Background: Epithelial sodium channels (ENaC) are activated by proteolytic cleavage. Several proteases including furin and prostasin cleave ENaC.
Results: Cathepsin B also cleaves and activates ENaC. Cathepsin B cleaves ENaC α but not β or γ subunits.
Conclusion: Cathepsin B is a secreted protease, so it may cleave ENaC at the cell surface.
Significance: Cathepsin B cleavage represents a novel ENaC regulatory mechanism.
Keywords: Cysteine Protease, ENaC, Epithelial Cell, Proteolytic Enzymes, Sodium Transport, Cathepsin B, ENaC Activation
Abstract
The epithelial sodium channel (ENaC) plays an important role in regulating sodium balance, extracellular volume, and blood pressure. Evidence suggests the α and γ subunits of ENaC are cleaved during assembly before they are inserted into the apical membranes of epithelial cells, and maximal activity of ENaC depends on cleavage of the extracellular loops of α and γ subunits. Here, we report that Xenopus 2F3 cells apically express the cysteine protease cathepsin B, as indicated by two-dimensional gel electrophoresis and mass spectrometry analysis. Recombinant GST ENaC α, β, and γ subunit fusion proteins were expressed in Escherichia coli and then purified and recovered from bacterial inclusion bodies. In vitro cleavage studies revealed the full-length ENaC α subunit fusion protein was cleaved by active cathepsin B but not the full-length β or γ subunit fusion proteins. Both single channel patch clamp studies and short circuit current experiments show ENaC activity decreases with the application of a cathepsin B inhibitor directly onto the apical side of 2F3 cells. We suggest a role for the proteolytic cleavage of ENaC by cathepsin B, and we suggest two possible mechanisms by which cathepsin B could regulate ENaC. Cathepsin B may cleave ENaC extracellularly after being secreted or intracellularly, while ENaC is present in the Golgi or in recycling endosomes.
Introduction
Epithelial sodium channels (ENaCs)2 are expressed within the apical membranes of epithelial cells, primarily in the collecting tubules of the kidney, lung, and colon. ENaC plays an important role in the regulation of renal sodium transport and helps maintain total body water and salt homeostasis. Specific mutations in ENaC lead to inherited forms of hypertension (e.g. Liddle's syndrome) and hypotension (e.g. pseudohypoaldosteronism type 1) (1–5).
ENaC functions most efficiently as a heteromultimeric protein complex formed from α, β, and γ subunits, but the α subunit alone may also form functional channels. Each of the three ENaC subunits consists of short intracellular NH2 and COOH domains, a large extracellular loop, and two transmembrane domains. Ion channels can be activated by selective ligands (6, 7), chemical compounds (8), hormones (6, 9), intrinsic signaling (6, 10), and by proteolytic cleavage (8, 11, 12). Several reports suggest the gating of ENaC is regulated by cations (13), anions (14), sodium (15), protons (16), and proteases (17–26). Recent evidence has shown that ENaC is activated when cleaved by several different tryptic proteases including channel-activating protease 1/2 (21, 28, 29), trypsin (30), furin (22, 32), and prostasin (32–34). The cleavage of ENaC was found to occur mainly in the Golgi apparatus before ENaC is inserted into the membrane. The ability of secreted proteases to cleave and activate ENaC has not been thoroughly investigated.
Various epithelial cells are known to secrete proteases. Cathepsin B is a lysosomal cysteine protease that is expressed in epithelial cells and tissues (35). The active enzyme is secreted upon lysosomal exocytosis or extracellular processing by surface activators (36). Cathepsin B is different from other cathepsins in that it functions as an endopeptidase at neutral pH and can be found outside of lysosomes including in the cytoplasm, at the plasma membrane, and in the extracellular space. Cathepsin B contributes to various biological processes including inflammation, cancer metastasis, apoptosis, and differentiation (37–41).
Like other integral membrane proteins, ENaC is susceptible to proteolysis by secreted proteases in response to physiological and pathophysiological conditions. Here we demonstrate that the commonly used Xenopus 2F3 subclone of the A6 cell line in the study of the regulation of ENaC secretes the protease cathepsin B exclusively on the apical side. Our findings also show that this protease can cleave ENaC in vitro and activate ENaC in situ.
MATERIALS AND METHODS
Cell Culture
2F3 cells were a kind gift of Dr. Dale Benos. The cells were seeded onto permeable polyester inserts and cultured in 50/50 Dulbecco's modified Eagle's medium/Ham's F-12 base media (Invitrogen) at pH 7.4 and supplemented with 5% fetal bovine serum (Invitrogen), 1.5 μm aldosterone, 0.6% penicillin, and 1% streptomycin (Irvine Scientific, Santa Ana, CA). Cells were maintained in a humidified incubator with 4% CO2 in air. Experiments were performed using cells between passages 98 and 106.
Two-dimensional Gel Electrophoresis
Apical and basolateral media was subject to a cleanup procedure using the ReadyPrep 2-D cleanup kit (Bio-Rad) to concentrate the sample and to remove contaminants that could interfere with isoelectric focusing. Two-hundred micrograms of the purified protein sample prepared in 125 μl of rehydration/sample buffer (8 m urea, 2% CHAPS, 50 mm dithiothreitol (DTT), 0.2% (w/v) Bio-lyte 3/10 ampholytes, and bromphenol blue) was applied to IPG strips (pH 4–7 ReadyStrip, 7-cm, Bio-Rad). The strips were allowed to rehydrate for 16 h at room temperature. The rehydrated IPG strips were transferred to an isoelectric focusing tray, and electrode wicks were used as receptacles for non-amphoteric constituents. The IPG strips were covered with mineral oil and then focused using a Protean IEF cell (Bio-Rad) according to the following parameters (250 V, linear ramp, 20 min; 4,000 V, linear ramp, 2 h; 4,000 V rapid ramp, 10,000 V-h; 20 °C, 50 μA/strip). Focused IPG strips were transferred to an equilibration tray and incubated with 2.5 ml of equilibration buffer 1 (6 m urea, 2% SDS, 0.375 m Tris-HCl (pH 8.8), 20% glycerol, and 2% (w/v) DTT) for 10 min on an orbital shaker at room temperature. The IPG strips were then incubated with 2.5 ml of equilibration buffer 2 (6 m urea, 2% SDS, 0.375 m Tris-HCl (pH 8.8), 20% glycerol, and with 2.5% (w/v) iodoacetamide) for 10 min on an orbital shaker at room temperature.
Coomassie Staining
To visualize the separated proteins, 4–15% gradient IPG gels were fixed with Colloidal Coomassie fixing solution (45% methanol, 1% acetic acid, 54% ultrapure H2O) for 1 h and then stained with Colloidal Coomassie staining solution (170 g/liter ammonium sulfate, 1 g/liter Coomassie G250, 0.5% acetic acid, 34% methanol) for 8 h and then destained in destaining solution (5% methanol, 95% ultrapure H2O) for 8 h.
