Background: YB-1 is a multifunctional protein that affects transcription, splicing, and translation.
Results: ΔNp63α, the main p63 protein isoform, interacts with YB-1 and affects YB-1 subcellular localization and regulation of cell survival and motility genes.
Conclusion: ΔNp63α and YB-1 interaction inhibits epithelial to mesenchymal transition and tumor cell motility.
Significance: This is the first demonstration of a physical and functional interaction between YB-1 and ΔNp63α oncoproteins.
Keywords: Epithelial Cell, Epithelial to Mesenchymal Transition, Gene Regulation, p63, Protein-Protein Interactions, Cold Shock Domain Superfamily, YB-1
Abstract
The Y-box binding protein 1 (YB-1) belongs to the cold-shock domain protein superfamily, one of the most evolutionarily conserved nucleic acid-binding proteins currently known. YB-1 performs a wide variety of cellular functions, including transcriptional and translational regulation, DNA repair, drug resistance, and stress responses to extracellular signals. Inasmuch as the level of YB-1 drastically increases in tumor cells, this protein is considered to be one of the most indicative markers of malignant tumors. Here, we present evidence that ΔNp63α, the predominant p63 protein isoform in squamous epithelia and YB-1, can physically interact. Into the nucleus, ΔNp63α and YB-1 cooperate in PI3KCA gene promoter activation. Moreover, ΔNp63α promotes YB-1 nuclear accumulation thereby reducing the amount of YB-1 bound to its target transcripts such as that encoding the SNAIL1 protein. Accordingly, ΔNp63α enforced expression was associated with a reduction of the level of SNAIL1, a potent inducer of epithelial to mesenchymal transition. Furthermore, ΔNp63α depletion causes morphological change and enhanced formation of actin stress fibers in squamous cancer cells. Mechanistic studies indicate that ΔNp63α affects cell movement and can reverse the increase of cell motility induced by YB-1 overexpression. These data thus suggest that ΔNp63α provides inhibitory signals for cell motility. Deficiency of ΔNp63α gene expression promotes cell mobilization, at least partially, through a YB-1-dependent mechanism.
Introduction
The Y-box binding protein 1 (YB-1), also known as NSEP1, CSBD, and MDR-NF1, is a member of the highly conserved Y-box family of proteins that regulate gene transcription by binding to the TAACC element (the Y-box) contained within many eukaryotic promoters (1). Transcriptional targets of YB-1 include genes associated with cell death, cell proliferation, and multidrug resistance (1, 2). YB-1 protein can also regulate gene transcription by binding to other transcription factors such as p53, AP1, and SMAD3 (2).
YB-1 is an important marker of tumorigenesis and is overexpressed in many malignant tissues, including breast cancer, non-small cell lung carcinoma, ovarian adenocarcinoma, human osteosarcomas, colorectal carcinomas, and malignant melanomas (1). Mostly cytosolic, YB-1 protein shuttles between the nucleus and cytoplasm. Cytoplasmic YB-1 acts as a translation factor of oncogenic/pro-metastatic genes (3). It binds to the mRNA cap structure and displaces the eukaryotic translation initiation factors eIF4E and eIF4G, thereby causing mRNA translational silencing (4). However, cytoplasmic YB-1 activates cap-independent translation of mRNA encoding SNAIL1, a transcription factor that promotes epithelial-mesenchymal transition by suppressing E-cadherin (5). According to those observations, the YB-1 protein level, in breast carcinoma, is positively correlated to the increase of SNAIL1 protein level and reduced expression of E-cadherin (6).
In normal conditions, YB-1 is located in the cytoplasm and transiently translocates to the nucleus at the G1/S transition of the cell cycle (7). Moreover, nuclear translocation of YB-1 was shown to occur in response to DNA-damaging agents, phosphorylation at Ser-102 by AKT kinase, UV irradiation, TGFβ stimulation, virus infection, hypothermia, and pharmacological compounds (1, 8, 9). Under stress stimuli, YB-1 nuclear translocation requires a physical interaction with wild type p53 (10).
Herein, we report that YB-1 directly interacts with ΔNp63α and accumulates in the nuclear compartment. ΔNp63α is encoded by the TP63 locus, a homologue of the p53 tumor suppressor gene and the most ancient member of the p53 family (11). Because of the presence of two promoters, the TP63 gene encodes two major classes of proteins as follows: those containing a transactivating domain homologous to the one present in p53 (i.e. TAp63), and those lacking it (i.e. ΔNp63) (12). In addition, alternate splicing at the C terminus generates at least three p63 variants (α, β, and γ) in each class. The TAp63γ isoform resembles most p53, whereas the α-isoforms include a conserved protein-protein interaction domain named the sterile α motif.
Gene knock-out mice demonstrated that p63 plays a critical role in the morphogenesis of organs/tissues developed by epithelial-mesenchymal interactions such as the epidermis, teeth, hair, and glands (13). In adults, ΔNp63α expression occurs in the basal cells of stratified epithelia. High expression of ΔNp63α is associated with the proliferative potential of epithelial cells (14) and is enhanced at the early stages of squamous cell carcinomas (SCCs)2 (15). Down-regulation of ΔNp63α by SNAIL transcription factor instead was found to reduce cell-cell adhesion and increase the migratory properties of squamous carcinoma cells (16). Moreover, a partial or complete loss of ΔNp63α expression was associated with tumor metastasis supporting the idea that ΔNp63α plays a relevant role in metastasis suppression (17, 18). Here, we provide experimental evidence that the YB-1 protein associates with ΔNp63α and such interaction plays a critical role in the control of genes involved in cell survival and motility.
EXPERIMENTAL PROCEDURES
Plasmids
cDNAs encoding human FLAG-tagged YB-1 were provided by Dr. Sandra Dunn (Research Institute for Children's and Women's Health, Vancouver, British Columbia, Canada). PIK3CA promoter luciferase plasmid was provided by Dr. Arezoo Astanehe (Research Institute for Children's and Women's Health, Vancouver, British Columbia, Canada). cDNA encoding human ΔNp63α and ΔNp63γ were previously described (19). For bacterial expression, ΔNp63γ and ΔNp63α cDNAs were inserted in pRSETA vector in XhoI/ClaI and XhoI/XbaI (filled) sites, respectively. GFP and YB-1-GFP vectors were provided by Dr. Paul R. Mertens (University Hospital Aachen, Germany). PET-YB-1 was from Dr. Jill Gershan (Medical College of Wisconsin, Milwaukee, WI).
Cell Lines, Transfection, and Antibodies
Squamous carcinoma cells (SCC011 and SCC022) were described previously and provided to us by Dr. C. Missero (20).
These cell lines were maintained in RPMI medium supplemented with 10% fetal bovine serum at 37 °C and 5% CO2. A431, H1299, and MDA-MB-231 cells were obtained from ATCC and maintained in DMEM supplemented with 10% fetal bovine serum at 37 °C and 5% CO2. Dox-inducible Tet-On H1299/ΔNp63α cells were described previously (21).
Lipofections were performed with Lipofectamine (Invitrogen), according to the manufacturer's recommendations. YB-1 transient silencing was carried out with ON-TARGET plus SMART pool YB-1-siRNA (Dharmacon) and RNAiMAX reagent (Invitrogen). ΔNp63α transient silencing in SCC011 cells was carried out with RIBOXX (IBONI p63-siRNA pool) and RNAiMAX reagent (Invitrogen), according to the manufacturer's recommendation. An si-RNA against luciferase (si-Luc) was used as a negative control. Anti-p63 (4A4) and anti-actin(1–19) were from Santa Cruz Biotechnology Inc., and anti-FLAG M2 was from Sigma. PARP, AKT, pAKT (Ser-473) and rabbit polyclonal YB-1 antibody (Ab12148) were from Cell Signaling Technology (Beverly, MA). Mouse monoclonal E-cadherin (ab1416) and SNAIL1 antibodies were from Abcam (Cambridge, UK). Mouse monoclonal N-cadherin (610921) was from BD Transduction Laboratories. Cy3-conjugated anti-mouse IgG and Cy5-conjugated anti-rabbit IgG were from Jackson ImmunoResearch. Doxycycline was from Clontech.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP was performed with chromatin from human MDA-MB-231 cells transfected with ΔNp63γ, ΔNp63α, and/or FLAG YB-1, as described previously (22). Real time PCR was performed with the 7500 Applied Biosystems apparatus and SYBR Green MasterMix (Applied Biosystems) using the following oligonucleotides: PIK3CA-For, CCCCCGAACTAATCTCGTTT, and PIK3CA-Rev, TGAGGGTGTTGTGTCATCCT.
Sequential ChIP was performed with chromatin from SCC011 cells. Method of chromatin extraction and controls used were previously described (22).
Chromatin samples were subjected to pre-clearing with 80 μl of salmon sperm DNA-saturated agarose A beads for 2 h at 4 °C with rotation. The first immunoprecipitation step was obtained with p63 antibodies (4A4). After several washes, the immunocomplexes were extracted from beads with 100 μl of 2% TE/SDS. The second immunoprecipitation step was obtained with YB-1 (Ab12148) antibodies after a 30-fold dilution in DB buffer (50 mm Tris-HCl, 5 mm EDTA, 200 mm NaCl, 0.5% Nonidet P-40, 15 mm DTT). Samples were incubated overnight at 4 °C with rotation. Immunocomplexes were extracted from beads as described above. Immunoprecipitated protein-DNA complexes were subjected to reverse cross-linking and proteinase K treatment. DNA was purified by phenol/chloroform extraction and resuspended in 40 μl of TE. 2 μl were used for PCR performed with the following primers: PIK3CA promoter For, ACAAACCCCTGGAATGTGAG, and PI3KCA promoter Rev, TGGAAAAGCGTAGGAGCAGT.
