Abstract
Background
Mantle cell lymphoma is a clinically heterogeneous disease characterized by overexpression of cyclin D1 protein. Blastoid morphology, high proliferation, and secondary genetic aberrations are markers of aggressive behavior. Expression profiling of mantle cell lymphoma revealed that predominance of the 3’UTR-deficient, short cyclin D1 mRNA isoform was associated with high cyclin D1 levels, a high “proliferation signature” and poor prognosis.
Design and Methods
Sixty-two cases of mantle cell lymphoma were analyzed for cyclin D1 mRNA isoforms and total cyclin D1 levels by real-time reverse transcriptase polymerase chain reaction, and TP53 alterations were assessed by immunohistochemistry and molecular analysis. Results were correlated with proliferation index and clinical outcome.
Results
Predominance of the short cyclin D1 mRNA was found in 14 (23%) samples, including four with complete loss of the standard transcript. TP53 alterations were found in 15 (24%) cases. Predominance of 3’UTR-deficient mRNA was significantly associated with high cyclin D1 mRNA levels (P=0.009) and more commonly found in blastoid mantle cell lymphoma (5/11, P=0.060) and cases with a proliferation index of >20% (P=0.026). Both blastoid morphology (11/11, P<0.001) and TP53 alterations (15/15, P<0.001) were significantly correlated with a high proliferation index. A proliferation index of 10% was determined to be a significant threshold for survival in multivariate analysis (P=0.01).
Conclusions
TP53 alterations are strongly associated with a high proliferation index and aggressive behavior in mantle cell lymphoma. Predominance of the 3’UTR-deficient transcript correlates with higher cyclin D1 levels and may be a secondary contributing factor to high proliferation, but failed to reach prognostic significance in this study.
Keywords: cyclin D1 mRNA, isoforms, mantle cell lymphoma, prognosis
Introduction
Mantle cell lymphoma (MCL) is an aggressive B-cell lymphoma genetically characterized by the t(11;14)(q13;q32) translocation leading to overexpression of cyclin D1 (CyD1), which is not normally expressed in lymphoid cells.1,2 Although aberrant expression of CyD1 is seen in almost 100% of MCL and a subset of other B-cell neoplasms, including 40% of multiple myelomas and hairy cell leukemias due to various mechanisms,3,4 the precise role of CyD1 overexpression in lymphomagenesis is still elusive. D-type cyclins, which act as growth factor sensors, are not per se sufficient to keep cells in cycle and need the cooperation of other oncogenes to induce malignant transformation.5–7
Although MCL is a quite homogenous disease based on phenotype and gene expression profile, the clinical course of MCL is highly variable, with survival times ranging from a few months to several years. Various studies have tried to identify prognostic markers that predict the clinical course and are able to distinguish aggressive variants from indolent MCL. Alterations in cell cycle regulators, including p53, CDK4 and p16/CDKN2, and DNA damage response pathways have been found to be associated with high proliferation index and aggressive clinical behavior.8–12 In particular, cases of MCL with inactivating TP53 mutations have a significantly shorter median survival compared to cases with wild-type TP53.9,11,13
The clinically aggressive blastoid variant of MCL, characterized by either lymphoblast-like or large cells (pleomorphic type), shows a higher frequency of these additional genetic aberrations, such as abnormalities of C-MYC, TP53, p16/CDKN2 and p14ARF, as well as, the common occurrence of hyperploidy or tetraploidy.14,15
Using global gene expression profiling, it was shown that MCL cases predominantly or exclusively expressing a short 3’UTR-deficient CyD1 mRNA isoform have higher CyD1 mRNA levels, a higher “proliferation signature” and a significantly poorer survival than cases which express the standard full-length transcript.16 The long 3’UTR contains AU-rich mRNA destabilizing elements, and the 3’UTR-deficient mRNA is thought to exhibit increased stability. In addition, the 3’UTR is targeted by microRNA (miRNA) of the miR-15/16 family and the miR17-92 cluster, indirectly reducing CyD1 protein.17 Loss of 3’UTR can be caused by genomic deletion or rearrangement of the region encoding the 3’UTR, or through mutations in the 3’UTR, which introduce alternative polyadenylation signal sites.18,19
Although the proliferation signature was suggested to represent an integrated measurement of different oncogenic events targeting the cell cycle in MCL, it was not correlated with the proliferation index as assessed by Ki-67 immunostaining, a conventional examination available in all pathology departments.16 Nonetheless, other studies using Ki-67 staining have confirmed that the proliferation index is one of the best prognostic indicators for MCL, even in the context of different novel therapy regimes.20,21 To date, no systematic correlative analysis of CyD1 mRNA isoform expression, alterations of TP53, and cell proliferation as determined by Ki-67 staining has been performed in MCL. The aim of our study was, therefore, to clarify the relationship and relative importance of total CyD1 expression, CyD1 isoform expression, genomic loss of the 3′ UTR, and p53 abnormalities with the proliferation index, and morphological subtypes of MCL.
Design and Methods
Case selection
Paraffin-embedded blocks from 62 patients with CyD1-positive MCL obtained at primary diagnosis were selected from the archives of the Institute of Pathology, Technical University of Munich, Germany and from the Tour de Pathologie, Liege, Belgium. Additional samples from sequential biopsies taken 7 months to 6 years later were available for four patients. Reactive lymph nodes and small intestinal mucosa samples were used as controls for quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) and fluorescent in situ hybridization (FISH) analysis. The diagnosis of MCL was based on morphological and immunophenotypic findings according to the World Health Organization (WHO) classification of Neoplastic Diseases of Hematopoietic and Lymphoid Tissue.22 The diagnosis of the morphological MCL subtype was based on cell morphology and blindly reviewed twice by two of the authors (LQ-M and FF). If discrepancies arose, the cases were discussed to achieve consensus. CyD1 positivity was confirmed by immunohistochemistry in all cases. The study was approved by the local ethics committee.
Immunohistochemical analysis
Immunohistochemistry for CyD1, Ki-67 (clone MIB-1) and p53 was performed on an automated immunostainer (Ventana Medical Systems, Inc., Tucson, AZ, USA) according to previously published procedures.23 CyD1 (rabbit monoclonal SP4), Ki-67 and p53 (DO-7 clone) antibodies were obtained from Dako (Copenhagen, Denmark). Appropriate positive controls were used to confirm the adequacy of the staining. The proliferation index was assessed by counting at least 300 tumor cells on Ki-67-stained sections, following the recommendations recently described in a consensus paper.24 The results were given as percentages of total tumor cells.24 If >30% of neoplastic cells showed Ki-67-positivity, the case was defined as highly proliferative. In addition Ki-67-positivity of 11–30% and 0–10% was defined as medium and low proliferation, respectively. Cases were evaluated as p53-positive if at least 20% of cells showed strong nuclear positivity. All p53-positive cases were screened for p53 mutations (see below).
