection.8–11 Considering that JAK2V617F allele burden at diagnosis is highly variable, the method chosen should be sensitive enough to detect low levels of mutant alleles, but also specific enough not to produce false positive results.
The JAK2V617F mutation can be present in a heterozygous state or there can be progression to homozygosity, most frequently by a mitotic recombination event resulting in uniparental disomy. Different studies have suggested that in essential thrombocythemia and polycythemia vera a higher allele burden is associated with disease progression to myelofibrosis,12–15 but, in contrast, in myelofibrosis, a low allele burden is related to poorer overall and leukemia-free survival.16 An estimation of the allele burden is, therefore, clinically relevant and can be an important marker in monitoring the molecular response to the novel JAK2 inhibitors.17,18
Several qualitative and quantitative molecular assays are currently available for the detection of the JAK2V617F mutation but each of them has some important limitations, such as low sensitivity and reproducibility and the requirement of labor-intensive procedures performed with expensive specialized equipment that may not always be readily available in clinical laboratories. Allele-specific oligonucleotide polymerase chain reaction (ASO-PCR) analysis is the most commonly used technique and allows the detection of an allele burden down to 1%, but it requires hands-on-time and care to achieve unambiguous results and is not a quantitative method.3 In contrast, amplification of exon 14 of JAK2, followed by enzymatic digestion, can provide a reliable evaluation of the JAK2V617F allele burden.19 Pyrosequencing has the advantage of providing sequence data in “real time”, without an additional post-PCR procedure, and it is also semi-quantitative. However, its sensitivity is between 5% and 10%.20 Finally, different quantitative assays have been described in the literature and some of them are commercially available but, as yet, accurate quantification of the JAK2V617F allele burden is still far from being a routine practice.21
Here we present a novel, non-PCR molecular method for the identification of the JAK2V617F mutation, based on the principle of loop-mediated isothermal amplification (LAMP).22 This technique, called allele-specific (AS)-LAMP, generates a large amount of amplified DNA within 1 h. The DNA is visible to the naked-eye and can also be monitored by real-time turbidimetry and does not require gel separation. This simple, easy to perform and rapid assay selectively detects the JAK2V617F mutated DNA with high sensitivity.
Design and Methods
Cell lines
Genomic DNA was extracted using a GENTRA kit (Qiagen, Hilden, Germany) from the following JAK2V617F-positive human cell lines: HEL (obtained from the German Collection of Microorganisms and Cell Culture, DSMZ, Braunschweig, Germany), UKE-1 (a kind gift from Walter Fiedler, Eppendorf Hospital, Hamburg, Germany), both harboring only JAK2V617F mutated alleles, and SET-2 (DSMZ), carrying wild-type and JAK2V617F mutated alleles. As negative controls, the following cell lines (all carrying only wild-type JAK2 alleles) were also used: HL60 (acute myeloid leukemia), TF1 (acute myeloid leukemia), GFD8 (acute myeloid leukemia with complex karyotype), KASU-MI-1 [a t(8;21) acute myeloid leukemia], K562 (a BCR/ABL-positive chronic myeloid leukemia in erythroid blast crisis), NB4 [a t(15;17)-positive acute promyelocytic leukemia], BJAB (a Burkitt, non-Hodgkin’s lymphoma), and U266B1 (a multiple myeloma). These cell lines were validated by analyzing the presence of specific rearrangements by PCR (K562 for BCR/ABL b3a2 transcript) (NB4 for PML/RARα BCR1 transcript) or cytogenetics (BJAB for c-myc amplification) (data not shown). In addition, all cell lines were tested by ASO-PCR, for the presence of JAK2V617F mutation and only HEL, UKE-1, and SET-2 proved positive. These latter cell lines were also tested to estimate the JAK2V617F allele burden: HEL and UKE-1 had only JAK2V617F mutated alleles whereas SET-2 had both wild-type and JAK2V617F mutated alleles (data not shown). In addition, the HEL, UKE-1, SET-2, BJAB and K562 cell lines were also tested for JAK2 allele copy number by fluorescence in situ hybridization using a bacterial artificial chromosome (RP11-927H16) encompassing the entire JAK2 gene (kindly provided by Prof. Cristina Mecucci, University of Perugia, Italy) and quantitative PCR with the following primers: JAK2 gene: forward primer: 5′-AAGCTTTCTCACAAGCATTTGGTTT-3′, reverse primer 5′-AGAAAGGCATTAGAAAGCCTGTAGTT-3′, probe FAM-5′-TGTGGAGACGAGAGTAAGT-3′-TAMRA; Albumin gene: forward primer : 5′–TGAAACATACGTTCCCAAAGAGTTT-3′, reverse primer: 5′–CTCTCCTCCTCAGAAGTGTGCATAT-3′, probe FAM -5′-GCTGAAACATTCACCTTCCATGCAGA-3′ TAMRA; β2 microglobulin gene forward primer: 5′-GGAATTGATTTGGGAGAGCATC-3′, reverse primer: 5′-AGTGTGACTGGGCAGATCATCCACCTTC-3′, probe: FAM-5′-AGTGTGACTGGGCAGATCATCCACCTTC3′-TAMRA. The JAK2 copy number was calculated as previously described.23 In keeping with published results,24 a concordant JAK2 copy number was defined by both approaches: UKE-1, two copies; HEL, 12 copies; SET-2, five copies; BJAB, two copies; and K562, two copies (data not shown).