Immunoblot Analyses and Co-immunoprecipitation
Immunoblots and co-immunoprecipitations were performed as described previously (19). To detect p63/YB-1 interaction in SCC011 cells, 5.0 × 105 cells were plated in 60-mm dishes. For co-immunoprecipitations, whole cell extracts, precleared with 30 μl of protein A-agarose (50% slurry; Roche Applied Science), were incubated overnight at 4 °C with anti-YB-1(3 μg) or α-rabbit IgG (3 μg). The reciprocal experiment was performed with anti-p63 (2 μg) or α-mouse IgG.
Far Western Assays
Far Western assays were conducted according to the protocol described by Cui et al. (23). Increasing amounts (0.2, 0.5, and 1.5 μg) of ΔNp63α or ΔNp63γ recombinant proteins were subjected to SDS-PAGE and transferred to PVDF membrane (IPVH00010 Immobilon Millipore, Milan, Italy). Bovine serum albumin (0.5 and 1.5 μg; BSA) was used as control of unspecific binding and equal loading of BSA, and recombinant YB-1 or p63 proteins in far Western blotting was verified by Coomassie staining (data not shown).
Luciferase Reporter Assay
MDA-MB-231 cells were co-transfected with ΔNp63α, PIK3CA luc promoter, and pRL-TK. YB-1 gene silencing was carried out 24 h before plasmid transfection. At 24 h after transfection, cells were harvested in 1× PLB buffer (Promega), and luciferase activity was measured using the Dual-Luciferase Reporter system (Promega) using pRL-TK activity as internal control. Firefly-derived luciferase activity was normalized for transfection efficiency. Successful transfection of p63 and silencing of YB-1 was confirmed by immunoblotting.
RNA Immunoprecipitation
1 × 106 MDA-MB-231 cells were seeded in 100-mm plates and transfected with pcDNA3.1 or ΔNp63α expression plasmids. 24 h after transfection, cells were fixed with 1% formaldehyde for 10 min and washed twice in ice-cold PBS. Cell extracts were prepared in RNA immunoprecipitation buffer (0.1% SDS, 1% Triton, 1 mm EDTA, 10 mm Tris, pH 7.5, 0.5 mm EGTA, 150 mm NaCl) supplemented with complete protease inhibitor mixture (Sigma), and sonication was carried out with BANDELINE SONOPULSE HD2200 instrument under the following conditions: 8 pulses of 4 s at 0.250% of intensity. Cell extracts were incubated with anti-YB-1 (3 μg) at 4 °C overnight. The RNA-protein immunocomplexes were precipitated with protein A beads (Roche Applied Science) saturated with tRNA and, after reverse cross-link, subjected to real time PCR.
Semi-quantitative and Real Time PCR
For PCR analysis, total RNA was isolated using the RNA mini extraction kit (Qiagen, GmbH, Hilden, Germany) according to the manufacturer's instructions. Total RNA (1 μg) was used to generate reverse-transcribed cDNA using SuperScript III (Invitrogen). PCR analysis was performed with the following primers: Twist (For), 5′-AGAAGTCTGCGGGCTGTG, and Twist (Rev), ′-TCTGCAGCTCCTCGTAAGACT; YB (For), 5′-CGCAGTGTAGGAGATGGAGAG, and YB (Rev), 5′-GAACACCACCAGGACCTGTAA; and HPRT (For), 5′-CCT GCT GGA TTA CAT TAA AGC, and HPRT (Rev), 5′ CTT CGT GGG GTC CTT TTC.
SNAIL1 amplification was performed using Quantitect Primers/Hs SNAIL1 from Qiagen (GmbH, Hilden, Germany). The amplification sequence consisted of 30 cycles of 94 °C for 1 min, 55 °C for 1min, and 72 °C for 1 min. PCR products were resolved by 2% agarose electrophoresis. RT-PCR amplification results were analyzed by Quantity One software (Bio-Rad). Real time PCR was performed with a 7500 RT-PCR Thermo Cycler (Applied Biosystem) as already described (22). HPRT was used for normalization. The results were expressed with the value relative to HPRT (set at 1) for each mRNA sample.
Immunofluorescence
A431, SCC011, and SCC022 cells (2.5 × 105) were plated in 35-mm dishes and grown on micro cover glasses (BDH). 24 h after seeding, cells were washed with cold phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde (Sigma) for 15 min at 4 °C. Cells were permeabilized with ice-cold 0.1% Triton X-100 for 10 min and then washed with PBS. p63 was detected using a 1:200 dilution of the monoclonal antibody 4A4.
YB-1 was detected using 1:100 dilution of the YB-1 antibody (Ab12148). After extensive washing in PBS, the samples were incubated with Cy3-conjugated anti-mouse IgGs and Cy5-conjugated anti-rabbit IgGs at room temperature for 30 min. After PBS washing, the cells were incubated with 10 mg/ml 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma) for 3 min. The glasses were mounted with Moviol (Sigma) and examined under a fluorescence microscope (Nikon). Images were digitally acquired and processed using Adobe Photoshop software CS.
H1299 and MDA-MB-231 cells (5.0 × 105) were plated in 35-mm dishes, grown on micro cover glasses (BDH), and transfected with 0.2 μg of pcDNAYB-1/GFP plasmid with or without 0.6 μg of ΔNp63α or ΔNp63γ plasmid. The GFP empty vector was used as control (data not shown). 24 h after transfection, cells were subjected to the immunofluorescence protocol as described above.
Cytoskeleton Analysis
Actin bundles were stained with TRITC-conjugated phalloidin. Cell fixation was performed with 4% paraformaldehyde for 20 min and then permeabilized with 0.1% Triton X-100 (Sigma) in PBS one time for 10 min. Actin staining was done by incubating sample with TRITC-phalloidin (Sigma) in PBS for 30 min at room temperature. Images of fluorescent cells were collected with a fluorescent inverted microscope (IX81, Olympus, Tokyo, Japan) equipped with an ORCA 2.8 digital camera (Hamamatsu Photonics, Japan).
Time-lapse Microscopy
SCC011 cells were cultured on 35-mm dishes (Corning, NY) at 2 × 104 cells/dish density. ΔNp63α transient silencing was performed as described under “Experimental Procedures.” At 48 h from the addition of ΔNp63α si-RNA, the culture dishes were placed in a mini-incubator connected to an automated stage of an optical microscope (Olympus Co., Japan). Images of selected positions of the cell culture were collected in bright field every 10 min for 12 h. The bright field images were mounted to obtain 72-frame time lapse video for each position. Cell trajectories were reconstructed from time lapse video using Metamorph software (Molecular Device, CA). Root mean square speed (S) and persistence time (P) were chosen as relevant parameters to describe the macroscopic features of cell migration in the different experimental conditions. These parameters were calculated according to the procedure reported by Dunn (24). Briefly, mean-squared displacement of each cell was calculated according to overlapping time interval methods (25). Subsequently, S and P were estimated by fitting the experimental data of the mean-squared displacement to a linear approximation of the persistent random walk model. The upper limit for the data fitting was set at approximately 200 min for each cell because of the deviation from linearity that was observed at higher time points. The fitting procedure provided (S, P) pairs for each cell and the statistical significance between S and P values of the different experimental setups was assessed by performing a nonparametric Kruskal-Wallis test in Matlab (MathWorks, MA). p values < 0.05 were considered significant.
Cell migration in H1299 cells was evaluated with a similar approach. Briefly, cells were cultured on 35-mm dishes (Corning, NY) at a density of 2 × 104 cells/dish. ΔNp63α expression was induced with 2 μg/ml doxycycline for 48 h. 16 h after transfection, cell dishes were placed in a mini-incubator connected to an automated stage of an optical microscope (Cell[caret]R, Olympus Co., Japan). Time 0 images of selected position were collected in fluorescence to localize transfected cells. Then, at the same positions, images were recorded in bright field every 10 min for 12 h. Data analysis was performed as described above.
