Background: ERα and p53 are transcription factors that play important roles in breast cancer.
Results: ERα transcriptionally regulates p53, which then modulates DNA damage-induced growth suppression.
Conclusion: p53 is a target of ERα and is responsible for the sensitivity of ERα-positive breast cancer cells to DNA damage.
Significance: Loss of ERα, causing a decrease in p53 expression, could lead to tumors resistant to both antiestrogen and chemotherapy.
Keywords: Breast Cancer, DNA Damage Response, Estrogen Receptor, p53, Transcription Regulation, p21
Abstract
In response to genotoxic stress, the p53 tumor suppressor induces target genes for cell cycle arrest, apoptosis, and DNA repair. Although p53 is the most commonly mutated gene in all human cancers, it is only mutated in about 20% of breast cancers. 70% of all breast cancer cases are estrogen receptor (ER)-positive and express ERα. ER-positive breast cancer generally indicates good patient prognosis and treatment responsiveness with antiestrogens, such as tamoxifen. However, ER-positive breast cancer patients can experience loss or a reduction in ERα, which is associated with aggressive tumor growth, increased invasiveness, poor prognosis, and loss of p53 function. Consistent with this, we found that p53 is a target gene of ERα. Specifically, we found that knockdown of ERα decreases expression of p53 and its downstream targets, MDM2 and p21. In addition, we found that ERα activates p53 transcription via binding to estrogen response element half-sites within the p53 promoter. Moreover, we found that loss of ERα desensitizes, whereas ectopic expression of ERα sensitizes, breast cancer cells to DNA damage-induced growth suppression in a p53-dependent manner. Altogether, this study provides an insight into a feedback loop between ERα and p53 and a biological role of p53 in the DNA damage response in ER-positive breast cancers.
Introduction
Estrogen receptors α (ERα)3 and β (ERβ) are members of the nuclear hormone receptor family and act as transcription factors when bound and activated by their ligand. In breast cells, ERα and ERβ are expressed and share 97% identity in their DNA-binding domains and 55% identity in their ligand-binding domains (1). Once activated by 17 β-estradiol (estrogen), ERα and ERβ form homo- and/or heterodimers that regulate expression of shared and unique target genes (2). An estimated 70% of breast cancer cases express estrogen receptors and thus are classified as ER-positive (3). However, ERα and ERβ have been shown to exhibit opposing effects in breast cancers. ERα expression is high in ER-positive breast cancers and is associated with tumor growth (4). On the other hand, ERβ is expressed at low levels in breast tissue and may play an inhibitory role in tumorigenesis (5). ER-positive breast cancer generally indicates good patient prognosis and treatment responsiveness with antiestrogens, such as tamoxifen. However, 30–50% of recurrent tumors are resistant to hormone therapy due to loss of ERα expression (6), which can occur by abnormal methylation of the ERα promoter, pathway inactivation, or spontaneous loss of ERα expression (7). Importantly, an ER-negative tumor status is associated with aggressive tumor growth, increased invasiveness, poor patient prognosis, and loss of p53 function (8).
The tumor suppressor p53 is activated by genotoxic stress to induce target genes for cell cycle arrest, DNA repair, and apoptosis (9). p21, a cyclin-dependent kinase inhibitor, is a major p53 target that blocks cell cycle progression at the G1/S transition to allow DNA repair (10). If damages are unrepairable, p53 induces several apoptotic target genes, such as PUMA, Bax, and Noxa, leading to programmed cell death (9). Under normal cellular conditions, p53 is kept inactive by its target, murine double minute 2 (MDM2), a RING finger E3 ubiquitin ligase (11). Indicative of its importance in genome stability, p53 is inactivated in more than 50% of all human cancers (12). Surprisingly, only 20% of breast cancers contain mutated p53, which suggests that other mechanisms are involved in inactivating p53 function (13).
Evidence has shown that the expression of ERα is correlated with the status of p53 (14, 15). It has been shown that knockdown of p53 decreases, whereas overexpression of p53 increases, ERα expression in ER-positive MCF7 breast cancer cells (14). Consistently, DNA damage increases ERα expression in mammary tumors in a p53-dependent manner (15). Indeed, it has been shown that p53 regulates ERα transcription by recruiting several transcription factors including cAMP-response element-binding protein (CREB)-binding protein (CBP) and Sp1 to the ERα promoter (15). However, physical interaction between p53 and ERα interferes with each other's activities to regulate gene expression (16, 17). Interestingly, it has been reported that upon treatment with estrogen, p53 expression is enhanced (18), suggesting that ERα may regulate p53 expression. In this study, we found that knockdown of ERα decreases, whereas ectopic expression of ERα increases, p53 transcription. In addition, we showed that ERα binds to and activates the p53 promoter via two ERE half-sites. Moreover, the p53 promoter is activated by estrogen. Finally, we showed that knockdown of ERα attenuates, whereas overexpression of ERα enhances, DNA damage-induced growth suppression in a p53-dependent manner. Taken together, our data suggest that p53 is a direct transcriptional target of ERα and modulates DNA damage-induced growth suppression in ERα-positive breast cancer cells.
EXPERIMENTAL PROCEDURES
Plasmids
To generate HA-tagged wild-type ERα in pCMV expression vector, an ERα cDNA fragment was amplified from MCF7 cDNA with forward primer 5′-GGACCACCATGTACCCATACGATGTTCCAGATTACGCTACCATGACCCTCCACACCAAAGCATC-3′ and reverse primer 5′-GAAGATCTCCACCATGCCCTCTAC-3′. Similarly, HA-tagged wild-type ERβ in pCMV was generated using forward primer 5′-GGACCACCATGTACCCATACGATGTTCCAGATTACGCTGATATAAAAAACTCACCATC-3′ and reverse primer 5′-CTCGAGTCACTGAGACTGTGGGTTCTGGG-3′. To generate untagged wild-type ERα in pcDNA4 for tetracycline-inducible expression (Invitrogen), the cDNA fragment was amplified from an ERα cDNA clone (EST clone no. 40128594; Open Biosystems) with forward primer 5′-AGGAATTCACCATGGAGCGGATCCCCAGCG-3′ and reverse primer 5′-AGTCTAGAAGGAAGGAAAGCAAAGCAG-3′. To generate a construct for the inducible expression of ERα shRNA, two oligonucleotides, 5′-GATCCCCAGTTTGTGTGCCTCAAATCTTCAAGAGAGATTTGAGGCACACAAACTTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAAGTTTGTGTGCCTCAAATCTCTCTTGAAGATTTGAGGCACACAAACTGGG-3′, were designed to target ERα exon 6 (in boldface). The oligonucleotides were annealed and cloned into pBabe-H1 as described previously (19). The resulting vector was designated pBabe-H1-siERα. The pBabe-U6-sip53 construct expressing p53 shRNA was described previously (20). To generate pGL2 luciferase reporters under control of the p53 promoter (nucleotides (nt) −1998 to +73 designated p53-P-2kb and nt −593 to +73 designated p53-P-593), genomic DNA fragments were amplified from MCF7 cells with forward primer 5′-ATGGGTACCAAGTGTAGGGCTAGGGCTG-3′ or 5′-TTGGTACCGCTTCAGACCTGTCTCCCTCATTC-3′ and reverse primer 5′-ACTCTCGAGTGGCTCTAGACTTTTGAGAAGCTC-3′. p53 promoter internal deletion mutants were generated by a PstI and PvuII (New England Biolabs) restriction enzyme digest and religation according to the manufacturer's instructions and designated p53-P-PstI and p53-P-PvuII, respectively. To generate individual wild-type or mutant estrogen response element (ERE) half-sites cloned upstream of the minimum c-fos promoter in the luciferase reporter OFLuc reporter vector (21), genomic DNA fragments were amplified from MCF7 cells with the following primer sets: −1828, forward primer 5′-GGGGAAGCTTTGAAAATCTCGGGGGTGGTCAG-3′ and reverse primer 5′-GGGGAGATCTTCGATTTCTCAGTGGTTCCTGGTCAG-3′; −1828M, forward primer 5′-GGGGAAGCTTTGAAAATCTCGGGGGTGTACAG-3′ and reverse primer 5′-GGGGAGATCTTCGATTTCTCAGTGGTTCCTGTACAG; −1611, forward primer 5′-GGGGAAGCTTAGGCCTGGAGAAGTGGGTCT-3′ and reverse primer 5′-GGGGAGATCTTAAGTGGTGATGGCAG-3′; −1611M, forward primer 5′-GGGAAGCTTAGGCCTGGAGAAGTGGTACTCAGGATT-3′ and reverse primer 5′-GGGGAGATCTTAGCTCCGGACTGCTGTACTTCAGTAC-3′; −1248, forward primer 5′-GGGGAAGCTTAGCCACAGGATCTGGGGACA-3′ and reverse primer 5′-GGGGAGATCTCACGCTTCCCCGATGA-3′; and −1224, forward primer 5′-GCGGAAGCTTCAGTTCAGAGTCC-3′ and reverse primer 5′-GGGCAGATCTTAGCTCCGGACTGCTG-3′. Added restriction enzyme sites are shown in italic. Wild-type and mutant ERE sites are shown in boldface.