Patients
Genomic DNA was obtained from the peripheral blood of patients with polycythemia vera (n=36), essential thrombocythemia (n=58), myelofibrosis (n=4) and unclassified myeloproliferative neoplasms (n=2). For control purposes, DNA was also extracted from normal healthy donors (n=73) and patients with acute lymphoblastic leukemia (n= 2), follicular non-Hodgkin’s lymphoma (n= 2), or B-cell chronic lymphocytic leukemia (n= 2). All these subjects are routinely followed for clinical reasons at Ospedali Riuniti di Bergamo. Sample preservation and genetic analysis required informed consent and approval by the Ethics Committee.
The allele-specific loop-mediated isothermal amplification assay
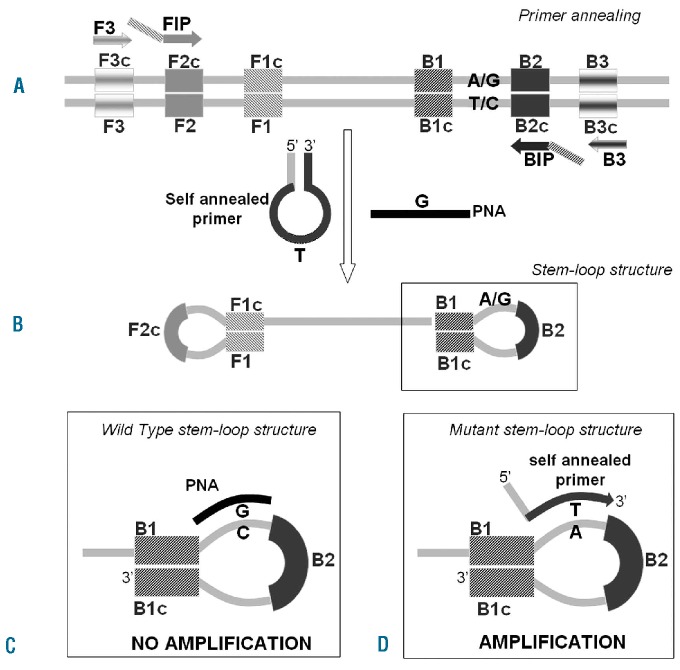
LAMP of DNA (see Online Supplementary Movie) was initially described by Notomi as an innovative tool to amplify DNA with high specificity, efficiency and rapidity under isothermal conditions.22 The AS-LAMP reaction (Figure 1) is based on the use of four primers specifically designed to recognize six distinct regions on the target gene: a pair of outer primers [F3 (GA231 5′ GCATCTTTATTATGGCAGAGAG 3′); and B3 (GA232 5′ TGCTCTGAGAAAGGCATTA 3′)] and a pair of inner primers [FIP (GA233 5′ GCTGCTTCAAAGAAAGACTAAGGAAATG-GACAACAGTCAAACAAC 3′) and BIP (GA234 5′GCTTTCTCACAAGCATTTGGTTTTAAATTAGCCTGTAGTTTTACTTACTCTC 3′)] presenting a tag complementary to a downstream region in the opposite strand of the target (F1 and B1). The method also includes an additional “self-annealed” primer (GA236 5′ GTCTCCACTGGAGTATGTTTCTGTGGAGAC 3′) complementary to the loop region in which the JAK2 T mutated base is located plus a peptide nucleic acid probe specific for the JAK2 G wild-type nucleotide. The reaction is conducted at a constant temperature in the presence of a DNA polymerase with strand displacement activity. The FIP and BIP primers anneal and are extended on the target DNA and the newly synthesized DNA chains are then displaced by extension of F3 and B3 (Figure 1A). The displaced product generates a “stem loop structure” which represents the starting structure for a classical LAMP reaction (Figure 1B). If the target gene is wild-type, the specific peptide nucleic acid forms a stable duplex with the dumbbell structure, thus interfering with the annealing and extension of the mutant self-annealed primer (Figure 1C). In the case of a few molecules not bound by the peptide nucleic acid, the amplification still does not proceed thanks to the auto-sequestration of the self-annealed primer, which cannot be extended by the polymerase since the melting temperature of the intra-molecular hairpin structure is higher than that of the primer-target hybridization one. If the target gene is mutated, the peptide nucleic acid does not anneal because of the single-base mismatch, while the self-annealed loop primer breaks its internal interaction to bind the target, consequently becoming extended (Figure 1D). The assay was performed at a constant temperature of 65°C for 1 h on an LA200 turbidimeter (Teramecs Co, Kyoto, Japan) for real-time monitoring. The variation of absorbance [in arbitrary units (a.u.)] can be analyzed to find the threshold time for each sample tested. The threshold time is the minute at which the sample absorbance, after baseline subtraction, reaches the a.u. value representing the threshold of 0.1 a.u., which coincides with the beginning of the exponential phase of the amplification process. It guarantees reliable results and is commonly adopted in LAMP applications used for the molecular diagnosis of infectious diseases.25 The threshold time [T(t)min] of each sample is correlated with the logarithm of DNA mutated copies per reaction. A LAMP control reaction to validate negative results is also performed in parallel by a mutant-insensitive amplification of the JAK2 gene using the same basic primer set described above and avoiding the peptide nucleic acid and self-annealed mutant loop primer in the reaction mixture. The amplification resulting from the control LAMP assay is indicative of correct annealing and extension of the LAMP primers as well as of the absence of inhibitors in the reaction solution.
