Background: Erythropoietin regulates the myogenic regulatory factor expression program and proliferation.
Results: Erythropoietin induces GATA-4 and TAL1 to retard differentiation via Sirt1 activity in skeletal myoblasts.
Conclusion: GATA-4, TAL1, and Sirt1 cross-talk with each other to mediate erythropoietin activity and negatively regulate myogenic differentiation.
Significance: These finding provide new insight into the molecular mechanism of erythropoietin function beyond erythropoiesis.
Keywords: Erythropoietin, GATA, Molecular Biology, Muscle, Transcription Factors, Myoblast Proliferation, Myogenic Differentiation
Abstract
Erythropoietin (EPO), the cytokine required for erythrocyte production, contributes to muscle progenitor cell proliferation and delay myogenic differentiation. However, the underlying mechanism is not yet fully understood. Here, we report that EPO changes the skeletal myogenic regulatory factor expression program and delays differentiation via induction of GATA-4 and the basic helix-loop-helix TAL1 and that knockdown of both factors promotes differentiation. EPO increases the Sirt1 level, a NAD+-dependent deacetylase, and also induces the NAD+/NADH ratio that further increases Sirt1 activity. Sirt1 knockdown reduced GATA-4 and TAL1 expression, impaired EPO effect on delayed myogenic differentiation, and the Sirt1 knockdown effect was abrogated when combined with overexpression of GATA-4 or TAL1. GATA-4 interacts with Sirt1 and targets Sirt1 to the myogenin promoter and represses myogenin expression, whereas TAL1 inhibits myogenin expression by decreasing MyoD binding to and activation of the myogenin promoter. Sirt1 was found to bind to the GATA-4 promoter to directly regulate GATA-4 expression and GATA-4 binds to the TAL1 promoter to regulate TAL1 expression positively. These data suggest that GATA-4, TAL1, and Sirt1 cross-talk each other to regulate myogenic differentiation and mediate EPO activity during myogenic differentiation with Sirt1 playing a role upstream of GATA-4 and TAL1. Taken together, our findings reveal a novel role for GATA-4 and TAL1 to affect skeletal myogenic differentiation and EPO response via cross-talk with Sirt1.
Introduction
Differentiation of the myogenic lineage is mainly regulated by the myogenic regulatory factors (MRFs)2 such as Myf5, MyoD, and myogenin. These DNA-binding proteins contain a basic helix-loop-helix (bHLH) domain and bind E-box motifs (CANNTG) (1). Expression of Myf5 and MyoD is required for the commitment of progenitor cells to the myogenic lineage, because disruption of both genes results in the absence of skeletal myoblasts (2). Myogenin is essential for the terminal differentiation of myoblasts, but is dispensable for establishing the myogenic lineage (3). MyoD can bind to the E-box region of the myogenin promoter to activate the expression of myogenin. Histone deacetylases have been reported to regulate muscle gene expression through modifying the MyoD acetylation state (4–6). The class III deacetylase, Sirt1, which is most homologous to yeast Sir2 and is a NAD+-dependent deacetylase (7, 8), targets many transcription factors, such as, p53, FOXO, PGC-1α, NF-κB, E2F1, and LXR to be involved in functions as diverse as cell fate determination, inflammatory responses, and energy metabolism (9). Importantly, Sirt1 has been found to negatively regulate muscle differentiation by deacetylating MyoD and forming a complex with the acetyltransferase PCAF and MyoD in a NAD+-dependent manner (10).
During erythroid differentiation of hematopoietic stem cells, erythropoietin (EPO) binds to its receptor (EpoR) located on the surface of early erythroid progenitor cells to promote cell survival, proliferation, and differentiation (11, 12). However, EPO signaling is not restricted to the erythroid lineage and can be found in many nonhematopoietic tissues including endothelial, neural, and muscle progenitor/precursor cells (13–15). The deacetylated PCAF and MyoD were found to inhibit muscle gene expression such as myogenin through binding at the myogenin promoter to retard myogenic differentiation (10). We previously reported that EPO up-regulates Myf5 and MyoD and contributes myoblast proliferation, but inhibits myogenin expression and retards myogenic differentiation and myotube formation (13). However, the detailed mechanism by which EPO retards myogenic differentiation and modifies expression of MRFs remains largely unknown. It is of interest to know if Sirt1 can take part in EPO action in the regulation of myogenic differentiation.
We previously demonstrated that EPO stimulates proliferation of myoblasts through binding to EpoR to expand the progenitor/precursor population during differentiation and may have a potential role in muscle maintenance or repair (13). Enhanced EpoR expression promotes donor cell survival in a mouse model for myoblast transplantation and increases the number of dystrophin expressing muscle fibers in mice with muscular dystrophy (16). EPO also increases the satellite cell number following muscle injury, improves myoblast proliferation and survival, and promotes repair and regeneration during muscle injury (17). Recently, a metabolic effect of EPO signaling in muscle was reported to provide protection against diet-induced obesity and increase glucose tolerance (18). It is important to understand how EPO exerts its activity in nonerythroid cells such as skeletal muscle myoblast to assess the activity of EPO in muscle maintenance, function, and repair.
In hematopoietic cells, EPO stimulation of erythropoiesis stimulates marked increases in erythroid transcription factors including GATA-1 and the bHLH transcription factor T-cell acute leukemia 1 (TAL1), which are required for erythroid maturation (19–21). These factors have been reported to express beyond erythroid cells. GATA factors have been largely reported to be crucial for development of other tissues. GATA-4 null mice die around E10 as a result of severe defects in the extra embryonic endoderm and display defects in heart and foregut morphogenesis (22, 23). During development, GATA-4 contributes importantly to myocardial anti-apoptosis and cell proliferation (24, 25) and mediates cardioprotective effects via regulating EpoR expression (26). TAL1 also plays important roles in other tissues such as endothelial cell specification and differentiation (27, 28), and endocardium morphogenesis (29). TAL1 was recently found decreased in Sirt1−/− embryonic stem cells that exhibit delayed hematopoietic differentiation (30), whereas forced expression of TAL1 in myoblasts was reported to block myogenic differentiation (31, 32). However, it has not yet been determined how endogenous GATA factors and TAL1 regulate myoblast differentiation in skeletal muscle.
In this current report, we describe that EPO modifies transcription factors expression including induction of GATA-4 and TAL1, both of which are observed to retard myogenic differentiation. Importantly, EPO activity changes the myoblast redox state as reflected in the increased NAD+/NADH ratio and stimulates Sirt1 activity resulting in inhibition of expression of myogenic differentiation required factors and myotube formation. Sirt1 can regulate GATA-4 and the TAL1 expression level. GATA-4 but not TAL1 interacts with Sirt1 and targets Sirt1 to the myogenin promoter to inhibit expression. TAL1 is positively regulated by GATA-4 and inhibits myogenin expression via repression of MyoD binding to and activation of the myogenin promoter. Our findings first demonstrate the new function of GATA-4 and TAL1 in regulating skeletal myogenic differentiation via cross-talk with Sirt1 and their targeted action and importance in myoblasts response to EPO.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
The murine C2C12 myoblast cell line (ATCC) was propagated in Dulbecco's modified Eagle's medium with 10% FBS (growth medium) in a humidified atmosphere of 5% CO2 at 37 °C (13). To induce differentiation, myoblasts were cultured to 80% confluence and then switched to differentiation medium (Dulbecco's modified Eagle's medium with 2% horse serum). When EPO (Epoetin α, Amgen) was added to the cell cultures, it was used at a final concentration of 5 units/ml unless otherwise indicated. MEL cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS. For proliferation assays, 106 C2C12 cells were seeded onto a T75 flask, trypsinized at specific time points, and counted with a hemocytometer. Human muscle primary cells (HMPC) (catalog number A12555, Invitrogen) were similarly cultured. Primary hematopoietic/erythroid progenitor cells were isolated from human peripheral blood using Ficoll-Hypaque (BioWhittaker) and cultured for 5 days in α-minimal essential medium with 10% FBS, 10% conditioned media from bladder carcinoma 5637 cultures, 1.5 mm glutamine, and 1 μg/ml of cyclosprin A as previously described (21).
Western Blotting
For Western blotting, cells were washed with cold PBS twice and then lysed in RIPA buffer (50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 0.25% sodium deoxycholate and protease inhibitors). Proteins were resolved on 4–12% NuPAGE bis-Tris gels (Invitrogen), transferred onto nitrocellulose membranes, incubated with specific antibodies or anti-β-actin antibody as a control followed by incubation with secondary antibody conjugated to horseradish peroxidase and developed by enhanced chemiluminescence (ECL) (GE Healthcare UK Ltd.).
Overexpression, RNAi, and Quantitative Real-time PCR
GATA-4 cDNA was a gift from Dr. Eric N. Olson (Southwestern Medical Center). GATA-4, TAL1, and PGC-1α cDNA expression vectors were transfected into C2C12 cells using the transfection reagent LipofectamineTM 2000 (Invitrogen). GATA-4, TAL1, Sirt1, Myf5, MyoD, EpoR, and negative control siRNAs (Thermo Scientific Dharmacon) were transfected into C2C12 cells. For differentiated cells, C2C12 cells were transfected with siRNA in growth medium and after 24 h the cultures were changed to differentiation medium and cultured for an additional required time prior to harvesting. Each experiment was carried out at least in triplicate. The quantitative real-time PCR assay was carried out using gene-specific primers (available upon request) and fluorescently labeled TaqMan probes or SYBR Green dye (Invitrogen) in a 7900 Sequence Detector (PE Applied Biosystems, Foster City, CA). Plasmid containing the cDNA of interest was used as template to generate a standard curve. S16 was used as an internal control.
BrdU Cell Proliferation Analysis
BrdU absorbance for cell proliferation was performed according to the manufacture's instructions provided for the BrdU cell proliferation assay kit (Cell Signaling Technology).