Cell Lines
MCF7 and ZR-75-1 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37 °C with 5% CO2. MCF7-TR-7, which expresses the tetracycline repressor, was generated in our laboratory (22). To generate cell lines that inducibly express wild-type ERα, MCF7-TR-7 cells were transfected with pcDNA4-ERα using Lipofectamine 2000 (Invitrogen) and selected with medium containing 200 μg/ml Zeocin. To generate cell lines in which ERα and/or p53 are inducibly knocked down, MCF7-TR-7 cells were transfected with pBabe-H1-siERα and/or pBabe-U6-sip53 and selected with 0.5 μg/ml puromycin. To generate cell lines that inducibly express wild-type ERα and in which p53 is knocked down, MCF7-TR-7 cells were transfected with pcDNA4-ERα and pBabe-U6-sip53 and selected with 200 μg/ml Zeocin and 0.5 μg/ml puromycin. The resulting cell lines were designated MCF7-ERα, MCF7-ERα-KD, MCF7(p53-KD)-ERα-KD, and MCF7(p53-KD)-ERα, respectively.
Luciferase Reporter Assay
The Dual Luciferase assay was performed in triplicate using MCF7 cells according to the manufacturer's instructions (Promega). Cells were mock-treated or treated with 17β-estradiol (Sigma) or ICI 182, 780 (Sigma) for 24 h prior to transfection. The fold change in relative luciferase activity was determined by the luciferase activity induced by ERα or ERβ divided by luciferase activity induced by an empty pcDNA3 vector.
Chromatin Immunoprecipitation (ChIP) Assay
A ChIP assay was performed as described previously (19). ERα protein binding to the p53 promoter at nt −1406 to −1111 (296-bp fragment) was detected with the forward primer 5′-TCAGAAAGTTCTTGCTCCTCG-3′ and the reverse primer 5′-CTTTGGAGACTCAACCGTTAGC-3′. The p53 promoter at nt −1741 to −1490 (252-bp fragment) was detected with forward primer 5′-CTGAACTCTGACCAGGAACCAC-3′ and reverse primer 5′-GGAAGATACCTCTGGGGAACC-3′. As a positive control, binding of ERα protein to the ERE within the pS2 promoter at nt −592 to −194 (399-bp fragment) was detected with the forward primer 5′-TCTATCAGCAAATCCTTCC-3′ and the reverse primer 5′-GTTGGGATTACAGCGTGAG-3′. Primers for the amplification of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) promoter were used as described previously (23).
Colony Formation Assay
Cells were seeded at 1000/well in 6-well plates with or without doxycycline in triplicate. 72 h postinduction, cells were mock-treated or treated with camptothecin (CPT) (250 nm) for 6 h or doxorubicin (Dox) (100 nm) for 2 h and maintained for 15 days. Colonies were fixed with a 7:1 mixture of methanol:glacial acetic acid, washed in H2O, and stained with 0.02% crystal violet.
Western Blot Analysis
Whole cell extracts were prepared with 1× SDS sample buffer and boiled for 5 min at 95 °C. Antibodies against ERα, p53, p21, MDM2, PUMA, PolH, and GAPDH were purchased from Santa Cruz Biotechnology. Antibody against MIC-1 was purchased from Upstate. Anti-actin was purchased from Sigma.
Reverse Transcription-PCR (RT-PCR)
A reverse transcription assay was performed as previously described (24). Transcripts were detected using the following primers: ERα (439-bp fragment), forward primer 5′-GGAGACATGAGAGCTGCCAAC-3′ and reverse primer 5′-CCAGCAGCATGTCGAAGATC-3′; p53 (309-bp fragment), forward primer 5′-GACCGGCGCACAGAGGAAGAGAATC-3′ and reverse primer 5′-GAGTTTTTTATGGCGGGAGGTAGAC-3′; and pS2 (209-bp fragment), forward primer 5′-TTGTGGTTTTCCTGGTGTC-3′ and reverse primer 5′-CCGAGCTCTGGGACTAATCA-3′. Primers for actin (225-bp fragment) were described previously (24).
Real Time PCR Analysis of p53 mRNA
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Reverse transcription of RNA was performed using Moloney murine leukemia virus reverse transcriptase (Promega) according to the manufacturer's instructions. p53 primers used for SYBR Green RT-quantitative PCR were 5′-GTTCCGAGAGCTGAATGAGG-3′ and 5′-TCTGAGTCAGGCCCTTCTGT-3′. Control primers for GAPDH were 5′-GAGTCAACGGATTTGGTCGT-3′ and 5′-GACAAGCTTCCCGTTCTCAG-3′. RT-quantitative PCR and relative quantification of mRNA were performed as described previously (25).
siRNA
To transiently knock down ERα and/or p53, cells were transfected with ERα siRNA (5′-GGAUUUGACCCUCCAUGAU-3′; Dharmacon) and/or p53 siRNA (5′-GGAAAUUUGCGUGUGGAGU-3′; Qiagen) using siLentFect (Bio-Rad) according to the manufacturers' instructions. A non-targeting scrambled siRNA (Dharmacon) was used as a control.
RESULTS
p53 Expression Is Regulated by ERα
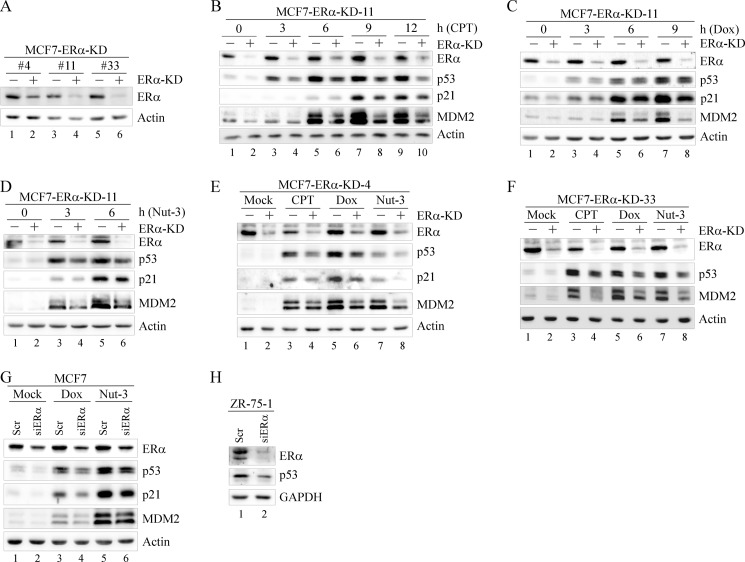
ER-positive breast cancer cells generally contain wild-type p53 and rely on estrogen for proliferation (26). Interestingly, high levels of estrogen are associated with increased p53 expression in breast cancer cells under a stress condition (27). Because ERα, the main estrogen receptor in breast tumors, is activated by estrogen to induce target gene expression (28), we determined whether ERα affects p53 expression. To test this, we generated multiple MCF7 cell lines in which ERα can be inducibly knocked down. The MCF7 cell line is known to express ERα and wild-type p53. As shown in Fig. 1A, ERα was efficiently knocked down in clones 4, 11, and 33. We found that the level of p53 protein was markedly decreased upon knockdown of ERα in clone 11 (Fig. 1B, p53 panel, compare lane 1 with lane 2). In addition, ERα knockdown (KD) inhibited stabilization of p53 induced by treatment with CPT (Fig. 1B, compare lanes 3, 5, 7, and 9 with lanes 4, 6, 8, and 10, respectively), Dox (Fig. 1C, compare lanes 3, 5, and 7 with lanes 4, 6, and 8, respectively), and Nutlin-3 (Nut-3) (Fig. 1D, compare lanes 3 and 5 with lanes 4 and 6, respectively). CPT and Dox are topoisomerase I and II inhibitors, respectively, and Nut-3 is an MDM2 antagonist; all three stabilize and activate p53 (29, 30). Moreover, we found that attenuation of p53 stabilization by ERα-KD led to decreased induction of the p53 targets p21 and MDM2 (Fig. 1, B–D). To confirm this, the same experiments were performed in clones 4 and 33, and similar results were observed (Fig. 1, E and F). To rule out potential off-target effects, ERα was transiently knocked down by another siRNA, which is different from that used for cell line generation. Consistently, we found that transient knockdown of ERα resulted in a decrease of p53, MDM2, and p21 regardless of treatment with Dox or Nut-3 (Fig. 1G, compare lanes 1, 3, and 5 with lanes 2, 4, and 6, respectively). Moreover, to rule out a potential cell type-specific effect, ERα was transiently knocked down in ZR-75-1, an ER-positive breast cancer cell line containing wild-type p53 (31). Again, we showed that knockdown of ERα led to a decreased level of p53 protein (Fig. 1H).
FIGURE 1.