RESULTS
YB-1 Interacts with ΔNp63α in Vitro and in Vivo
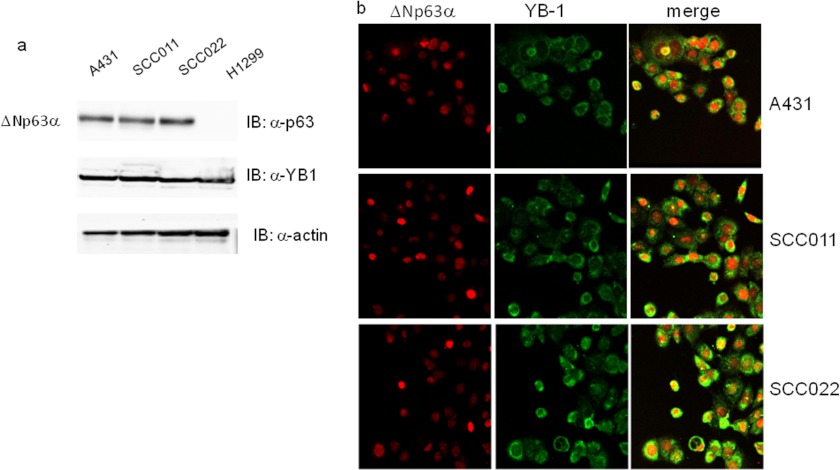
As a result of a comprehensive screening for ΔNp63α interactors, using affinity purification followed by mass spectrometry, we identified a set of proteins with DNA/RNA binding activity, including YB-1 (26). We decided to confirm the interaction between ΔNp63α and YB-1 in SCC as in this cellular context the TP63 gene is often amplified and/or overexpressed (27). Immunoblots and immunofluorescence assays performed in A431, SCC022, and SCC011 cell lines showed intense signals for both proteins thus confirming that endogenous YB-1 and ΔNp63α are abundantly co-expressed in human squamous carcinoma cells (Fig. 1, a and b).
FIGURE 1.
ΔNp63α and YB-1 are co-expressed in squamous carcinoma cell lines. a, whole cell lysates were obtained from H1299 (non-small cell lung carcinoma), A431 (epidermoid carcinoma cell line), and SCC011 and SCC022 (keratinocyte-derived SCC) cells. 30 μg of total protein extracts were separated by SDS-PAGE and subjected to immunoblot (IB). Proteins were detected with specific antibodies as indicated. Images were acquired with CHEMIDOC (Bio-Rad) and analyzed with the Quantity-ONE software. b, A431, SCC011, and SCC022 cells were seeded (2.5 × 105) on a 35-mm dish and grown on micro cover glasses (BDH). After 24 h at seeding, cells were fixed and subjected to double immunofluorescence using rabbit primary YB-1 antibody and Fitch-conjugated secondary antibodies (green). p63 protein was detected using mouse anti-p63 and Cy3-conjugated secondary antibodies (red). Images of merge (yellow) show the co-expression of two proteins.
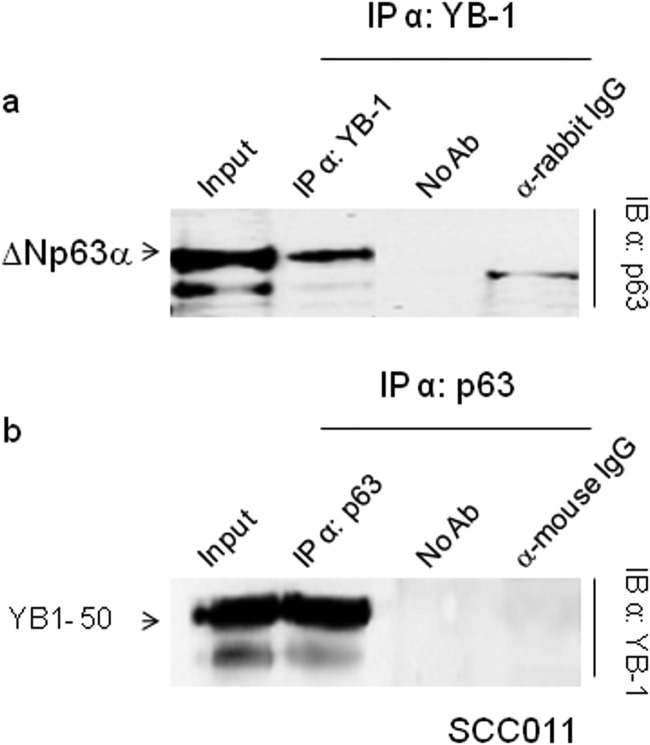
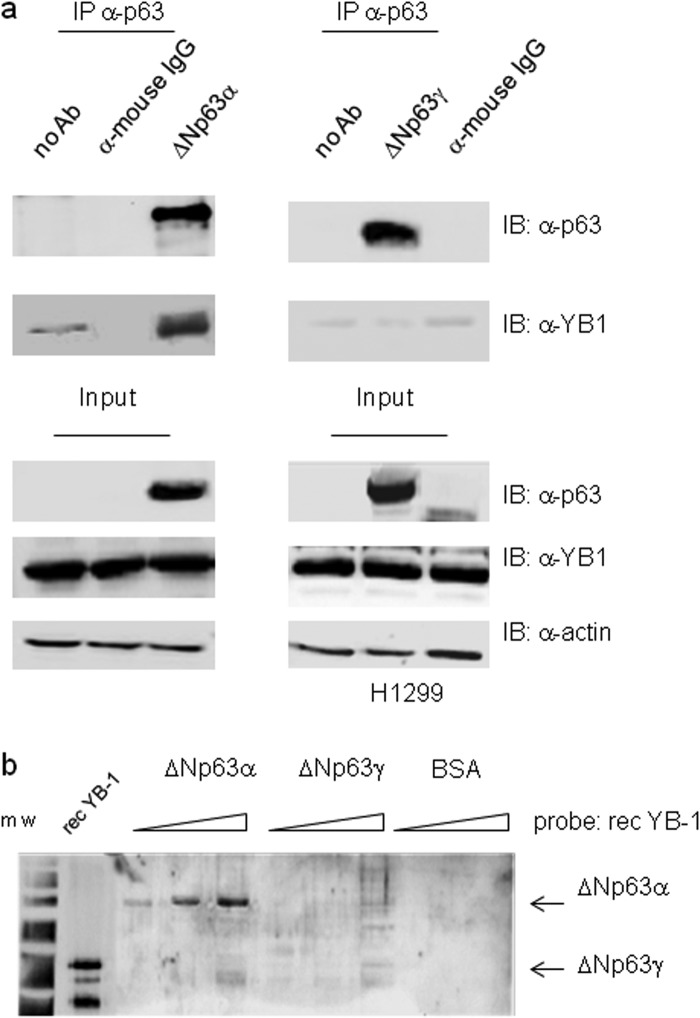
We next performed co-immunoprecipitation experiments in SCC011 cells. Immunocomplexes containing ΔNp63α and YB-1 were isolated from total cell lysates, using either antibodies against YB-1 or antibodies against p63 thus confirming the interaction inferred by mass spectrometry (Fig. 2, a and b). To determine whether YB-1 interacts specifically with the α-isoform of ΔNp63, we transfected p63 null H1299 cells with ΔNp63α or ΔNp63γ to perform co-immunoprecipitation assays. As shown in Fig. 3a, YB-1 associates with ΔNp63α but not ΔNp63γ. This result was also confirmed by far Western blot analysis using ΔNp63α, ΔNp63γ, and YB-1 recombinant proteins purified by affinity chromatography (Fig. 3b). The obtained results indicate that the α-isoform C-terminal region is involved in the association with YB-1, and the interaction between the two proteins requires no additional factors.
FIGURE 2.
ΔNp63α interacts with YB-1. a, extracts from SCC011 cells were immunoprecipitated (IP) with anti-YB-1 antibodies, and the immunocomplexes were blotted and probed with anti-p63 as indicated. b, extracts from SCC011 cells were immunoprecipitated with anti-p63 antibodies, and the immunocomplexes were blotted and probed with anti-YB-1. Samples with no antibody (no Ab) or irrelevant α-mouse and α-rabbit antibodies were included as controls. IB, immunoblot.
FIGURE 3.
ΔNp63α but not ΔNp63γ interacts with YB-1. a, H1299 cells were transiently transfected with 5 μg of ΔNp63α (left panel) or ΔNp63γ (right panel) expression vectors. Equal amounts (1 mg) of extracts were immunoprecipitated (IP) with anti-p63 antibodies (4A4) or unrelated α-mouse IgG. The immunocomplexes were blotted and probed with anti-p63 and anti-YB-1 antibodies, as indicated. b, far Western analysis. Increasing amounts of purified recombinant ΔNp63α (0.2, 0.5, and 1.5), ΔNp63γ (0.2, 0.5, and 1.5), or BSA (0.5 and 1.5) were subjected to SDS-PAGE. After Coomassie staining to monitor equal loading, the proteins were transferred to a PVDF membrane, and the filter was incubated with purified YB-1 recombinant protein (0.8 μg/ml). After extensive washing, the membrane was subjected to immunoblotting (IB) with YB-1 antibodies followed by ECL detection. Recombinant YB-1 protein (0.1 μg) was used as positive control. m.w., molecular weight markers.