Immunofluorescence Staining
C2C12 myoblasts were cultured in cell culture chamber slides (NUNC international) and transfected with siRNA or treated with EPO. After washing with PBS and treating with 4% paraformaldehyde for 10 min. Slides were washed with PBS for 3 times, treated with 5% Triton X-100 for 5–10 min, and blocked with 3% BSA for 30 min. Cell culture slides were incubated with primary antibody for MHC overnight at 4 °C. After staining with fluorescence-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA), slides were mounted using vector DAPI mounting medium (Vector Laboratories Inc., Burlingame, CA). Samples were examined, captured, and quantified under an Inverted Axio Observer.Z1 confocal microscope equipped with a Zeiss LSM 5 Live DuoScan laser scanning system (Carl Zeiss MicroImaging). The fusion index of myotubes was measured according to the Ref. 33.
NAD+/NADH Assay
NAD+ and NAD+/NADH levels were determined according to the manufacture's instructions provided for the NAD+/NADH assay (BIOVISION).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP analysis was carried out as described (34). In brief, cells were lysed, nuclei were isolated and cross-linked with 1% formaldehyde, lysed in lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris, pH 8.1), and cross-linked chromatin DNA was sheared by sonication on ice to an average length around 500 bp. The sonicated supernatant was diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.1, and 167 mm NaCl). Pre-cleared chromatin using protein G-agarose (Upstate) was incubated with specific antibody or IgG as the control at 4 °C overnight. Immunoprecipitations were recovered by incubation with protein G-agarose (Upstate) at 4 °C for 2 h, followed by low-speed centrifugation. The washed pellets were reverse cross-linked. DNA was extracted with phenol-chloroform/isoamyl alcohol (25:24:1), precipitated with ethanol, and used for quantitative PCR analysis. The primer sequences for ChIP assay are shown in the supplemental Table S1.
Co-immunoprecipitation Assay
Cell nuclear extract prepared in RIPA buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 2 mm EDTA, 1% Nonidet P-40, 1 mm EGTA, protease inhibitor) and 100 mg of protein extract was pre-cleared with control IgG (Santa Cruz) and protein A/G beads (Upstate). Pre-cleared extract was then incubated with specific antibody or preimmune serum and protein A/G beads in 1.5 ml of immunoprecipitation buffer (0.5% Nonidet P-40, 10 mm Tris-HCl, 150 mm NaCl, 2 mm EDTA, 10% glycerol, protease inhibitor) at 4 °C for 4–6 h. After a brief centrifugation, the pellet was washed in immunoprecipitation buffer 4–5 times at 4 °C for 10 min, and the immunoprecipitated protein complexes were analyzed by Western blot analysis with specific antibodies.
Reporter Gene Analysis
A mouse myogenin promoter-Luc reporter gene assay was performed as described (10). Briefly, cells were harvested and luciferase activities were determined by the Dual Luciferase Assay System (Promega, Madison, WI).
Antibodies
GATA-3, GATA-4, TAL1, Myf5, MyoD, myogenin, Sirt1, and EpoR antibodies (Santa Cruz), STAT-5 and phosphorylated STAT-5 (Cell Signaling), and histone-3 acetylation, PGC-1α antibodies (Millipore) were obtained commercially.
Statistical Analyses
Data are presented as mean ± S.D. Statistical analyses were performed using analysis of variance.
RESULTS
EPO Induces GATA-4 and TAL1 Expression in Myoblasts
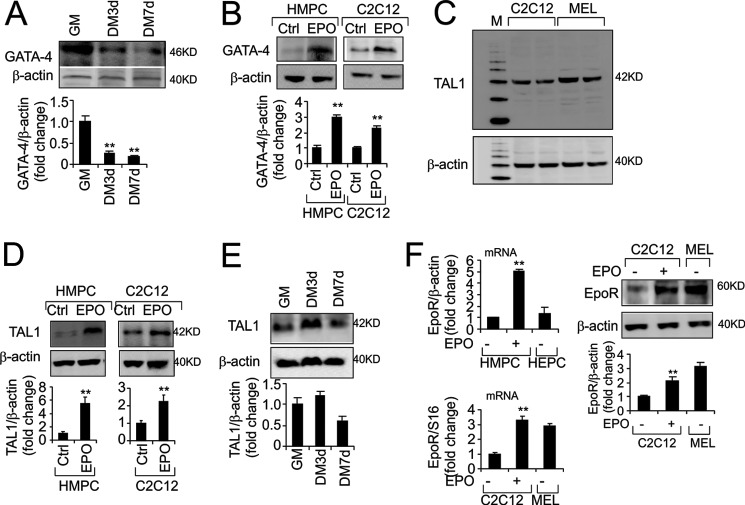
EPO stimulates GATA transcription factor response in erythroid, myoblast, and neural cells. To determine which of the six GATA-like family members respond to EPO stimulation in myoblasts, we examined expression of GATA-like transcription factors in C2C12 myoblasts. GATA-1, GATA-2, and GATA-5 were not detectable in C2C12 cells. We found GATA-4 to be expressed at the highest level in myoblasts compared with GATA-3, and GATA-6 appeared to be an order of magnitude lower (supplemental Fig. S1A). The GATA-4 protein level was also found to decrease with myoblast differentiation (Fig. 1A). EPO induction of GATA-4 protein was readily detected in C2C12 cells cultured in growth media and also in differentiated C2C12 cells that exhibit the decrease in GATA-4 to a low level with differentiation (supplemental Fig. S1B). HMPC also respond to EPO stimulation with increased proliferation (supplemental Fig. S1C) and induction of GATA-4 expression (Fig. 1B). EPO retards HMPC differentiation as indicated by reduced immunoreactivity to myosin heavy chain (MHC) (supplemental Fig. S1C).
FIGURE 1.
EPO induces GATA-4 and TAL1 in HMPC and C2C12 myoblasts. A, GATA-4 protein level was assessed using Western blotting and quantified in C2C12 cells cultured in growth medium (GM) and differentiation medium (DM) for 3 and 7 days as indicated. β-Actin was used as a loading control. B, GATA-4 protein expression was induced by EPO stimulation in primary HMPC and C2C12 myoblasts. C, TAL1 protein in C2C12 cells, compared with MEL cells was assessed by Western blotting using β-actin as loading control. D, EPO induction of TAL1 in HMPC and C2C12 cells was determined by Western blotting normalized to β-actin and quantified. E, TAL1 protein level was assessed using Western blotting with myoblasts differentiation and quantified. F, EPO induction of EpoR mRNA and comparison between HMPC and primary human hematopoietic/erythroid progenitor cells (HEPC) are shown. EPO induction of EpoR mRNA and protein levels in C2C12 myoblasts and comparison between C2C12 cells and MEL cells are also shown. For quantification, the mean values from 3 experiments are shown. Error bars represent S.D.; * indicates p < 0.05; **, p < 0.01.
EPO stimulation of erythroid progenitor cells induces expression of erythroid transcription factors including GATA-1 and TAL1. Because TAL1 is known to be expressed in both hematopoietic and nonhematopoietic tissue, we examined TAL1 expression in myoblasts. Surprisingly, TAL1 was readily detected by Western blotting in C2C12 myoblast cells compared with no expression in HeLa cells (Fig. 1C). The expression level was comparable with murine erythroleukemia MEL cells that can be induced to undergo further erythroid differentiation. In addition, EPO induced TAL1 expression in C2C12 cells (Fig. 1D). We also detected EPO induction of TAL1 and to a greater extent in HMPC, suggesting HMPC is more sensitive to EPO than the C2C12 cell line (Fig. 1D), possibly reflecting a stronger EPO response in primary cells. In contrast to GATA-4, there is not a marked decrease in TAL1 protein expression with differentiation compared with proliferating myoblasts (Fig. 1, A and E). EPO response is determined by EpoR expression. Unexpectedly, we observed that the EpoR mRNA expression level in HMPC from skeletal muscle was comparable with expression in primary human hematopoietic/erythropoietic progenitor cells without EPO stimulation (Fig. 1F). The EpoR mRNA level in HMPC was further increased by 4-fold with EPO stimulation. In the C2C12 cell line, the EpoR mRNA level was about ⅓ that of the erythroleukemia MEL cell line and increased by about 3-fold with EPO stimulation at the mRNA level (Fig. 1F). The increase in EpoR protein in C2C12 myoblasts is consistent with the increase in mRNA level (Fig. 1F).