MALDI-TOF Mass Spectrometry
Selected protein spots were excised from Coomassie-stained gels and then transferred to keratin-free tubes. The gel spots were subject to in-gel tryptic digestion followed by liquid extraction of the gel fragments. The peptides were extracted and concentrated under vacuum centrifugation. A nanoflow liquid chromatograph (Easy-nLC, Proxeon, Odense, Denmark) coupled to an electrospray ion trap mass spectrometer (LTQ, Thermo, San Jose, CA) was used for tandem mass spectrometry peptide sequencing experiments. Spectra were analyzed by searching with the publicly available search algorithm Mascot using the National Center for Biotechnology Information and SwissProt databases.
SDS-PAGE and Immunoblot Analysis
2F3 cells were washed with 1× PBS and lysed with mammalian protein extraction reagent (Thermo Scientific; Rockford, IL) containing protease and phosphatase inhibitors (Thermo Scientific) followed by passing the lysate 5 times through a 23-gauge needle with a syringe before being incubated on ice for 1 h. Protein concentration was determined using the BCA assay (Thermo Scientific) according to the manufacturer's instructions. Samples were prepared in Laemmli sample buffer (Bio-Rad) supplemented with 5% β-mercaptoethanol. One hundred micrograms of protein was loaded and resolved on 7.5% Tris-HCl polyacrylamide precast gels using either the Criterion or Protean Western blotting system (Bio-Rad). The separated proteins were electrophoretically transferred to Hybond C-extra nitrocellulose membranes (GE Healthcare) and then blocked in 5% (w/v) dry milk in 1× TBS (Bio-Rad) for 1 h at room temperature. Immunoblotting was performed by first incubating the blocked membranes with peroxidase-conjugated anti-GST antibody (Sigma) or anti-cathepsin B antibody (Cell Signaling Technologies) at a 1:1000 dilution prepared in 5% BSA, 1× TBS for 8 h at 4 °C. After a series of three 5-min washes with 1× TBS, the membranes were incubated with HRP-conjugated goat anti-rabbit secondary antibody at a dilution of 1:3000 prepared in blocking solution. The immunoreactive bands were visualized using a SuperSignal Dura chemiluminescent substrate kit (Pierce) according to the manufacturer's instructions. The membranes were exposed using the Kodak Gel Logic 2200 Imager and Molecular Imaging software system (Carestream Health, Rochester, NY), and relative molecular masses of the immunoreactive bands were assessed using Precision Plus Protein Standards (Bio-Rad).
Densitometric Analysis
Densitometry was conducted using ImageJ software (National Institutes of Health). Protein levels are presented as arbitrary units of a ratio between the band intensity for the protein of interest and the band intensity of actin.
Recombinant Protein Production
The entire coding sequence of each ENaC subunit was subcloned into the pGEX4T-2 expression vector. The constructs were transformed into competent Escherichia coli cells, and expression was induced with isopropyl 1-thio-β-d-galactopyranoside. The fusion proteins were purified and recovered from bacterial inclusion bodies as previously described by Alli and Gower (42, 43).
Electrophysiological Transepithelial Measurements
The voltage and resistance across confluent monolayers were measured with an epithelial Voltohmeter (EVOM; World Precision Instrument, Sarasota, FL), and transepithelial amiloride-sensitive current was calculated per unit area according to Ohm's law before and after application of 10 μm cathepsin B inhibitor (CA-74) (Calbiochem) or vehicle dimethyl sulfoxide (DMSO) (Sigma) at time points between 5 and 60 min. The cells were harvested for protein for biochemical studies as described above.
Cell-attached Single Channel Patch Clamp Studies
Micropipettes were pulled (two-stage vertical puller; Tokyo, Japan) from filamented borosilicate glass capillaries (TW-150F, World Precision Instruments). The basolateral and apical membranes of 2F3 cells subcultured on Lucite rings were exposed to pipette solution (96 mm NaCl, 3.4 mm KCl, 0.8 mm MgCl2, 0.8 mm CaCl2, and 10 mm HEPES). The micropipettes were filled with the same patch solution and had a resistance between 5 and 10 ohms. The product of the number of channels (N) and the single-channel open probability (Po) was used to calculate ENaC activity within a patch. The number of current levels or the number of peaks detected in current amplitude histograms were used to estimate the total number of channels in a patch. The open probability of a single channel in a patch was calculated by dividing NPo by the number of channels in a patch. Single channel currents from gigaohm seals were filtered at 100 Hz and recorded at 500 Hz using the Clampfit 10.1 software (Molecular Devices).
In Vitro Cathepsin B Cleavage Assay
Active cathepsin B protease purified from human liver was purchased from Calbiochem. Five micrograms of freshly prepared recombinant ENaC GST fusion proteins in elution buffer was incubated with 5 μg of cathepsin B protease at 37 °C for 8 h. The reactions were terminated by adding sample buffer containing SDS. The samples were analyzed by SDS-PAGE and Coomassie staining.
siRNA-mediated Knockdown of Cathepsin B
siRNA targeting cathepsin B (sense, 5′-AGUCUAUGGCAGUGACAAAUU-3′; antisense, 5′-UUUGUCACUGCCAUAGACUUU-3′) was designed using the siDESIGN tool and purchased from Thermo Scientific. As a control, non-targeting siRNA was used (siGENOME non-targeting siRNA #2; Thermo Scientific). Cells were transfected with siRNA using Xfect transfection reagent (Clontech; Mountain View, CA) while following the manufacturer's instructions.
Cell Surface Biotinylation
Confluent cells grown on transwell-permeable support were treated with drug or vehicle for 3 h in an incubator. The apical side of the inserts was washed twice with sterile ice-cold PBS and then incubated for 30 min in ice-cold borate buffer (85 mm NaCl, 4 mm KCl, 15 mMNa2B4O7, pH 9.0) containing biotin (ThermoScientific) (0.5 mg/ml). The cells were then washed with lysine solution (18.3 g/liter complete growth medium) and then incubated for 15 min in the same solution. The cells were washed with sterile ice-cold PBS and lysed in mammalian protein extraction reagent containing protease and phosphatase inhibitors. The lysate was incubated with prewashed avidin beads and incubated for 6 h at 4 °C. The complex was washed 3 times with sterile PBS, and proteins were eluted by boiling for 10 min at 100 °C in SDS sample buffer. Thirty microliters of the prepared sample was loaded and resolved on 7.5% Tris-HCl polyacrylamide gels and subject to SDS-PAGE and immunoblotting as mentioned above but with anti-ENaC α antibody (44) at a dilution of 1:1000.
Statistical Analysis
The values obtained from experiments performed in triplicate (n = 3) are presented as the mean ± S.E. Statistical significance was determined by using the Student's t test at p < 0.05.