Knockdown of ERα inhibits p53 expression. A, generation of MCF7 cell lines in which ERα is inducibly knocked down. Western blots were prepared with extracts from MCF7 cells uninduced (−) or induced (+) to express ERα shRNA for 72 h. B–D, knockdown of ERα decreased the expression of p53. Western blots were prepared with MCF7-ERα-KD-11 cells that were uninduced (−) or induced (+) to knock down ERα followed by mock treatment or treatment with CPT (250 nm) for 0, 3, 6, 9, or 12 h (B); treatment with Dox (400 nm) for 0, 3, 6, or 9 h (C); and treatment with Nut-3 (7 μm) for 0, 3, or 6 h (D). E and F, Western blots were prepared with MCF7-ERα-KD-4 (E) or MCF7-ERα-KD-33 (F) cells that were uninduced (−) or induced (+) to knock down ERα followed by mock treatment or treatment with CPT (250 nm), Dox (400 nm) for 9 h, and Nut-3 (7 μm) for 6 h. G, Western blots were prepared with MCF7 cells that were transiently transfected with scrambled (Scr) or ERα siRNA (siERα) for 72 h and then mock-treated or treated with Dox (400 nm) or Nut-3 (7 μm) for 6 h. H, Western blots were prepared with ZR-75-1 cells that were transiently transfected with scrambled or ERα siRNA for 72 h. ERα, p53, MDM2, p21, GAPDH, and actin were detected by their respective antibodies.
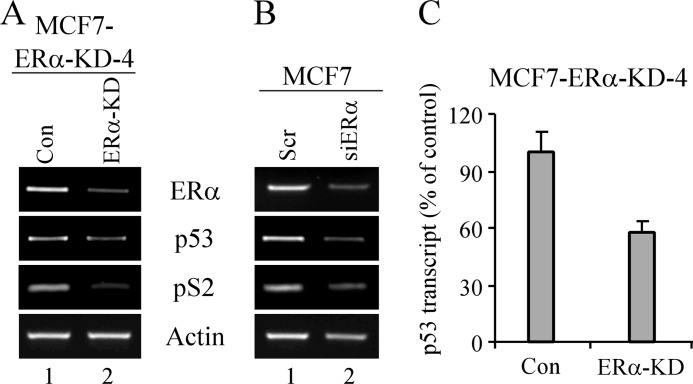
Next, we examined whether the decrease in p53 protein was due to a decrease in p53 transcript. As a positive control, we tested pS2, which is known to be transcriptionally regulated by the estrogen receptor (32). We found that knockdown of ERα resulted in a decrease of ERα, pS2, and p53 transcripts in clone 4 (Fig. 2A). Similarly, transient knockdown of ERα by another siRNA also resulted in a decrease of ERα, pS2, and p53 transcripts (Fig. 2B). In addition, quantitative real time RT-PCR was performed and confirmed that knockdown of ERα decreased p53 transcript in clone 4 (Fig. 2C). Together, these data suggest that p53 is transcriptionally regulated by ERα.
FIGURE 2.
Knockdown of ERα reduces p53 transcription. A, the levels of transcripts for ERα, p53, pS2, and actin were measured by RT-PCR with total RNA purified from MCF7 cells uninduced (−) or induced (+) to express ERα shRNA for 72 h. B, RT-PCR was performed with total RNA from MCF7 cells transiently transfected with scrambled (Scr) or ERα siRNA (siERα) for 72 h. C, the level of p53 transcripts was analyzed by quantitative real time RT-PCR with cDNAs from A. Results were normalized to GAPDH (error bars represent S.D.; n = 3). Con, control.
ERα Binds to ERE Half-sites on the p53 Promoter to Induce p53 Expression
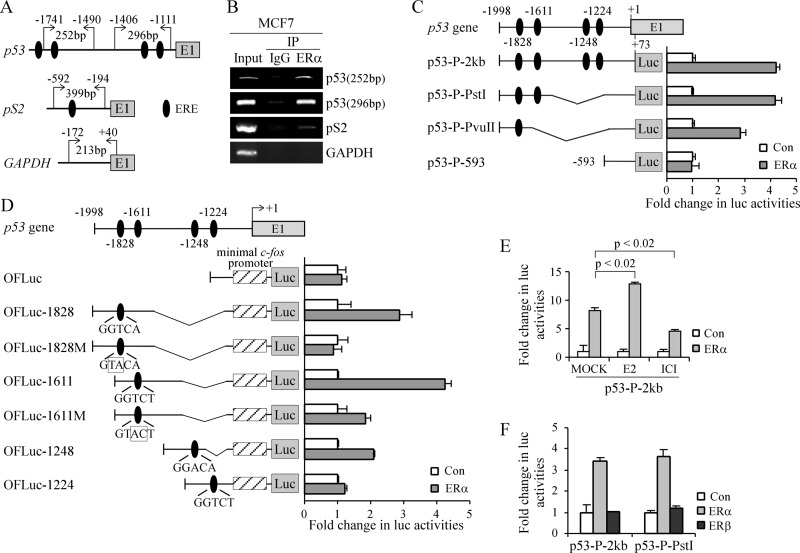
ERα, a nuclear hormone receptor, regulates gene expression by binding to consensus and non-consensus EREs on target gene promoters. A consensus ERE is composed of two palindromic half-sites separated by 3 nt, 5′-GGTCANNNTGACC-3′ (where N represents any nucleotide) (33). However, ERα is also able to bind to and activate target gene promoters containing imperfect or truncated ERE sites (34). In addition, ERα can activate gene expression by binding to enhancer sites located at a distance of >100 kb from the transcriptional start site of estrogen-regulated target genes (35). Thus, we analyzed the genome-wide ChIP-ENCODE database, which contains a comprehensive library of transcription factor interactions on the human genome (36). Probing of this data set showed an ERα interaction on the p53 proximal promoter region. A closer look at the p53 promoter sequence revealed four potential ERE half-sites (Fig. 3A, p53 panel). To determine whether ERα binds to the p53 promoter in vivo, ChIP assay was performed using chromatin collected from MCF7 cells. The binding of ERα to the pS2 gene, a well defined target of ERα, served as a positive control (37). The binding of ERα to the GAPDH promoter was measured as a nonspecific binding control. We showed that ERα bound to the p53 and pS2 promoters but not the GAPDH promoter (Fig. 3B).
FIGURE 3.
p53 is a transcriptional target of ERα. A, schematic presentation of p53, pS2, and GAPDH promoters with the location of potential EREs and primers used for ChIP assays. B, ERα binds to the p53 promoter in vivo. MCF7 chromatin was immunoprecipitated (IP) with anti-ERα or a control IgG. EREs on the p53 and pS2 promoters were amplified by PCR. C, left panel, schematic representation of luciferase reporter constructs. Right panel, luciferase (Luc) activity measured in the presence or absence of ERα. D, left panel, schematic representation of OFLuc luciferase reporter constructs. Right panel, luciferase activity measured in the presence or absence of ERα. E, luciferase activity measured in the presence or absence of ERα along with mock treatment or treatment with estrogen (E2) or ICI 182,780 (ICI). F, luciferase activity measured in the presence or absence of ERα or ERβ. Error bars represent S.D.; n = 3. Con, control.
Next, to determine which of the four potential ERE half-sites is responsive to ERα, a luciferase reporter under the control of the p53 promoter (nt −1998 to +73), which contains all four ERE half-sites (at nt −1224, −1248, −1611, and −1828), was constructed and designated p53-P-2kb (Fig. 3C, left panel). In addition, luciferase reporters containing one, two, or none of the ERE half-sites were constructed and designated p53-P-PvuII, p53-P-PstI, and p53-P-593, respectively (Fig. 3C, left panel). We found that ERα induced a 4-fold increase in luciferase activity for p53-P-2kb and p53-P-PstI and a 2.7-fold increase for p53-P-PvuII but no increase for p53-P-593 (Fig. 3C, right panel). These results suggest that the ERE half-sites at nt −1611 and −1828 are important for ERα activation of p53 transcription. To confirm this, the four potential ERE half-sites were individually cloned into the OFLuc reporter vector (21), which contains a minimal c-fos promoter, and the resulting constructs were designated OFLuc−1828, OFLuc−1611, OFLuc−1248, and OFLuc−1224 (Fig. 3D, left panel). We showed that upon expression of ERα luciferase activity was increased 4-fold for OFLuc−1611, 3-fold for OFLuc−1828, 2-fold for OFLuc−1248, and less than 50% for OFLuc−1224 (Fig. 3D, right panel). To further test the ERE half-sites at nt −1828 and −1611, the ERE consensus sequence for each half-site was mutated, and the resulting reporters carrying a mutant ERE were designated as OFLuc−1828M and OFLuc−1611M, respectively (Fig. 3D, left panel). We showed that the luciferase activity was not increased by ERα for OFLuc−1828M, and there was little if any increase for OFLuc−1611M (Fig. 3D, right panel). Taken together, these results indicate that the ERE half-sites located at nt −1611 and −1828 on the p53 promoter are primarily responsible for ERα activation of p53 transcription.