GATA-4 and TAL1 Regulate Myoblast Proliferation, Expression of MRFs, and Myogenic Differentiation
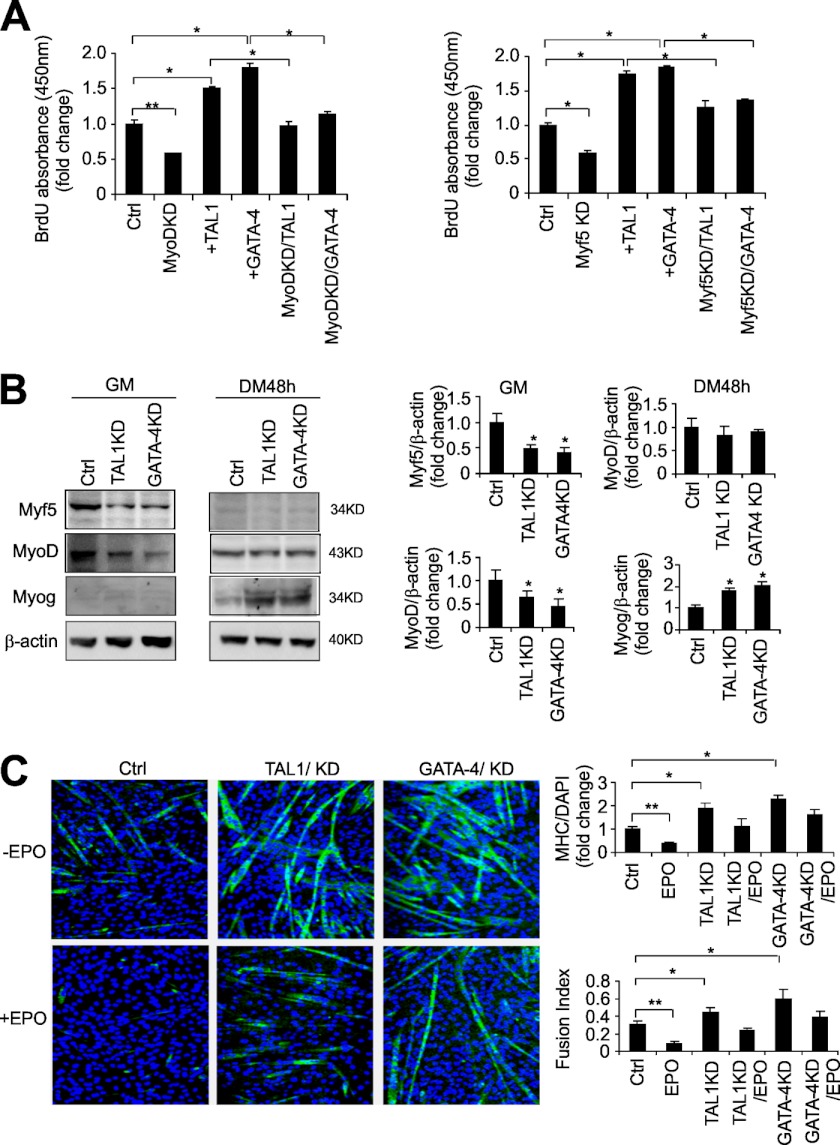
We investigated the activity of GATA-4 and TAL1 in myoblasts by overexpression and knockdown of the respective transcription factors (supplemental Fig. S1, D and E). Transfection of a GATA-4 expression vector into C2C12 cells promoted myoblast proliferation as shown in the increased BrdU absorbance (Fig. 2A) and cell number (supplemental Fig. S1F). The increased proliferation obtained with EPO treatment was further enhanced with increased GATA-4 expression, suggesting a synergistic effect between EPO stimulation and overexpression of GATA-4 (supplemental Fig. S1, F and G). Similarly, overexpression of TAL1 increased myoblast proliferation and the combination of EPO treatment and increased expression of TAL1 further increased proliferation compared with EPO treatment alone as shown by increased BrdU incorporation (Fig. 2A) and cell number (supplemental Fig. S1, F and G). Also, increased EpoR expression was observed with the overexpression of TAL1 and GATA-4 (supplemental Fig. S1H).
FIGURE 2.
GATA-4 and TAL1 increase myoblast proliferation, change MRF expression patterns in C2C12 myoblasts, and delay myogenic differentiation. A, cell proliferation was determined by BrdU cell uptake in C2C12 myoblasts with Myf5 and MyoD knockdown and TAL1, GATA-4 overexpression. B, changes in Myf5, MyoD, and myogenin protein in control (Ctrl) and knockdown (KD) of GATA-4 and TAL1 in C2C12 cells cultured in growth medium (GM) and differentiation medium (DM) for 48 h was determined by Western blotting. C, MHC staining, quantification, and fusion index measurement in control (Ctrl), TAL1, and GATA-4 knockdown (KD) without or with EPO treatment in C2C12 myoblasts cultured in differentiated medium for 36 h. For quantification, the mean values from 3 experiments are shown. Error bars represent S.D.; * indicates p < 0.05 and ** indicates p < 0.01.
EPO stimulation increases Myf5 and MyoD. To examine a possible regulation by GATA-4 and TAL1 on expression of MRFs, we used siRNA to knockdown GATA-4 or TAL1 expression (supplemental Fig. S1, D and E). We observed a decrease in Myf5 and MyoD in undifferentiated myoblasts (Fig. 2B) that may be mediated via direct or indirect GATA-4 or TAL1 activity on gene expression of MRFs. In contrast, in differentiating myoblasts, expression of myogenin, which is expressed only at minimal levels in undifferentiated myoblasts, is significantly increased by knockdown of GATA-4 or TAL1 (Fig. 2B). With myoblast differentiation, the decrease expression in Myf5 to a minimal or not detectable level and the level of MyoD are not affected by knockdown of GATA-4 and TAL1 (Fig. 2B). Therefore, knocking down GATA-4 or TAL1 in undifferentiated myoblasts reduced expression of Myf5 and MyoD, factors required for specification of the myoblast lineage and progenitor status in growth medium, whereas in differentiating myoblasts knocking down GATA-4 or TAL1 induces myogenin. These data demonstrate that GATA-4 and TAL1 affect myoblast proliferation/differentiation independent of EPO stimulation.
MyoD and Myf5 have been reported to play a role in regulating myoblast proliferation (35, 36). We knocked down MyoD and Myf5 to determine whether the effect of GATA-4 and TAL1 on cell proliferation is due to the altered MRFs level. Knocking down Myf5 and MyoD (supplemental Fig. S1I) significantly attenuated the effect of GATA-4 and TAL1 on cell proliferation (Fig. 2A), suggesting that GATA-4 and TAL1 increase myoblast proliferation in part through changing MRF expression. These results indicate that GATA-4 and TAL1 are able to alter the programmed expression of the MRF transcription family members usually correlated with muscle myogenesis.
To investigate how myoblast differentiation to myotube formation is affected by GATA-4 and TAL1, we performed immunostaining of MHC in differentiating myoblasts at 36 h. Knocking down GATA-4 increases MHC expression and myotube formation (Fig. 2C). Knocking down TAL1 also increases MHC expression and myotube formation (Fig. 2C). Although EPO inhibits myoblast differentiation (Fig. 2C), both knocking down of GATA-4 and TAL1 impaired the EPO effect on myogenic differentiation (Fig. 2C, bottom panels). These observations are consistent with the increased expression of myogenin in differentiating myoblasts with knockdown of GATA-4 or TAL1 and indicate that decreasing GATA-4 and TAL1 promotes myogenic differentiation and that both GATA-4 and TAL1 may mediate EPO effect.
GATA-4 and TAL1 Stimulate STAT-5 Activation and Cell Cycle Progression
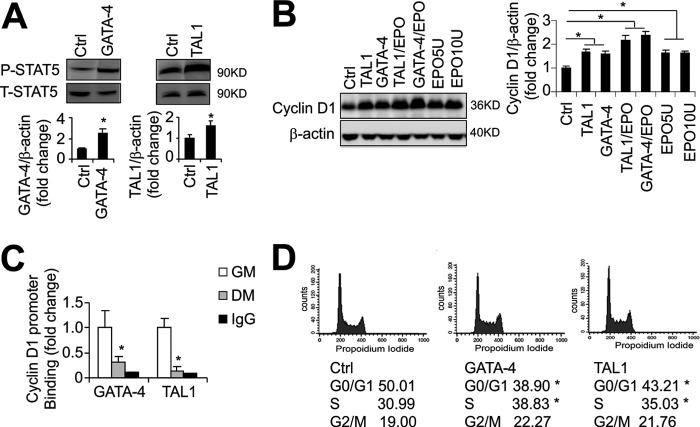
We previously demonstrated STAT-5 activation with EPO stimulation in myoblasts (13). Here, we found that in addition to increased proliferation of myoblasts with overexpression of GATA-4, STAT-5 phosphorylation is also increased in the absence of EPO treatment (Fig. 3A). Similarly, overexpression of TAL1 in undifferentiated myoblasts increases proliferation and STAT-5 phosphorylation (Fig. 3A). These data provide further evidence that myoblast induction of GATA-4 and TAL1 mediates in part the EPO response.
FIGURE 3.
GATA-4 and TAL1 retard myogenic differentiation, increase EPO signal, and facilitate cell cycle transition. A, STAT5 phosphorylation (p-STAT5) level relative to total STAT5 (T-STAT5) was determined in C2C12 cells with GATA-4 and TAL1 overexpression as indicated using Western blotting. B, Cyclin D1 protein level in C2C12 myoblasts was determined by Western blotting (relative to Control) and quantified, with overexpression of TAL1 (T1), without and with EPO treatment (5 units/ml), with overexpression of GATA-4 (G4), without and with EPO treatment, and with EPO treatment at 5 (E5) and 10 units/ml (E10). Cyclin D1 quantification was normalized to β-actin and indicated as fold-change relative to control culture. C, ChIP was used to analyze the GATA-4 and TAL1 binding at the Cyclin D1 promoter region during C2C12 differentiation. D, cell cycle was determined by flow cytometry for C2C12 cells, cells with overexpression of GATA-4 and cells with overexpression of TAL1 as indicated. The percentage of cells in G0/G1, S, and G2/M were quantified, and are representative of 3 independent experiments. The mean values from 3 experiments are shown. Error bars represent S.D.; * indicates p < 0.05.
Cell cycle transition has been reported to be involved in myoblast proliferation (37). To determine whether cell cycle transition was involved in EPO response as well as the effect of GATA-4 and TAL1 on myoblast proliferation, we examined expression of the cell cycle G1/S transition factor, Cyclin D1. During myogenic differentiation, proliferation decreases and Cyclin D1 expression is reduced (supplemental Fig. S1J), and myoblasts fuse to form myotubes. We observed that myoblasts stimulated with EPO or subjected to forced expression of GATA-4 and TAL1 exhibit a modest increase in Cyclin D1 (Fig. 3B). The increase in Cyclin D1 is further enhanced by the combination of EPO plus overexpression of GATA-4 or EPO plus overexpression of TAL1 (Fig. 3B), analogous to the increase in proliferation with the combination of EPO plus overexpression of GATA-4 or TAL1 (supplemental Fig. S1, F and G). Using ChIP analysis, we also observed that GATA-4 and TAL1 bind to the Cyclin D1 promoter region (Fig. 3C), and with differentiation, the binding decreased, suggesting that GATA-4 and TAL1 regulate Cyclin D1 expression directly to modify cell cycle progression in the myoblasts and with differentiation, GATA-4 and TAL1 decrease binding at the Cyclin D1 promoter concomitant myoblast release from the cell cycle to differentiate. FACS analysis of cell cycle distribution in proliferation media revealed that GATA-4 and TAL1 overexpression modestly facilitate cell cycle progression, especially G1/S cell cycle progression via reduction of the G0/G1 cell percentage by 7–11% and increase of G1/S cell percentage by 4–8% (Fig. 3D). These significant but modest effects of GATA-4 and TAL1 on cell cycle progression suggest that other activities in addition to cell cycle transition contribute to the EPO response to regulate myogenic differentiation.