ΔNp63α Induces YB-1 Nuclear Accumulation
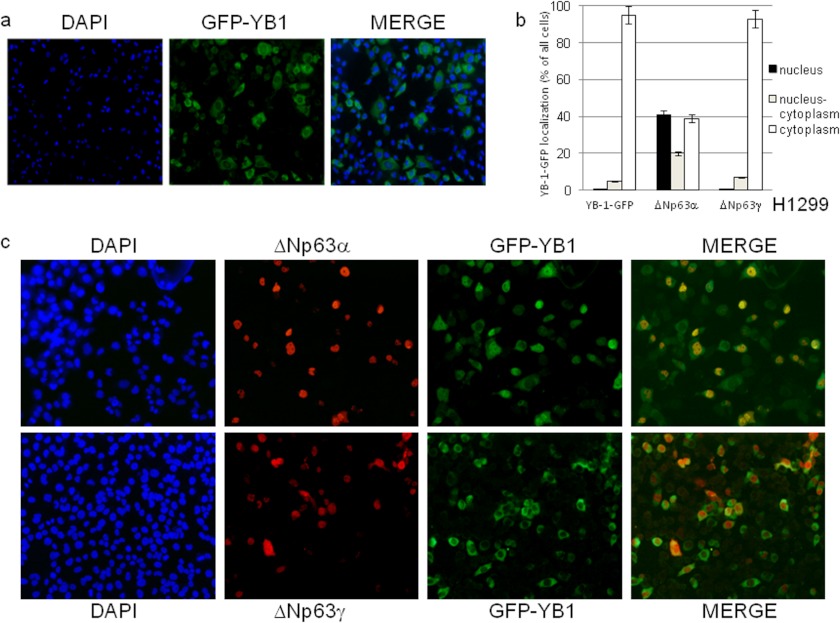
It has been reported that YB-1 is ubiquitously expressed and predominantly localized to the cytoplasm (2), ΔNp63α is mainly expressed by basal and myoepithelial cells in skin and glands and is almost completely restricted to the nuclear compartment (29). Because we noticed some overlapping between the p63 and YB-1 immunofluorescence signals in the nucleus of squamous carcinoma cell lines, we sought to determine whether ΔNp63α has an effect on YB-1 subcellular localization. Fusion of YB-1 with the green fluorescent protein (YB-1-GFP) was transfected in H1299 cells. H1299 cells are p63 null and express moderate levels of endogenous YB-1. GFP-positive cells were counted, and 95% showed a strong cytoplasmic signal, although in the remaining 5%, the signal was nuclear or distributed between the nucleus and cytoplasm (Fig. 4, a and b). A very similar result was observed following ΔNp63γ co-transfection (Fig. 4, b and c, lower panel). Remarkably, combined expression of YB-1-GFP and ΔNp63α caused a dramatic change of YB-1-GFP subcellular localization. In fact, among the GFP-positive cells, more than 40% showed an intense nuclear signal (Fig. 4, b and c, upper panel); an additional 20% exhibited YB-1-GFP almost equally distributed between the nucleus and cytoplasm, and the remaining 40% exhibited exclusively cytoplasmic YB-1-GFP (Fig. 4b). As expected, ΔNp63α and ΔNp63γ immunofluorescence signals were almost exclusively nuclear (Fig. 4c).
FIGURE 4.
ΔNp63α induces YB-1 nuclear accumulation. a, H1299 cells (2.3 × 105) were seeded at 60% confluency in 24 × 24-mm sterile coverglasses placed in 60-mm dishes and transiently transfected with GFP-YB-1 expression vector (0.3 μg). GFP-YB-1 was detected by direct immunofluorescence. b, H1299 cells (2.3 × 105) were seeded as described in a and transiently transfected with GFB-YB-1 (0.3 μg) along with 1 μg of ΔNp63α or ΔNp63γ expression vectors. Plot shows the percentage of nuclear (dark gray), nucleo-cytoplasmic (gray), and cytoplasmic (white) GFP-YB-1 in cells transfected with ΔNp63α or ΔNp63γ expression vectors. Each experimental point is the average of counts performed on 100 cells in five independent fields. Each histogram bar represents the mean and standard deviation of values from three biological replicates. c, H1299 cells (2.3 × 105) were transfected as described in b. GFP-YB-1 was detected by direct immunofluorescence (green), and p63 was detected using anti-p63 antibodies and secondary anti-mouse Cy3-conjugated (red). DAPI was used to stain nuclei.
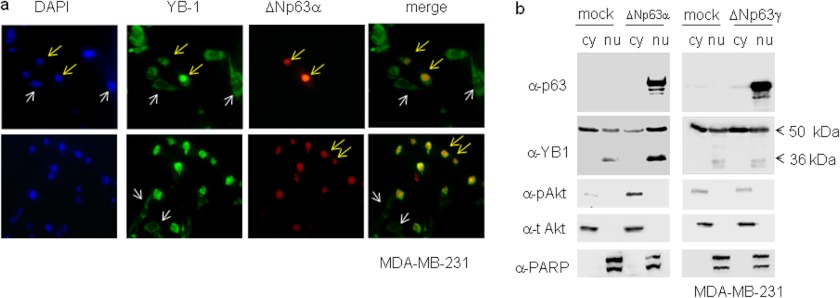
ΔNp63α is a selective nuclear marker of human breast myoepithelial cells. ΔNp63α persists in benign lesions of the breast, although it consistently disappears in invasive carcinomas (30). We transfected ΔNp63α in MDA-MB-231 cells, human metastatic breast carcinoma cells lacking p63 but expressing high level of endogenous YB-1. As shown in Fig. 5, in cells with no detectable p63 signal, YB-1 exhibited a largely prevalent cytoplasmic localization (Fig. 5a, white arrows), although 95% of ΔNp63α-expressing cells showed a strong YB-1 nuclear staining (Fig. 5a, yellow arrows). We verified this observation by nuclear/cytoplasmic fractionation of MDA-MB-231 cells transfected with ΔNp63α or ΔNp63γ. Endogenous YB-1 protein, in MDA-MB-231 cells, was detectable as 50- and 36-kDa immunoreactive forms. Both forms of YB-1 appear to accumulate in the nuclear compartment of cells transfected with ΔNp63α but not ΔNp63γ (Fig. 5b). Immunoblots with antibodies against PARP-1 (nuclear) and AKT (cytoplasmic) were performed to check for the quality of the fractionation. Interestingly, although the amount of total AKT was comparable, phosphorylated AKT, at serine 473, was enhanced in ΔNp63α but not ΔNp63γ-transfected cells (Fig. 5b). This observation was in line with previous data showing the specific ability of the ΔNp63α isoform to promote PI3K/AKT pathway activation (31).
FIGURE 5.
Endogenous YB-1 accumulates in the nucleus of breast cancer cells expressing ΔNp63α. a, MDA-MB-231 cells were seeded at 60% confluency (2.3 × 105) on 24 × 24-mm sterile coverglasses placed in 60-mm dishes and transiently transfected with 1 μg of ΔNp63α expression vector. Cells were fixed and subjected to double indirect immunofluorescence using rabbit primary YB-1 antibody and Fitch-conjugated secondary antibodies (green). p63 protein was detected using mouse anti-p63 and Cy3-conjugated secondary antibodies (red). DAPI was used to stain nuclei (blue). A representative image is given of a cell expressing ΔNp63α and showing nuclear endogenous YB-1 (yellow arrows). A representative image is given showing YB-1 cytoplasmic localization in cells bearing no detectable ΔNp63α expression (white arrows). b, MB-MDA-231 cells were transiently transfected with a fixed amount (5 μg) of an empty vector (mock) and ΔNp63α or ΔNp63γ expression vector in 100-mm dishes. 24 h after transfection, cell lysates were fractionated to obtain cytoplasmic (cy) and nuclear (nu) fractions. 20 μg of nuclear and cytoplasmic extracts were separated by SDS-PAGE and subjected to immunoblot. Proteins were detected with specific antibodies, as indicated. PARP and total AKT were used as nuclear and cytoplasmic controls, respectively, to check for cross-contamination. Images were acquired with CHEMIDOC (Bio-Rad) and analyzed with the Quantity-ONE software.
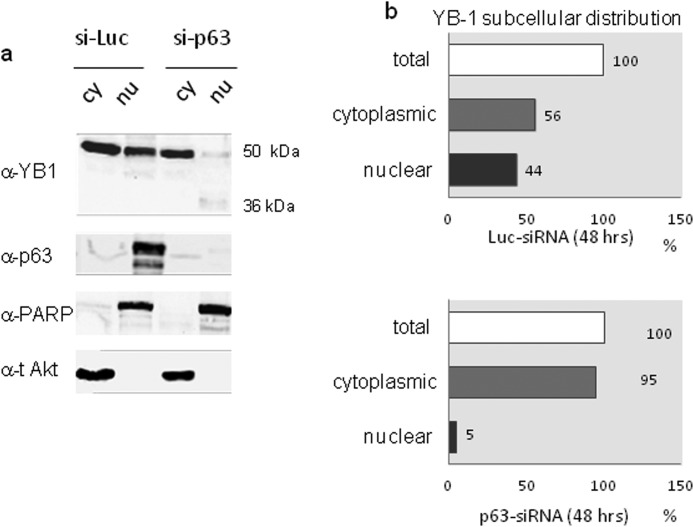
We then evaluated the contribution of ΔNp63α to YB-1 subcellular distribution in SCC011 cells expressing both proteins endogenously. To this aim we analyzed YB-1 subcellular distribution by immunoblot on nuclear/cytoplasmic fractions after depletion of ΔNp63α by siRNA. As shown in Fig. 6, a and b, following p63 knockdown the YB-1 nuclear pool was dramatically reduced, and the 36-kDa form of YB-1 became detectable (Fig. 6a).
FIGURE 6.