Figure 1.
The principle of AS-LAMP. (A) At a constant reaction temperature FIP and BIP are extended on the target DNA and the newly synthesized DNA chains are then displaced by extension of F3 and B3. (B) The displaced product generates a “stem-loop structure” which represents the starting structure for a LAMP reaction. (C) If the target in the reaction is wild-type, the peptide nucleic acid (PNA) forms a stable duplex with the stem-loop structure, preventing the annealing and extension of the mutant self-annealed primer and, therefore, suppressing the amplification. (D) If the target in solution is mutated, the PNA does not anneal, while the self-annealed primer breaks its internal interaction to bind its target and finally it is extended. The amplification can, therefore, proceed.
Evaluation of the JAK2V617F allele burden by allele-specific loop-mediated isothermal amplification
In order to quantify allele burden, a standard curve was prepared using three calibrators containing genomic DNA from the UKE-1 cell line (homozygous for the JAK2V617F mutation) diluted (100%, 10% and 1%) into DNA from the BJAB cell line (wild-type JAK2). Each calibrator was used in triplicate. The threshold time of unknown samples can be plotted in the standard curve to calculate the amount of JAK2V617F mutated allele. Samples with a T(t)min between 10% and 100% of the calibrators’ T(t)min are further analyzed after 1:5 dilution in Tris with yeast RNA. If the amount of JAK2V617F mutant allele exceeds 50%, the amplification of both the undiluted and the 1:5 diluted samples occurs before the threshold time of the 10% calibrator. In the case of a JAK2V617F mutant allele between 10% and 50%, the T(t)min of the 1:5 dilution occurs after that of the 10% calibrator. Samples with an allele burdens below 10% can be directly defined as low positive (<10%).
Identification of JAK2V617F by polymerase chain reaction amplification
The ASO-PCR for the JAK2V617F mutation was performed as previously described.3 Positivity was confirmed and the allele burden estimated by amplification of exon 14 and subsequent digestion with BsaXI (New England Biolabs, Ipswich, MA, USA).26
Polymerase chain reaction-clamping and sequencing
In case of discordant results obtained by AS-LAMP and ASO-PCR on clinical samples, a PCR amplification of the JAK2 region was also performed in the presence of the peptide nucleic acid, which inhibits amplification of the wild-type allele thus leading to increased amplification of the mutated allele. The PCR reaction was performed in the presence of 1x reaction buffer, 2.5 mM MgCl2, 200 μM dNTP, 500 nM primers (F3 forward and B3 reverse, used for LAMP), 1.5 μM peptide nucleic acid (the same as that used in the LAMP), 1.25U Taq Gold (Applied Biosystem, Carlsbad, CA, USA) and 100 ng DNA in a final volume of 50 μL. The resulting solution was amplified using the following program: 10 min at 95°C; 35 cycles of 30 sec at 94°C, 40 sec at 62°C, 30 sec at 58°C and 30 sec at 72°C; and a final extension for 10 min at 72°C. If the mutated region is enriched at least to a level of 20% of the amplified product, it can be detected by direct sequencing. This technique was validated by amplifying and sequencing UKE-1 DNA diluted into BJAB DNA (10%, 1%, 0.9%, 0.8%, 0.7%, 0.6%, 0.5%, 0.4%, 0.3%, 0.2% and 0.1%) and negative controls (wild-type cell lines, 11 replicates).