Sirt1 Contributes to EPO Activity
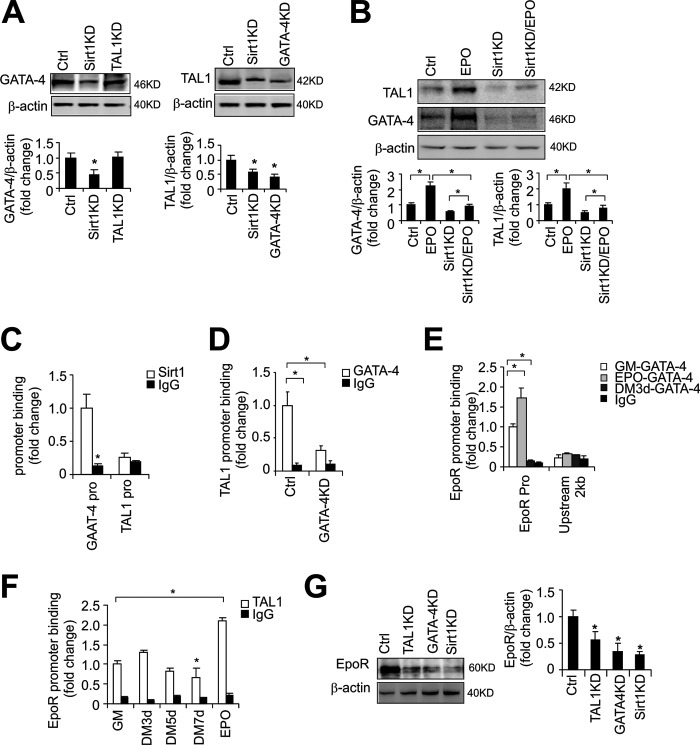
We previously showed that EPO decreases myogenin in C2C12 myoblasts to delay myogenic differentiation (13). Sirt1 inhibits myogenin expression to retard myogenic differentiation. To investigate a possible relationship between EPO stimulation and Sirt1 activity in myogenic differentiation, we examined the effect of EPO on Sirt1 expression. We found that EPO treatment in C2C12 cells up-regulated the Sirt1 protein level (Fig. 4A) whereas knockdown of GATA-4 or TAL1 in C2C12 cells did not significantly affect the Sirt1 level (Fig. 4A), indicating that EPO induction of Sirt1 is independent of EPO stimulation of and upstream of GATA-4 or TAL1 expression.
FIGURE 4.
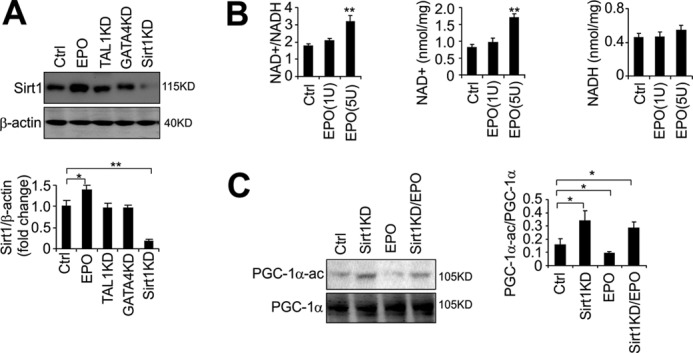
EPO increases the NAD+/NADH ratio and Sirt1 activity. A, Sirt1 protein level in C2C12 myoblasts was determined by Western blotting in C2C12 control cells (Ctrl), or with EPO stimulation (5units/ml), Sirt1 knockdown (KD), TAL1 knockdown, and GATA-4 knockdown. B, NAD+/NADH ratio, NAD+, and NADH levels were determined in C2C12 myoblasts. C2C12 myoblasts were treated with different dosages of EPO (1 and 5 units/ml) or PBS (Ctrl) as indicated. C, Western blots and quantification of acetylated PGC-1α (PGC-1α-ac) in C2C12 myoblasts. PGC-1α was overexpressed in C2C12 myoblasts with EPO treatment (5 units/ml) or PBS treatment with or without Sirt1 knockdown. PGC-1α was immunoprecipitated and blotted with acetylated lysine antibody. T-PGC-1α indicates total PGC-1α. All data are averages of three independent experiments. *, p < 0.05; **, p < 0.01.
Sirt1 is an NAD+-dependent histone deacetylase that can sense an elevated NAD+/NADH ratio to deacetylate its targets (8). We found that EPO stimulation increases the cellular NAD+/NADH ratio significantly (p ≤ 0.01) at 5 units/ml (Fig. 4B). This change was also reflected in the cellular NAD+ level. The NAD+/NADH ratio decreases as muscle cells differentiate (38), it is also possible that EPO signaling negatively regulates myogenic differentiation through regulating cellular NAD+/NADH redox status. To assess Sirt1 activity, we monitored the acetylation status of PGC-1α, a factor reported to be deactylated uniquely by Sirt1 (39). We overexpressed PGC-1α in C2C12 myoblasts to increase the PGC-1α protein level (supplemental Fig. S2A) to increase our ability to detect changes in PGC-1α acetylation. Knocking down Sirt1 (Fig. 4A) increased the extent of PGC-1α acetylation (Fig. 4C). Conversely, EPO treatment decreased PGC-1α acetylation. However, knocking down Sirt1 abolished the decrease in PGC-1α acetylation by EPO (Fig. 4C), suggesting that EPO action on PGC-1α is through increasing Sirt1 activity in myoblasts. We also determined another Sirt1 target, P53 acetylation, and Sirt1 knockdown impaired the EPO effect on decreased P53 acetylation (supplemental Fig. S2B).
Sirt1 Activity Is Involved in EPO Effect of Retarded Myogenic Differentiation
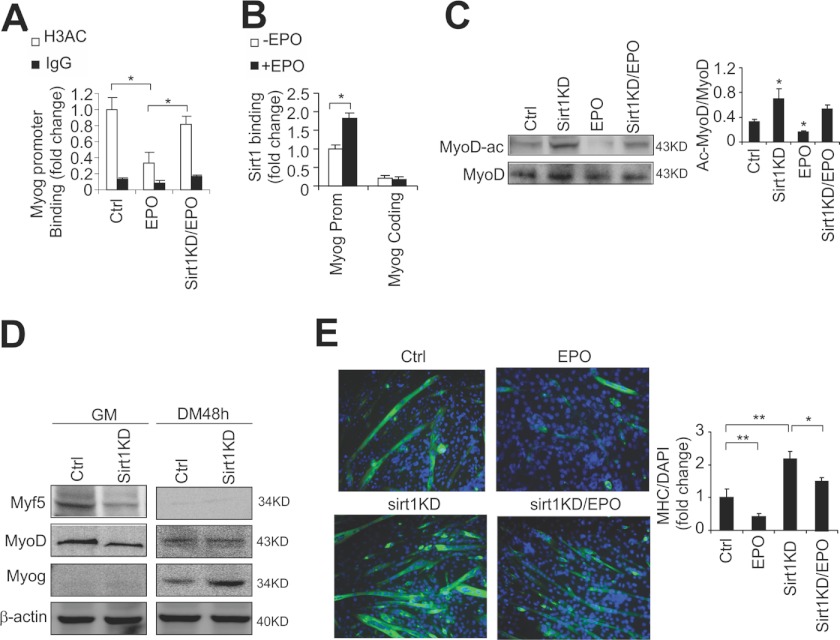
EPO inhibition of myogenin expression is also reflected in changes in histone 3 acetylation (H3AC) in the myogenin promoter that includes a MyoD E-box binding region for activation of myogenin expression. Chromatin regions permissive for gene expression are associated with enriched H3AC. In differentiating C2C12, EPO-treated (48 h) cells significantly decreased H3AC associated with the myogenin promoter (Fig. 5A). Conversely, knocking down Sirt1 reversed the decrease by EPO in H3AC associated with the myogenin promoter (Fig. 5A). It has been proposed that Sirt1 can associate with and deacetylate both PCAF and MyoD and together they form a complex to inhibit muscle gene expression by histone hypoacetylation (10). We determined that Sirt1 bound to the myogenin promoter preferentially compared with the coding region, and that Sirt1 binding to the myogenin promoter was significantly increased with EPO treatment (Fig. 5B). Furthermore, we speculate that EPO can increase Sirt1 expression and, importantly, induce Sirt1 deacetylation enzymatic activity to deacetylate MyoD, which also represses MyoD transcriptional activity of myogenin in differentiated myoblasts. Indeed, we found that MyoD acetylation was decreased with EPO treatment (Fig. 5C). Conversely, knocking down Sirt1 increased MyoD acetylation and impaired the EPO effect in decreasing of MyoD acetylation (Fig. 5C), suggesting that Sirt1 is involved in EPO response and impairment of myogenic differentiation. Sirt1 knockdown also decreased Myf5 and MyoD in undifferentiated myoblasts but not in differentiating myoblasts (Fig. 5D). In contrast, expression of myogenin, which is expressed only at minimal levels in undifferentiated myoblasts, is significantly increased by knockdown of Sirt1 (Fig. 5D).
FIGURE 5.