ΔNp63α depletion reduces the pool of nuclear YB-1. a, SCC011 cells were seeded at 60% confluency (1.5 × 106) in 100-mm dishes. 24 h after seeding, cells were transiently silenced with IBONI p63-siRNA pool (20 nm final concentration). 48 h after p63-silencing, cell lysates were fractionated to obtain cytoplasmic (cy) and nuclear (nu) fractions. Nuclear and cytoplasmic extracts were separated by SDS-PAGE and subjected to immunoblot. Proteins were detected with specific antibodies, as indicated. PARP and total AKT were used as nuclear and cytoplasmic control respectively, to check for cross-contamination. Images were acquired with CHEMIDOC (Bio-Rad) and analyzed with the Quantity-ONE software. b, plots show the percentage of YB-1 subcellular distribution in control sample (upper panel) and in p63-silenced sample (lower panel) considering the total amount of YB-1 between the nucleus and cytoplasm as 100%.
ΔNp63α and YB-1 Bind to the PIK3CA Gene Promoter and Cooperate in Its Transcriptional Activation
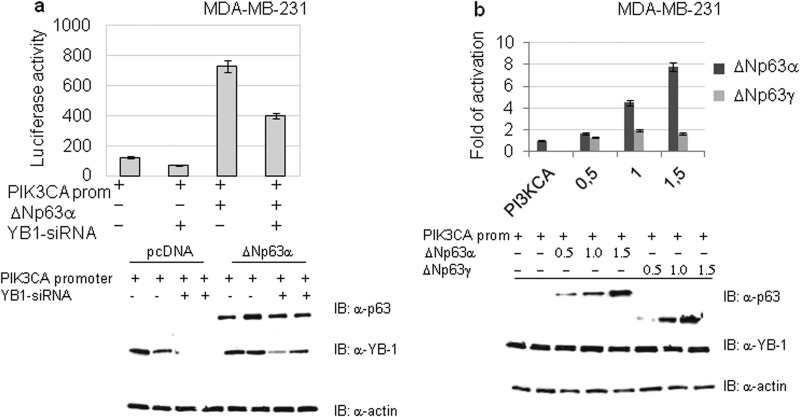
The PIK3CA gene encodes the p110α catalytic subunit of the PI3K and is a well characterized YB-1 direct transcriptional target (32). We speculated that ΔNp63α should increase PIK3CA promoter activity because of its ability to cause YB-1 nuclear accumulation. We tested this hypothesis by luciferase assays. As shown in Fig. 7a, ΔNp63α transfection in MDA-MB-231 cells caused a relevant increase of the PIK3CA promoter activity that was consistently attenuated by YB-1 knockdown. In addition, we observed that ΔNp63α was able to transactivate the PIK3CA promoter in a dose-dependent manner, although ΔNp63γ was completely ineffective (Fig. 7b).
FIGURE 7.
ΔNp63α and YB-1 activates the PIK3CA gene promoter. a, MB-MDA− cells were transfected with 1 μg of PIK3CA promoter luciferase reporter plasmid. The luciferase activity upon siRNA-mediated knockdown of endogenous YB-1 was measured in the presence or absence of ΔNp63α expression. Values are shown as mean ± S.D. of three biological replicates. The extent of YB-1 knockdown was documented by Western blotting as shown in the lower panel. b, MB-MDA-231 cells were seeded at 60% confluency (1.2 × 106) in 100-mm dishes and transiently transfected with 1 μg of PIK3CA promoter luciferase reporter plasmid and the indicated amounts of ΔNp63α or ΔNp63γ plasmid. Values are the mean ± S.D. of three independent experimental points. Lower panel, immunoblotting (IB) with indicated antibodies to detect proteins in samples transfected with different amounts of ΔNp63α or ΔNp63γ.
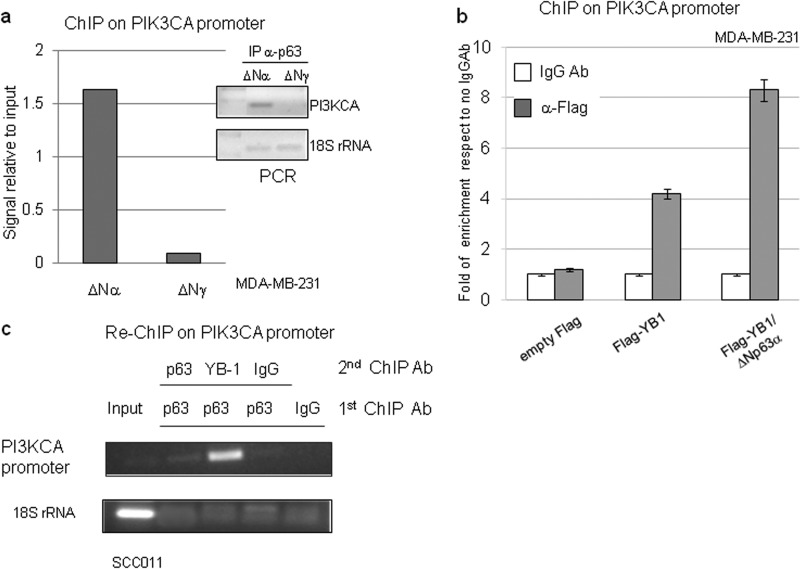
The role of ΔNp63α and YB-1 in PIK3CA promoter activation was thoroughly investigated by chromatin immunoprecipitation (ChIP). We first performed ChIP assays in MDA-MB-231 cells transfected with ΔNp63α or ΔNp63γ using anti-p63 (4A4) antibodies and PIK3CA promoter-specific primers. As shown in Fig. 8a, ΔNp63α, but not ΔNp63γ, binds to the genomic sequence of the PIK3CA promoter.
FIGURE 8.
ΔNp63α increases YB-1 binding to the PIK3CA gene promoter. a, MDA-MB-231 cells were seeded at 60% confluency (1.2 × 106) in 100-mm dishes and transiently transfected with ΔNp63α or ΔNp63γ plasmid (5 μg). After formaldehyde cross-linking, the DNA-proteins complexes were immunoprecipitated (IP) with anti-p63 (4A4) antibody. Immunoprecipitated DNA was PCR-amplified with PIK3CA promoter oligonucleotides and 18 S rRNA oligonucleotides (right). The data obtained from the ChIP assay were measured by densitometry and are presented as signal relative to the input (left). b, MDA-MB-231 cells were seeded at 60% confluency (1.2 × 106) in 100-mm dishes and transiently transfected with 3×FLAG empty vector or 3×FLAG-YB-1 (5 μg) plasmid with or without ΔNp63α (2.5 μg) expression vector. The cells were cross-linked with formaldehyde, and DNA-protein complexes were immunoprecipitated with anti-FLAG antibody or irrelevant IgG antibody as negative control. The DNA immunoprecipitates were analyzed by quantitative PCR using PIK3CA or GAPDH promoter oligonucleotides. Quantitative RT-PCR results were analyzed with the ΔΔCT method and expressed as fold of enrichment with respect to the IgGAb control samples. Values are represented as the mean of three independent experiments. c, SCC011 cells at 85% confluency were fixed with formaldehyde, and the DNA-protein complexes were subjected to sequential ChIP with anti-p63 (4A4), anti-YB-1(Ab12148), or irrelevant IgG antibody as described under “Experimental Procedures.” Immunoprecipitated DNA was PCR-amplified with PIK3CA promoter primers and 18 S rRNA primers to check the quality of the input chromatin and the cleaning of the other samples.
Then we evaluated the effect of ΔNp63α expression on the binding of YB-1 to the PIK3CA promoter. FLAG-YB-1 was transfected, in MB-MDA-231 cells, with or without ΔNp63α plasmid. ChIP was carried out with anti-FLAG antibodies and coupled with quantitative real time PCR. As show in Fig. 8b, FLAG-YB-1 efficiently binds to the PIK3CA promoter, and its binding was substantially increased by ΔNp63α. Finally, by using a sequential chromatin immunoprecipitation assay (Re-ChIP) in SCC011 cells, expressing endogenous ΔNp63α and YB-1, we found that both proteins are recruited to the PIK3CA proximal promoter suggesting that they cooperate in PIK3CA gene transactivation as a complex (Fig. 8c).