Estrogen, an ERα ligand, induces a conformational change of ERα and then promotes ERα dimerization and binding to ERE sites (38). To test whether the p53 promoter is estrogen-responsive, MCF7 cells were pretreated with estrogen or the antiestrogen ICI 182,780 (Fulvestrant). We showed that estrogen enhanced, but ICI 182,780 suppressed, the ability of ERα to increase the luciferase activity under the control of the p53 promoter (Fig. 3E).
Although ERα and ERβ recognize the same EREs on target gene promoters, they are capable of regulating both common and distinct sets of target genes (34, 39). Thus, we examined whether ERβ regulates p53 transcription. Surprisingly, we found that, unlike ERα, ERβ had no effect on luciferase activity for p53-P-2kb (Fig. 3F). Altogether, we concluded that ERα regulates p53 transcription through multiple ERE half-sites on the p53 promoter.
Knockdown of ERα Desensitizes Cells to DNA Damage-induced Growth Suppression in a p53-dependent Manner
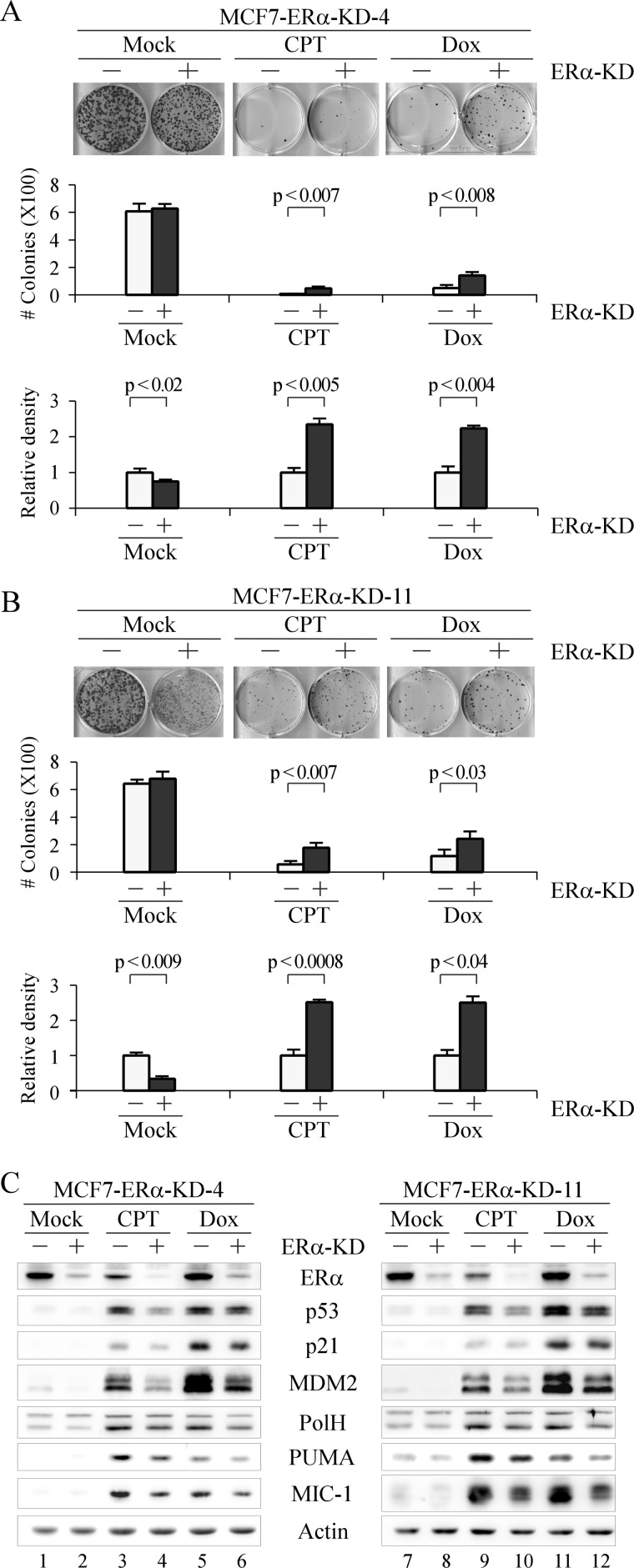
It is well established that, when activated by DNA damage, p53 induces target genes for cell cycle arrest and apoptosis (9). In contrast, it is well known that ERα induces target genes that promote cell growth (40). To determine the biological function of p53 expression induced by ERα, a colony formation assay was performed to examine the effect of ERα-KD on cell proliferation in MCF7 cells in which ERα can be inducibly knocked down. We found that knockdown of ERα decreased the size of colonies formed by MCF7-ERα-KD-4 cells (Fig. 4A, top panel, mock treatment column). The overall number of colonies and cell density were quantified to measure the level of cell proliferation. We found that ERα-KD did not change the number of colonies but inhibited cell proliferation (Fig. 4A, compare middle panel with bottom panel, mock treatment column). In addition, we found that when MCF7 cells were treated with CPT and Dox, the number of colonies and overall cell density were markedly decreased (Fig. 4A, top panel, compare CPT and Dox columns with mock treatment column). However, the number of colonies and overall cell density formed by ERα-KD cells were significantly increased compared with that formed by control cells upon treatment with CPT or Dox (Fig. 4A, compare middle panel with bottom panel, CPT and Dox columns). Similar results were obtained with MCF7-ERα-KD-11 cells (Fig. 4B).
FIGURE 4.
Knockdown of ERα decreases cell sensitivity to DNA damage-inducing growth suppression. A and B, top panel, a colony formation assay was performed in triplicate with MCF7-ERα-KD-4 (A) or -11 (B) cells uninduced (−) or induced (+) to knock down ERα for 72 h followed by mock treatment or treatment with CPT (250 nm) for 6 h or Dox (100 nm) for 2 h and then maintained for 15 days. Middle panel, the number of colonies was counted using the UVP VisionWorksLS software (error bars represent S.D.; n = 3). Bottom panel, all stained cells in a well were scanned using the UVP VisionWorksLS software to determine total cell density. The density of MCF7 cells without ERα-KD was arbitrarily set at 1.0 regardless of mock treatment or treatment with CPT and Dox. The -fold change in cell density by ERα-KD was calculated in triplicate (error bars represent S.D.; n = 3). C, Western blots were prepared with extracts from MCF7-ERα-KD-4 (left panel) or -11 (right panel) cells were uninduced (−) or induced (+) to knock down ERα followed by mock treatment or treatment with CPT (250 nm) or Dox (400 nm) for 9 h. ERα, p53, p21, MDM2, PolH, PUMA, MIC-1, and actin were detected by their respective antibodies.
To further determine the cellular response to DNA damage when ERα is knocked down, we analyzed several p53 target genes in cell cycle arrest and cell death including p21, MDM2, PolH, PUMA, and MIC-1 (41–45). We found that knockdown of ERα led to a decrease in the protein levels of p21, MDM2, PolH, PUMA, and MIC-1 in MCF7-ERα-KD-4 and -11 cells treated with CPT and Dox (Fig. 4C, compare lanes 3, 5, 9, and 11 with lanes 4, 6, 10, and 12, respectively).
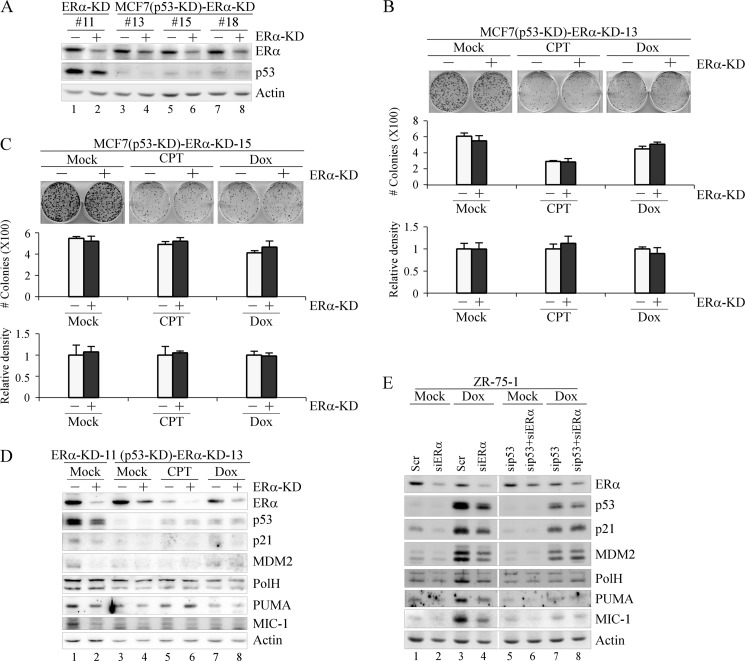
To determine whether the decreased sensitivity of ERα-KD cells to DNA damage is due to p53, we generated multiple MCF7 cell lines, designated MCF7(p53-KD)-ERα-KD, in which p53 was stably knocked down and ERα can be inducibly knocked down. As shown in Fig. 5A, both ERα and p53 were knocked down in clones 13, 15, and 18. The levels of ERα and p53 were also measured in ERα-KD clone 11 (Fig. 5A), which was used as a control. Next, a colony formation assay was performed with MCF7(p53-KD)-ERα-KD cell lines (clones 13 and 15). We found that upon knockdown of p53, ERα-KD had no effect on the size and number of colonies regardless of DNA damage (Fig. 5, B and C). In addition, we showed that unlike in p53-proficient cells (Fig. 4C), the expression of p53 target genes was not affected by ERα-KD in p53-deficient cells treated with CPT and Dox (Fig. 5D). Similarly, we showed that expression of p53 along with its targets, p21, MDM2, PolH, PUMA, and MIC-1, was decreased by ERα-KD (Fig. 5E, compare lane 3 with lane 4), which was diminished if not abrogated by p53-KD in ZR-75-1 cells treated with Dox (Fig. 5E, compare lane 7 with lane 8). Altogether, we conclude that knockdown of ERα desensitizes MCF7 cells to DNA damage-induced growth suppression in a p53-dependent manner.