Results
Sensitivity and specificity of the allele-specific loop-mediated isothermal amplification assay
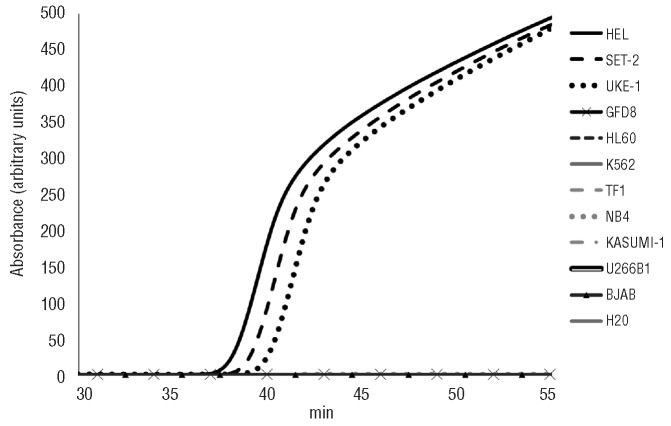
The AS-LAMP method was initially tested on genomic DNA from three JAK2V617F mutant (UKE-1, SET-2 and HEL) and eight wild-type JAK2 cell lines (HL60, TF1, GFD8, KASUMI-1, K562, NB4, BJAB and U266B1). After reaction for 1 h, no amplification was observed in the cell lines with wild-type JAK2, while all the JAK2V617F mutants proved positive. All the negative results were validated by a control reaction performed in parallel to exclude inhibitors or incorrect reaction conditions (Figure 2).
Figure 2.
Specificity of AS-LAMP on cell lines. Eight JAK2 wild-type (GFD8, HL60, K562, TF1, NB4, KASU-MI-1, U266B1, BJAB) and three JAK2V617F mutant cell lines (HEL, SET-2 and UKE-1) were analyzed by AS-LAMP. Within 60 min, only DNA samples obtained from mutant cell lines showed an amplified product, while no amplification was detected in any of the wild-type samples.
The AS-LAMP assay was further validated on 66 samples obtained from patients with polycythemia vera (n=36), essential thrombocythemia (n=24), myelofibrosis (n=4) or unclassified myeloproliferative neoplasms (n=2) who had been previously analyzed and found to be JAK2V617F-positive by conventional ASO-PCR. There was a 100% concordance between the results of the two assays for all samples. In addition, 80 DNA samples obtained from the peripheral blood of healthy donors (n=73) or patients with acute lymphoblastic leukemia (n=2), follicular non-Hodgkin’s lymphoma (n=2) or B-cell chronic lymphocytic leukemia (n=2) proved consistently negative (data not shown).
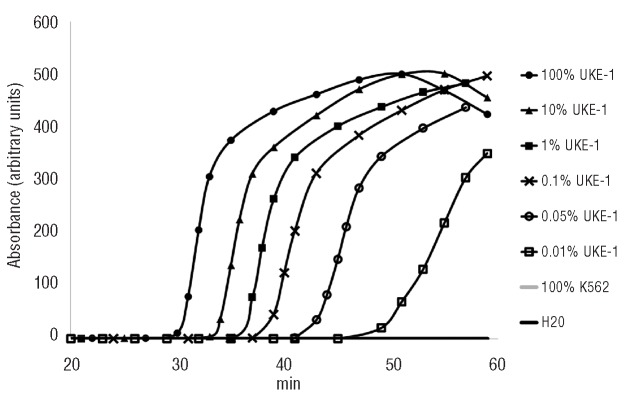
To evaluate the ability of the AS-LAMP to detect low amounts of JAK2V617F mutated DNA, we optimized the assay on a JAK2V617F-containing plasmid serially diluted into a wild-type JAK2 plasmid. This enabled us to reach a sensitivity of 0.05% (70 mutated copies/350000 total plasmid copies) (data not shown). Furthermore, we performed the assay using genomic DNA from the UKE-1 cell line serially diluted (100, 10, 1, 0.1, 0.05, 0.01 and 0%) into wild-type DNA from K562 cells. The reproducible sensitivity of AS-LAMP was 0.1%, detected in 100% of cases. The 0.05% dilution was revealed in 50% of cases, while the 0.01% dilution was revealed in 38% of cases, this being the level of maximal sensitivity of the assay (Figure 3). A linear relationship (R2=0.99) between the threshold time and the logarithm-dilution factor of the mutant DNA was observed between the 100% and 0.1% doses. We also used AS-LAMP to test clinical samples from 34 patients with essential thrombocythemia who were negative for the JAK2V617F mutation according to ASO-PCR. Interestingly, the AS-LAMP assay led to six of these patients being reclassified as JAK2V617F-positive, albeit with a low tumor allele burden (≤1%). In addition, a patient with post-polycythemia vera myelofibrosis who achieved complete hematologic remission after allogeneic transplantation proved molecularly negative by repeated ASO-PCR assays, but low positive by AS-LAMP. This result was confirmed by PCR-clamping followed by direct sequencing, thus proving the superior ability of the AS-LAMP assay to detect minute amounts of the mutated allele. To assess and validate the sensitivity of the PCR-clamping analysis, we amplified and sequenced several dilutions of UKE-1 DNA into wild-type BJAB DNA (10%, 1%, 0.9%, 0.8%, 0.7%, 0.6%, 0.5%, 0.4%, 0.3%, 0.2% and 0.1%). With this approach, the JAK2V617F mutation could be detected by sequencing down to 0.6% (maximal sensitivity of the test). Using this approach, no mutant sequences were detected in DNA from wild-type cell lines used as negative controls (11 replicates). All in all, these findings confirm that discordant results obtained in clinical samples can be solved by PCR clamping.