Analysis of total cyclin D1 and cyclin D1 isoform expression
To measure total CyD1 mRNA expression levels and CyD1 mRNA isoforms, real-time qRT-PCR was performed. Total RNA was obtained from formalin-fixed, paraffin-embedded tissues and was either isolated from entire sections or after macro-dissection, to enrich the neoplastic population in cases with an irregular pattern of infiltration. RNA isolation, DNase digestion and cDNA synthesis were performed as described previously. 25,26 Total CyD1 expression levels were given relative to the housekeeping gene TATA box binding protein (TBP). The cut-off for total CyD1 mRNA was set at 8.6, defined as the mean ratio of CyD1/TBP + three standard deviations (SD) for the normal lymphoid control samples. Expression levels of CyD1 mRNA isoforms were determined using two different qRT-PCR assays, one spanning the exon 1–2 junction (CyD1 total assay) and the other located in the proximal 3’UTR (CyD1 3’UTR assay). The amount of short CyD1 isoform was calculated by the ΔΔCt method.25,27 Reactive lymphoid tissue was used as the control for background CyD1 levels, and small intestinal samples, which show higher physiological CyD1 expression, served as a control to assess the normal range of the CyD1 isoform ratio (CyD1 isoform ratio=2−ΔΔCt, ΔΔCt=[Ct-value CyD1 3’UTR(tumor)–Ct-value CyD1 total(tumor)]-[(mean Ct-value CyD1 3’UTR(controls) – (mean Ct-value CyD1 total (controls)]. A ratio (proximal 3’UTR/exon1-2) of 0.6 or lower was regarded as evidence of significant expression of the short Δ3’UTR-deficient CyD1 isoform. No RT controls were used to control for DNA contamination in the 3’UTR assay. The sequences for primers and probes (CyD1 total, CyD1 3’UTR and TBP) used in the study have been published previously.27 Using 300 nmol/L of primers and 200 nmol/L of the corresponding probes, we obtained very similar amplification efficiencies of 3’UTR amplicons in relation to the exon 1–2 junction, allowing direct gene dosage estimation without using calibration standards.28 TaqMan assays were performed in duplicate using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Foster City, CA, USA).
Cyclin D1 3’UTR genomic deletion
Genomic DNA was obtained from 62 MCL cases and extracted either from entire or macrodissected sections, as described above. At least three replicates of 40 ng genomic DNA were subjected to qRT-PCR analysis to study CyD1 3’UTR gene dosage. The primers and probe sequences for the CyD1 3’UTR region were identical to those used for the expression analysis. Since MCL commonly shows complex karyotypes with aneuploidy or tetraploidy, we used several genomic regions on different chromosomes as endogenous controls, including Beta-globin, SOCS 1, Beta-2-microglobulin and Albumin, as described previously.27 For each sample, CyD1 3’UTR was compared to these endogenous control genes, using normal lymphoid tissues as reference. We obtained very similar amplification efficiencies of 3’UTR genomic amplicons in relation to endogenous controls, allowing direct gene dosage estimation without using calibration standards.28
Mutation analysis
Screening for mutations of exons 5 to 8 of the TP53 gene, including the intron/exon boundaries, was performed in a subset of MCL cases by denaturing high performance liquid chromatography (DHPLC) on an automated DHPLC analysis system (WAVE, Transgenomic, Omaha, NE, USA) as described previously.29 Chromatograms were read manually and evaluated independently by three observers (GK, JSH and LQM). All PCR products showing suspicious chromatograms were subjected to direct sequencing analysis.
Exons 4 and 5 of the CCND1 gene, including the proximal CyD1 3’UTR region, were sequenced in cases with predominant expression of the short CyD1 mRNA variant, as previously described.27 Automated fluorescent sequencing was performed with the BigDye Terminator Cycle Sequencing kit (PE Applied Biosystems, Foster City, CA, USA) and ABI Prism 377 automated sequencer (PE Applied Biosystems). Comparison with the wild-type sequences was performed with DNASIS 2.6 software (Hitachi Software Engineering, Yokohama, Japan).
CCND1/cyclin D1 gene analysis by fluorescent in situ hybridization
FISH analysis was performed using the commercially available LSI IgH/CCND1 and LSI Cyclin D1/CEP 11 probes according to previously published procedures.30 Probes were purchased from Vysis (Vysis Inc, Downers Grove, IL, USA) and used according to the manufacturer’s instructions. The nuclei were counterstained with Hoechst 33342 (Molecular Probes, Leiden) and slides were mounted with Vectashield medium (Vector Laboratories, USA). Imaging was performed with a LSM-510 NLO laser microscope (Zeiss, Jena, Germany). Image processing was carried out with Zeiss computer software (AIM 3.2). Control specimens were prepared from reactive lymph nodes to determine cut-off levels for aberrant signals.
Follow-up
The patients were followed up for a range of 1–105 months. Data were collected from at least one of the following sources: the hospitals’ inpatient and outpatient records (periodically reviewed for updated information), primary physicians’ offices, and direct contact with the patient or family. The primary endpoint was overall survival duration from the date of MCL diagnosis to the date of death. Patients who were alive at the end of the study period were censored on the last date of follow-up. The end of the observation period was the end of April 2011.
Statistical analysis
SPSS statistical software (SPSS Inc., Chicago IL, USA) was used for the statistical analyses. Associations in 2 × 2 tables were evaluated with Fisher’s exact test. Correlations were assessed by Pearson’s or Spearman’s correlation analysis. Continuous variables between different groups were compared using the Mann-Whitney U test. Survival was analyzed using Kaplan-Meier estimates and the curves were compared by the log-rank test. P levels <0.05 were considered statistically significant. In addition, maximally selected log-rank statistics were used to determine the optimal cut-off for proliferation. To consider multiple testing in this setting, the R-function ‘maxstat.test’ was employed.
Results
Clinico-pathological features and proliferation index
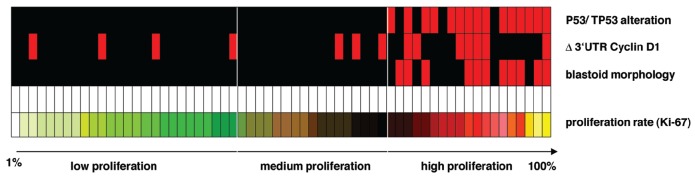
Of the 62 patients included in the study, 42 were males and 14 females, with a median age of 67 years at diagnosis (range, 34 to 89 years). No personal data were available for six patients. Follow-up information was obtained from the medical records for 51 patients (38 males and 13 females). Most tissue samples were lymph nodes biopsies (n=44), whereas the remainder were derived from various extranodal sites (ten from the naso- and oropharynx, four from soft tissue, three from the intestine and one from prostate). All cases showed strong CyD1 immunostaining and a phenotype and morphology consistent with MCL. Fifty-one (82%) of 62 cases were classified as classical and 11/62 (18%) as blastoid subtype of MCL, including two pleomorphic variants (Figure 1). Among the four patients with classical MCL for whom sequential biopsies were available, blastoid transformation was evident in one case (#21) after a period of 16 months, accompanied by an increase of proliferation index from 3% to 26% (Ki-67). The median proliferation index, as assessed by Ki-67 staining, was 15%, ranging from 1% to 100% of tumor cells (Figure 1). The high proliferation group included 19 MCL cases (31%), the medium proliferation group contained 17 cases (27%), and the low proliferation group contained 26 cases (42%), with a median proliferation index of 36%, 21% and 7%, respectively. All 11 cases (100%) of blastoid MCL were in the high proliferative group as opposed to 8/51 (16%) of the classic variant (P<0.001) (Figure 2A).
Figure 1.