FIGURE 5.
The effect of ERα-KD on cell proliferation is p53-dependent. A, generation of MCF7 cell lines in which p53 is stably knocked down and ERα can be inducibly knocked down. Western blots were prepared with extracts from MCF7-ERα-KD-11, MCF7(p53-KD)-ERα-KD-13, -15, and -18 cells uninduced (−) or induced (+) to knock down ERα. ERα, p53, and actin were detected by their respective antibodies. B and C, top panel, colony formation assay was performed in triplicate with MCF7(p53-KD)-ERα-KD-13 (B) or -15 (C) cells uninduced (−) or induced (+) to knock down ERα for 72 h followed by mock treatment or treatment with CPT (250 nm) for 6 h or Dox (100 nm) for 2 h and then maintained for 15 days. Middle panel, the number of colonies was counted using the UVP VisionWorksLS software (error bars represent S.D.; n = 3). Bottom panel, all stained cells in a well were scanned using the UVP VisionWorks LS software to determine total cell density. The density of MCF7 cells without ERα-KD was arbitrarily set at 1.0 regardless of mock treatment or treatment with CPT and Dox. The -fold change in cell density by ERα-KD was calculated in triplicate (error bars represent S.D.; n = 3). D, Western blots were prepared with extracts from MCF7(p53-KD)-ERα-KD-13 cells that were uninduced (−) or induced (+) to knock down ERα followed by mock treatment or treatment with CPT (250 nm) or Dox (400 nm) for 9 h. ERα, p53, p21, MDM2, PolH, PUMA, MIC-1, and actin were detected by their respective antibodies. MCF7-ERα-KD-11 cells that were uninduced (−) or induced (+) to knock down ERα were used as a control. E, Western blots were prepared with extracts from ZR-75-1 cells that were transiently transfected with scrambled (Scr) and/or ERα siRNA (siERα) (left panel) and p53 (sip53) and/or ER siRNA (right panel) for 72 h followed by mock treatment or treatment with Dox (400 nm) for 9 h. ERα, p53, p21, MDM2, PolH, PUMA, MIC-1, and actin were detected by their respective antibodies.
Ectopic Expression of ERα Sensitizes MCF7 Cells to DNA Damage-induced Growth Suppression in a p53-dependent Manner
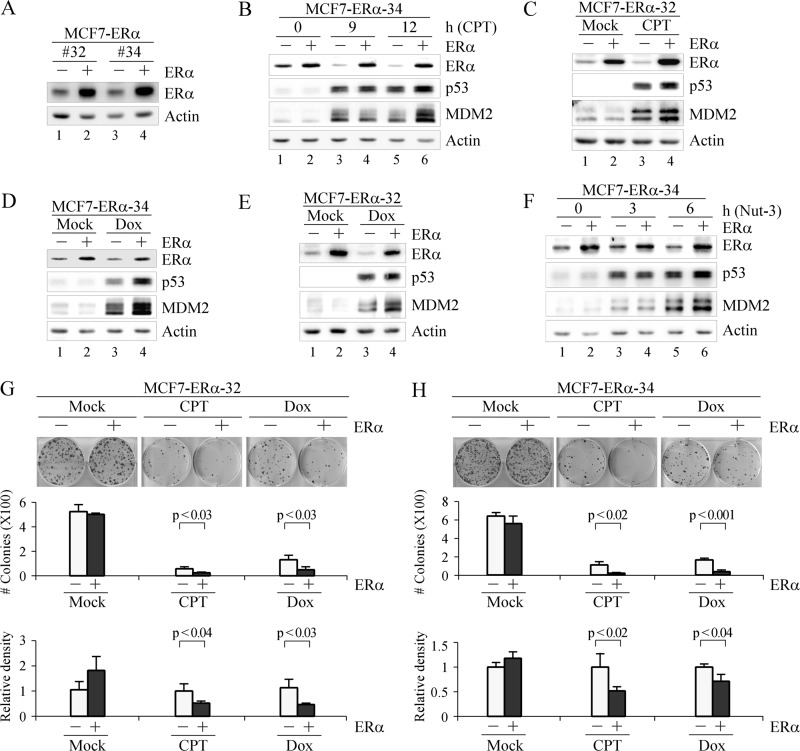
Overexpression of ERα is a hallmark of ER-positive breast cancer (46). Interestingly, overexpression of ERα is also correlated with higher levels of p53 (47, 48) and a favorable prognosis (6). To test whether p53 plays a role in the favorable prognosis of ERα-positive breast cancer patients, we generated multiple MCF7 cell lines that can inducibly express ERα, designated MCF7-ERα (Fig. 6A, ERα panel, compare lanes 1 and 3 with lanes 2 and 4, respectively). We showed that ectopic expression of ERα led to increased accumulation of p53 in MCF7 cells upon treatment with CPT (Fig. 6, B, p53 panel, compare lanes 3 and 5 with lanes 4 and 6, respectively, and C, p53 panel, compare lane 3 with lane 4), Dox (Fig. 6, D and E, p53 panel, compare lane 3 with lane 4), or Nut-3 (Fig. 6F, p53 panel, compare lanes 3 and 5 with lanes 4 and 6, respectively). In addition, the enhanced level of p53 by ERα overexpression was transcriptionally active as MDM2, a p53 target, was also increased (Fig. 6, B and F, MDM2 panel, compare lanes 5 with lane 6, and C–E, MDM2 panel, compare lanes 3 with lanes 4). Next, a colony formation assay was performed to examine the effect of ERα overexpression on cell growth. We found that overexpression of ERα in MCF7 cells enhanced cell proliferation (Fig. 6, G and H, mock treatment panels), consistent with previous results (49). However, ERα-overexpressing cells were more sensitive to treatment with CPT and Dox compared with control cells (Fig. 6, G and H, CPT and Dox panels).
FIGURE 6.
Overexpression of ERα increases p53 levels and MCF7 cell sensitivity to DNA damage-induced growth suppression. A, generation of MCF7 cell lines in which ERα can be inducibly expressed. B, Western blots were prepared with extracts from MCF7-ERα-34 cells that were uninduced (−) or induced (+) to express ERα for 24 h followed by mock treatment or treatment with CPT (250 nm) for 9 and 12 h. C, Western blots were prepared with extracts from MCF7-ERα-32 cells that were uninduced (−) or induced (+) to express ERα for 24 h followed by mock treatment or treatment with CPT (250 nm) for 9 h. D and E, extracts for Western blots were prepared as in C except that Dox (150 nm) was used to treat MCF7-ERα-34 (D) and -32 (E) cells for 9 h. F, extracts for Western blots were prepared as in B except that cells were treated with Nut-3 (7 μm) for 3 and 6 h. ERα, p53, MDM2, and actin were detected by their respective antibodies. G and H, top panel, a colony formation assay was performed in triplicate with MCF7-ERα-32 (G) or -34 (H) cells uninduced (−) or induced (+) to express ERα for 48 h followed by mock treatment or treatment with CPT (250 nm) for 6 h or Dox (100 nm) for 2 h and then maintained for 15 days. Middle panel, the number of colonies was counted using the UVP VisionWorksLS software (error bars represent S.D.; n = 3). Bottom panel, all stained cells in a well were scanned using the UVP VisionWorksLS software to determine total cell density. The density of MCF7 cells without ERα overexpression was arbitrarily set at 1.0 regardless of mock treatment or treatment with CPT and Dox. The -fold change in cell density by ERα overexpression was calculated in triplicate (error bars represent S.D.; n = 3).