Figure 3.
Sensitivity of AS-LAMP on genomic DNA. Serial dilutions of mutant UKE-1 DNA in wild-type K562 DNA at concentrations of 100, 10, 1, 0.1, 0.05, 0.01 and 0% (100% K562) were analyzed by AS-LAMP. Mutant alleles could be reproducibly detected down to a mutant-to-wild-type ratio of 0.1% within 1 h. The 0.05% dilution is detectable with a probability of 50%, while the 0.01% dilution with a probability of 38%. Negative samples did not generate specific amplification signals.
By the addition of an intercalating dye, the AS-LAMP reaction was also monitored on a real-time instrument with a comparable rapid response and sensitivity (data not shown).
Evaluation of JAK2V617F allele burden in clinical samples by allele-specific loop-mediated isothermal amplification
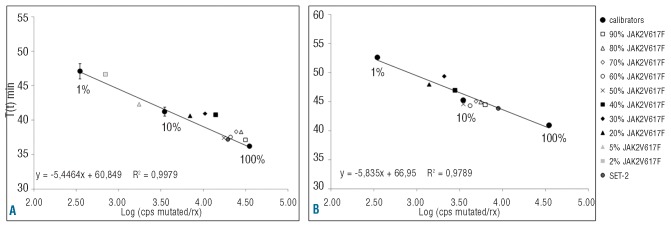
To test the ability of the assay to define the JAK2V617F allele burden, experiments were planned using genomic DNA from UKE-1 cells serially diluted into wild-type DNA from BJAB cells. Samples containing a known amount of JAK2V617F-mutated DNA (from 90% to 2%) were analyzed and compared to the standard curve prepared using three calibrators containing JAK2V617F allele burdens of 100%, 10% and 1% (Figure 4A). Samples with a detected T(t)min between those of the 10% and 1% calibrators could be directly defined as low positive (allele burden <10%). For samples containing a JAK2V617F allele burden above 10%, the efficiency and speed of AS-LAMP do not allow immediate definition of the tumor allele burden (Figure 4A). After 1:5 dilution, samples with an allele burden below 50% showed a T(t)min higher than the 10% calibrator while samples with an allele burden between 90% and 60% showed a T(t)min between those of the 100% and 10% calibrators. DNA from SET-2 cells showed an allele burden above 50% (Figure 4B). We then tested the AS-LAMP assay on DNA obtained from freshly isolated peripheral blood leukocytes from 19 patients with polycythemia vera in whom the JAK2V617F allele burden had been previously defined by a conventional enzymatic digestion method and in all cases AS-LAMP properly defined a tumor allele burden above or below 50% (data not shown).
Figure 4.
Allele burden determination. (A) A standard curve of the AS-LAMP assay was created using three calibrators (1%-10%-100% JAK2V617F allele) and the indicated DNA dilutions of UKE-1 into BJAB (from 90% to 2%) were tested. DNA samples containing an allele burden of 2% and 5% could be directly defined as low positive (allele burden between 1% and 10%). DNA samples containing a JAK2V617F allele burden between 20% and 90% were appropriately defined as having an allele burden between 10% and 100%. (B) A further 1:5 dilution of these latter samples allowed discrimination of samples into those with a JAK2V617F allele burden above or below 50% (see text).