RESULTS
Cathepsin B Is Found in the Apical but Not Basolateral Medium from Confluent Xenopus 2F3 Cells
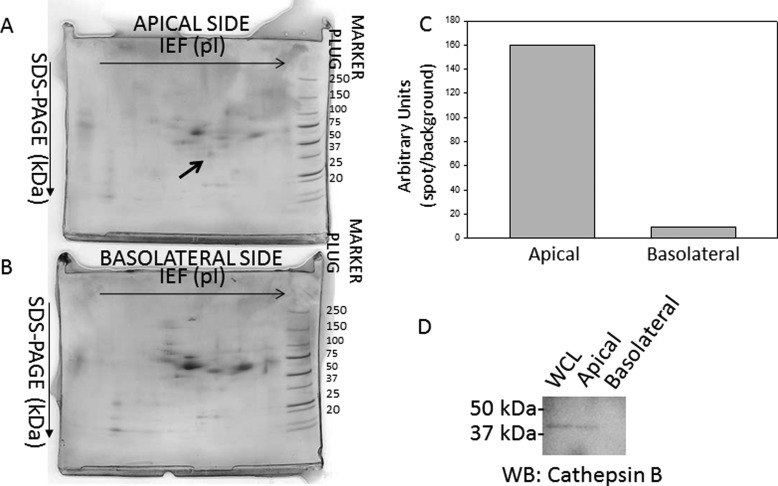
The 2F3 cell line remains an excellent model system to study the regulation of ENaC using biochemical and electrophysiological approaches. ENaC is known to be activated by various proteases. To determine if 2F3 cells secrete any proteases for which ENaC could be a substrate, media from the apical and basolateral sides of these cells were concentrated and then subjected to two-dimensional gel electrophoresis. Because the samples were processed extensively to remove contaminants that could potentially minimize resolution and cause vertical and horizontal streaking, spot profiles from the apical and basolateral samples could be analyzed, and the protein spots could be identified by mass spectrometry. The proteins were separated by their isoelectric point in the horizontal direction and then by their molecular weight in the vertical direction. Several spots corresponding to proteins unique to either the apical or basolateral sides were excised and subject to mass spectrometry sequencing for protein identification. Fig. 1 shows one such protein spot that was found on the apical side but not the basolateral side. Mass spectrometry analysis of this protein spot revealed multiple signature peptides corresponding to cathepsin B.
FIGURE 1.
Two-dimensional gel electrophoresis analysis of proteins located on the apical and basolateral side of 2F3 cells. Coomassie-stained gel of proteins from media collected and concentrated from the apical side (A) or basolateral side (B) of Xenopus 2F3 cells that were subcultured on permeable supports and allowed to form tight junctions and generate a measurable transepithelial resistance and voltage. Some proteins were found to be exclusively on the apical or basolateral side. The protein spot in A, labeled with an arrow, was excised and identified as cathepsin B after mass spectrometry protein sequencing. C, densitometric analysis of the protein spot indicated by an arrow in A. D, shown is a representative Western blot of cathepsin B expression in the cell lysate (whole cell lysate (WCL)) and in the media collected and concentrated from the apical and basolateral side of Xenopus 2F3 cells. IEF, isoelectric focusing.
ENaC Is Cleaved in Vitro in the Presence of Active Cathepsin B
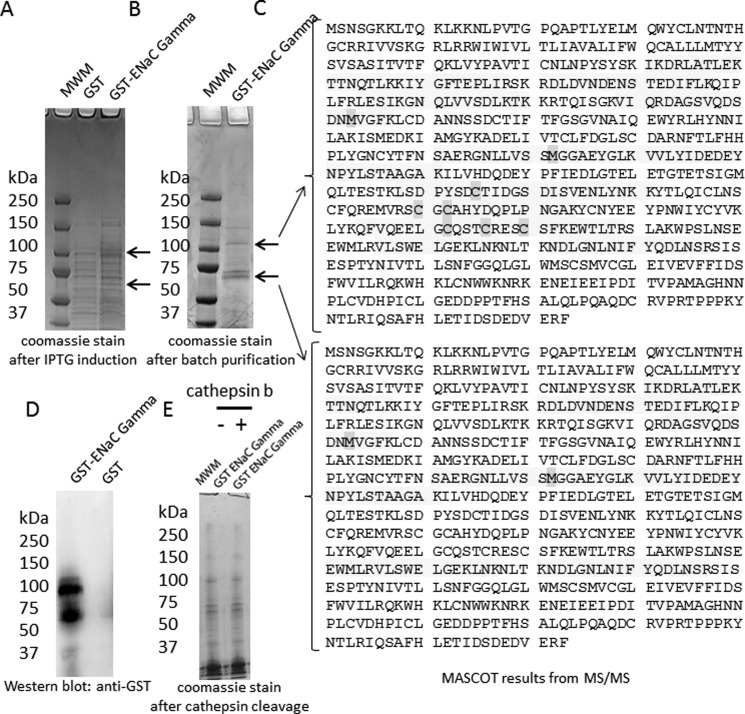
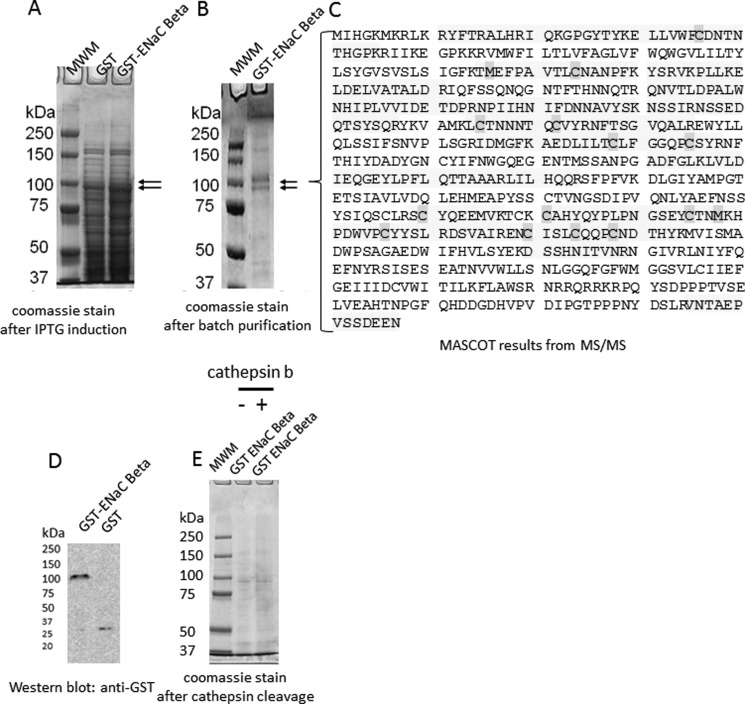
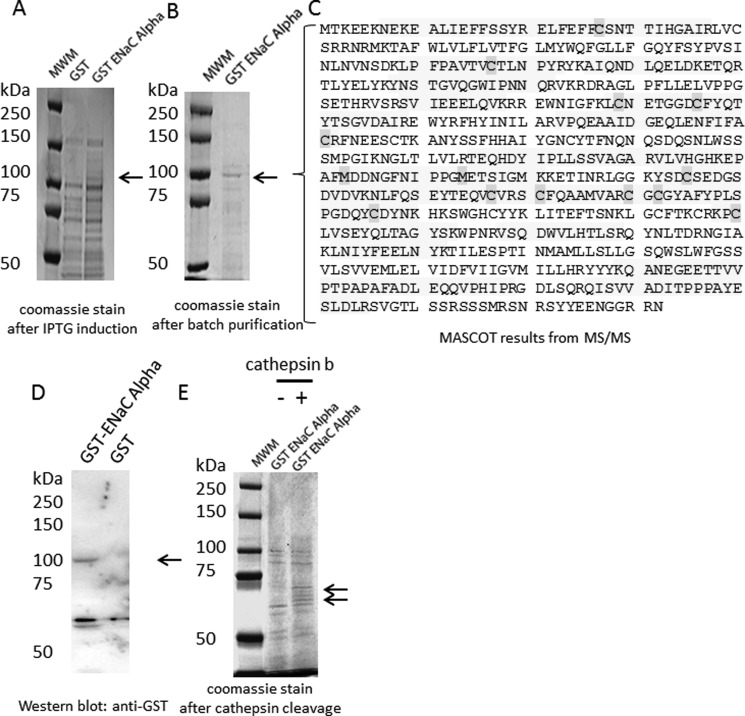
To determine whether each of the three structurally similar but distinct ENaC subunits could be substrates of cathepsin B, we first subcloned the full Xenopus coding sequence of each subunit into a bacterial expression vector comprising a GST tag. Recombinant fusion proteins can be valuable resources in many different applications because they can serve as a renewable source of purified protein and they can be obtained in large quantities. However, expression of foreign proteins in bacterial systems and particularly those proteins with hydrophobic domains often pose problems because they tend to aggregate in bacterial inclusion bodies. Each of the ENaC subunits contains two transmembrane domains and aggregated in inclusion bodies. Using a system previously described by Alli and Gower (42), we expressed each ENaC subunit as a GST fusion protein and then purified them from bacterial inclusion bodies. The expression of foreign proteins in a bacterial system may result in the protein being prematurely cleaved by host proteases, and the purification of those proteins often results in copurification of host proteins. Therefore, we excised protein bands that corresponded to each of the three ENaC subunits from Coomassie-stained gels and confirmed the identity and coverage of the proteins by mass spectrometry. Mascot reports of the mass spectral data revealed multiple large signature peptides corresponding to ENaC γ (Fig. 2C), ENaC β (Fig. 3C), and ENaC α (Fig. 4C). Of the two proteins bands excised from Coomassie-stained gels after SDS-PAGE analysis of recombinant GST ENaC γ (Fig. 2B), the lower molecular weight band represents ENaC γ without the GST moiety, whereas the higher molecular weight band represents ENaC γ with the GST moiety (Fig. 2C). The mass spectrometry sequence coverage did not reveal any mutations or sequence coverage for the GST moiety of any of the ENaC subunit fusion proteins. The fusion proteins were used to perform in