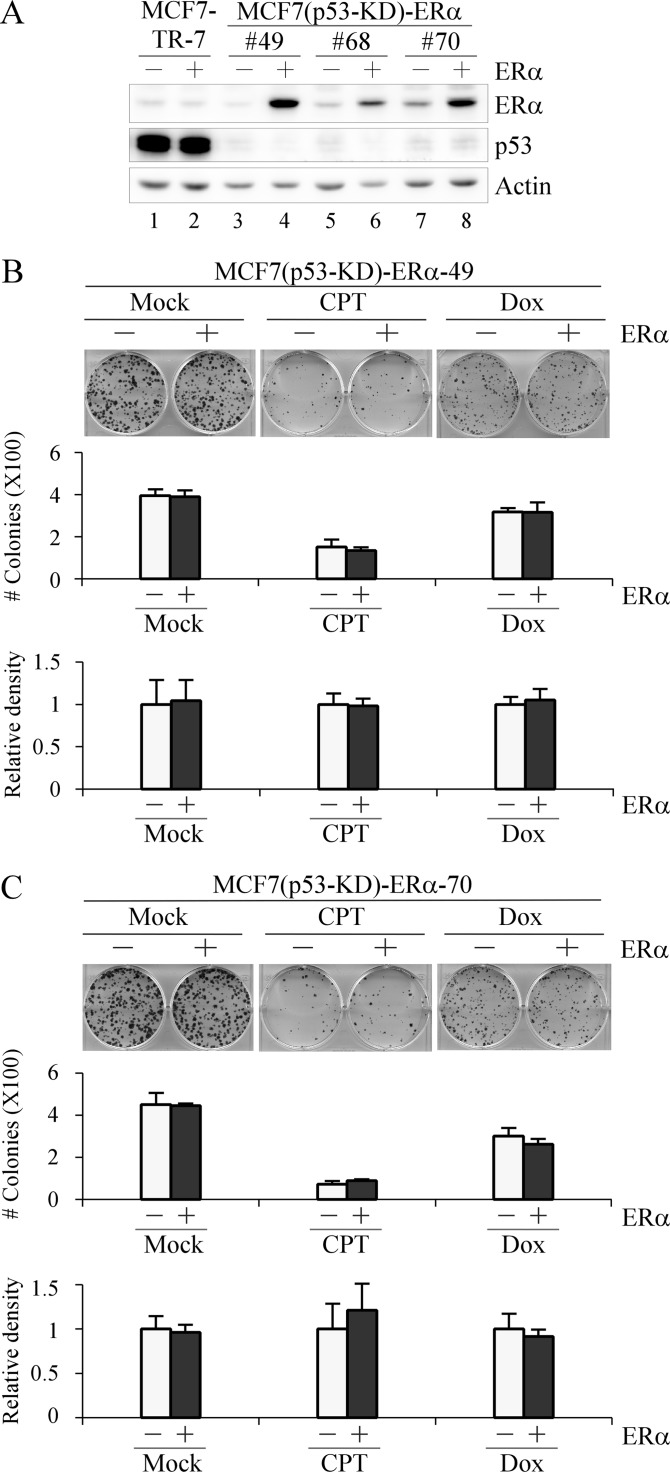
To determine whether the increased sensitivity of ERα-overexpressing cells to DNA damage was dependent on p53, we generated multiple MCF7 cell lines, designated MCF7(p53-KD)-ERα, in which p53 was stably knocked down and ERα can be inducibly expressed. Western blot analysis showed that p53 was undetectable, whereas ERα was inducibly expressed in MCF7(p53-KD)-ERα (clones 49 and 70) compared with that in MCF7-TR-7 cells (Fig. 7A). Next, a colony formation assay was performed and showed that the increased sensitivity of ERα-overexpressing cells to DNA damage was abrogated by p53-KD (Fig. 7, B and C).
FIGURE 7.
The effect of ERα overexpression on cell sensitivity to DNA damage-induced growth suppression is p53-dependent. A, generation of MCF7 cell lines in which p53 is stably knocked down and ERα can be inducibly expressed. Western blots were prepared with extracts from MCF7(p53-KD)-ERα-49, -68, or -70 cells that were uninduced (−) or induced (+) to express ERα for 24 h. B and C, top panel, a colony formation assay was performed in triplicate with MCF7(p53-KD)-ERα-49 (B) and -70 (C) cells uninduced (−) or induced (+) to express ERα for 48 h followed by mock treatment or treatment with CPT (250 nm) for 6 h or Dox (100 nm) for 2 h and then maintained for 15 days. Middle panel, the number of colonies was counted using the UVP VisionWorksLS software (error bars represent S.D.; n = 3). Bottom panel, all stained cells in a well were scanned using the UVP VisionWorksLS software to determine total cell density. The density of MCF7 cells without ERα overexpression was arbitrarily set at 1.0 regardless of mock treatment or treatment with CPT and Dox. The -fold change in cell density by ERα overexpression was calculated in triplicate (error bars represent S.D.; n = 3).
DISCUSSION
The p53 tumor suppressor is commonly mutated in over 50% of human cancers (12). However, the overall frequency of p53 mutations in breast cancers is significantly lower than in other type of cancers (13). Inactivation of p53 in cancers maintaining a wild-type p53 allele could be achieved via alterations in upstream regulators and downstream effectors (50). For example, positive regulators of p53, such as ataxia telangiectasia mutated (ATM)/Chk2 and p19ARF, have been found to be inactivated, whereas negative regulators of p53, such as MDM2, have been found to be overexpressed (51, 52). In addition, p53 target genes, such as PIG8 (a proapoptotic factor) and 14-3-3σ (a G2/M arrest regulator), are often inactivated in breast cancers (53, 54). Importantly, it has been shown that a reduced basal level of p53 mRNA due to loss of HoxA5 expression is correlated with primary breast carcinomas (55). Indeed, p53 is a direct target of HoxA5 (55). Reports also showed that p53 is transcriptionally activated by c-Jun and c-Fos through the AP-1 site, p50NF-κB1 and p65RelA through the NF-κB motif, and c-Myc/Max/upstream stimulatory factor through the E-box element (56). During the time we were preparing this manuscript, one report showed that Oldenlandia diffusa extract stimulates ERα association with Sp1 at the GC-rich motif in the proximal p53 promoter (57). However, whether ERα directly regulates p53 transcription is unclear. Here, we found that knockdown of ERα results in reduced expression of p53 protein and mRNA (Figs. 1 and 2). In addition, we found that ERα activates p53 transcription via binding to ERE half-sites on the p53 promoter (Fig. 3). The antiestrogen ICI 182,780 competes with estrogen for binding to ERα, promotes ERα degradation, disrupts nuclear localization and dimerization of ERα, and reduces ERα binding to ERE sites (38, 58). Thus, ICI 182,780 attenuates ERα transcriptional activity and reduces steady-state levels of ERα (58, 59). Consistently, we showed that activation of the p53 promoter by ERα is enhanced by estrogen but reduced by ICI 182,780 (Fig. 3E). These data indicate that p53 is a direct target of ERα. Importantly, it has been shown that ERα is regulated by p53 (14, 15). Therefore, our findings provide new insights into a positive feedback loop between p53 and ERα.
ERα, a nuclear hormone receptor and a transcription factor, is involved in several physiological processes, such as development of the female reproductive system, metabolism, and bone homeostasis (60). However, enhanced proliferation of ER-positive cells by ERα contributes to mammary tumorigenesis (61). As a result, ERα inhibitors, such as tamoxifen, have been successfully developed for breast cancer treatment (62). Importantly, the mortality for ER-positive breast cancer when treated with antiestrogen and adjuvant chemotherapy is markedly decreased (62, 63). However, the underlying mechanism is not clear. As a master mediator of DNA damage signals to induce cell cycle arrest and apoptosis, the status of p53 predicts a good outcome following chemotherapy (65, 66). However, the extent of p53 involvement in endocrine therapy and chemotherapy in breast cancers is still uncertain (67). Here, we found that ERα-KD cells with impaired p53 expression are more resistant (Fig. 4), whereas ERα-overexpressing cells with elevated p53 expression are more sensitive (Fig. 6) to DNA damage-induced growth suppression as compared with controls. We also showed that knockdown of p53 diminishes, if not abrogates, the prosurvival activity of ERα-KD and the antisurvival activity of ERα overexpression upon DNA damage (Figs. 5 and 7). Moreover, we showed that expression of p53 targets in cell cycle arrest (p21) and in cell death (PolH, PUMA, and MIC-1) was decreased by ERα-KD in p53-proficient (Figs. 4C and 5E) but not in p53-deficient cells (Fig. 5, D and E). Thus, our data indicate that in addition to hormone therapy, ERα is implicated in chemotherapy via regulating the p53 pathway.
In clinical studies, ER-positive breast cancer is defined as having a statistically significant chance of responding to hormone therapy with a positive outcome. ER-positive cases make up 70% of all invasive breast cancer diagnoses (68). However, 30–40% of ER-positive breast cancer patients can experience a decrease in ERα activity from hormone treatment (69). Interestingly, evidence showed that p53 alteration has been found to be correlated with ER-negative and high grade breast tumors (64). Altogether, we hypothesized that due to a positive feedback regulatory loop between ERα and p53 loss of ERα would lead to a decrease in p53 expression, which in turn could lead to the formation of a more aggressive tumor refractory to both antiestrogen and chemotherapy. Therefore, further exploration of the relationship between ERα and p53 in breast cancers will improve our understanding and practical management of different types of breast tumors.