Discussion
Here we have described a new molecular assay for the identification of the JAK2V617F mutation. This assay is a non-PCR DNA amplification method based on the LAMP principle and is called AS-LAMP. LAMP is an innovative technique that rapidly amplifies DNA or RNA targets under isothermal conditions in which reagents react fast with high specificity and efficiency.27 LAMP is performed using strand-displacement polymerase and does not require Taq DNA polymerase or thermal cycling. The potential diagnostic applications of this technology are remarkable and rapidly expanding, particularly in the field of infectious diseases.28–30 The simplicity of its use is such that LAMP may be applied in low income, developing countries facing significant problems in healthcare.31 The AS-LAMP we developed allowed rapid and robust identification of DNA samples harboring the JAK2V617F mutation. No false positive or negative results, compared to those obtained by ASO-PCR or direct sequencing, were recorded. This new assay also proved remarkably sensitive since the mutated JAK2V617F DNA could be detected down to 0.01% with a reproducible sensitivity of 0.1%. Finally, the assay allows accurate definition of whether the JAK2V617F allele burden is higher or lower than 50%. The method we have described is rapid, easy to perform and does not require either a thermal cycler or gel separation, thus reducing the risk of contamination due to post-amplification manipulation and the costs of the procedure. In fact, the proceeding reaction is visible to the naked eye and can be monitored by real-time turbidimetry thanks to pyrophosphate salts generated as a by-product of the DNA amplification process.32 When compared to other molecular methods currently employed to detect the JAK2V617F mutation, AS-LAMP proved more sensitive than ASO-PCR, more accurate than enzymatic digestion of the PCR products and did not require the use of real-time PCR instruments. All these characteristics make this method feasible in routine diagnostic laboratories even in the absence of major technical infrastructure or highly qualified personnel. The simplicity and rapidity of AS-LAMP (less than 1 h compared to at least 4 h for ASO-PCR) enables the clinical sample to be processed immediately after sampling without the support of a specialized molecular laboratory. Considering the widespread request for molecular profiling of patients with a suspected chronic myeloproliferative disorder, this assay may represent a substantial improvement.
When compared to ASO-PCR, this method significantly increases our ability to detect a low tumor allele burden, which is important when monitoring patients treated with the aim of eradicating the disease, such as those undergoing allogeneic transplantation.33
The evaluation of tumor allele burden is clinically relevant since various studies have shown that a high allele burden is associated with a faster transformation to myelofibrosis and acute myeloid leukemia12–15 and a higher risk of thrombotic events.34–36 In its present form AS-LAMP is capable of reproducible detection of very small numbers of JAK2V617F-positive hematopoietic cells, but it cannot be used as a truly quantitative assay since the high efficiency of the DNA amplification limits per se the possibility of quantifying the amount of the mutated alleles precisely. This technique may, therefore, still have limitations if the purpose of its application is the clinical evaluation of new drugs, which may affect tumor allele burden only slightly.37 On the other hand, the current AS-LAMP assay may allocate each sample to discrete intervals of positivity, comparable to other molecular methods.38 Further development of this technique is currently ongoing: this development, based on the use of fluorochromes, will enable a pure quantitative approach.
Acknowledgments
The authors would like to thank Professor Cristina Mecucci of the University of Perugia for providing the probe for the FISH analysis.
Footnotes
The online version of this article has a Supplementary Appendix.
Funding: this work was supported in part by grants from the Associazione Italiana per la Ricerca contro il Cancro (AIRC, Milano) “Special Program Molecular Clinical Oncology 5×1000” to AGIMM (AIRC-Gruppo Italiano Malattie Mieloproliferative), project number #1005 and Associazione Italiana Lotta alla Leucemia (AIL), sezione Paolo Belli.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Tefferi A, Skoda R, Vardiman JW. Myeloproliferative neoplasms: contemporary diagnosis using histology and genetics. Nat Rev Clin Oncol. 2009;6(11):627–37. doi: 10.1038/nrclinonc.2009.149. [DOI] [PubMed] [Google Scholar]