EPO retards myogenic differentiation via Sirt1. A, ChIP was used to analyze the myogenin promoter histone 3 acetylation modification in differentiated C2C12 cells with EPO treatment, and EPO treatment combined with Sirt1 knockdown (KD). B, ChIP was used to analyze the Sirt1 binding at the myogenin promoter region in C2C12 cells with or without EPO treatment (5 units/ml). C, Western blot and quantification of acetylated MyoD(Ac-Lys) in C2C12 myoblasts with EPO treatment (5 units/ml) or PBS treatment with or without Sirt1 knockdown. MyoD was immunoprecipitated and blotted with acetylated lysine antibody. D, Myf5, MyoD, and myogenin expression in control (Ctrl) and knocking down of Sirt1 in C2C12 cells cultured in growth medium (GM) and differentiation medium (DM) for 48 h were determined by Western blotting. E, MHC staining and quantification in control (Ctrl) and Sirt1 knockdown in C2C12 myoblasts with or without EPO treatment. All data are averages of three independent experiments. *, p < 0.05; **, p < 0.01.
EPO inhibition of myogenic differentiation is further illustrated by the decrease in MHC staining with EPO treatment in differentiating C2C12 cells for 36 h, whereas knocking down Sirt1 promotes differentiation indicated by increased MHC staining (Fig. 5E). However, knocking down Sirt1 impaired but not completely abolished the EPO effect on decreasing myogenic differentiation, providing further evidence that EPO may impair myogenic differentiation partially through Sirt1 although we cannot exclude the possibility that knocking down Sirt1 is not complete. Taken together, these data suggest that Sirt1 is involved in EPO activity in myoblasts through an elevated NAD+ level and NAD+/NADH ratio to negatively regulate myogenic differentiation.
Relationship of Sirt1, GATA-4, and TAL1 in Myogenic Differentiation
To determine how Sirt1 takes part in EPO signaling in myogenic differentiation, we first performed BrdU analysis and observed that knocking down Sirt1 decreased C2C12 proliferation as did knockdown of GATA-4 and TAL1 (supplemental Fig. S2C). Also knocking down of Sirt1, GATA-4, and TAL1 all attenuated the EPO effect on C2C12 myoblasts (supplemental Fig. S2C). We then detected the expression of GATA factors and TAL1 in C2C12 myoblast after Sirt1 was knocked down. Knocking down Sirt1 decreased GATA-4 and TAL1 expression (Fig. 6A). Furthermore, GATA-4 knockdown also decreased TAL1 expression but TAL1 knockdown had no effect on GATA-4 expression (Fig. 6A), indicating that GATA-4 exerts its role upstream of TAL1, and that Sirt1 functions upstream of both GATA-4 and TAL1. Of note, GATA-3 levels were not affected by knocking down Sirt1 (supplemental Fig. S2D). However, we observed an increased Cyclin D1 expression in C2C12 myoblasts and decreased myogenin expression in C2C12 cultured in differentiation medium (supplemental Fig. S2E), suggesting that GATA-3 may affect myogenic differentiation through cell cycle regulation but not Sirt1 activity. Also, the effect of EPO on GATA-4 and TAL1 induction was attenuated but not completely abolished by Sirt1 knockdown, suggesting EPO regulates GATA-4 and TAL1 expression in part through Sirt1 (Fig. 6B).
FIGURE 6.
Sirt1 regulates GATA-4, TAL1, and EpoR, and GATA-4 regulates TAL1 and EpoR expression. A, GATA-4 and TAL1 protein levels in C2C12 myoblasts were determined by Western blotting in C2C12 control cells (Ctrl) without and with TAL1 knockdown (KD), GATA-4 knockdown, and Sirt1 knockdown. B, C2C12 cells with Sirt1 knockdown and EPO treatment were differentiated for 24 h and GATA-4 and TAL1 protein level were determined by Western blotting. C, ChIP was used to analyze the Sirt1 binding at the GATA-4 and TAL1 promoter regions in C2C12 myoblasts. D, ChIP was used to analyze the GATA-4 binding at TAL1 promoter region in C2C12 myoblasts. E and F, ChIP was used to analyze the binding status of GATA-4 (E) and TAL1 (F) at the EpoR promoter in C2C12 cells cultured in growth media (GM) or differentiation media (DM) as indicated. The ChIP data are normalized to results from Input and then expressed as fold-change relative to control without EPO. G, EpoR protein levels in C2C12 myoblasts were determined by Western blotting in C2C12 control cells or with TAL1 knockdown, GATA-4 knockdown, or Sirt1 knockdown.
To determine whether Sirt1 directly regulates GATA-4 and TAL1 expression, we performed ChIP analysis using anti-Sirt1 antibody. Sirt1 binds to the GATA-4 promoter but not the TAL1 promoter (Fig. 6C). However, GATA-4 was observed to bind to the TAL1 promoter (Fig. 6D). Taken together, these data suggest that Sirt1 directly regulates GATA-4 expression and GATA-4 regulates TAL1 expression by binding to the TAL1 promoter. ChIP analysis also showed that GATA-4 (Fig. 6E) and TAL1 (Fig. 6F) both can bind to the EPO receptor (EpoR) promoter region, and with EPO stimulation, the binding of GATA-4 and TAL1 at the EpoR promoter was increased (Fig. 6, E and F). Hence, as observed in erythroid progenitor cells, EPO regulates its own promoter in myoblasts and induction of EpoR can increase EPO sensitivity, whereas down-regulation of EpoR with terminal differentiation abrogates EPO response. It is notable that GATA-4 binding at EpoR decreased with myoblast differentiation (Fig. 6E), but TAL1 binding at the EpoR promoter did not decrease until the final differentiation at 7 days (Fig. 6F). Considering that TAL1 may also recruit some repressors to negatively regulate target gene expression (40) and that EpoR expression declined with myoblast differentiation, it is possible that with myoblast differentiation TAL1 may recruit some co-repressors to inhibit EpoR expression. Although knocking down Sirt1, GATA-4, or TAL1 all decreased EpoR expression (Fig. 6G), Sirt1 did not show any binding at the EpoR promoter region. Therefore, GATA-4 and TAL1 both may regulate EpoR expression that is required for EPO response by directly binding to the EpoR promoter. Although Sirt1 affects EpoR expression, it is likely that Sirt1 regulates EpoR via indirect mechanisms such as changing the cellular environment or through regulating GATA-4 and TAL1 factors.
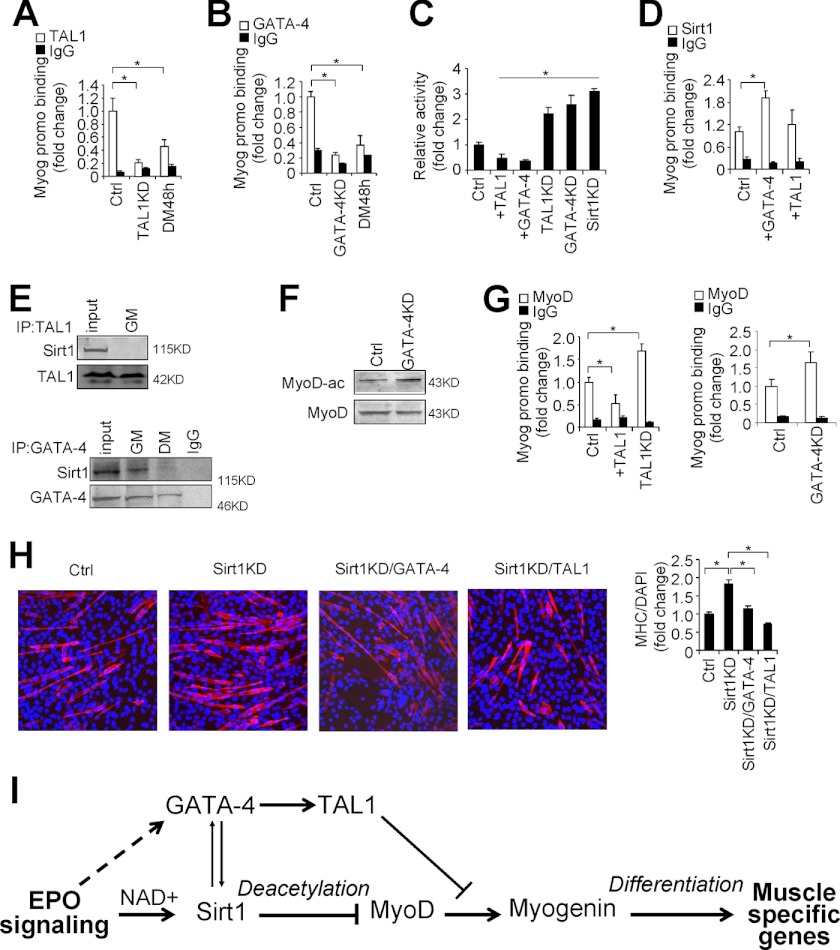
Sirt1 Interacts with GATA-4 to Regulate Myogenin Expression, and TAL1 Represses MyoD Binding to the Myogenin Promoter
We examined transcription factor binding to the myogenin promoter to determine the extent that Sirt1, GATA-4, and TAL1 regulate myoblast differentiation via changing myogenin expression. ChIP analysis demonstrated that TAL1 and GATA-4 bound directly to the myogenin promoter region containing a MEF2 binding site and E-box site (Fig. 7, A and B, and supplemental Fig. S2F). Knocking down TAL1 and GATA-4 decreased binding of these two factors to the myogenin promoter. Sirt1 has been shown to bind to the myogenin promoter to repress myogenin expression (Fig. 7, A and B). Consistently, the myogenin promoter activity was reduced by overexpression of TAL1 and GATA-4, but elevated by knocking down of TAL1, GATA-4, and Sirt1 (Fig. 7C), suggesting an inhibitory effect of TAL1, GATA-4, and Sirt1 on the myogenin promoter. To determine whether GATA-4 and TAL1 facilitate Sirt1 binding to the myogenin promoter, we overexpressed GATA-4 and TAL1 in the C2C12 myoblast and found that only increased GATA-4 but not TAL1 increased Sirt1 binding at the myogenin promoter region (Fig. 7D). Furthermore, immunoprecipitation analysis suggested a direct interaction between Sirt1 and GATA-4 but not between Sirt1 and TAL1 (Fig. 7E). Also an increased MyoD acetylation was observed with GATA-4 knocked down (Fig. 7F), supporting the hypothesis that GATA-4 can target Sirt1 to the myogenin promoter to repress myogenin expression.