vitro cathepsin B cleavage experiments. The GST ENaC γ fusion protein was subject to treatment with or without active cathepsin B protease. Coomassie-stained gels and Western blot analysis demonstrated a stained and immunoreactive band, respectively, of 100 kDa for ENaC γ fusion protein (Fig. 2). Further analysis revealed the GST ENaC γ subunit fusion was not cleaved by cathepsin B (Fig. 2). Similar to GST ENaC γ, Coomassie-stained gels and Western blot analysis demonstrated a stained and immunoreactive band, respectively, of 100 kDa for ENaC β fusion protein (Fig. 3). Also, further analysis revealed the GST ENaC β subunit fusion was not cleaved by cathepsin B. Coomassie-stained gels and Western blot analysis demonstrated a stained and immunoreactive band, respectively, of 100 kDa for ENaC α fusion protein (Fig. 4). Unlike for recombinant GST ENaC γ and GST ENaC β fusion proteins, lower molecular bands were present when GST ENaC α fusion was exposed to cathepsin B (Fig. 4), suggesting cathepsin B cleavage of ENaC α.
FIGURE 2.
Characterization and proteolytic analysis of full-length ENaC γ subunit. A, a Coomassie-stained gel shows the expression of the entire γ subunit of ENaC as a GST fusion protein (GST-ENaC γ) after isopropyl 1-thio-β-d-galactopyranoside (IPTG) induction. B, a Coomassie-stained gel shows the double band corresponding to GST-ENaC γ or ENaC γ after batch purification. C, MASCOT mass spectrometry peptide sequence coverage of several signature peptides corresponding to Xenopus ENaC γ subunit is shown. The shaded residues represent oxidation of methionine residues or alkylation of cysteine residues. D, shown is Western blot analysis of GST-ENaC using a peroxidase-conjugated antibody against GST to detect a 100-kDa immunoreactive band corresponding to GST-ENaC γ. E, a Coomassie-stained gel indicates GST-ENaC γ is not cleaved by cathepsin B in vitro. MWM, molecular weight markers.
FIGURE 3.
Characterization and proteolytic analysis of full-length ENaC β subunit. A, a Coomassie-stained gel shows the expression of the entire β subunit of ENaC as a GST fusion protein (GST-ENaC β) after isopropyl 1-thio-β-d-galactopyranoside (IPTG) induction. B, a Coomassie-stained gel shows the double band corresponding to GST-ENaC β after batch purification. C, MASCOT mass spectrometry peptide sequence coverage of several signature peptides corresponding to Xenopus ENaC β subunit is shown. The shaded residues represent oxidation of methionine residues or alkylation of cysteine residues. D, shown is Western blot analysis of GST-ENaC using a peroxidase-conjugated antibody against GST to detect a 100-kDa immunoreactive band corresponding to GST-ENaC β. E, a Coomassie-stained gel indicates GST-ENaC β is not cleaved by cathepsin B in vitro. MWM, molecular weight markers.
FIGURE 4.
Characterization and proteolytic analysis of full-length ENaC α subunit. A, a Coomassie-stained gel shows the expression of the entire α subunit of ENaC as a GST fusion protein (GST-ENaC α) after isopropyl 1-thio-β-d-galactopyranoside (IPTG) induction. B, a Coomassie-stained gel shows a band corresponding to GST-ENaC α after batch purification. C, shown is MASCOT mass spectrometry peptide sequence coverage of several signature peptides corresponding to the Xenopus ENaC α subunit. The shaded residues represent oxidation of methionine residues or alkylation of cysteine residues. D, shown is a Western blot analysis of GST-ENaC using a peroxidase-conjugated antibody against GST to detect a 100-kDa immunoreactive band corresponding to GST-ENaC α. E, shown is a Coomassie-stained gel indicating GST-ENaC α is cleaved by cathepsin B in vitro. MWM, molecular weight markers.
The Extracellular Loop of ENaC α Contains Putative Cathepsin B Cleavage Sites
The bioinformatics tool PEPS (Prediction of Endopeptidase Substrates) was used to predict putative cathepsin B cleavage sites within the entire coding sequence of Xenopus ENaC subunits. PEPS overcomes limitations of other bioinformatics tools and has been found to offer reliable endopeptidase substrate predictions (45). The sequence of Homo sapiens cathepsin B that was used to perform the in vitro cathepsin B cleavage assays was compared with that of Xenopus laevis and found to be identical, as shown in Fig. 5A. At least one putative cathepsin B cleavage site lies within the extracellular loop of ENaC α (Fig. 5B). However, the β and γ subunits of ENaC lack putative cathepsin b cleavage sites.