- 2.James C, Ugo V, Casadevall N, Constantinescu SN, Vainchenker W. A JAK2 mutation in myeloproliferative disorders: pathogenesis and therapeutic and scientific prospects. Trends Mol Med. 2005;11(12):546–54. doi: 10.1016/j.molmed.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365(9464):1054–61. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 4.Dusa A, Staerk J, Elliott J, Pecquet C, Poirel HA, Johnston JA, et al. Substitution of pseudokinase domain residue Val-617 by large non-polar amino acids causes activation of JAK2. J Biol Chem. 2008;283(19):12941–8. doi: 10.1074/jbc.M709302200. [DOI] [PubMed] [Google Scholar]
- 5.Kota J, Caceres N, Constantinescu SN. Aberrant signal transduction pathways in myeloproliferative neoplasms. Leukemia. 2008;22(10):1828–40. doi: 10.1038/leu.2008.236. [DOI] [PubMed] [Google Scholar]
- 6.Vainchenker W, Dusa A, Constantinescu SN. JAKs in pathology: role of Janus kinases in hematopoietic malignancies and immunodeficiencies. Semin Cell Dev Biol. 2008;19(4):385–93. doi: 10.1016/j.semcdb.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Tefferi A, Vardiman JW. Classification and diagnosis of myeloproliferative neoplasms: the 2008 World Health Organization criteria and point-of-care diagnostic algorithms. Leukemia. 2008;22(1):14–22. doi: 10.1038/sj.leu.2404955. [DOI] [PubMed] [Google Scholar]
- 8.Konstantou JK, Iliadi AC, Ioannou PC, Christopoulos TK, Anagnostopoulos NI, Kanavakis E, et al. Visual screening for JAK2V617F mutation by a disposable dipstick. Anal Bioanal Chem. 2010;397(5):1911–6. doi: 10.1007/s00216-010-3747-z. [DOI] [PubMed] [Google Scholar]
- 9.Steensma DP. JAK2 V617F in myeloid disorders: molecular diagnostic techniques and their clinical utility: a paper from the 2005 William Beaumont Hospital Symposium on Molecular Pathology. J Mol Diagn. 2006;8(4):397–411. doi: 10.2353/jmoldx.2006.060007. quiz 526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denys B, El Housni H, Nollet F, Verhasselt B, Philippe J. A real-time polymerase chain reaction assay for rapid, sensitive, and specific quantification of the JAK2V617F mutation using a locked nucleic acid-modified oligonucleotide. J Mol Diagn. 2010;12(4):512–9. doi: 10.2353/jmoldx.2010.090137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huijsmans CJ, Poodt J, Savelkoul PH, Hermans MH. Sensitive detection and quantification of the JAK2V617F allele by real-time PCR blocking wild-type amplification by using a peptide nucleic acid oligonucleotide. J Mol Diagn. 2011;13(5):558–64. doi: 10.1016/j.jmoldx.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vannucchi AM, Antonioli E, Guglielmelli P, Pardanani A, Tefferi A. Clinical correlates of JAK2V617F presence or allele burden in myeloproliferative neoplasms: a critical reappraisal. Leukemia. 2008;22(7):1299–307. doi: 10.1038/leu.2008.113. [DOI] [PubMed] [Google Scholar]
- 13.Tefferi A. JAK and MPL mutations in myeloid malignancies. Leuk Lymphoma. 2008;49(3):388–97. doi: 10.1080/10428190801895360. [DOI] [PubMed] [Google Scholar]
- 14.Barosi G, Bergamaschi G, Marchetti M, Vannucchi AM, Guglielmelli P, Antonioli E, et al. JAK2 V617F mutational status predicts progression to large splenomegaly and leukemic transformation in primary myelofibrosis. Blood. 2007;110(12):4030–6. doi: 10.1182/blood-2007-07-099184. [DOI] [PubMed] [Google Scholar]
- 15.Tiedt R, HaoShen H, Sobas MA, Looser R, Dirnhofer S, Schwaller J, et al. Ratio of mutant JAK2-V617F to wild-type Jak2 determines the MPD phenotypes in transgenic mice. Blood. 2008;111(8):3931–40. doi: 10.1182/blood-2007-08-107748. [DOI] [PubMed] [Google Scholar]
- 16.Guglielmelli P, Barosi G, Specchia G, Rambaldi A, Lo Coco F, Antonioli E, et al. Identification of patients with poorer survival in primary myelofibrosis based on the burden of JAK2V617F mutated allele. Blood. 2009;114(8):1477–83. doi: 10.1182/blood-2009-04-216044. [DOI] [PubMed] [Google Scholar]
- 17.Verstovsek S, Kantarjian H, Mesa RA, Pardanani AD, Cortes-Franco J, Thomas DA, et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med. 2010;363(12):1117–27. doi: 10.1056/NEJMoa1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pardanani A, Gotlib JR, Jamieson C, Cortes JE, Talpaz M, Stone RM, et al. Safety and efficacy of TG101348, a selective JAK2 inhibitor, in myelofibrosis. J Clin Oncol. 2011;29(7):789–96. doi: 10.1200/JCO.2010.32.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell PJ, Scott LM, Baxter EJ, Bench AJ, Green AR, Erber WN. Methods for the detection of the JAK2 V617F mutation in human myeloproliferative disorders. Methods Mol Med. 2006;125:253–64. doi: 10.1385/1-59745-017-0:253. [DOI] [PubMed] [Google Scholar]