FIGURE 7.
GATA-4 interacts with and targets Sirt1 to the myogenin promoter and TAL1 inhibits MyoD binding to the myogenin promoter. A and B, ChIP was used to analyze TAL1 (A) and GATA-4 (B) binding at the myogenin promoter in C2C12 cells and C2C12 cells with TAL1 knockdown (KD) and GATA-4 knockdown and differentiated myoblasts (48 h). C, relative myogenin promoter activity was determined using the luciferase reporter gene assay system in C2C12 with overexpression of TAL1 and GATA-4 and knocking down of TAL1, GATA-4, and Sirt1. D, C2C12 myoblast were differentiated and ChIP was used to analyze Sirt1 binding at the myogenin promoter in C2C12 cells and C2C12 cells with GATA-4 (+GATA-4) and TAL1 (+TAL1) overexpression. E, C2C12 cell nuclear extract was immunoprecipitated with GATA-4- and TAL1-specific antibodies and analyzed by Western blotting for GATA-4, Sirt1, and TAL1. IgG was used for nonspecific binding. F, Western blot of acetylated MyoD(Ac-Lys) in C2C12 myoblasts with GATA-4 knockdown. MyoD was immunoprecipitated and blotted with acetylated lysine antibody. G, C2C12 cells with TAL1 overexpression (+TAL1), TAL1 knockdown, and GATA-4 knockdown were differentiated for 48 h and ChIP was used to analyze MyoD binding at the myogenin promoter region. All ChIP data are normalized to results from Input and then expressed as fold-change relative to control (Ctrl). H, C2C12 myoblasts were differentiated for 36 h and MHC staining and quantification were checked in control, and Sirt1 knockdown without and with TAL1 (+TAL1) and GATA-4 (+GATA-4) overexpression. I, schematic diagram showing EPO activation of Sirt1 by increasing the cellular NAD+/NADH ratio, induction of GATA-4 and TAL1 expression. The pathway converges on regulation of myogenin expression, and, ultimately, on myogenic differentiation. A known pathway is that Sirt1 deacetylates MyoD and inhibits myogenin expression and myogenic differentiation (heavy lines). A predicted pathway is that EPO increases Sirt1 activity, Sirt1 directly regulates GATA-4 expression, and GATA-4 targets Sirt1 to the myogenin promoter (thin lines). TAL1 can be regulated by GATA-4 directly; however, TAL1 inhibits myogenic differentiation via repressing MyoD binding to the myogenin promoter (thin lines). EPO may also regulate GATA-4 expression through a Sirt1 independent way (dashed line). The mean values from 3 experiments are shown. Error bars represent S.D.; * indicates p < 0.05.
Although TAL1 overexpression did not increase Sirt1 binding to the myogenin promoter (Fig. 7D), we found that increased TAL1 decreased MyoD binding to the myogenin promoter and knocking down TAL1 enhanced the binding of MyoD to the myogenin promoter (Fig. 7G), suggesting that TAL1 inhibits myogenin expression possibly by blocking MyoD binding to the myogenin promoter and, therefore, inhibits MyoD transcription activation of myogenin expression, which is required for myogenic differentiation. Also, MyoD binding at the myogenin promoter was increased when GATA-4 was knocked down (Fig. 7G, right panel). Although it is possible that knocking down GATA-4 reduced TAL1 expression resulting in decreased inhibition of MyoD binding at the myogenin promoter, we cannot exclude that GATA-4 may inhibit MyoD binding directly. Finally, we found that increased myogenic differentiation by Sirt1 knockdown was blocked by overexpression of GATA-4 and TAL1 (Fig. 7H), providing further evidence that whereas Sirt1 can directly or indirectly increase GATA-4 and TAL1 expression, GATA-4 and TAL1 can also act downstream and independent of Sirt1 to block myoblast differentiation. Taken together, our data suggest the following hypothesis for EPO response during myogenic differentiation (Fig. 7I): EPO signaling increases Sirt1 activity via the NAD+ pathway to increase MyoD deacetylation to inhibit myogenin expression and myogenic differentiation. Sirt1 activity also regulates GATA-4 expression and increased GATA-4 is able to target more Sirt1 to the myogenin promoter to inhibit its expression and myogenic differentiation. Although TAL1 is not involved in targeting Sirt1 to the myogenin promoter, increased GATA-4 by Sirt1 regulates TAL1 expression that is able to inhibit MyoD binding to the myogenin promoter to decrease myogenin expression and myogenic differentiation, providing an additional possible mechanism by which GATA-4 and TAL1 can inhibit myogenic differentiation independent of Sirt1.
DISCUSSION
Satellite cells or muscle progenitor cells are involved in the growth and regeneration of skeletal muscle (41). We previously demonstrated that EPO stimulates myoblast proliferation and delays differentiation in culture and contributes to myoblast survival in vivo (13, 16). EPO exerts its effect by binding to its cell surface receptor, EpoR, and the extent of EpoR expression determines EPO response. Here, we observed for the first time that EpoR expression in primary human muscle precursor cells is comparable with that on primary human hematopoietic/erythroid progenitor cells, providing support for a potential role of EPO signaling in primary myoblasts. We determined that EPO promoted expression of GATA-4 and TAL1 in addition to Myf5 and MyoD in primary human muscle precursor cells and in murine C2C12 myoblasts. Overexpression of GATA-4 or TAL1 promotes myoblast proliferation and knocking down GATA4 or TAL1 increased myogenic differentiation. EPO signaling was also found to be involved in Sirt1 activity through increasing the NAD+ level and NAD+/NADH ratio and elevating Sirt1 expression to retard myogenic differentiation, linking the EPO response to Sirt1 regulation of myoblast differentiation and metabolism (42). Furthermore, GATA-4 was demonstrated to be involved in the Sirt1 effect on myogenic differentiation through interacting and targeting Sirt1 to the promoter of the myogenin gene that is required for Sirt1 activity to inhibit myogenic differentiation. Our data also indicates that whereas Sirt1 is required for EPO induction of GATA-4 and TAL1, both GATA-4 and TAL1 can inhibit myogenic differentiation independently of Sirt1. For example, TAL1 plays an inhibition role in myogenic differentiation by repressing MyoD binding to the myogenin promoter and GATA-4 can induce TAL1.
In cardiomyocytes GATA-4 regulates cell cycle genes such as Cyclin D2 and Cyclin A2 to affect proliferation and cell cycle (43). TAL1 in myeloid precursor cells regulates cell proliferation and entry into and traversal through the cell cycle (44). Here, we showed that either GATA-4 or TAL1 overexpression stimulates proliferation of myoblasts and their induction by EPO stimulation provides an explanation for the resultant proliferative EPO response. We also demonstrated that decreased MyoD and Myf5 impaired the effect of GATA-4 and TAL1 on increased cell proliferation, suggesting that the changes in MRFs expression contribute to the effect of GATA-4 and TAL1 on myoblast proliferation. This is consistent with the reported role of Myf5 and MyoD in regulating myoblast proliferation (35, 36). Also, whereas we showed that GATA-4 associates with the myogenin promoter, further experiments are needed to address if the GATA-4 and TAL1 effects on the other MRFs are direct or indirect. The increased proliferation by TAL1 and GATA-4 could also result in part from regulation of the cell cycle. Cyclin D1 is required for progression of the cell cycle from the G1 to S phase (45). Skeletal myoblasts from insulin-like growth factor-1 expressing transgenic mice exhibit prolonged proliferative reserves and increased Cyclin D1 with enhanced G1 to S phase cell cycle progression and blocking insulin-like growth factor-1 signaling caused a G1/S cell cycle arrest and induced a senescent-like state (37). Similarly, we observed that skeletal myoblasts overexpressing EpoR increase proliferation (16), whereas myoblasts from mice with restricted EpoR expression exhibit decreased proliferative capacity (17). Our observations of increased Cyclin D1 by TAL1 and GATA-4 overexpression and direct binding of GATA-4 and TAL1 at Cyclin D1 promoter suggest that TAL1 and GATA-4 may also facilitate the G1 to S phase cell cycle progression via regulating cell cycle molecules to affect skeletal muscle cell proliferation and differentiation partially. However, the modest change of Cyclin D1 and cell cycle progression raises the possibility that the cell cycle may not be the only EPO-regulated activity during myogenic differentiation.