FIGURE 5.
Sequence analysis of cathepsin B and ENaC. A, shown is multiple sequence alignment of H. sapiens and X. laevis cathepsin B. The amino acid sequence of cathepsin B is highly conserved between H. sapiens and X. laevis, as it is for several other species. B, shown is the predicted cleavage site of cathepsin B in the extracellular loop of Xenopus ENaC α subunit. Based on known cleavage sites of other substrates of cathepsin B, the amino acid region underlined and in bold italics is the predicted cathepsin B cleavage site within the extracellular loop of the α subunit of ENaC.
Inhibition and siRNA-mediated Knockdown of Cathepsin B Decrease Amiloride-sensitive Transepithelial Current in 2F3 Cells
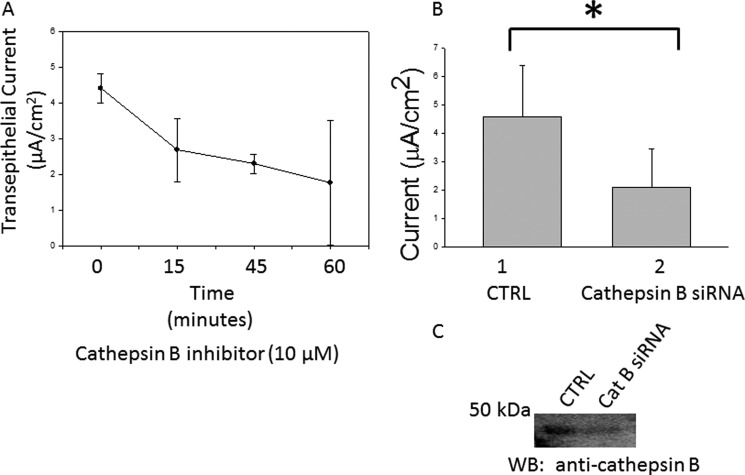
Amiloride-sensitive ENaC are responsible for transepithelial sodium transport. It is possible to correlate changes in ENaC activity at the apical membrane with changes in transepithelial current measured by an electrode voltage ohmmeter because it measures net transepithelial current. The correlation between changes in transepithelial current and ENaC activity (based on electrode voltage ohmmeter measurements) are supported because an increase in transepithelial current can be attenuated with the addition of amiloride. As shown in Fig. 6A, application of a selective cathepsin B inhibitor significantly decreased ENaC activity in a dose- and time-dependent manner. The resistance measured before and after application of the inhibitor did not decrease but instead slightly increased after application of the inhibitor (consistent with a decrease in apical ENaC conductance). The effect of the cathepsin B inhibitor on transepithelial current was corroborated by siRNA-mediated knockdown of cathepsin B. A statistically significant decrease in transepithelial current was observed 48 h after transient transfection of siRNA targeting cathepsin B when compared with transfection of nontargeting siRNA in Xenopus 2F3 cells (Fig. 6B). Western blot analysis showed cathepsin B decreased at the protein level after siRNA transfection (Fig. 6C).
FIGURE 6.
Measurements of amiloride-sensitive transepithelial current in 2F3 cells. A, transepithelial sodium current decreased with time after treating Xenopus 2F3 cells with 10 μm cathepsin B inhibitor. B, transepithelial sodium current decreased after siRNA-mediated knockdown of cathepsin B in Xenopus 2F3 cells. Values are presented as the means ± S.E., n = 3 for each data point. *, p < 0.05. CTRL, control. C, a representative Western blot (WB) shows a decrease in cathepsin B at the protein level after siRNA-mediated knockdown as presented in B.
Inhibition of Cathepsin B Decreases the Number of Active α ENaCs in 2F3 Cells
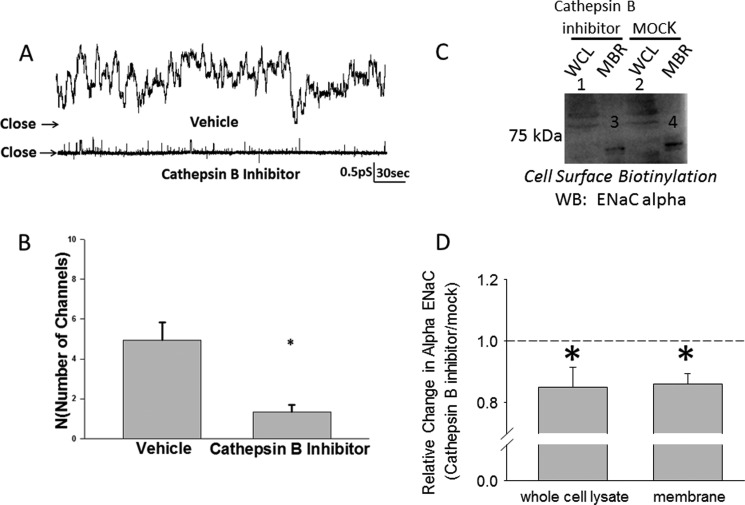
To determine if inhibiting cathepsin B affects the open probability or the number of active channels in the apical membrane or both, we performed single channel patch clamp studies. Fig. 7 shows representative traces of eight patches with active ENaC channels in which 2F3 cells were pretreated with vehicle or the cathepsin B inhibitor for 1 h before acquiring a seal. The cathepsin B inhibitor significantly decreased the number of active channels at the membrane; however, the open probability of the remaining channels was unchanged from that before treatment. To further confirm cathepsin B affects the density of ENaC at the membrane, cell surface biotinylation studies were performed. As shown in Fig. 7, C and D, representative Western blots show a statistically significant decrease in the density of α ENaC both in the cell lysate and at the membrane after treatment with the cathepsin B inhibitor.
FIGURE 7.
Single channel recording from Xenopus 2F3 cells showing an effect on ENaC activity after application of a cathepsin B inhibitor. A, shown are representative traces of Xenopus 2F3 cells pretreated with vehicle (DMSO) or cathepsin B inhibitor (CA-074) for 1 h before acquiring a patch. B, results of eight independent patch clamp studies show the cathepsin B inhibitor significantly decreased the number of active channels in a patch of Xenopus 2F3 cells. *, p < 0.001. Empty patches were included in the calculation of N, but the inhibitor did not affect Po. C, a representative Western blot (WB) of three independent experiments shows a decrease in ENaC α protein in the cell lysate and at the membrane after treating Xenopus 2F3 cells with the cathepsin B inhibitor. WCL represents whole cell lysate, and MBR represents membrane fractions. D, a densitometric analysis of immunoreactive bands in C shows a statistically significant decrease in ENaC α protein in either the whole cell lysate or membrane fractions after treatment with the cathepsin B inhibitor relative to treatment with the vehicle (MOCK). n = 3; *, p < 0.05.