Acknowledgments
We thank Colleen Sweeney (University of California, Davis, CA) for generously providing ZR-75-1 cells and for constructive advice. We also thank Ariane Scoumanne and Seong-Jun Cho for providing technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant CA076069.
- ER
- estrogen receptor
- ERE
- estrogen response element
- KD
- knockdown
- Dox
- doxorubicin
- CPT
- camptothecin
- MDM2
- murine double minute 2
- nt
- nucleotides
- PolH
- polymerase η
- Nut-3
- Nutlin-3
- PUMA
- p53-upregulated modulator of apoptosis.
REFERENCES
- 1. Mosselman S., Polman J., Dijkema R. (1996) ERβ: identification and characterization of a novel human estrogen receptor. FEBS Lett. 392, 49–53 [DOI] [PubMed] [Google Scholar]
- 2. Kuiper G. G., Carlsson B., Grandien K., Enmark E., Häggblad J., Nilsson S., Gustafsson J. A. (1997) Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors α and β. Endocrinology 138, 863–870 [DOI] [PubMed] [Google Scholar]
- 3. Stanford J. L., Szklo M., Brinton L. A. (1986) Estrogen receptors and breast cancer. Epidemiol. Rev. 8, 42–59 [DOI] [PubMed] [Google Scholar]
- 4. Leygue E., Dotzlaw H., Watson P. H., Murphy L. C. (1998) Altered estrogen receptor α and β messenger RNA expression during human breast tumorigenesis. Cancer Res. 58, 3197–3201 [PubMed] [Google Scholar]
- 5. Lazennec G., Bresson D., Lucas A., Chauveau C., Vignon F. (2001) ERβ inhibits proliferation and invasion of breast cancer cells. Endocrinology 142, 4120–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Allegra J. C., Lippman M. E. (1980) Estrogen receptor status and the disease-free interval in breast cancer. Recent Results Cancer Res. 71, 20–25 [DOI] [PubMed] [Google Scholar]
- 7. Ottaviano Y. L., Issa J. P., Parl F. F., Smith H. S., Baylin S. B., Davidson N. E. (1994) Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res. 54, 2552–2555 [PubMed] [Google Scholar]
- 8. Parl F. F., Schmidt B. P., Dupont W. D., Wagner R. K. (1984) Prognostic significance of estrogen receptor status in breast cancer in relation to tumor stage, axillary node metastasis, and histopathologic grading. Cancer 54, 2237–2242 [DOI] [PubMed] [Google Scholar]
- 9. Vousden K. H., Prives C. (2009) Blinded by the light: the growing complexity of p53. Cell 137, 413–431 [DOI] [PubMed] [Google Scholar]
- 10. el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 11. Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J. (1992) The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 12. Hollstein M., Sidransky D., Vogelstein B., Harris C. (1991) p53 mutations in human cancers. Science 253, 49–53 [DOI] [PubMed] [Google Scholar]
- 13. Pharoah P. D., Day N. E., Caldas C. (1999) Somatic mutations in the p53 gene and prognosis in breast cancer: a meta-analysis. Br. J. Cancer 80, 1968–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Angeloni S. V., Martin M. B., Garcia-Morales P., Castro-Galache M. D., Ferragut J. A., Saceda M. (2004) Regulation of estrogen receptor-α expression by the tumor suppressor gene p53 in MCF-7 cells. J. Endocrinol. 180, 497–504 [DOI] [PubMed] [Google Scholar]
- 15. Shirley S. H., Rundhaug J. E., Tian J., Cullinan-Ammann N., Lambertz I., Conti C. J., Fuchs-Young R. (2009) Transcriptional regulation of estrogen receptor α by p53 in human breast cancer cells. Cancer Res. 69, 3405–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu C. L., Driggers P., Barrera-Hernandez G., Nunez S. B., Segars J. H., Cheng S. (1997) The tumor suppressor p53 is a negative regulator of estrogen receptor signaling pathways. Biochem. Biophys. Res. Commun. 239, 617–620 [DOI] [PubMed] [Google Scholar]
- 17. Sayeed A., Konduri S. D., Liu W., Bansal S., Li F., Das G. M. (2007) Estrogen receptor α inhibits p53-mediated transcriptional repression: implications for the regulation of apoptosis. Cancer Res. 67, 7746–7755 [DOI] [PubMed] [Google Scholar]
- 18. Qin C., Nguyen T., Stewart J., Samudio I., Burghardt R., Safe S. (2002) Estrogen up-regulation of p53 gene expression in MCF-7 breast cancer cells is mediated by calmodulin kinase IV-dependent activation of a nuclear factor κB/CCAAT-binding transcription factor-1 complex. Mol. Endocrinol. 16, 1793–1809 [DOI] [PubMed] [Google Scholar]
- 19. Liu G., Xia T., Chen X. (2003) The activation domains, the proline-rich domain, and the C-terminal basic domain in p53 are necessary for acetylation of histones on the proximal p21 promoter and interaction with p300/CREB-binding protein. J. Biol. Chem. 278, 17557–17565 [DOI] [PubMed] [Google Scholar]
- 20. Yan W., Chen X. (2006) GPX2, a direct target of p63, inhibits oxidative stress-induced apoptosis in a p53-dependent manner. J. Biol. Chem. 281, 7856–7862 [DOI] [PubMed] [Google Scholar]
- 21. Johansen F. E., Prywes R. (1994) Two pathways for serum regulation of the c-fos serum response element require specific sequence elements and a minimal domain of serum response factor. Mol. Cell. Biol. 14, 5920–5928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harms K. L., Chen X. (2007) Histone deacetylase 2 modulates p53 transcriptional activities through regulation of p53-DNA binding activity. Cancer Res. 67, 3145–3152 [DOI] [PubMed] [Google Scholar]
- 23. Liu G., Chen X. (2005) The C-terminal sterile α motif and the extreme C terminus regulate the transcriptional activity of the α isoform of p73. J. Biol. Chem. 280, 20111–20119 [DOI] [PubMed] [Google Scholar]
- 24. Qian Y., Jung Y. S., Chen X. (2011) ΔNp63, a target of DEC1 and histone deacetylase 2, modulates the efficacy of histone deacetylase inhibitors in growth suppression and keratinocyte differentiation. J. Biol. Chem. 286, 12033–12041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qian Y., Chen X. (2008) ID1, inhibitor of differentiation/DNA binding, is an effector of the p53-dependent DNA damage response pathway. J. Biol. Chem. 283, 22410–22416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gretarsdottir S., Tryggvadottir L., Jonasson J. G., Sigurdsson H., Olafsdottir K., Agnarsson B. A., Ogmundsdottir H., Eyfjörd J. E. (1996) TP53 mutation analyses on breast carcinomas: a study of paraffin-embedded archival material. Br. J. Cancer 74, 555–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fernández-Cuesta L., Anaganti S., Hainaut P., Olivier M. (2011) Estrogen levels act as a rheostat on p53 levels and modulate p53-dependent responses in breast cancer cell lines. Breast Cancer Res. Treat. 125, 35–42 [DOI] [PubMed] [Google Scholar]
- 28. Holst F., Stahl P. R., Ruiz C., Hellwinkel O., Jehan Z., Wendland M., Lebeau A., Terracciano L., Al-Kuraya K., Jänicke F., Sauter G., Simon R. (2007) Estrogen receptor α (ESR1) gene amplification is frequent in breast cancer. Nat. Genet. 39, 655–660 [DOI] [PubMed] [Google Scholar]
- 29. Fornari F. A., Randolph J. K., Yalowich J. C., Ritke M. K., Gewirtz D. A. (1994) Interference by doxorubicin with DNA unwinding in MCF-7 breast tumor cells. Mol. Pharmacol. 45, 649–656 [PubMed] [Google Scholar]
- 30. Tovar C., Rosinski J., Filipovic Z., Higgins B., Kolinsky K., Hilton H., Zhao X., Vu B. T., Qing W., Packman K., Myklebost O., Heimbrook D. C., Vassilev L. T. (2006) Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc. Natl. Acad. Sci. U.S.A. 103, 1888–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Troester M. A., Herschkowitz J. I., Oh D. S., He X., Hoadley K. A., Barbier C. S., Perou C. M. (2006) Gene expression patterns associated with p53 status in breast cancer. BMC Cancer 6, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jakowlew S. B., Breathnach R., Jeltsch J. M., Masiakowski P., Chambon P. (1984) Sequence of the pS2 mRNA induced by estrogen in the human breast cancer cell line MCF-7. Nucleic Acids Res. 12, 2861–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klein-Hitpass L., Ryffel G. U., Heitlinger E., Cato A. C. (1988) A 13 bp palindrome is a functional estrogen responsive element and interacts specifically with estrogen receptor. Nucleic Acids Res. 16, 647–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Driscoll M. D., Sathya G., Muyan M., Klinge C. M., Hilf R., Bambara R. A. (1998) Sequence requirements for estrogen receptor binding to estrogen response elements. J. Biol. Chem. 273, 29321–29330 [DOI] [PubMed] [Google Scholar]