In mammals, Sirt1 has been reported to take part in different metabolic processes. For example, Sirt1 can repress peroxisome proliferator-activated receptor-γ expression and increase lipolytic rates in white fat tissue (46), and Sirt1 can deacetylate PGC-1α and activate PGC-1α to promote mitochondrial fatty acid oxidation and hepatic glucose output (39, 47). Sirt1 was also found to interact with pCAF and GCN5 to prevent their acetylation effect on MyoD, which was shown to inhibit skeletal muscle differentiation (10, 48). First, we report that EPO can influence Sirt1 activity through changing the cellular NAD+ level and NAD+/NADH ratio and by elevating Sirt1 expression. It is possible that EPO increases cell oxidative capacity to increase NADH oxidation leading to an increased NAD level and NAD+/NADH ratio. It is also likely that EPO increases NAD production to increase the NAD+/NADH ratio. Interestingly, the NAD+/NADH ratio decreases as muscle cells differentiate (38), and EPO signaling may negatively regulate myogenic differentiation through regulating the cellular NAD+/NADH redox status. Sirt1 can deacetylate MyoD to inhibit MyoD transcription activation of myogenin, which is NAD+ dependent (10), raising the possibility that this process is also involved in EPO action. Our data demonstrate that EPO treatment can increase PGC-1α deacetylation, a marker of Sirt1 enzymatic activity, suggesting EPO impacts on the Sirt1 enzymatic deacetylation activity. Importantly, EPO treatment increased MyoD deacetylation and this effect was decreased when Sirt1 was knocked down, suggesting that EPO exerts its function through Sirt1 to affect myogenic differentiation. Our data further confirmed this view. EPO treatment increased Sirt1 binding at the promoter of the MyoD target gene, myogenin. EPO also decreased H3AC at the myogenin promoter, whereas Sirt1 knockdown impaired the effect of EPO on myogenin promoter deacetylation. Finally, the decreased MHC staining by EPO was reversed by Sirt1 knockdown, providing further evidence that Sirt1 activity is involved the role of EPO in retarding myogenic differentiation. Interestingly, although TAL1 and GATA-4 can decrease myogenin expression, no effect was found on Sirt1 expression with TAL1 or GATA-4 knockdown. However, Sirt1 knockdown decreased GATA-4 and TAL1 expression, suggesting an upstream function of Sirt1 on GATA-4 and TAL1. Furthermore, GATA-4 knockdown decreases TAL1 expression, indicating that EPO can stimulate Sirt1 activity to increase GATA-4 expression and that increased GATA-4 can induce TAL1 expression, and collectively these effects contribute to inhibition of myogenic differentiation.
In Sirt1 deleted mouse embryonic stem cells, GATA-4 was found to be regulated by Sirt1 and GATA-4 expression was decreased (30, 49). Here we demonstrated that Sirt1 directly binds to the GATA-4 promoter to regulate GATA-4 expression in myoblasts. It is possible that Sirt1 deacetylates some inhibitors of GATA-4 expression to active GATA-4 expression. Although we did not find Sirt1 association with the TAL1 promoter, Sirt1 knockdown decreased TAL1 expression. Also GATA-4 directly binds to the TAL1 promoter and GATA-4 knockdown decreased TAL1 expression, suggesting that GATA-4 specifically regulates TAL1 in skeletal myoblasts and that Sirt1 may regulate TAL1 expression through GATA-4. TAL1 regulation by GATA family members has been observed in hematopoietic and other cell types. TAL1 is regulated by GATA-1 and GATA-2 in hematopoietic lineages (50, 51) and TAL1 expression is also GATA factor dependent in the central nervous system (52). It is notable that GATA-4 and TAL1 were both found to bind at the myogenin promoter, and that GATA-4 interacts with Sirt1 and targets Sirt1 to the myogenin promoter to inhibit the myogenic required myogenin gene expression. On the other hand, overexpression of TAL1 negatively regulates MyoD binding to the myogenin promoter, suggesting that TAL1 is not directly involved in GATA-4 inhibition of myogenin, but rather inhibits MyoD activation of myogenin by competing with MyoD for binding to the myogenin promoter. As a member of the bHLH family, the E-box binding activity of TAL1 is thought to be critical for either repression or activation of target genes during normal and malignant hematopoiesis (53, 54). The binding region of TAL1 on the myogenin promoter includes one E-box for MyoD binding that contributes to myogenin promoter activation. Our data indicate that TAL1 may share the same E-box sequence as MyoD on the myogenin promoter as an overlapped binding site to inhibit myogenin promoter activity.
Although Sirt1, GATA-4, and TAL1 may regulate myogenic differentiation via suppression of myogenin expression, Sirt1, GATA-4, and TAL1 positively regulate EpoR expression. GATA-4 and TAL1, but not Sirt1, bind directly to the EpoR promoter and increase expression during myoblast proliferation. GATA-4 association with the EpoR promoter decreased, concomitant with decrease in the level of GATA-4, whereas TAL1 binding was found to persist during myoblast differentiation. In erythroid cells, TAL1 acts as repressor and activator through association with different co-repressors or co-activators during erythroid differentiation (55). Our data suggest that TAL1 represses myogenin expression but activates EpoR expression before myogenic differentiation. It is possible that TAL1 associates with different cofactors to exert its dual function in myoblast.
Transcription factor response suggests that EPO increases Sirt1 and downstream induction of GATA-4 and TAL1 and alters expression of the MRF program to promote proliferation and inhibit differentiation and myotube fusion. In addition, EPO increases the NAD+ level and NAD+/NADH ratio that increases Sirt1 activity reflected in reduced PGC-1α acetylation and reduced histone 3 acetylation associated with the myogenin promoter during myoblast differentiation. Overexpression and knockdown of GATA-4 or TAL1 showed that these transcription factors can affect myoblast differentiation independent of EPO and that overexpression of GATA-4 or TAL1 can overcome the ability of Sirt1 knockdown to promote myoblast differentiation. GATA-4, which was induced by Sirt1, interacted with the myogenin promoter to increase Sirt1 binding, whereas TAL1, which was induced by GATA-4, associated with the myogenin promoter and inhibited MyoD binding and activation. GATA-4 and TAL1 activities were mediated in part by MRF expression, but they also associated with the Cyclin D1 promoter and modified Cyclin D1 expression. Beyond MRF expression, we found that Sirt1, GATA-4, and TAL1 increased EpoR expression mediated via direct binding to the EpoR promoter region by GATA-4 and TAL1. Conversely, knocking down Sirt1, GATA-4, or TAL1 decreased EpoR expression and decreased myoblast response to EPO. These data provide evidence for GATA-4 and TAL1 to affect myoblast differentiation via cross-talk with and induction by Sirt1 and suggest a new mechanism for EPO inhibition of myoblast differentiation.
Supplementary Material
Acknowledgments
We thank Dr. Alan Schechter for helpful discussions. We also thank Dr. Vittorio Sartorelli for the Myogenin-promoter-Luc reporter vector.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program of the NIDDK.

This article contains supplemental Figs. S1 and S2 and Table S1.
- MRF
- myogenic regulatory factor
- bHLH
- basic helix-loop-helix
- EPO
- erythropoietin
- TAL1
- T-cell acute leukemia 1
- HMPC
- human muscle primary cell
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- H3AC
- histone 3 acetylation
- PGC-1α
- peroxisome proliferator-activated receptor γ-coactivator 1α.
REFERENCES
- 1. Weintraub H., Davis R., Tapscott S., Thayer M., Krause M., Benezra R., Blackwell T. K., Turner D., Rupp R., Hollenberg S. (1991) The myoD gene family. Nodal point during specification of the muscle cell lineage. Science 251, 761–766 [DOI] [PubMed] [Google Scholar]
- 2. Rudnicki M. A., Schnegelsberg P. N., Stead R. H., Braun T., Arnold H. H., Jaenisch R. (1993) MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75, 1351–1359 [DOI] [PubMed] [Google Scholar]
- 3. Nabeshima Y., Hanaoka K., Hayasaka M., Esumi E., Li S., Nonaka I., Nabeshima Y. (1993) Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature 364, 532–535 [DOI] [PubMed] [Google Scholar]
- 4. Lu J., McKinsey T. A., Zhang C. L., Olson E. N. (2000) Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell 6, 233–244 [DOI] [PubMed] [Google Scholar]
- 5. Mal A., Sturniolo M., Schiltz R. L., Ghosh M. K., Harter M. L. (2001) A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD. Inhibition of the myogenic program. EMBO J. 20, 1739–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Puri P. L., Iezzi S., Stiegler P., Chen T. T., Schiltz R. L., Muscat G. E., Giordano A., Kedes L., Wang J. Y., Sartorelli V. (2001) Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol. Cell 8, 885–897 [DOI] [PubMed] [Google Scholar]
- 7. Landry J., Sutton A., Tafrov S. T., Heller R. C., Stebbins J., Pillus L., Sternglanz R. (2000) The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. U.S.A. 97, 5807–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Imai S., Armstrong C. M., Kaeberlein M., Guarente L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 [DOI] [PubMed] [Google Scholar]
- 9. Michan S., Sinclair D. (2007) Sirtuins in mammals. Insights into their biological function. Biochem. J. 404, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fulco M., Schiltz R. L., Iezzi S., King M. T., Zhao P., Kashiwaya Y., Hoffman E., Veech R. L., Sartorelli V. (2003) Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell 12, 51–62 [DOI] [PubMed] [Google Scholar]
- 11. Lin C. S., Lim S. K., D'Agati V., Costantini F. (1996) Differential effects of an erythropoietin receptor gene disruption on primitive and definitive erythropoiesis. Genes Dev. 10, 154–164 [DOI] [PubMed] [Google Scholar]
- 12. Wu H., Liu X., Jaenisch R., Lodish H. F. (1995) Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell 83, 59–67 [DOI] [PubMed] [Google Scholar]
- 13. Ogilvie M., Yu X., Nicolas-Metral V., Pulido S. M., Liu C., Ruegg U. T., Noguchi C. T. (2000) Erythropoietin stimulates proliferation and interferes with differentiation of myoblasts. J. Biol. Chem. 275, 39754–39761 [DOI] [PubMed] [Google Scholar]