DISCUSSION
Accumulating evidence suggests cleavage of the extracellular loop of α and γ ENaC subunits by several tryptic proteases enhances the activity of the channel. This cleavage is thought to occur principally during channel assembly within the trans-Golgi network before ENaC is inserted into the apical membrane. It has been shown that there are at least two cleavage sites within the extracellular loop of the α and γ subunits of mammalian ENaC. Besides there being other cleavage sites by other proteases than those that have been previously reported, ENaC may be cleaved at different points as it traffics to the apical membrane. It is also possible that a population of uncleaved or partially cleaved ENaC at the membrane may be subject to further cleavage, or a population of ENaC may be cleaved while being recycled. ENaC can be internalized through a clathrin-dependent pathway and is subsequently localized to early endosomes. A population of the channels is targeted for lysosomal degradation, whereas another population is deubiquitinated to allow recycling.
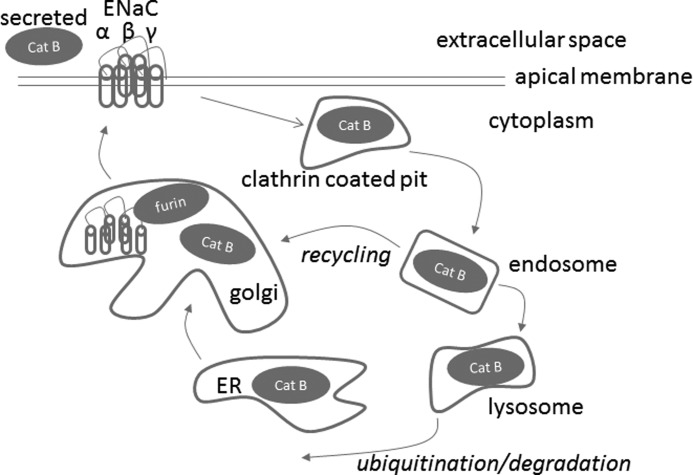
Cathepsin B is post-translationally processed in the rough endoplasmic reticulum, Golgi, endosomal, and lysosomal compartments (Fig. 8). Cathepsin B is also secreted into the extracellular space of various tumor epithelial cells (46, 47). Taking this into consideration along with our findings that cathepsin B is secreted on the apical side of 2F3 cells, we considered several possibilities: cathepsin B cleaving ENaC within the Golgi during ENaC assembly, extracellular cleavage while ENaC is in the apical membrane, or cleavage within endosomes while ENaC is being recycled (Fig. 8).
FIGURE 8.
Proposed model depicting the cathepsin B cleavage-dependent activation of ENaC. The cysteine protease cathepsin B (Cat B) is synthesized as a latent preproenzyme and post-translationally processed in the rough endoplasmic reticulum, Golgi, endosomal, and lysosomal compartments. Cathepsin B is secreted or found in the cytoplasm of certain epithelial cells. Unlike other cysteine proteases, cathepsin B exhibits both endopeptidase and exopeptidase activities. Cathepsin B is shown to potentially cleave the extracellular loop of the α subunit of ENaC either within the Golgi, extracellularly, or within endosomes.
We used a non-permeable inhibitor of cathepsin B in our patch clamp studies to investigate the role of cathepsin B secreted into the extracellular space. Because there is secreted cathepsin B on the apical side of our 2F3 cells, we predicted application of the inhibitor would not have an immediate effect as ENaC would already be cleaved. Therefore, before patching we pretreated our cells for at least 1 h with the non-permeable form of the cathepsin B inhibitor to allow for a new population of uncleaved or partially cleaved ENaC to insert into the membrane. Both single-channel patch clamp studies and cell surface biotinylation studies suggest cathepsin B affects the density of ENaC at the membrane. In other experiments, when we applied active cathepsin B protease directly onto transporting 2F3 cells we did not observe a significant effect. This is likely because a majority, if not all, of the functional ENaC channels were already cleaved to produce maximal activity.
Our studies are consistent with those of others in that the α subunit of ENaC is subject to proteolytic cleavage. Cathepsin B appears to be a highly conserved protease, as suggested by multiple sequence alignments of at least 12 different organisms. Because it could be obtained in an active and purified form, human cathepsin B from human liver was used to determine if recombinant full-length Xenopus ENaC α, β, and γ subunit fusion proteins could be cleaved by cathepsin B in vitro. We choose to perform our in vitro proteolytic cleavage experiments at optimal conditions to achieve maximum protease activity while minimizing potential degradation and nonspecific proteolytic activity. Therefore, our studies involved digesting each of the three recombinant GST ENaC subunit fusion proteins with or without cathepsin B at 37 °C for 8 h.
2F3 cells are a clone of the A6 cell line, and both cell lines are excellent models that are commonly utilized in electrophysiology to investigate the regulation of ENaC. Our findings of cathepsin B being secreted on the apical side of 2F3 cells are novel, because previous reports of cathepsin B being secreted by certain epithelial cells were limited to cancer cells. Cathepsin B can be released in the cytosol and has been found to contribute to various cellular processes including caspase activation (48, 49) and arachidonic acid release (46). Foghsgaard et al. (46) showed cathepsin B regulates TNF-induced arachidonic acid release by cleaving cPLA2. Because Wei et al. (31) has already shown arachidonic acid decreases ENaC activity, we investigated if cathepsin B could differentially regulate ENaC when it is expressed in the cytoplasm or when it is expressed in the extracellular space after being secreted.
Another possible mechanism by which cathepsin B could increase ENaC activity is through indirect proteolytic processing of other proteins, which could subsequently activate ENaC. Others have shown cathepsin B is involved in the transcriptional and post-translational processing of various proteinases. For example, cathepsin B is also a candidate for the regulation of TNF-α vesicle trafficking through the regulation of SNARE components (27). Because cathepsin B is such a versatile protease, future studies may look to investigate if proteins known to regulate the insertion, gating, recycling, or degradation of ENaC are also substrates for cathepsin B.
We screened the entire coding sequence of each Xenopus ENaC subunit using the PEPS algorithm to identify putative cathepsin B cleavage sites. The analysis revealed at least one site that might be recognized and cleaved by cathepsin B and that is within the extracellular loop domain of Xenopus ENaC α but is not present in the extracellular loop of either Xenopus ENaC β or γ. This finding is consistent with our hypothesis that the extracellular loop of at least one ENaC subunit is cleaved after cathepsin B is secreted into the extracellular space. However, despite the algorithm identifying a putative cleavage site, the consensus sequence for cathepsin B remains ambiguous and substrate prediction algorithms are often not definitive. In fact, we observed multiple bands after digesting our purified ENaC α recombinant protein with active cathepsin B, which suggests that there could be multiple cathepsin B cleavage sites. Our initial attempt to confirm the cathepsin B consensus sequence using mass spectrometry was unsuccessful because we could not generate enough protein from the bands excised from Coomassie-stained gels after digesting recombinant Xenopus ENaC protein with active cathepsin B. Confirming that cathepsin B cleaves ENaC α at the site predicted by PEPS, and identifying the exact amino residues around the cleavage site will require extensive studies because there could be multiple cleavage sites. Our future plans are to mutate the putative cathepsin B cleavage site by site-directed mutagenesis, produce recombinant Xenopus ENaC α protein, and perform in vitro cathepsin B assay studies.