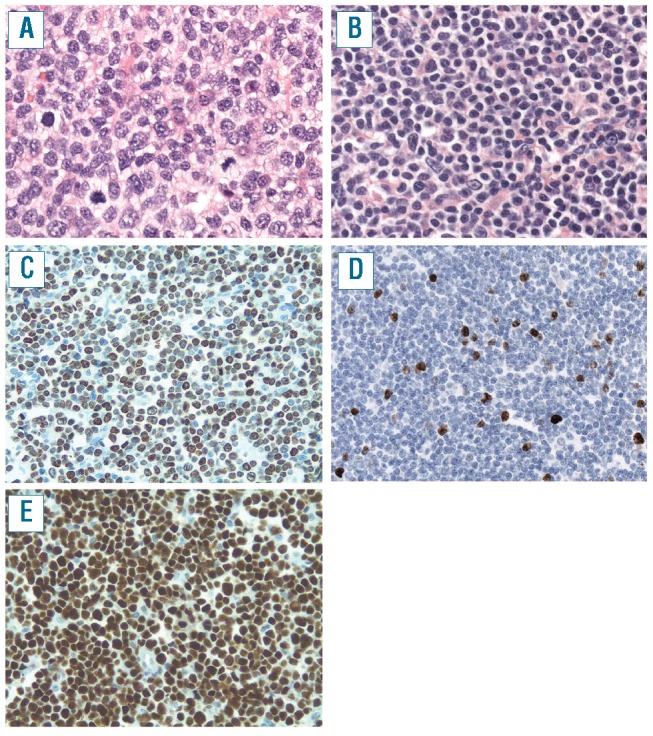
Blastoid (A, C, E) and classic MCL (B, D). (A) Blastoid MCL shows large tumor cells with irregular large nuclei with open chromatin and a high mitotic rate (original magnification 400x) (C) MIB1 staining shows 80% positive tumor cells (case 3, original magnification 200x) (E) Virtually all tumor cells show strong expression of p53 protein (case 49, original magnification 200x) (B) Classical MCL shows cells with scant cytoplasm and small irregular nuclei (original magnification 400x) (D) MIB1 shows nuclear positivity in 13% of tumor cells (case 42, original magnification 200x).
Figure 2.
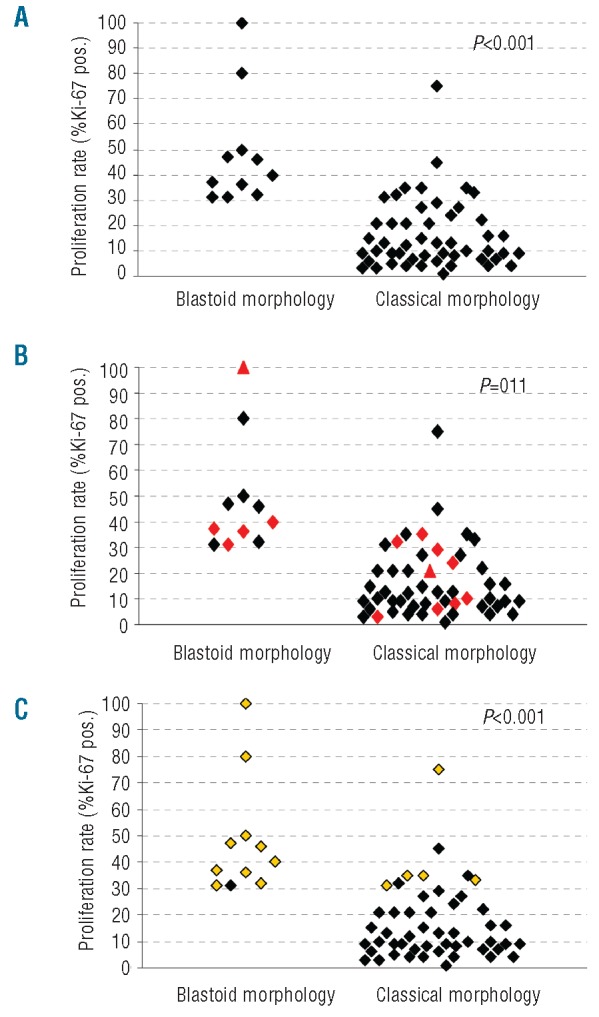
Scatter plots show the correlation between (A) proliferation index and blastoid morphology, (B) proliferation, morphological subtype and CyD1 isoform expression (red diamonds: predominant Δ3’UTR expression) and (C) proliferation, morphological subtype and p53 alteration (yellow diamonds: p53 overexpression/mutation).
Cyclin D1 mRNA isoform expression and cyclin D1 3’UTR alterations
Total CyD1 mRNA expression levels in MCL samples ranged widely from 9 to 215 (median CyD1/TBP=56). The long standard CyD1 transcript was the predominant isoform expressed in 48/62 MCL cases (77%). These cases expressed only low levels of the short Δ3’UTR mRNA. In contrast, 14 MCL cases (23%) predominantly expressed the short Δ3’UTR CyD1 mRNA, indicated by a 3’UTR/exon1-2 ratio of less than 0.6, including four cases (6%) with a complete loss of the 3’UTR of CyD1 mRNA. In order to investigate the cause of complete loss of the 3’UTR of CyD1 mRNA, quantitative genomic PCR was performed. Two of the four cases showed genomic deletion of the 3’UTR locus whereas in the remaining two cases, neither deletions nor mutations of exons 4 and 5 including the 3’UTR were found. The 14 MCL cases with predominant expression of the Δ3’UTR CyD1 isoform showed significantly higher total CyD1 mRNA expression levels than the MCL cases with the long standard transcript (median CyD1/TBP ratio 93 versus 50, interquartile range (IQR) 56–117 versus 29–79, P=0.009). The two MCL cases with genomic 3’UTR deletion had total CyD1 levels of 95.3 and 68.3 and did not differ from other MCL cases with predominance of the short CyD1 isoform. The sequential samples from four MCL patients showed similar CyD1 mRNA levels and CyD1 mRNA isoform expression.
p53 overexpression and TP53 mutational analysis
p53 overexpression was observed in 13 (21%) of 62 MCL cases; eight cases had blastoid morphology (8/11; 73%) and five cases corresponded to the classical MCL variant (5/51; 10%). Ten of these 13 cases showed p53 overexpression in more than 50% of the tumor cells (Figure 1). To complete the TP53 status analysis, mutational screening of exons 5 to 8 of the TP53 gene was performed by DHPLC followed by sequence analysis in MCL cases with a proliferation index >30% for which enough DNA was available (seven blastoid and five classical MCL). These cases included eight p53-positive cases and four p53-negative cases. In six cases with p53 overexpression and in two highly proliferative cases negative for p53 by immunohistochemistry, missense mutations leading to amino acid substitutions were detected (five in exon 5 and one each in exon 6, exon 7 and exon 8). In the remaining two p53-positive cases DHPLC analysis showed an abnormal DHPLC pattern suggestive of mutations, one in exon 6 and one in exon 8, but not enough DNA was available for sequence analysis (Table 1). As a control, mutational screening of exons 5 to 8 of the TP53 gene was performed by DHPLC in 16 MCL negative for p53 and with a proliferation index <30%. In all cases normal patterns, indicative of wild-type TP53, were observed. In summary, p53/TP53 alterations were found in 15 MCL cases, ten with blastoid morphology and five with classical morphology.
Table 1.
Clinicopathological features of highly proliferative mantle cell lymphomas.