- 20.Jones AV, Kreil S, Zoi K, Waghorn K, Curtis C, Zhang L, et al. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood. 2005;106(6):2162–8. doi: 10.1182/blood-2005-03-1320. [DOI] [PubMed] [Google Scholar]
- 21.Lippert E, Girodon F, Hammond E, Jelinek J, Reading NS, Fehse B, et al. Concordance of assays designed for the quantification of JAK2V617F: a multicenter study. Haematologica. 2009;94(1):38–45. doi: 10.3324/haematol.13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Quentmeier H, MacLeod RA, Zaborski M, Drexler HG. JAK2 V617F tyrosine kinase mutation in cell lines derived from myeloproliferative disorders. Leukemia. 2006;20(3):471–6. doi: 10.1038/sj.leu.2404081. [DOI] [PubMed] [Google Scholar]
- 25.Nakao R, Stromdahl EY, Magona JW, Faburay B, Namangala B, Malele I, et al. Development of loop-mediated isothermal amplification (LAMP) assays for rapid detection of Ehrlichia ruminantium. BMC Microbiol. 2010;10:296. doi: 10.1186/1471-2180-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerini V, Barbui V, Spinelli O, Salvi A, Dellacasa C, Carobbio A, et al. The histone deacetylase inhibitor ITF2357 selectively targets cells bearing mutated JAK2(V617F) Leukemia. 2008;22(4):740–7. doi: 10.1038/sj.leu.2405049. [DOI] [PubMed] [Google Scholar]
- 27.Parida M, Sannarangaiah S, Dash PK, Rao PVL, Morita K. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev Med Virol. 2008;18(6):407–21. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon LLM, Wong BWY, Ma EHT, Chan KH, Chow LMC, Abeyewickreme W, et al. Sensitive and inexpensive molecular test for falciparum malaria: detecting plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal Amplification. Clin Chem. 2006;52(2):303–6. doi: 10.1373/clinchem.2005.057901. [DOI] [PubMed] [Google Scholar]
- 29.Iwamoto T, Sonobe T, Hayashi K. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J Clin Microbiol. 2003;41(6):2616–22. doi: 10.1128/JCM.41.6.2616-2622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Njiru ZK, Mikosza ASJ, Armstrong T, Enyaru JC, Ndung’u JM, Thompson ARC. Loop-mediated isothermal amplification (LAMP) method for rapid detection of Trypanosoma brucei rhodesiense. PLoS Negl Trop Dis. 2008;2(2):e147. doi: 10.1371/journal.pntd.0000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boehme CC, Nabeta P, Henostroza G, Raqib R, Rahim Z, Gerhardt M, et al. Operational feasibility of using loop-mediated isothermal amplification for diagnosis of pulmonary tuberculosis in microscopy centers of developing countries. J Clin Microbiol. 2007;45(6):1936–40. doi: 10.1128/JCM.02352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mori Y, Kitao M, Tomita N, Notomi T. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J Biochem Biophys Methods. 2004;59(2):145–57. doi: 10.1016/j.jbbm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Kroger N, Badbaran A, Holler E, Hahn J, Kobbe G, Bornhauser M, et al. Monitoring of the JAK2-V617F mutation by highly sensitive quantitative real-time PCR after allogeneic stem cell transplantation in patients with myelofibrosis. Blood. 2007;109(3):1316–21. doi: 10.1182/blood-2006-08-039909. [DOI] [PubMed] [Google Scholar]
- 34.De Stefano V, Za T, Rossi E, Vannucchi AM, Ruggeri M, Elli E, et al. Increased risk of recurrent thrombosis in patients with essential thrombocythemia carrying the homozygous JAK2 V617F mutation. Ann Hematol. 2010;89(2):141–6. doi: 10.1007/s00277-009-0788-5. [DOI] [PubMed] [Google Scholar]
- 35.Carobbio A, Finazzi G, Antonioli E, Guglielmelli P, Vannucchi AM, Dellacasa CM, et al. JAK2V617F allele burden and thrombosis: a direct comparison in essential thrombocythemia and polycythemia vera. Exp Hematol. 2009;37(9):1016–21. doi: 10.1016/j.exphem.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barbui T, Carobbio A, Cervantes F, Vannucchi AM, Guglielmelli P, Antonioli E, et al. Thrombosis in primary myelofibrosis: incidence and risk factors. Blood. 2010;115(4):778–82. doi: 10.1182/blood-2009-08-238956. [DOI] [PubMed] [Google Scholar]
- 37.Antonioli E, Carobbio A, Pieri L, Pancrazzi A, Guglielmelli P, Delaini F, et al. Hydroxyurea does not appreciably reduce JAK2 V617F allele burden in patients with polycythemia vera or essential thrombocythemia. Haematologica. 2010;95(8):1435–8. doi: 10.3324/haematol.2009.021444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiladjian JJ, Cassinat B, Chevret S, Turlure P, Cambier N, Roussel M, et al. Pegylated interferon-alfa-2a induces complete hematologic and molecular responses with low toxicity in polycythemia vera. Blood. 2008;112(8):3065–72. doi: 10.1182/blood-2008-03-143537. [DOI] [PubMed] [Google Scholar]