ΔNp63α Affects YB-1 Binding to SNAIL and YB-1 mRNAs
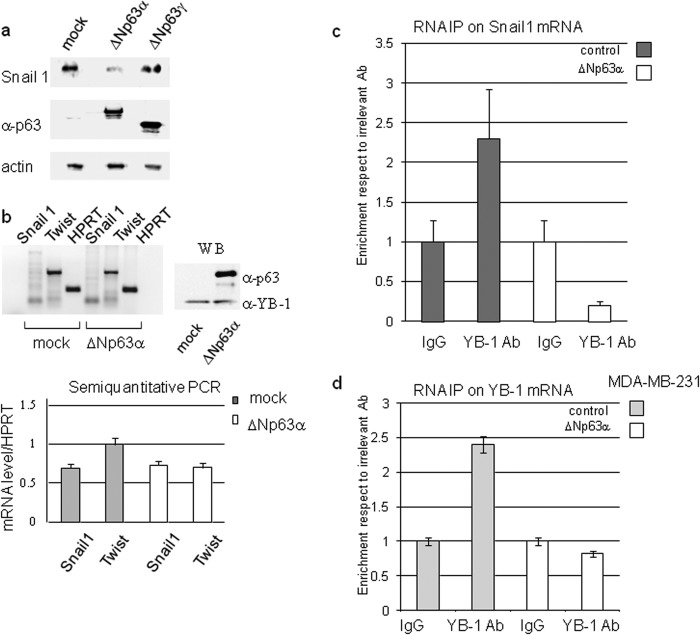
Cytoplasmic localization of YB-1 was associated with binding and translational activation of SNAIL1 mRNA (33). Snail1 protein is a transcriptional repressor of ΔNp63α and E-cadherin and promotes the epithelial-mesenchymal transition in several epithelium-derived cancer cell lines (34). We first observed that ΔNp63α overexpression in MB-MDA-231 caused a reduction of SNAIL1 protein level (Fig. 9a). Such a reduction was not associated with a decrease of SNAIL1 mRNA levels (Fig. 9b) thereby suggesting that the SNAIL1 protein was down-regulated at the translational level. We hypothesized that the specific association of ΔNp63α with YB-1 would reduce the amount of YB-1 bound to the SNAIL1 transcript. To test this hypothesis, we performed RNA-immunoprecipitation coupled with real time PCR to quantitate the amount of SNAIL1 mRNA bound to endogenous YB-1, in mock and ΔNp63α-transfected MDA-MB-231 cells. The obtained results confirm that ΔNp63α reduces the amount of YB-1 bound to SNAIL1 transcript (Fig. 9c). A similar analysis performed on the YB-1 transcript gave the same result (Fig. 9d).
FIGURE 9.
ΔNp63α affects SNAIL1 protein level and YB-1 binding to SNAIL1 and YB-1 mRNAs in MDA-MB-231 cells. a, MDA-MB-231 cells were transfected with an empty vector (mock) or a fixed amount (1 μg in 60-mm dishes) of ΔNp63α or ΔNp63γ expression vector. 24 h after transfection, cells were harvested, and total extracts were prepared. 20 μg of each extract were loaded on SDS-PAGE and subjected to immunoblot with the indicated antibodies. b, MB-MDA-231 cells were transfected with an empty vector or ΔNp63α encoding plasmid. 24 h after transfection, total RNA was purified and retrotranscribed as described under “Experimental Procedures.” The expression level of ΔNp63α was checked by immunoblot (right panel). WB, Western blot. PCR was performed with primers designed to specifically amplify SNAIL1, TWIST, or HPRT transcripts and analyzed by agarose gel electrophoresis (left panel). Plot showing the level of SNAIL1 and TWIST transcripts was normalized respect to HPRT. Values are the mean of three independent experiments (lower panel). Nuclear extracts from ΔNp63α transfected MB-MDA-231 cells were immunoprecipitated with anti-YB-1 antibody. After reverse cross-linking, the YB-1-bound RNA was purified, retrotranscribed, and subjected to quantitative RT-PCR analysis using oligonucleotides designed to specifically amplify SNAIL1 transcript (c) or YB-1 transcript (d). Plots represent the % of enrichment of SNAIL1/YB-1 transcript normalized as indicated under “Experimental Procedures.” 1:50 of the input extract was loaded as control. The values are the means ± S.D. of three biological replicates.
ΔNp63α Affects Cell Shape and Motility
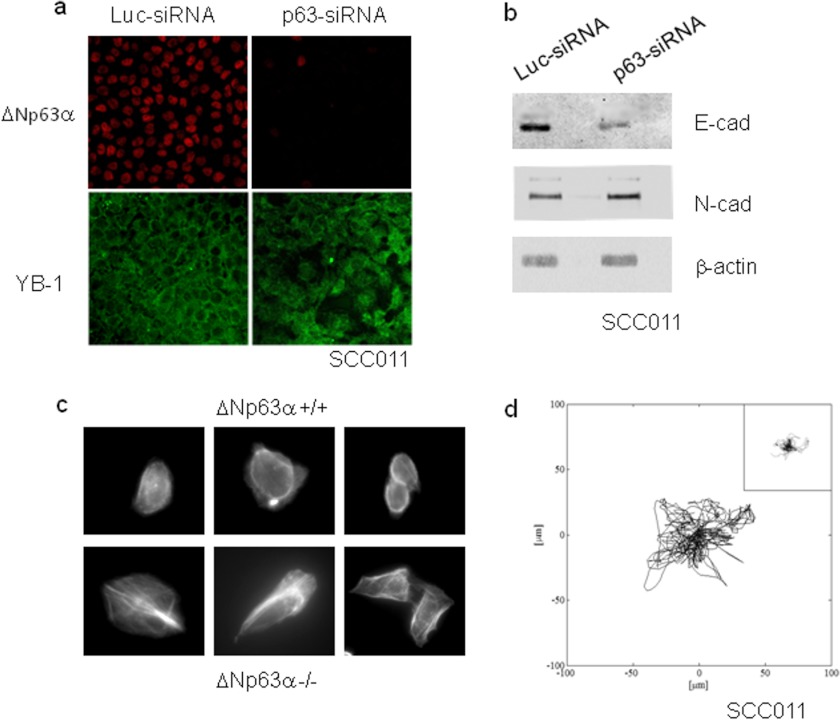
So far, little attention has been focused on the effects of ΔNp63α on epithelial cancer cell morphology and motility. We have unexpectedly noted a profound change in cell morphology when SCC011 cells were depleted of ΔNp63α by siRNA. SCC011 cells are round in shape, and under high density culture conditions, they appear orderly arranged with tight cell-cell contacts. Conversely, ΔNp63α-depleted cells tend to lose their contacts and exhibited an unusual extended phenotype that was distinct from that of the control cells cultured at the same density (Fig. 10a). The unusual change in morphology of ΔNp63α-depleted cells indicated possible alterations in actin cytoskeleton organization. Therefore, we examined the status of actin stress fibers at the level of individual cells, in control and ΔNp63α-depleted cells, with TRITC-phalloidin followed by fluorescent microscopy. As shown in Fig. 10b, the control cells exhibited a diffuse pattern of actin staining, although ΔNp63α-depleted cells displayed a more substantial enhancement of actin stress fibers suggesting an inhibitory effect of ΔNp63α on cancer cell motility. Remarkably, compared with the control cells, ΔNp63α-depleted SCC011 cells express higher levels of N-cadherin and lower amounts of E-cadherin, suggesting that they were undergoing epithelial to mesenchymal trans-differentiation (Fig. 10c).
FIGURE 10.
ΔNp63α affects cell shape and motility. a, SCC011 cells were transiently silenced with IBONI p63-siRNA pool (20 nm final concentration). Cells were fixed and subjected to double immunofluorescence 48 h after silencing, as already described. b, protein extracts from control and p63-depleted cells were separated by SDS-PAGE and subjected to immunoblot. Proteins were detected with specific antibodies. c, actin staining with TRITC-conjugated phalloidin of control and ΔNp63α-depleted SCC011 cells (see “Experimental Procedures”). Cell expressing ΔNp63α displayed a round morphology. Conversely, ΔNp63α-depleted cells had a polarized morphology with more evident lamellipodia and trailing edge. d, windrose plot of trajectories described by ΔNp63α depleted SCC011 or SCC011 control cells (inset) in 12-h time lapse video. Only 15 cell trajectories were reported in each plot for graphical clarity. E-cad, E-cadherin; N-cad, N-cadherin.
We determined the influence of ΔNp63α on cell motility by time-lapse microscopy using siRNA-based silencing of endogenous ΔNp63α in SCC011 cells. Silencing efficiency was about 90% as determined by fluorescent staining for p63 protein (data not shown). SCC011 migration was characterized by oscillations of the cells around their initial adhesion site. These oscillations occurred at random directions in space, as reported in the windrose plots of the trajectories (Fig. 10d, inset). Conversely, ΔNp63α silenced cells display much wider oscillations (Fig. 10d). These observations were confirmed by the analysis of the migration parameters “Speed (S)” and “persistence time (P)” that are computed from cell tracks and are a measure of the frequency of cell steps and of the minimum time that is necessary for a cell to significantly change direction. According to these parameters, SCC011 exhibit lower speeds (p < 0.05) and shorter persistence times (0.26 ± 0.03 μm/min and 6.5 ± 0.7 min) with respect to ΔNp63α-depleted cells (0.44 ± 0.05 μm/min and 8.2 ± 2.3 min). These data indicate a role for ΔNp63α in affecting cell motility. In particular, the absence of ΔNp63α causes the cells to migrate faster with less frequent direction changes, which enables the cells to explore larger areas in a fixed time interval.