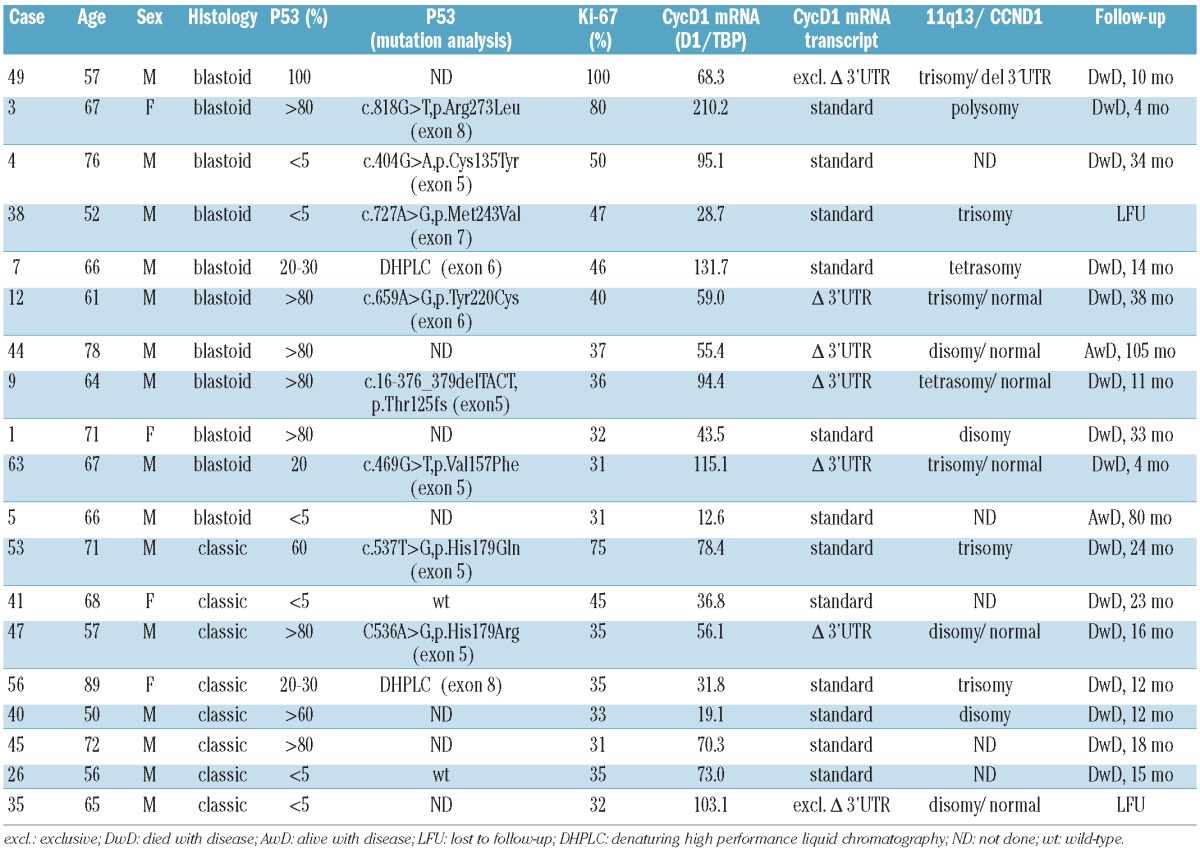
Correlation between proliferation index and histological subtype, cyclin D1 expression levels and p53 alterations
The correlations between histological subtype, CyD1 isoform expression, p53 alterations and the proliferation index are illustrated in Figures 2 and 3. Median proliferation indices were 40% versus 10% in blastoid versus classical cases (IQR 50-32 versus 22-7, P<0.001), 30% versus 13% in cases with Δ3’UTR CyD1 versus standard CyD1 (IQR 36-10 versus 30-7, P=0.11) and 37% versus 10% in MCL cases with p53 alterations versus wild-type p53 (IQR 50-33 versus 21-7, P<0.001). All cases with blastoid morphology (11/11, 100%) showed a high proliferation index (median MIB1=40%, IQR 32–50%) whereas only eight of the cases with classical morphology (8/51, 16%) belonged to the high proliferation group (median MIB1=35%, P<0.001). Total CyD1 expression levels did not differ significantly between high, medium and low proliferation groups (median CyD1/TBP: 68 versus 83 versus 51, IQR 37–95 versus 36–107 versus 28–63, P=0.116), but there was a tendency for cases with predominant Δ3’UTR CyD1 mRNA to be more frequently found in the group with blastoid morphology (5/11) than in the classical subtype (9/51) (P=0.060) (Figure 2B). Although, total CyD1 levels were higher in blastoid MCL than in the classical MCL subtype, the difference was not statistically significant (median CyD1/TBP 68 versus 53, IQR 44–115 versus 32–85, P=0.253). MCL cases with predominant Δ3’UTR CyD1 mRNA were found in the high, medium and low proliferation groups (7/19=37% versus 3/17=18% versus 4/26=15%, P=0.20) with over-representation in the highly proliferative group. However, only if a Ki67 index of 20% was used as a single cut-off were both total CyD1mRNA levels (P=0.022) and Δ3’UTR CyD1 mRNA (P=0.026) significantly associated with high proliferation. Complete 3’UTR loss at the mRNA level, as well as, genomic 3’UTR deletion were found in both morphological subtypes. Alterations of p53/TP53 (overexpression and/or mutations) were found exclusively in the highly proliferative group (15/19), but in none of the MCL cases with medium or low proliferation index (0/43) (P<0.001). Similarly, ten (91%) of the 11 blastoid MCL cases showed alterations of TP53, whereas only five (10%) of the 51 typical MCL cases (10%) did (P<0.001). p53 alterations remained highly statistically significant when cases were separated into two groups, irrespective of the cut-off points for proliferation (10%, 20%, or 30%). Of interest, the MCL case with the highest proliferation index of 100% showed both strong nuclear p53 positivity in >90% of the tumor cells and predominance of the Δ3’UTR transcript.
Figure 3.
Relationship between p53 overexpression/mutation, Δ3’UTR cyclin D1 isoform expression, blastoid morphology and the proliferation index measured by MIB-1 immunostaining. Sixty-two MCL cases are ordered by MIB1 positivity (1–100% from left to right). Red squares in the upper three bars represent cases with p53 alteration, predominant Δ3’UTR cyclin D1 isoform expression and blastoid morphology, respectively.
The characteristics of the highly proliferative cases are summarized in Table 1. Concurrent predominant expression of short ΔCyD1 isoform and p53/TP53 alterations was observed in six cases, p53/TP53 alterations alone in nine cases and Δ3’UTR CyD1 expression without evidence of TP53 alterations was observed in one case, whereas neither was present in the remaining three cases. Thus, in the high proliferation group, all but one of the cases with predominance of the Δ3’UTR CyD1 mRNA additionally showed overexpression and/or mutation of p53. However, there were no significant differences in the proliferation index irrespective of whether none, any or both of the above features were present in the tumor cells.