Acknowledgment
We thank B. J. Duke for concentrating the media and for maintaining 2F3 cells in culture. We also thank the Proteomics Core Facility at the Moffitt Cancer Center, Tampa, FL for mass spectrometry services.
This work was supported, in whole or in part, by National Institutes of Health Grants F32 DK093255–01 (to Abdel A. Alli), T32 DK007771 (to C. A. Parkos), 5R01 DK067110 (to H.-P. M.), and R37 DK037963 (to D. C. E.).
- ENaC
- epithelial sodium channel.
REFERENCES
- 1. Bertog M., Cuffe J. E., Pradervand S., Hummler E., Hartner A., Porst M., Hilgers K. F., Rossier B. C., Korbmacher C. (2008) Aldosterone responsiveness of the epithelial sodium channel (ENaC) in colon is increased in a mouse model for Liddle's syndrome. J. Physiol. 586, 459–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu C., Pribanic S., Debonneville A., Jiang C., Rotin D. (2007) The PY motif of ENaC, mutated in Liddle syndrome, regulates channel internalization, sorting, and mobilization from subapical pool. Traffic 8, 1246–1264 [DOI] [PubMed] [Google Scholar]
- 3. Konstas A. A., Mavrelos D., Korbmacher C. (2000) Conservation of pH sensitivity in the epithelial sodium channel (ENaC) with Liddle's syndrome mutation. Pflugers Arch. 441, 341–350 [DOI] [PubMed] [Google Scholar]
- 4. Prince L. S., Welsh M. J. (1999) Effect of subunit composition and Liddle's syndrome mutations on biosynthesis of ENaC. Am. J. Physiol. 276, C1346–C1351 [DOI] [PubMed] [Google Scholar]
- 5. Schild L. (1996) The ENaC channel as the primary determinant of two human diseases. Liddle syndrome and pseudohypoaldosteronism. Nephrologie 17, 395–400 [PubMed] [Google Scholar]
- 6. Krsek J., Dittert I., Hendrych T., Hník P., Horák M., Petrovic M., Sedlácek M., Susánková K., Svobodová L., Tousová K., Ujec E., Vlachová V., Vyklický L., Vyskocil F., Vyklický L., Jr. (2004) Activation and modulation of ligand-gated ion channels. Physiol. Res. 53, S103–S113 [PubMed] [Google Scholar]
- 7. Zimmermann I., Dutzler R. (2011) Ligand activation of the prokaryotic pentameric ligand-gated ion channel ELIC. PLoS. Biol. 9, e1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lester H. A. (1995) Activation of ion channels by acetylcholine. Two contrasting transduction pathways. Harvey Lect. 91, 79–98 [PubMed] [Google Scholar]
- 9. Furukawa T., Kurokawa J. (2007) Regulation of cardiac ion channels via non-genomic action of sex steroid hormones. Implication for the gender difference in cardiac arrhythmias. Pharmacol. Ther. 115, 106–115 [DOI] [PubMed] [Google Scholar]
- 10. Haddock R. E., Hill C. E. (2002) Differential activation of ion channels by inositol 1,4,5-trisphosphate (IP3)- and ryanodine-sensitive calcium stores in rat basilar artery vasomotion. J. Physiol 545, 615–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clark E. B., Jovov B., Rooj A. K., Fuller C. M., Benos D. J. (2010) Proteolytic cleavage of human acid-sensing ion channel 1 by the serine protease matriptase. J. Biol. Chem. 285, 27130–27143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grant A., Amadesi S., Bunnett N. W. (2007) in TRP Ion Channel Function in Sensory Transduction and Cellular Signaling Cascades (Liedtke W. B., Heller S., eds) pp. 421–434, CRC Press, Inc, Boca Raton, FL: [PubMed] [Google Scholar]
- 13. Palmer L. G., Andersen O. S. (1989) Interactions of amiloride and small monovalent cations with the epithelial sodium channel. Inferences about the nature of the channel pore. Biophys. J. 55, 779–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Collier D. M., Snyder P. M. (2009) Extracellular chloride regulates the epithelial sodium channel. J. Biol. Chem. 284, 29320–29325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Knight K. K., Wentzlaff D. M., Snyder P. M. (2008) Intracellular sodium regulates proteolytic activation of the epithelial sodium channel. J. Biol. Chem. 283, 27477–27482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Collier D. M., Snyder P. M. (2009) Extracellular protons regulate human ENaC by modulating Na+ self-inhibition. J. Biol. Chem. 284, 792–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adebamiro A., Cheng Y., Johnson J. P., Bridges R. J. (2005) Endogenous protease activation of ENaC. Effect of serine protease inhibition on ENaC single channel properties. J. Gen. Physiol 126, 339–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carattino M. D., Hughey R. P., Kleyman T. R. (2008) Proteolytic processing of the epithelial sodium channel γ subunit has a dominant role in channel activation. J. Biol. Chem. 283, 25290–25295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chraïbi A., Vallet V., Firsov D., Hess S. K., Horisberger J. D. (1998) Protease modulation of the activity of the epithelial sodium channel expressed in Xenopus oocytes. J. Gen. Physiol. 111, 127–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Diakov A., Bera K., Mokrushina M., Krueger B., Korbmacher C. (2008) Cleavage in the γ-subunit of the epithelial sodium channel (ENaC) plays an important role in the proteolytic activation of near-silent channels. J. Physiol. 586, 4587–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. García-Caballero A., Dang Y., He H., Stutts M. J. (2008) ENaC proteolytic regulation by channel-activating protease 2. J. Gen. Physiol. 132, 521–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harris M., Garcia-Caballero A., Stutts M. J., Firsov D., Rossier B. C. (2008) Preferential assembly of epithelial sodium channel (ENaC) subunits in Xenopus oocytes. Role of furin-mediated endogenous proteolysis. J. Biol. Chem. 283, 7455–7463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hughey R. P., Bruns J. B., Kinlough C. L., Harkleroad K. L., Tong Q., Carattino M. D., Johnson J. P., Stockand J. D., Kleyman T. R. (2004) Epithelial sodium channels are activated by furin-dependent proteolysis. J. Biol. Chem. 279, 18111–18114 [DOI] [PubMed] [Google Scholar]
- 24. Hughey R. P., Carattino M. D., Kleyman T. R. (2007) Role of proteolysis in the activation of epithelial sodium channels. Curr. Opin. Nephrol. Hypertens. 16, 444–450 [DOI] [PubMed] [Google Scholar]