- 14. Anagnostou A., Liu Z., Steiner M., Chin K., Lee E. S., Kessimian N., Noguchi C. T. (1994) Erythropoietin receptor mRNA expression in human endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 91, 3974–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morishita E., Masuda S., Nagao M., Yasuda Y., Sasaki R. (1997) Erythropoietin receptor is expressed in rat hippocampal and cerebral cortical neurons, and erythropoietin prevents in vitro glutamate-induced neuronal death. Neuroscience 76, 105–116 [DOI] [PubMed] [Google Scholar]
- 16. Jia Y., Warin R., Yu X., Epstein R., Noguchi C. T. (2009) Erythropoietin signaling promotes transplanted progenitor cell survival. Faseb J. 23, 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jia Y., Suzuki N., Yamamoto M., Gassmann M., Noguchi C. T. (2012) Endogenous erythropoietin signaling facilitates skeletal muscle repair and recovery following pharmacologically induced damage. FASEB J. 26, 2847–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hojman P., Brolin C., Gissel H., Brandt C., Zerahn B., Pedersen B. K., Gehl J. (2009) Erythropoietin overexpression protects against diet-induced obesity in mice through increased fat oxidation in muscles. PLoS One 4, e5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ravet E., Reynaud D., Titeux M., Izac B., Fichelson S., Roméo P. H., Dubart-Kupperschmitt A., Pflumio F. (2004) Characterization of DNA-binding-dependent and -independent functions of SCL/TAL1 during human erythropoiesis. Blood 103, 3326–3335 [DOI] [PubMed] [Google Scholar]
- 20. Kassouf M. T., Chagraoui H., Vyas P., Porcher C. (2008) Differential use of SCL/TAL-1 DNA-binding domain in developmental hematopoiesis. Blood 112, 1056–1067 [DOI] [PubMed] [Google Scholar]
- 21. Rogers H. M., Yu X., Wen J., Smith R., Fibach E., Noguchi C. T. (2008) Hypoxia alters progression of the erythroid program. Exp. Hematol. 36, 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Molkentin J. D., Lin Q., Duncan S. A., Olson E. N. (1997) Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 11, 1061–1072 [DOI] [PubMed] [Google Scholar]
- 23. Kuo C. T., Morrisey E. E., Anandappa R., Sigrist K., Lu M. M., Parmacek M. S., Soudais C., Leiden J. M. (1997) GATA-4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 11, 1048–1060 [DOI] [PubMed] [Google Scholar]
- 24. Suzuki Y. J., Evans T. (2004) Regulation of cardiac myocyte apoptosis by the GATA-4 transcription factor. Life Sci. 74, 1829–1838 [DOI] [PubMed] [Google Scholar]
- 25. Xin M., Davis C. A., Molkentin J. D., Lien C. L., Duncan S. A., Richardson J. A., Olson E. N. (2006) A threshold of GATA4 and GATA6 expression is required for cardiovascular development. Proc. Natl. Acad. Sci. U.S.A. 103, 11189–11194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salisch S., Klar M., Thurisch B., Bungert J., Dame C. (2011) Gata4 and Sp1 regulate expression of the erythropoietin receptor in cardiomyocytes. J. Cell. Mol. Med. 15, 1963–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sumanas S., Gomez G., Zhao Y., Park C., Choi K., Lin S. (2008) Interplay among Etsrp/ER71, Scl, and Alk8 signaling controls endothelial and myeloid cell formation. Blood 111, 4500–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ema M., Faloon P., Zhang W. J., Hirashima M., Reid T., Stanford W. L., Orkin S., Choi K., Rossant J. (2003) Combinatorial effects of Flk1 and Tal1 on vascular and hematopoietic development in the mouse. Genes Dev. 17, 380–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bussmann J., Bakkers J., Schulte-Merker S. (2007) Early endocardial morphogenesis requires Scl/Tal1. PLoS Genet. 3, e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiang C., Qu A., Matsubara T., Chanturiya T., Jou W., Gavrilova O., Shah Y. M., Gonzalez F. J. (2011) Disruption of hypoxia-inducible factor 1 in adipocytes improves insulin sensitivity and decreases adiposity in high-fat diet-fed mice. Diabetes 60, 2484–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goldfarb A. N., Lewandowska K. (1995) Inhibition of cellular differentiation by the SCL/tal oncoprotein. Transcriptional repression by an Id-like mechanism. Blood 85, 465–471 [PubMed] [Google Scholar]
- 32. Hofmann T. J., Cole M. D. (1996) The TAL1/Scl basic helix-loop-helix protein blocks myogenic differentiation and E-box dependent transactivation. Oncogene 13, 617–624 [PubMed] [Google Scholar]
- 33. Das M., Wilson K., Molnar P., Hickman J. J. (2007) Differentiation of skeletal muscle and integration of myotubes with silicon microstructures using serum-free medium and a synthetic silane substrate. Nat. Protoc. 2, 1795–1801 [DOI] [PubMed] [Google Scholar]
- 34. Wen J., Huang S., Pack S. D., Yu X., Brandt S. J., Noguchi C. T. (2005) Tal1/SCL binding to pericentromeric DNA represses transcription. J. Biol. Chem. 280, 12956–12966 [DOI] [PubMed] [Google Scholar]
- 35. Ustanina S., Carvajal J., Rigby P., Braun T. (2007) The myogenic factor Myf5 supports efficient skeletal muscle regeneration by enabling transient myoblast amplification. Stem Cells 25, 2006–2016 [DOI] [PubMed] [Google Scholar]
- 36. Zhang K., Sha J., Harter M. L. (2010) Activation of Cdc6 by MyoD is associated with the expansion of quiescent myogenic satellite cells. J. Cell Biol. 188, 39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chakravarthy M. V., Abraha T. W., Schwartz R. J., Fiorotto M. L., Booth F. W. (2000) Insulin-like growth factor-1 extends in vitro replicative life span of skeletal muscle satellite cells by enhancing G1/S cell cycle progression via the activation of phosphatidylinositol 3′-kinase/Akt signaling pathway. J. Biol. Chem. 275, 35942–35952 [DOI] [PubMed] [Google Scholar]
- 38. MacDonald M. J., Marshall L. K. (2000) Mouse lacking NAD+-linked glycerol phosphate dehydrogenase has normal pancreatic beta cell function but abnormal metabolite pattern in skeletal muscle. Arch. Biochem. Biophys. 384, 143–153 [DOI] [PubMed] [Google Scholar]
- 39. Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 40. Hu X., Li X., Valverde K., Fu X., Noguchi C., Qiu Y., Huang S. (2009) LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. Proc. Natl. Acad. Sci. U.S.A. 106, 10141–10146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shea K. L., Xiang W., LaPorta V. S., Licht J. D., Keller C., Basson M. A., Brack A. S. (2010) Sprouty1 regulates reversible quiescence of a self-renewing adult muscle stem cell pool during regeneration. Cell Stem Cell 6, 117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fulco M., Cen Y., Zhao P., Hoffman E. P., McBurney M. W., Sauve A. A., Sartorelli V. (2008) Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of NAMPT. Dev. Cell 14, 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rojas A., Kong S. W., Agarwal P., Gilliss B., Pu W. T., Black B. L. (2008) GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol. Cell Biol. 28, 5420–5431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dey S., Curtis D. J., Jane S. M., Brandt S. J. (2010) The TAL1/SCL transcription factor regulates cell cycle progression and proliferation in differentiating murine bone marrow monocyte precursors. Mol. Cell Biol. 30, 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Santra M. K., Wajapeyee N., Green M. R. (2009) F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature 459, 722–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M. W., Guarente L. (2004) Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature 429, 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gerhart-Hines Z., Rodgers J. T., Bare O., Lerin C., Kim S. H., Mostoslavsky R., Alt F. W., Wu Z., Puigserver P. (2007) Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J. 26, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brunet A., Sweeney L. B., Sturgill J. F., Chua K. F., Greer P. L., Lin Y., Tran H., Ross S. E., Mostoslavsky R., Cohen H. Y., Hu L. S., Cheng H. L., Jedrychowski M. P., Gygi S. P., Sinclair D. A., Alt F. W., Greenberg M. E. (2004) Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 49. Calvanese V., Lara E., Suárez-Alvarez B., Abu Dawud R., Vázquez-Chantada M., Martínez-Chantar M. L., Embade N., López-Nieva P., Horrillo A., Hmadcha A., Soria B., Piazzolla D., Herranz D., Serrano M., Mato J. M., Andrews P. W., López-Larrea C., Esteller M., Fraga M. F. (2010) Sirtuin 1 regulation of developmental genes during differentiation of stem cells. Proc. Natl. Acad. Sci. U.S.A. 107, 13736–13741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lecointe N., Bernard O., Naert K., Joulin V., Larsen C. J., Romeo P. H., Mathieu-Mahul D. (1994) GATA- and SP1-binding sites are required for the full activity of the tissue-specific promoter of the tal-1 gene. Oncogene 9, 2623–2632 [PubMed] [Google Scholar]
- 51. Göttgens B., Nastos A., Kinston S., Piltz S., Delabesse E. C., Stanley M., Sanchez M. J., Ciau-Uitz A., Patient R., Green A. R. (2002) Establishing the transcriptional program for blood. The SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 21, 3039–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sinclair A. M., Göttgens B., Barton L. M., Stanley M. L., Pardanaud L., Klaine M., Gering M., Bahn S., Sanchez M., Bench A. J., Fordham J. L., Bockamp E., Green A. R. (1999) Distinct 5′ SCL enhancers direct transcription to developing brain, spinal cord, and endothelium. Neural expression is mediated by GATA factor binding sites. Dev. Biol. 209, 128–142 [DOI] [PubMed] [Google Scholar]
- 53. Kassouf M. T., Hughes J. R., Taylor S., McGowan S. J., Soneji S., Green A. L., Vyas P., Porcher C. (2010) Genome-wide identification of TAL1's functional targets. Insights into its mechanisms of action in primary erythroid cells. Genome Res 20, 1064–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Palii C. G., Perez-Iratxeta C., Yao Z., Cao Y., Dai F., Davison J., Atkins H., Allan D., Dilworth F. J., Gentleman R., Tapscott S. J., Brand M. (2011) Differential genomic targeting of the transcription factor TAL1 in alternate hematopoietic lineages. EMBO J. 30, 494–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hu X., Ybarra R., Qiu Y., Bungert J., Huang S. (2009) Transcriptional regulation by TAL1. A link between epigenetic modifications and erythropoiesis. Epigenetics 4, 357–361 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.