Survival
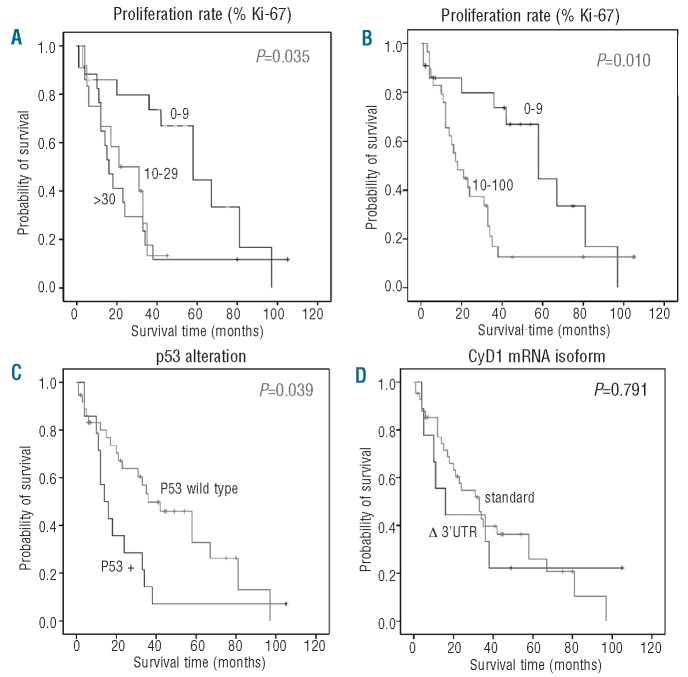
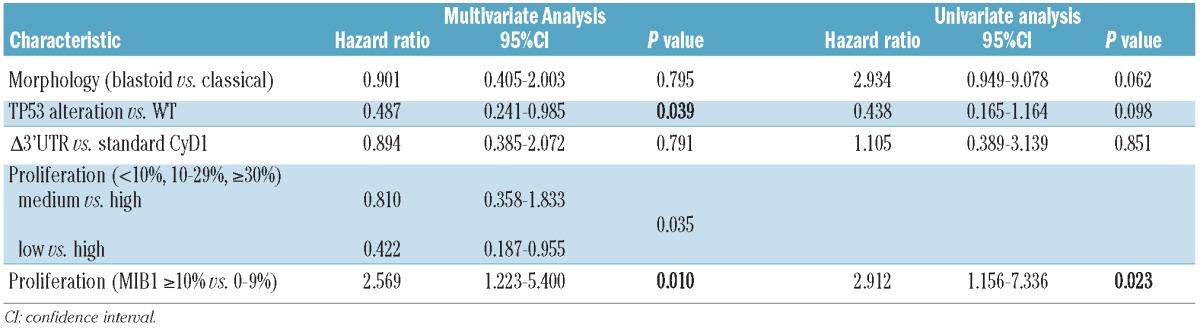
Follow-up data were available for 51 patients. Thirty-five patients (69%) died of progressive disease with a mean period from the date of diagnosis to death of 39.8 months (range, 1–97 months). Sixteen patients are still alive with disease (follow-up period from 2–105 months). Univariate analysis of the prognostic significance of the different parameters was performed for overall survival by the Kaplan-Meier method (Figure 4). Using the described cut-offs, overall survival differed significantly between the three proliferation groups with a median survival time of 16 months in the high proliferation group (n=17), 21 months in the medium (n=12) and 58 months in the low proliferation group (n=22) (P=0.035, Figure 4A). Notably, overall survival did not differ between the medium and the high proliferation groups in our set of cases. Using maximally selected log rank statistics, proliferation was highly significant for overall survival in multivariate analysis (P=0.023) with a determined optimal cut-off of 10% (Figure 4B). Results of uni- and multivariate analyses are summarized in Table 2. Patients with p53 overexpression/TP53 alteration had a significantly worse prognosis (median survival, 14 months) compared to patients with wild-type p53 (median survival, 36 months) (P=0.039, Figure 4C). In contrast, there was no statistically significant influence of Δ3’UTR CyD1 expression on overall survival (P=0.791, Figure 4D), as patients with the standard CyD1 isoform had a median survival of 33 months compared to the 24 months of patients with predominant Δ3’UTR transcript. In order to control for confounding factors, multivariate analysis was performed after leaving out either Ki67 index or p53 alterations. In both instances, proliferation and p53 (P=0.008), respectively, were highly significant for overall survival, whereas CyD1 mRNA levels and Δ3’UTR transcript were not. This indicates that p53 and proliferation index are interdependent factors concerning survival.
Figure 4.
Overall survival analysis (Kaplan-Meier curves) of 51 MCL patients. (A) according to separation into three groups with high (>30% MIB1-positivity), medium (11–30% MIB1-positivity) and low proliferation index (0–10% MIB1-positivity), (B) separation into two groups with a cut-off of 10% as determined by maximally selected log-rank statistics (C) according to p53 status and (D) according to predominant cyclin D1 (CyD1) isoform expression.
Table 2.
Uni- and multivariate analyses of effects of prognostic factors on overall survival.
Discussion
The proliferation index is an important prognostic parameter in MCL. The impact of different genetic alterations on proliferation index has not yet been analyzed systematically, although several secondary alterations associated with a poor prognosis have been described. In this study, we demonstrate that alterations of the TP53 tumor suppressor gene are strongly associated with a high Ki67 index and blastoid morphology, representing major biological determinants of aggressive behavior. Expression of the truncated Δ3’UTR CyD1 mRNA variant was associated with higher CyD1 mRNA levels and occurred more often in cases with a proliferation index >20%, but lacked independent prognostic significance.
MCL is an aggressive subtype of B-cell non-Hodgkin’s lymphoma with a median survival of 3–4 years with conventional therapy. Nevertheless, the range of clinical behavior is broad, and a subset of patients with indolent disease experience long-term survival.2 A variety of tumor-related risk factors for poor outcome have been identified, namely blastoid morphology and high proliferation index, as assessed by MIB1 staining.20,21,31–34 Poor prognostic features are commonly associated with alterations of TP53, C-MYC and CDK4, indicating that secondary genetic events contribute to the aggressive behavior of the lymphoma and may provide independent prognostic information.10–12,14,35 Whereas blastoid MCL invariably has a high proliferation index, the growth fraction of classical MCL shows a broad range, ranging from less than 5% to more than 70% MIB-1-positive tumor cells. In our series, 16% of classical MCL belonged to the highly proliferative group with >30% MIB-1 positivity, similar to the findings of other authors.20,21 Separating MCL according to growth fractions of <10%, 10–29% and >30% discerned patients with median overall survivals of 112, 59 and 30 months, respectively, in a large multicenter study. The difference between these groups was preserved when rituximab was part of the treatment regimen.20 The recognition of the paramount importance of the proliferation index has led the European MCL group to adopt guidelines for quantifying MIB1 staining in MCL, in order to increase reproducibility of the proliferation index.24
A distinct approach for looking at prognostic markers in MCL has been taken by Rosenwald et al.16 Using gene expression profiling of a series of 92 CyD1-positive MCL, they identified a set of overexpressed genes involved in cell proliferation, the so-called proliferation signature, which was strongly associated with poor survival. Patients in the lowest quartile of the expression signature had a median survival of 6.71 years, as compared to only 0.83 years of the patients in the highest quartile. Of interest, MCL cases with a high proliferation signature and shorter disease-related survival were more likely to express, predominantly or exclusively, the short isoform of CyD1 mRNA. Since cases with a truncated CyD1 transcript showed higher levels of total CyD1 mRNA, and mRNA and protein levels are closely correlated, the authors postulated that CyD1 level was a major determinant of the proliferation signature and, ultimately, prognosis, because of the crucial role of CyD1 protein levels for cell cycle progression.