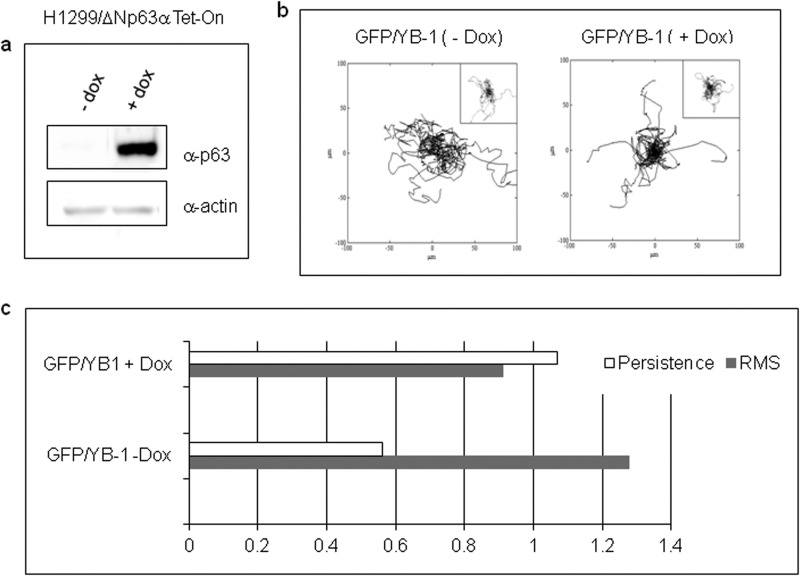
To gain a better insight into the effects of the interaction of ΔNp63α and YB-1 on cell migration, we used the genetically modified Tet-On-H1299 cells expressing ΔNp63α upon Dox addition (Fig. 11a). Cells were transfected with GFP or GFP-YB-1 plasmid, and unfluorescent and fluorescent cells were tracked separately. Cells migrated describing trajectories that are randomly distributed in plane, as depicted in plots of Fig. 10b. In Dox-free medium (−Dox), GFP-YB-1-expressing cells were characterized by a significantly higher S and lower P (0.69 ± 0.04 μm/min and 8.4 ± 0.5 min) than cells expressing GFP alone (0.54 ± 0.05 μm/min and 15 ± 2.1 min). Remarkably, in Dox-supplemented medium (+Dox), GFP-YB-1-expressing cells exhibited S and P values (0.62 ± 0.05 μm/min and 9.2 ± 1.0 min), which were not significantly different from those of cells transfected with GFP alone (0.68 ± 0.05 μm/min and 8.6 ± 0.9 min) (Fig. 10c). We also noticed that unfluorescent cells were characterized by the highest S values suggesting that expression of the green fluorescent protein alters cell migration to some extent (data not shown). However, S of unfluorescent cells with or without Dox was comparable. Overall, these data indicate that ΔNp63α expression can revert the increase of cell motility induced by YB-1-enforced expression.
FIGURE 11.
Effects of ΔNp63α and YB-1 on cell migration. a, after being subjected to the migration experiment, extracts from Tet-On H1299 cells, induced (+dox) or not (−dox) with doxycycline, were prepared and subjected to immunoblot analysis to monitor ΔNp63α induced expression. b, trajectories of GFP/YB-1- or GFP (insets)-transfected cells. Inducible H1299/ΔNp63α cells were transfected with a fixed amount of GFP-YB-1(1 μg) or GFP empty vector (1 μg). 4 h after transfection, 2 μg/ml doxycycline was added to induce ΔNp63α expression, and 16 h later uninduced (−Dox) and induced (+Dox) cells were tracked with optical microscopy. Plots represent data from the following number of cells (YB-1/−dox 79; YB−1/+dox 69; control/−dox 44; control/+dox 49). c, bar chart of speed (S; gray bars) and persistence ratios (P; white bars). The ratios are computed by dividing the population average values of speed and persistence for H1299 control cells (− Dox) and ΔNp63α-expressing cells (+ Dox).
DISCUSSION
In normal epithelium, ΔNp63α protein expression is abundant in basal cells and decreases with differentiation (35). Transient overexpression of ΔNp63α was shown to enhance cell proliferation, inhibit differentiation, and promote malignant conversion of primary keratinocytes (36, 37). However, enhanced and stable expression of ΔNp63α, as seen in human squamous cell carcinomas, is believed to alter skin homeostasis and be directly implicated in the etiology of human cutaneous carcinomas. In normal cells, YB-1 is mainly detected in the cytosol, particularly at the perinuclear region, with a minor pool located in the nucleus (38). Nuclear localization of YB-1 appears to be critical for its role in promoting proliferation. In fact, in the nucleus YB-1 may not only act as a transcription factor of various genes that are closely associated with DNA replication, cell proliferation, and multidrug resistance but also exert SOS signaling against genotoxic factors that could undermine the integrity of highly proliferative basal cells.
This report unveils a novel protein-protein association involving ΔNp63α and YB-1. This specific association results in the accumulation of nuclear YB-1, as shown by immunofluorescence assays and nuclear-cytoplasmic fractionation. Moreover, we have demonstrated that ΔNp63α and YB-1 cooperate in PIK3CA gene activation with both being recruited to the PIK3CA proximal promoter, thereby supporting the hypothesis that this molecular association can function as a pro-survival mechanism. It's worth mentioning, however, that additional mechanisms have been described to activate and translocate YB-1 to the nucleus (8).
Into the cytoplasm, YB-1 is known to act as a positive translational regulator of SNAIL1, a potent epithelial to mesenchymal transition inducer able to reduce cell-cell adhesion and increase migratory properties of cancer cells (33). Accordingly, we have observed that enforced expression of YB-1 in tumor cells increases their motility and migration speed. Remarkably, we have shown that ΔNp63α-enforced expression reduces the activating binding of YB-1 to the SNAIL1 transcript and restores a normal migratory behavior to YB-1-overexpressing cells. However, depletion of endogenous ΔNp63α in SCC cells results in SNAIL1 up-regulation, E-cadherin repression, and increased cell motility.
Finally, cytoplasmic YB-1 is known to bind to actin filaments and causes actin fibers to bundle in vitro (39). It was also demonstrated that YB-1 plays a relevant role in the organization of the actin cytoskeleton by binding to F-actin and microtubules (40, 41). In normal conditions, SCC011 cells are round in shape and exhibit a more diffuse actin distribution, as determined by phalloidin staining. Here, we show that ΔNp63α depletion in SCC cells promotes morphological changes and causes a profound cytoskeleton reorganization. ΔNp63α-depleted cells had a polarized morphology with more evident lamellipodia and trailing edge. They also exhibited mature stress fibers suggesting a pro-migratory behavior. The alterations in cell morphology and motility of ΔNp63α-depleted SCC cells closely resemble the process of epithelial to mesenchymal transition (28). We can speculate that the absence of ΔNp63α can promote F-actin polymerization by YB-1 thereby promoting the formation of stress fibers.
All together, our data indicate that ΔNp63α-YB-1 association into the nucleus can act as a pro-proliferative/survival mechanism, although the loss of p63 predisposes the acquisition of mesenchymal characteristics at least in part by restoring YB-1 cytoplasmic functions. Moreover, we suggest that the balance between ΔNp63α and YB-1 protein levels could be critical for the transition to a mesenchyme-like phenotype explaining, at least in part, why ΔNp63α depletion promotes cancer cell invasion and spreading (35).
Acknowledgments
We thank Sandra Dunn, Arezoo Astanehe, Karsten Jurchott, and Jill Gershan for kindly providing expression and reporter constructs. We acknowledge Dr. Caterina Missero for providing SCC cell lines.
This work was supported by MIUR Grant PRIN 2009KFS94X_003 (to V. C.).
- SCC
- squamous cell carcinoma
- For
- forward
- Rev
- reverse
- PARP
- poly(ADP-ribose) polymerase
- Dox
- doxycycline
- TRITC
- tetramethylrhodamine isothiocyanate
- S
- speed
- P
- persistence time
- Ab
- antibody
- HPRT
- hypoxanthine-guanine phosphoribosyltransferase.