- 25. Korbmacher C. (2009) Proteolytic activation of the epithelial sodium channel (ENaC) in health and disease. J. Med. Invest 56, 306–307 [DOI] [PubMed] [Google Scholar]
- 26. Vuagniaux G., Vallet V., Jaeger N. F., Pfister C., Bens M., Farman N., Courtois-Coutry N., Vandewalle A., Rossier B. C., Hummler E. (2000) Activation of the amiloride-sensitive epithelial sodium channel by the serine protease mCAP1 expressed in a mouse cortical collecting duct cell line. J. Am. Soc. Nephrol. 11, 828–834 [DOI] [PubMed] [Google Scholar]
- 27. Ha S. D., Martins A., Khazaie K., Han J., Chan B. M., Kim S. O. (2008) Cathepsin B is involved in the trafficking of TNF-α-containing vesicles to the plasma membrane in macrophages. J. Immunol. 181, 690–697 [DOI] [PubMed] [Google Scholar]
- 28. Planès C., Randrianarison N. H., Charles R. P., Frateschi S., Cluzeaud F., Vuagniaux G., Soler P., Clerici C., Rossier B. C., Hummler E. (2010) ENaC-mediated alveolar fluid clearance and lung fluid balance depend on the channel-activating protease 1. EMBO Mol. Med. 2, 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vallet V., Pfister C., Loffing J., Rossier B. C. (2002) Cell-surface expression of the channel activating protease xCAP-1 is required for activation of ENaC in the Xenopus oocyte. J. Am. Soc. Nephrol. 13, 588–594 [DOI] [PubMed] [Google Scholar]
- 30. Nesterov V., Dahlmann A., Bertog M., Korbmacher C. (2008) Trypsin can activate the epithelial sodium channel (ENaC) in microdissected mouse distal nephron. Am. J. Physiol. Renal Physiol. 295, F1052–F1062 [DOI] [PubMed] [Google Scholar]
- 31. Wei Y., Lin D. H., Kemp R., Yaddanapudi G. S., Nasjletti A., Falck J. R., Wang W. H. (2004) Arachidonic acid inhibits epithelial Na+ channel via cytochrome P450 (CYP) epoxygenase-dependent metabolic pathways. J. Gen. Physiol. 124, 719–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bruns J. B., Carattino M. D., Sheng S., Maarouf A. B., Weisz O. A., Pilewski J. M., Hughey R. P., Kleyman T. R. (2007) Epithelial Na+ channels are fully activated by furin- and prostasin-dependent release of an inhibitory peptide from the γ-subunit. J. Biol. Chem. 282, 6153–6160 [DOI] [PubMed] [Google Scholar]
- 33. Wakida N., Kitamura K., Tuyen D. G., Maekawa A., Miyoshi T., Adachi M., Shiraishi N., Ko T., Ha V., Nonoguchi H., Tomita K. (2006) Inhibition of prostasin-induced ENaC activities by PN-1 and regulation of PN-1 expression by TGF-β1 and aldosterone. Kidney Int. 70, 1432–1438 [DOI] [PubMed] [Google Scholar]
- 34. Adachi M., Kitamura K., Miyoshi T., Narikiyo T., Iwashita K., Shiraishi N., Nonoguchi H., Tomita K. (2001) Activation of epithelial sodium channels by prostasin in Xenopus oocytes. J. Am. Soc. Nephrol. 12, 1114–1121 [DOI] [PubMed] [Google Scholar]
- 35. Mort J. S., Buttle D. J. (1997) Cathepsin B. Int. J. Biochem. Cell Biol. 29, 715–720 [DOI] [PubMed] [Google Scholar]
- 36. Podgorski I., Sloane B. F. (2003) Cathepsin B and its role(s) in cancer progression. Biochem. Soc. Symp. 70, 263–276 [DOI] [PubMed] [Google Scholar]
- 37. Eykelbosh A. J., Van Der Kraak G. (2010) A role for the lysosomal protease cathepsin B in zebrafish follicular apoptosis. Comp Biochem. Physiol A Mol. Integr. Physiol. 156, 218–223 [DOI] [PubMed] [Google Scholar]
- 38. Foghsgaard L., Wissing D., Mauch D., Lademann U., Bastholm L., Boes M., Elling F., Leist M., Jäättelä M. (2001) Cathepsin B acts as a dominant execution protease in tumor cell apoptosis induced by tumor necrosis factor. J. Cell Biol. 153, 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guenette R. S., Mooibroek M., Wong K., Wong P., Tenniswood M. (1994) Cathepsin B, a cysteine protease implicated in metastatic progression, is also expressed during regression of the rat prostate and mammary glands. Eur. J. Biochem. 226, 311–321 [DOI] [PubMed] [Google Scholar]
- 40. Jane D. T., Morvay L., Dasilva L., Cavallo-Medved D., Sloane B. F., Dufresne M. J. (2006) Cathepsin B localizes to plasma membrane caveolae of differentiating myoblasts and is secreted in an active form at physiological pH. Biol. Chem. 387, 223–234 [DOI] [PubMed] [Google Scholar]
- 41. Martel-Pelletier J., Cloutier J. M., Pelletier J. P. (1990) Cathepsin B and cysteine protease inhibitors in human osteoarthritis. J. Orthop. Res. 8, 336–344 [DOI] [PubMed] [Google Scholar]
- 42. Alli A. A., Gower W. R., Jr. (2010) Molecular approaches to examine the phosphorylation state of the C type natriuretic peptide receptor. J. Cell. Biochem. 110, 985–994 [DOI] [PubMed] [Google Scholar]
- 43. Alli A. A., Gower W. R., Jr. (2009) The C type natriuretic peptide receptor tethers AHNAK1 at the plasma membrane to potentiate arachidonic acid-induced calcium mobilization. Am. J. Physiol. Cell Physiol. 297, C1157–C1167 [DOI] [PubMed] [Google Scholar]
- 44. Alli A. A., Bao H. F., Alli A. A., Aldrugh Y., Song J. Z., Ma H. P., Yu L., Al-Khalili O., Eaton D. C. (2012) Phosphatidylinositol phosphate-dependent regulation of Xenopus ENaC by MARCKS protein. Am. J. Physiol. Renal Physiol, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lohmüller T., Wenzler D., Hagemann S., Kiess W., Peters C., Dandekar T., Reinheckel T. (2003) Toward computer-based cleavage site prediction of cysteine endopeptidases. Biol. Chem. 384, 899–909 [DOI] [PubMed] [Google Scholar]
- 46. Foghsgaard L., Lademann U., Wissing D., Poulsen B., Jaattela M. (2002) Cathepsin B mediates tumor necrosis factor-induced arachidonic acid release in tumor cells. J. Biol. Chem. 277, 39499–39506 [DOI] [PubMed] [Google Scholar]
- 47. Mort J. S., Recklies A. D., Poole A. R. (1985) Release of cathepsin B precursors from human and murine tumors. Prog. Clin. Biol. Res. 180, 243–245 [PubMed] [Google Scholar]
- 48. Joy B., Sivadasan R., Abraham T. E., John M., Sobhan P. K., Seervi M., T. R. S. (2010) Lysosomal destabilization and cathepsin B contributes for cytochrome c release and caspase activation in embelin-induced apoptosis. Mol. Carcinog. 49, 324–336 [DOI] [PubMed] [Google Scholar]
- 49. Guicciardi M. E., Deussing J., Miyoshi H., Bronk S. F., Svingen P. A., Peters C., Kaufmann S. H., Gores G. J. (2000) Cathepsin B contributes to TNF-α-mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J. Clin. Invest. 106, 1127–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]