The reasons for increased levels of CyD1 mRNA in cases with predominance of the truncated variant probably lie in the destabilizing function of the 3’UTR, which contains AU-rich elements, which are crucial for regulating mRNA stability.36 Genes with long 3’UTR such as CyD1 usually encode for short-lived proteins involved in proliferation and other tightly regulated, transient cellular functions.37 In addition, truncation of the 3’UTR of the CyD1 mRNA results in loss of miR-16-1 binding sites and may change regulation of CyD1 protein expression.38
Although the predominance of the short CyD1 isoform in MCL is common – 23% in our series, similar to the 18% observed by Wiestner et al. - the reasons for this are evident only in a minority of cases.19 Only 6% of MCL lacked the long canonical transcript completely, and in only half of these cases were we able to demonstrate a genomic deletion of the 3’UTR explaining the complete loss of expression. Since previously described mutations introducing stop codons were not observed in our series,19 translocations with rearrangement of the 3’UTR without loss of genomic material (and, therefore, not detectable by q-PCR) or epigenetic alterations might play a role in these cases.18
Another potential reason for a high percentage of short, Δ3’UTR transcripts would be the predominant generation of the CyD1b isoform, which lacks exon 5 and the conventional 3’UTR region because of alternative splicing, supposedly due to a common splice site polymorphism A870G at the exon 4/intron 4 boundary.39,40 The protein encoded by this splice variant lacks the PEST sequence important for protein turn-over, but retains catalytic function and has been found to be potently transforming in vitro.41,42 Despite the potentially increased oncogenicity of this mRNA variant, we did not find any correlation between the splice site A/G polymorphism and the percentage of CyD1b transcript, which made up less than 10% of the total CyD1 mRNA in most studied cases, on the one hand, and levels of total CyD1 mRNA or proliferation index on the other hand (data not shown). More importantly, we failed to find a correlation between the levels of the short CyD1 transcript and CyD1b transcript. These negative results are in line with more recent findings by several groups, which also failed to identify a major role for the CyD1b transcript or the splice site polymorphism in MCL.43–46
Irrespective of the reasons for the predominance of the Δ3’UTR transcript, the initial finding of a correlation with high total CyD1 levels, high proliferation index and shorter survival induced us to investigate the interplay between this and other known factors of poor prognosis in more detail, given the paucity of comparative data.47 Of note, Rosenwald et al. did not correlate their findings with conventional assessment of MIB1/Ki67 staining. Furthermore, they studied alterations of p53 only by quantitative PCR, without mutational analysis or immunostaining, and failed to identify a correlation between the proliferation signature and p53 deletion.16 However, the high frequency of TP53 alterations in highly proliferative cases including the blastoid variant in our study is not surprising and confirms previous findings, which clearly indicate that further perturbations of cell cycle regulatory mechanisms are necessary for uncontrolled proliferation.10–12,14,35 TP53 gene mutations in lymphoid neoplasms have been associated with progression, high grade transformation and poor prognosis. In MCL TP53 mutations/deletions and p53 overexpression are frequently detected in the terminal leukemic phase and may be among the mechanisms involved in the development of aggressive forms of this lymphoma.9,11,13,48–50
The role of CyD1, which acts as a growth factor sensor under physiological circumstances, in the oncogenesis of B-cell neoplasms is far from clear. Both observations in human tumors as well as in CyD1-transgenic animals indicate that a simple mechanistic model – high CyD1 levels result in increased proliferation –is insufficient to explain aggressive behavior in MCL. In humans, B-cell neoplasms with overexpression of CyD1 show a broad spectrum of clinical behavior. In multiple myeloma (MM), we previously failed to identify a correlation between predominant expression of the Δ3’UTR CyD1 and higher proliferation, although t(11;14)-positive MM cases predominantly expressing the truncated mRNA variant tended to show higher levels of total CyD1 transcript, similar to MCL.27 A potential explanation for this lack of correlation between CyD1 levels and proliferation in t(11;14)-positive MM was proposed by Ely et al., who demonstrated that CyD1 alone is not sufficient to induce retinoblastoma protein phosphorylation and cell cycle progression in CyD1-positive MM, unless CDK4 is also elevated.51 Of note, average CyD1 mRNA levels in t(11;14)-positive MM are significantly higher than in MCL, probably because of the higher transcriptional activity of the translocated IgH locus in neoplastic plasma cells.30,52 Hairy cell leukemia, another B-cell non-Hodgkin’s lymphoma expressing CyD1 due to an unknown mechanism, usually has a very low proliferation index. Furthermore, the t(11;14) translocation is detected in a significant number of cases of monoclonal gammopathy of unknown significance without having a negative prognostic influence,53 and, recently, cases of in situ MCL with indolent behavior have been observed.54
On the other hand, overexpression of CyD1 is unable to induce B-cell lymphomas without the help of cooperating oncogenes in transgenic mouse models. Furthermore, we have recently shown that siRNA-mediated knockdown of CyD1 in MCL cell lines has little influence on cell proliferation, because of a compensatory up-regulation of cyclin D2.55
Despite this heterogeneous picture concerning the role of CyD1 in lymphoma and although both high total CyD1 mRNA levels and predominance of the short mRNA transcript were found in all proliferation groups, we did observe a significant correlation for both parameters with a proliferation index >20%, and blastoid MCL cases were more likely to express the truncated Δ3’UTR; a phenomenon which has also been observed by others.56 However, we failed to identify an association between high CyD1 levels or Δ3’UTR transcript predominance and overall survival. This might be due to the small cohort, or alternatively indicates that the Δ3’UTR mRNA acts only as a secondary factor for proliferation. Accordingly, concomitant occurrence of p53 alterations and short CyD1 mRNA expression was common in highly proliferative cases, which could indicate that these two phenomena can collaborate to induce an aggressive phenotype in MCL. Furthermore, it should be pointed out that MIB1 staining and assessment of the “proliferation signature” including CyD1 isoforms as one parameter, as performed by Rosenwald et al., are two different approaches to assess cell proliferation.
In summary, our data indicate that high CyD1 levels alone, irrespective of the causative mechanism, do not necessarily cause a high proliferation index in MCL. We also confirm that expression of the short Δ3’UTR variant CyD1 transcript occurs in a significant minority of cases of MCL, may be associated with genomic deletions of the 3’UTR, and results on average in higher CyD1 mRNA levels. Furthermore, the very high frequency of TP53 alterations in MCL cases with a high Ki67 index and poor prognosis underlines the importance of secondary genetic alterations in the further disruption of cell cycle control promoting high proliferation and an aggressive clinical behavior.
Footnotes
Funding: this work was supported by a grant from the Mantle Cell Lymphoma Consortium, Lymphoma Research Foundation, NY (to LQM) and a grant for female graduate students from the Technical University Munich to JSH.
Authorship and Disclosures
The information provided by the authors about contributions from persons listed as authors and in acknowledgments is available with the full text of this paper at www.haematologica.org.
Financial and other disclosures provided by the authors using the ICMJE (www.icmje.org) Uniform Format for Disclosure of Competing Interests are also available at www.haematologica.org.