- 35. Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R., Fox E. A., Silver P. A., Brown M. (2005) Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43 [DOI] [PubMed] [Google Scholar]
- 36. Rosenbloom K. R., Dreszer T. R., Pheasant M., Barber G. P., Meyer L. R., Pohl A., Raney B. J., Wang T., Hinrichs A. S., Zweig A. S., Fujita P. A., Learned K., Rhead B., Smith K. E., Kuhn R. M., Karolchik D., Haussler D., Kent W. J. (2010) ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 38, D620–D625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brown A. M., Jeltsch J. M., Roberts M., Chambon P. (1984) Activation of pS2 gene transcription is a primary response to estrogen in the human breast cancer cell line MCF-7. Proc. Natl. Acad. Sci. U.S.A. 81, 6344–6348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Parker M. G., Arbuckle N., Dauvois S., Danielian P., White R. (1993) Structure and function of the estrogen receptor. Ann. N.Y. Acad. Sci. 684, 119–126 [DOI] [PubMed] [Google Scholar]
- 39. Huang J., Li X., Maguire C. A., Hilf R., Bambara R. A., Muyan M. (2005) Binding of estrogen receptor α to estrogen response element in situ is independent of estradiol and impaired by its amino terminus. Mol. Endocrinol. 19, 2696–2712 [DOI] [PubMed] [Google Scholar]
- 40. Shiozawa T., Miyamoto T., Kashima H., Nakayama K., Nikaido T., Konishi I. (2004) Estrogen-induced proliferation of normal endometrial glandular cells is initiated by transcriptional activation of cyclin D1 via binding of c-Jun to an AP-1 sequence. Oncogene 23, 8603–8610 [DOI] [PubMed] [Google Scholar]
- 41. Liu G., Chen X. (2006) DNA Polymerase η, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol. Cell. Biol. 26, 1398–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tan M., Wang Y., Guan K., Sun Y. (2000) PTGF-β, a type β transforming growth factor (TGF-β) superfamily member, is a p53 target gene that inhibits tumor cell growth via TGF-β signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 97, 109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu J., Zhang L., Hwang P. M., Kinzler K. W., Vogelstein B. (2001) PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell 7, 673–682 [DOI] [PubMed] [Google Scholar]
- 44. Nakano K., Vousden K. H. (2001) PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 7, 683–694 [DOI] [PubMed] [Google Scholar]
- 45. Qian Y., Jung Y. S., Chen X. (2012) Differentiated embryo-chondrocyte expressed gene 1 regulates p53-dependent cell survival versus cell death through macrophage inhibitory cytokine-1. Proc. Natl. Acad. Sci. U.S.A. 109, 11300–11305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sluyser M., Van Nie R. (1974) Estrogen receptor content and hormone-responsive growth of mouse mammary tumors. Cancer Res. 34, 3253–3257 [PubMed] [Google Scholar]
- 47. Ivshina A. V., George J., Senko O., Mow B., Putti T. C., Smeds J., Lindahl T., Pawitan Y., Hall P., Nordgren H., Wong J. E., Liu E. T., Bergh J., Kuznetsov V. A., Miller L. D. (2006) Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 66, 10292–10301 [DOI] [PubMed] [Google Scholar]
- 48. Kao K. J., Chang K. M., Hsu H. C., Huang A. (2011) Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer 11, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Aitken S. C., Lippman M. E. (1985) Effect of estrogens and antiestrogens on growth-regulatory enzymes in human breast cancer cells in tissue culture. Cancer Res. 45, 1611–1620 [PubMed] [Google Scholar]
- 50. Gasco M., Shami S., Crook T. (2002) The p53 pathway in breast cancer. Breast Cancer Res. 4, 70–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Agrawal A., Yang J., Murphy R. F., Agrawal D. K. (2006) Regulation of the p14ARF-Mdm2-p53 pathway: an overview in breast cancer. Exp. Mol. Pathol. 81, 115–122 [DOI] [PubMed] [Google Scholar]
- 52. Sullivan A., Yuille M., Repellin C., Reddy A., Reelfs O., Bell A., Dunne B., Gusterson B. A., Osin P., Farrell P. J., Yulug I., Evans A., Ozcelik T., Gasco M., Crook T. (2002) Concomitant inactivation of p53 and Chk2 in breast cancer. Oncogene 21, 1316–1324 [DOI] [PubMed] [Google Scholar]
- 53. Gentile M., Ahnström M., Schön F., Wingren S. (2001) Candidate tumour suppressor genes at 11q23-q24 in breast cancer: evidence of alterations in PIG8, a gene involved in p53-induced apoptosis. Oncogene 20, 7753–7760 [DOI] [PubMed] [Google Scholar]
- 54. Ferguson A. T., Evron E., Umbricht C. B., Pandita T. K., Chan T. A., Hermeking H., Marks J. R., Lambers A. R., Futreal P. A., Stampfer M. R., Sukumar S. (2000) High frequency of hypermethylation at the 14-3-3σ locus leads to gene silencing in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 97, 6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Raman V., Martensen S. A., Reisman D., Evron E., Odenwald W. F., Jaffee E., Marks J., Sukumar S. (2000) Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature 405, 974–978 [DOI] [PubMed] [Google Scholar]
- 56. Kirch H. C., Flaswinkel S., Rumpf H., Brockmann D., Esche H. (1999) Expression of human p53 requires synergistic activation of transcription from the p53 promoter by AP-1, NF-κB and Myc/Max. Oncogene 18, 2728–2738 [DOI] [PubMed] [Google Scholar]
- 57. Gu G., Barone I., Gelsomino L., Giordano C., Bonofiglio D., Statti G., Menichini F., Catalano S., Andò S. (2012) Oldenlandia diffusa extracts exert antiproliferative and apoptotic effects on human breast cancer cells through ERα/Sp1-mediated p53 activation. J. Cell. Physiol. 227, 3363–3372 [DOI] [PubMed] [Google Scholar]
- 58. Dauvois S., White R., Parker M. G. (1993) The antiestrogen ICI 182780 disrupts estrogen receptor nucleocytoplasmic shuttling. J. Cell Sci. 106, 1377–1388 [DOI] [PubMed] [Google Scholar]
- 59. Howell A., Osborne C. K., Morris C., Wakeling A. E. (2000) ICI 182,780 (FaslodexTM). Cancer 89, 817–825 [DOI] [PubMed] [Google Scholar]
- 60. Barkhem T., Nilsson S., Gustafsson J. A. (2004) Molecular mechanisms, physiological consequences and pharmacological implications of estrogen receptor action. Am. J. Pharmacogenomics 4, 19–28 [DOI] [PubMed] [Google Scholar]
- 61. Ariazi E. A., Ariazi J. L., Cordera F., Jordan V. C. (2006) Estrogen receptors as therapeutic targets in breast cancer. Curr. Top. Med. Chem. 6, 181–202 [PubMed] [Google Scholar]
- 62. Early Breast Cancer Trialists' Collaborative Group (EBCTCG), Peto R., Davies C., Godwin J., Gray R., Pan H. C., Clarke M., Cutter D., Darby S., McGale P., Taylor C., Wang Y. C., Bergh J., Di Leo A., Albain K., Swain S., Piccart M., Pritchard K. (2012) Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet 379, 432–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hubay C. A., Pearson O. H., Manni A., Gordon N. H., McGuire W. L. (1985) Adjuvant endocrine therapy, cytotoxic chemotherapy and immunotherapy in stage II breast cancer: 6-year result. J. Steroid Biochem. 23, 1147–1150 [DOI] [PubMed] [Google Scholar]
- 64. Sasa M., Kondo K., Komaki K., Morimoto T., Monden Y. (1994) p53 alteration correlates with negative ER, negative PgR, and high histologic grade in breast cancer. J. Surg. Oncol. 56, 46–50 [DOI] [PubMed] [Google Scholar]
- 65. Petitjean A., Achatz M. I., Borresen-Dale A. L., Hainaut P., Olivier M. (2007) TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene 26, 2157–2165 [DOI] [PubMed] [Google Scholar]
- 66. Velculescu V. E., El-Deiry W. S. (1996) Biological and clinical importance of the p53 tumor suppressor gene. Clin. Chem. 42, 858–868 [PubMed] [Google Scholar]
- 67. Thompson A. M., Lane D. P. (2009) p53 transcriptional pathways in breast cancer: the good, the bad and the complex. J. Pathol. 220, 401–403 [DOI] [PubMed] [Google Scholar]
- 68. Harvey J. M., Clark G. M., Osborne C. K., Allred D. C. (1999) Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J. Clin. Oncol. 17, 1474–1781 [DOI] [PubMed] [Google Scholar]
- 69. Kuukasjärvi T., Kononen J., Helin H., Holli K., Isola J. (1996) Loss of estrogen receptor in recurrent breast cancer is associated with poor response to endocrine therapy. J. Clin. Oncol. 14, 2584–2589 [DOI] [PubMed] [Google Scholar]