References
- 1.Bosch F, Jares P, Campo E, Lopez-Guillermo A, Piris MA, Villamor N, et al. PRAD-1/cyclin D1 gene overexpression in chronic lymphoproliferative disorders: a highly specific marker of mantle cell lymphoma. Blood. 1994;84(8):2726–32. [PubMed] [Google Scholar]
- 2.Campo E, Raffeld M, Jaffe ES. Mantle-cell lymphoma. Semin Hematol. 1999;36(2):115–27. [PubMed] [Google Scholar]
- 3.Bergsagel PL, Kuehl WM. Critical roles for immunoglobulin translocations and cyclin D dysregulation in multiple myeloma. Immunol Rev. 2003;194:96–104. doi: 10.1034/j.1600-065x.2003.00052.x. [DOI] [PubMed] [Google Scholar]
- 4.de Boer CJ, Kluin-Nelemans JC, Dreef E, Kester MG, Kluin PM, Schuuring E, et al. Involvement of the CCND1 gene in hairy cell leukemia. Ann Oncol. 1996;7(3):251–6. doi: 10.1093/oxfordjournals.annonc.a010568. [DOI] [PubMed] [Google Scholar]
- 5.Bodrug SE, Warner BJ, Bath ML, Lindeman GJ, Harris AW, Adams JM. Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the myc gene. Embo J. 1994;13(9):2124–30. doi: 10.1002/j.1460-2075.1994.tb06488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lovec H, Grzeschiczek A, Kowalski MB, Moroy T. Cyclin D1/bcl-1 cooperates with myc genes in the generation of B-cell lymphoma in transgenic mice. EMBO J. 1994;13(15):3487–95. doi: 10.1002/j.1460-2075.1994.tb06655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18(22):2699–711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 8.Dreyling MH, Bullinger L, Ott G, Stilgenbauer S, Muller-Hermelink HK, Bentz M, et al. Alterations of the cyclin D1/p16-pRB pathway in mantle cell lymphoma. Cancer Res. 1997;57(20):4608–14. [PubMed] [Google Scholar]
- 9.Greiner TC, Dasgupta C, Ho VV, Weisenburger DD, Smith LM, Lynch JC, et al. Mutation and genomic deletion status of ataxia telangiectasia mutated (ATM) and p53 confer specific gene expression profiles in mantle cell lymphoma. Proc Natl Acad Sci USA. 2006;103(7):2352–7. doi: 10.1073/pnas.0510441103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez L, Bea S, Pinyol M, Ott G, Katzenberger T, Rosenwald A, et al. CDK4 and MDM2 gene alterations mainly occur in highly proliferative and aggressive mantle cell lymphomas with wild-type INK4a/ARF locus. Cancer Res. 2005;65(6):2199–206. doi: 10.1158/0008-5472.CAN-04-1526. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez L, Fest T, Cazorla M, Teruya-Feldstein J, Bosch F, Peinado MA, et al. p53 gene mutations and protein overexpression are associated with aggressive variants of mantle cell lymphomas. Blood. 1996;87(8):3351–9. [PubMed] [Google Scholar]
- 12.Pinyol M, Hernandez L, Cazorla M, Balbin M, Jares P, Fernandez PL, et al. Deletions and loss of expression of p16INK4a and p21Waf1 genes are associated with aggressive variants of mantle cell lymphomas. Blood. 1997;89(1):272–80. [PubMed] [Google Scholar]
- 13.Louie DC, Offit K, Jaslow R, Parsa NZ, Murty VV, Schluger A, et al. p53 overexpression as a marker of poor prognosis in mantle cell lymphomas with t(11;14)(q13;q32) Blood. 1995;86(8):2892–9. [PubMed] [Google Scholar]
- 14.Fernandez V, Hartmann E, Ott G, Campo E, Rosenwald A. Pathogenesis of mantle-cell lymphoma: all oncogenic roads lead to dysregulation of cell cycle and DNA damage response pathways. J Clin Oncol. 2005;23(26):6364–9. doi: 10.1200/JCO.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 15.Ott G, Kalla J, Ott MM, Schryen B, Katzenberger T, Muller JG, et al. Blastoid variants of mantle cell lymphoma: frequent bcl-1 rearrangements at the major translocation cluster region and tetraploid chromosome clones. Blood. 1997;89(4):1421–9. [PubMed] [Google Scholar]
- 16.Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3(2):185–97. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 17.Deshpande A, Pastore A, Deshpande AJ, Zimmermann Y, Hutter G, Weinkauf M, et al. 3’UTR mediated regulation of the cyclin D1 proto-oncogene. Cell Cycle. 2009;8(21):3584–92. doi: 10.4161/cc.8.21.9993. [DOI] [PubMed] [Google Scholar]
- 18.Rimokh R, Berger F, Bastard C, Klein B, French M, Archimbaud E, et al. Rearrangement of CCND1 (BCL1/PRAD1) 3′ untranslated region in mantle-cell lymphomas and t(11q13)-associated leukemias. Blood. 1994;83(12):3689–96. [PubMed] [Google Scholar]
- 19.Wiestner A, Tehrani M, Chiorazzi M, Wright G, Gibellini F, Nakayama K, et al. Point mutations and genomic deletions in cyclin D1 create stable truncated mRNAs that are associated with increased proliferation rate and shorter survival in mantle cell lymphoma. Blood. 2007;109(11):4599–606. doi: 10.1182/blood-2006-08-039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Determann O, Hoster E, Ott G, Wolfram Bernd H, Loddenkemper C, Leo Hansmann M, et al. Ki-67 predicts outcome in advanced-stage mantle cell lymphoma patients treated with anti-CD20 immunochemotherapy: results from randomized trials of the European MCL Network and the German Low Grade Lymphoma Study Group. Blood. 2008;111(4):2385–7. doi: 10.1182/blood-2007-10-117010. [DOI] [PubMed] [Google Scholar]
- 21.Tiemann M, Schrader C, Klapper W, Dreyling MH, Campo E, Norton A, et al. Histopathology, cell proliferation indices and clinical outcome in 304 patients with mantle cell lymphoma (MCL): a clinicopathological study from the European MCL Network. Br J Haematol. 2005;131(1):29–38. doi: 10.1111/j.1365-2141.2005.05716.x. [DOI] [PubMed] [Google Scholar]
- 22.Swerdlow SH, Campo E, Seto M, Mueller-Hermelink HK. Mantle cell lymphoma. In: Swerdlow SH, Campo E, Harris NL, et al., editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: IARC Press; 2008. [Google Scholar]
- 23.Quintanilla-Martinez L, Kremer M, Specht K, Calzada-Wack J, Nathrath M, Schaich R, et al. Analysis of signal transducer and activator of transcription 3 (Stat 3) pathway in multiple myeloma: Stat 3 activation and cyclin D1 dysregulation are mutually exclusive events. Am J Pathol. 2003;162(5):1449–61. doi: 10.1016/S0002-9440(10)64278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klapper W, Hoster E, Determann O, Oschlies I, van der Laak J, Berger F, et al. Ki-67 as a prognostic marker in mantle cell lymphoma-consensus guidelines of the pathology panel of the European MCL Network. J Hematop. 2009;2(2):103–111. doi: 10.1007/s12308-009-0036-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koch I, Slotta-Huspenina J, Hollweck R, Anastasov N, Hofler H, Quintanilla-Martinez L, et al. Real-time quantitative RT-PCR shows variable, assay-dependent sensitivity to for-malin fixation: implications for direct comparison of transcript levels in paraffin-embedded tissues. Diagn Mol Pathol. 2006;15(3):149–56. doi: 10.1097/01.pdm.0000213450.99655.54. [DOI] [PubMed] [Google Scholar]
- 26.Salaverria I, Espinet B, Carrio A, Costa D, Astier L, Slotta-Huspenina J, et al. Multiple recurrent chromosomal breakpoints in mantle cell lymphoma revealed by a combination of molecular cytogenetic techniques. Genes Chromosomes Cancer. 2008;47(12):1086–97. doi: 10.1002/gcc.20609. [DOI] [PubMed] [Google Scholar]
- 27.Slotta-Huspenina J, Koch I, Richter M, Bink K, Kremer M, Specht K, et al. Cyclin D1 positive multiple myeloma: predominance of the short, 3’UTR-deficient transcript is associated with high cyclin D1 mRNA levels in cases with t(11;14) translocation, but does not correlate with proliferation rate or genomic deletions. Leuk Res. 2008;32(1):79–88. doi: 10.1016/j.leukres.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔ C(T)) Method. methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Quintanilla-Martinez L, Kremer M, Keller G, Nathrath M, Gamboa-Dominguez A, Meneses A, et al. p53 Mutations in nasal natural killer/T-cell lymphoma from Mexico: association with large cell morphology and advanced disease. Am J Pathol. 2001;159(6):2095–105. doi: 10.1016/S0002-9440(10)63061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Specht K, Haralambieva E, Bink K, Kremer M, Mandl-Weber S, Koch I, et al. Different mechanisms of cyclin D1 overexpression in multiple myeloma revealed by fluorescence in situ hybridization and quantitative analysis of mRNA levels. Blood. 2004;104(4):1120–6. doi: 10.1182/blood-2003-11-3837. [DOI] [PubMed] [Google Scholar]
- 31.Velders GA, Kluin-Nelemans JC, De Boer CJ, Hermans J, Noordijk EM, Schuuring E, et al. Mantle-cell lymphoma: a population-based clinical study. J Clin Oncol. 1996;14(4):1269–74. doi: 10.1200/JCO.1996.14.4.1269. [DOI] [PubMed] [Google Scholar]
- 32.Argatoff LH, Connors JM, Klasa RJ, Horsman DE, Gascoyne RD. Mantle cell lymphoma: a clinicopathologic study of 80 cases. Blood. 1997;89(6):2067–78. [PubMed] [Google Scholar]
- 33.Bosch F, Lopez-Guillermo A, Campo E, Ribera JM, Conde E, Piris MA, et al. Mantle cell lymphoma: presenting features, response to therapy, and prognostic factors. Cancer. 1998;82(3):567–75. doi: 10.1002/(sici)1097-0142(19980201)82:3<567::aid-cncr20>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 34.Katzenberger T, Petzoldt C, Holler S, Mader U, Kalla J, Adam P, et al. The Ki67 proliferation index is a quantitative indicator of clinical risk in mantle cell lymphoma. Blood. 2006;107(8):3407. doi: 10.1182/blood-2005-10-4079. [DOI] [PubMed] [Google Scholar]
- 35.Salaverria I, Zettl A, Bea S, Moreno V, Valls J, Hartmann E, et al. Specific secondary genetic alterations in mantle cell lymphoma provide prognostic information independent of the gene expression-based proliferation signature. J Clin Oncol. 2007;25(10):1216–22. doi: 10.1200/JCO.2006.08.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lebwohl DE, Muise-Helmericks R, Sepp-Lorenzino L, Serve S, Timaul M, Bol R, et al. A truncated cyclin D1 gene encodes a stable mRNA in a human breast cancer cell line. Oncogene. 1994;9(7):1925–9. [PubMed] [Google Scholar]
- 37.Espel E. The role of the AU-rich elements of mRNAs in controlling translation. Semin Cell Dev Biol. 2005;16(1):59–67. doi: 10.1016/j.semcdb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 38.Chen RW, Bemis LT, Amato CM, Myint H, Tran H, Birks DK, et al. Truncation in CCND1 mRNA alters miR-16-1 regulation in mantle cell lymphoma. Blood. 2008;112(3):822–9. doi: 10.1182/blood-2008-03-142182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Betticher CD, Thatcher N, Altermatt HJ, Hoban P, Ryder WDJ, Heighway J. Alternative spling produces a novel cyclin D1 transcript. Oncogene. 1995;11(5):1005–11. [PubMed] [Google Scholar]
- 40.Hosokawa Y, Gadd M, Smith AP, Koerner FC, Schmidt EV, Arnold A. Cyclin D1 (PRAD1) alternative transcript b: full-length cDNA cloning and expression in breast cancers. Cancer Lett. 1997;113(1–2):123–30. doi: 10.1016/s0304-3835(97)04605-3. [DOI] [PubMed] [Google Scholar]
- 41.Alt JR, Cleveland JL, Hannink M, Diehl JA. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 2000;14(24):3102–14. doi: 10.1101/gad.854900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu F, Gladden AB, Diehl JA. An alternatively spliced cyclin D1 isoform, cyclin D1b, is a nuclear oncogene. Cancer Res. 2003;63(21):7056–61. [PubMed] [Google Scholar]
- 43.Carrere N, Belaud-Rotureau MA, Dubus P, Parrens M, de Mascarel A, Merlio JP. The relative levels of cyclin D1a and D1b alternative transcripts in mantle cell lymphoma may depend more on sample origin than on CCND1 polymorphism. Haematologica. 2005;90(6):854–6. [PubMed] [Google Scholar]
- 44.Howe D, Lynas C. The cyclin D1 alternative transcripts [a] and [b] are expressed in normal and malignant lymphocytes and their relative levels are influenced by the polymorphism at codon 241. Haematologica. 2001;86(6):563–9. [PubMed] [Google Scholar]
- 45.Krieger S, Grunau C, Sabbah M, Sola B. Cyclin D1 gene activation in human myeloma cells is independent of DNA hypomethylation or histone hyperacetylation. Exp Hematol. 2005;33(6):652–9. doi: 10.1016/j.exphem.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 46.Marzec M, Kasprzycka M, Lai R, Gladden AB, Wlodarski P, Tomczak E, et al. Mantle cell lymphoma cells express predominantly Cyclin D1a isoform and are highly sensitive to selective inhibition of CDK4 kinase activity. Blood. 2006;108(5):1744–50. doi: 10.1182/blood-2006-04-016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ek S, Bjorck E, Porwit-MacDonald A, Nordenskjold M, Borrebaeck CA. Increased expression of Ki-67 in mantle cell lymphoma is associated with de-regulation of several cell cycle regulatory components, as identified by global gene expression analysis. Haematologica. 2004;89(6):686–95. [PubMed] [Google Scholar]
- 48.Greiner TC, Moynihan MJ, Chan WC, Lytle DM, Pedersen A, Anderson JR, et al. p53 mutations in mantle cell lymphoma are associated with variant cytology and predict a poor prognosis. Blood. 1996;87(10):4302–10. [PubMed] [Google Scholar]
- 49.Solenthaler M, Matutes E, Brito-Babapulle V, Morilla R, Catovsky D. p53 and mdm2 in mantle cell lymphoma in leukemic phase. Haematologica. 2002;87(11):1141–50. [PubMed] [Google Scholar]
- 50.Zoldan MC, Inghirami G, Masuda Y, Vandekerckhove F, Raphael B, Amorosi E, et al. Large-cell variants of mantle cell lymphoma: cytologic characteristics and p53 anomalies may predict poor outcome. Br J Haematol. 1996;93(2):475–86. doi: 10.1046/j.1365-2141.1996.5421085.x. [DOI] [PubMed] [Google Scholar]
- 51.Ely S, Di Liberto M, Niesvizky R, Baughn LB, Cho HJ, Hatada EN, et al. Mutually exclusive cyclin-dependent kinase 4/cyclin D1 and cyclin-dependent kinase 6/cyclin D2 pairing inactivates retinoblastoma protein and promotes cell cycle dysregulation in multiple myeloma. Cancer Res. 2005;65(24):11345–53. doi: 10.1158/0008-5472.CAN-05-2159. [DOI] [PubMed] [Google Scholar]
- 52.Specht K, Kremer M, Muller U, Dirnhofer S, Rosemann M, Hofler H, et al. Identification of cyclin D1 mRNA overexpression in B-cell neoplasias by real-time reverse transcription-PCR of microdissected paraffin sections. Clin Cancer Res. 2002;8(9):2902–11. [PubMed] [Google Scholar]
- 53.Fonseca R, Barlogie B, Bataille R, Bastard C, Bergsagel PL, Chesi M, et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 2004;64(4):1546–58. doi: 10.1158/0008-5472.can-03-2876. [DOI] [PubMed] [Google Scholar]
- 54.Nodit L, Bahler DW, Jacobs SA, Locker J, Swerdlow SH. Indolent mantle cell lymphoma with nodal involvement and mutated immunoglobulin heavy chain genes. Hum Pathol. 2003;34(10):1030–4. doi: 10.1053/s0046-8177(03)00410-6. [DOI] [PubMed] [Google Scholar]
- 55.Klier M, Anastasov N, Hermann A, Meindl T, Angermeier D, Raffeld M, et al. Specific lentiviral shRNA-mediated knockdown of cyclin D1 in mantle cell lymphoma has minimal effects on cell survival and reveals a regulatory circuit with cyclin D2. Leukemia. 2008;22(11):2097–105. doi: 10.1038/leu.2008.213. [DOI] [PubMed] [Google Scholar]
- 56.Shakir R, Ngo N, Naresh KN. Correlation of cyclin D1 transcript levels, transcript type and protein expression with proliferation and histology among mantle cell lymphoma. J Clin Pathol. 2008;61(8):920–7. doi: 10.1136/jcp.2008.057455. [DOI] [PubMed] [Google Scholar]