REFERENCES
- 1. Matsumoto K., Bay B. H. (2005) Significance of the Y-box proteins in human cancers. J. Mol. Genet. Med. 1, 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Homer C., Knight D. A., Hananeia L., Sheard P., Risk J., Lasham A., Royds J. A., Braithwaite A. W. (2005) Y-box factor YB-1 controls p53 apoptotic function. Oncogene 24, 8314–8325 [DOI] [PubMed] [Google Scholar]
- 3. Wu J., Stratford A. L., Astanehe A., Dunn S. (2007) YB-1 is a transcription/translation factor that orchestrates the oncogenome by hardwiring signal transduction to gene expression. Translational Oncogenomics 2, 49–65 [PMC free article] [PubMed] [Google Scholar]
- 4. Evdokimova V., Ruzanov P., Anglesio M. S., Sorokin A. V., Ovchinnikov L. P., Buckley J., Triche T. J., Sonenberg N., Sorensen P. H. (2006) Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Mol. Cell. Biol. 26, 277–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sou P. W., Delic N. C., Halliday G. M., Lyons J. (2010) Snail transcription factors in keratinocytes. Enough to make your skin crawl. Int. J. Biochem. Cell Biol. 42, 1940–1944 [DOI] [PubMed] [Google Scholar]
- 6. Evdokimova V., Tognon C., Ng T., Ruzanov P., Melnyk N., Fink D., Sorokin A., Ovchinnikov L. P., Davicioni E., Triche T. J., Sorensen P. H. (2009) Translational activation of Snail and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell 15, 402–415 [DOI] [PubMed] [Google Scholar]
- 7. Jurchott K., Bergmann S., Stein U., Walther W., Janz M., Manni I., Piaggio G., Fietze E., Dietel M., Royer H. D. (2003) YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. J. Biol. Chem. 278, 27988–27996 [DOI] [PubMed] [Google Scholar]
- 8. Sutherland B. W., Kucab J., Wu J., Lee C., Cheang M. C., Yorida E., Turbin D., Dedhar S., Nelson C., Pollak M. (2005) Akt phosphorylates the Y-box binding protein 1 at Ser-102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene 24, 4281–4292 [DOI] [PubMed] [Google Scholar]
- 9. Higashi K., Tomigahara Y., Shiraki H., Miyata K., Mikami T., Kimura T., Moro T., Inagaki Y., Kaneko H. (2011) A novel small compound that promote nuclear translocation of YB-1 ameliorates experimental hepatic fibrosis in mice. J. Biol. Chem. 286, 4485–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Y. F., Homer C., Edwards S. J., Hananeia L., Lasham A., Royds J., Sheard P., Braithwaite A. W. (2003) Nuclear localization of Y-box factor YB-1 requires wild type p53. Oncogene 22, 2782–2794 [DOI] [PubMed] [Google Scholar]
- 11. Moll U. M. (2004) p63 and p73. Roles in development and tumor formation. Mol. Cancer Res. 2, 371–386 [PubMed] [Google Scholar]
- 12. Yang A., Kaghad M., Wang Y., Gillett E., Fleming M. D., Dötsch V., Andrews N. C., Caput D., McKeon F. (1998) p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death inducing, and dominant-negative activities. Mol. Cell 2, 305–316 [DOI] [PubMed] [Google Scholar]
- 13. Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R. T., Tabin C., Sharpe A., Caput D., Crum C., McKeon F. (1999) p63 is essential for regenerative proliferation in limb, craniofacial, and epithelial development. Nature 398, 714–718 [DOI] [PubMed] [Google Scholar]
- 14. Parsa R., Yang A., McKeon F., Green H. (1999) Association of p63 with proliferative potential in normal and neoplastic human keratinocyte. J. Invest. Dermatol. 113, 1099–1105 [DOI] [PubMed] [Google Scholar]
- 15. Fukunishi N., Katoh I., Tomimori Y., Tsukinoki K., Hata R., Nakao A., Ikawa Y., Kurata S. (2010) Induction of ΔNp63 by the newly identified keratinocyte-specific transforming growth factor β-signaling pathway with Smad2 and IkB kinase α in squamous cell carcinoma. Neoplasia 12, 969–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Higashikawa K., Yoneda S., Tobiume K., Taki M., Shigeishi H., Kamata N. (2007) Snail induced down-regulation of ΔNp63α acquires invasive phenotype of human squamous cell carcinoma. Cancer Res. 67, 9207–9213 [DOI] [PubMed] [Google Scholar]
- 17. Graziano V., De Laurenzi V. (2011) Role of p63 in cancer development. Biochim. Biophys. Acta 1816, 57–66 [DOI] [PubMed] [Google Scholar]
- 18. Adorno M., Cordenonsi M., Montagner M., Dupont S., Wong C., Hann B., Solari A., Bobisse S., Rondina M. B., Guzzardo V., Parenti A. R., Rosato A., Bicciato S., Balmain A., Piccolo S. (2009) A mutant-p53/Smad complex opposes p63 to empower TGFβ-induced metastasis. Cell 137, 87–98 [DOI] [PubMed] [Google Scholar]
- 19. Di Costanzo A., Festa L., Duverger O., Vivo M., Guerrini L., La Mantia G., Morasso M. I., Calabrò V. (2009) Homeodomain protein Dlx3 induces phosphorylation-dependent p63 degradation. Cell Cycle 8, 1185–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lefort K., Mandinova A., Ostano P., Kolev V., Calpini V., Kolfschoten I., Devgan V., Lieb J., Raffoul W., Hohl D., Neel V., Garlick J., Chiorino G., Dotto G. P. (2007) Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKα kinases. Genes Dev. 21, 562–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lo Iacono M., Di Costanzo A., Calogero R. A., Mansueto G., Saviozzi S., Crispi S., Pollice A., La Mantia G., Calabrò V. (2006) The Hay Wells syndrome-derived TAp63αQ540L mutant has impaired transcriptional and cell growth regulatory activity. Cell Cycle 5, 78–87 [DOI] [PubMed] [Google Scholar]
- 22. Radoja N., Guerrini L., Lo Iacono N., Merlo G. R., Costanzo A., Weinberg W. C., La Mantia G., Calabrò V., Morasso M. I. (2007) Homeobox gene Dlx3 is regulated by p63 during ectoderm development. Relevance in the pathogenesis of ectodermal dysplasia. Development 134, 13–18 [DOI] [PubMed] [Google Scholar]
- 23. Cui S., Arosio D., Doherty K. M., Brosh R. M., Jr., Falaschi A., Vindigni A. (2004) Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein. Nucleic Acid Res. 32, 2158–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dunn G. A. (1983) Characterizing a kinesis response. Time averaged measures of cell speed and directional persistence. Agents Actions Suppl. 12, 14–33 [DOI] [PubMed] [Google Scholar]
- 25. Dickinson R. B., Tranquillo R. T. (1993) Optimal estimation of cell movement indices from the statistical analysis of cell tracking data. AIChE J. 39, 1995–2010 [Google Scholar]
- 26. Amoresano A., Di Costanzo A., Leo G., Di Cunto F., La Mantia G., Guerrini L., Calabrò V. (2010) Identification of ΔNp63α protein interactions by mass spectrometry. J. Proteome Res. 9, 2042–2048 [DOI] [PubMed] [Google Scholar]
- 27. Hibi K., Trink B., Patturajan M., Westra W. H., Caballero O. L., Hill D. E., Ratovitski E. A., Jen J., Sidransky D. (2000) AIS is an oncogene amplified in squamous cell carcinoma. Proc. Natl. Acad. Sci U.S.A. 97, 5462–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee J. M., Dedhar S., Kalluri R., Thompson E. W. (2006) The epithelial-mesenchymal transition. New insights in signaling, development, and disease. J. Cell Biol. 172, 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Di Como C. J., Urist M. J., Babayan I., Drobnjak M., Hedvat C. V., Teruya-Feldstein J., Pohar K., Hoos A., Cordon-Cardo C. (2002) p63 expression profiles in human normal and tumor tissues. Clin. Cancer Res. 8, 494–501 [PubMed] [Google Scholar]
- 30. Barbareschi M., Pecciarini L., Cangi M. G., Macrì E., Rizzo A., Viale G., Dogliosi C. (2001) p63, a p53 homologue, is a selective nuclear marker of myoepithelial cells of the human breast. Am. J. Surg. Pathol. 25, 1054–1060 [DOI] [PubMed] [Google Scholar]
- 31. Danilov A. V., Neupane D., Nagaraja A. S., Feofanova E. V., Humphries L. A., DiRenzo J., Korc M. (2011) ΔNp63α-mediated induction of epidermal growth factor receptor promotes pancreatic cancer cell growth and chemoresistance. PLoS One 6, e26815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Astanehe A., Finkbeiner M. R., Hojabrpour P., To K., Fotovati A., Shadeo A., Stratford A. L., Lam W. L., Berquin I. M., Duronio V., Dunn S. E. (2009) The transcriptional induction of PIK3CA in tumor cells is dependent on the oncoprotein Y-box binding protein-1. Oncogene 28, 2406–2418 [DOI] [PubMed] [Google Scholar]
- 33. Evdokimova V., Tognon C., Ng T., Ruzanov P., Melnyk N., Fink D., Sorokin A., Ovchinnikov L. P., Davicioni E., Triche T. J., Sorensen P. H. (2009) Reduced proliferation and enhanced migration: Two sides of the same coin? Molecular mechanisms of metastatic progression by YB-1. Cancer Cell 15, 402–415 [DOI] [PubMed] [Google Scholar]
- 34. Peiró S., Escrivà M., Puig I., Barberà M. J., Dave N., Herranz N., Larriba M. J., Takkunen M., Francí C., Muñoz A., Virtanen I., Baulida J., García de Herreros A. (2006) Snail1 transcriptional repressor binds to its own promoter and controls its expression. Nucleic Acids Res. 34, 2077–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barbieri C. E., Tang L. J., Brown K. A., Pietenpol J. A. (2006) Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res. 66, 7589–7597 [DOI] [PubMed] [Google Scholar]
- 36. King K. E., Ponnamperuma R. M., Yamashita T., Tokino T., Lee L. A. (2003) ΔNp63α functions as a positive and negative transcriptional regulator and blocks in vitro differentiation of murine keratinocytes. Oncogene 22, 3635–3644 [DOI] [PubMed] [Google Scholar]
- 37. Ha L., Ponnamperuma R. M., Jay S., Ricci M. S., Weinberg W. C. (2011) Dysregulated ΔNp63α inhibits expression of Ink4a/arf, blocks senescence, and promotes malignant conversion of keratinocytes. PLoS One 6, e21877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koike K., Uchiumi T., Ohga T., Toh S., Wada M., Kohno K., Kuwano M. (1997) Nuclear translocation of the Y-box binding protein by ultraviolet irradiation. FEBS Lett. 417, 390–394 [DOI] [PubMed] [Google Scholar]
- 39. Ruzanov P. V., Evdokimova V. M., Korneeva N. L., Hershey J. W., Ovchinnikov L. P. (1999) Interaction of the universal mRNA-binding protein, p50, with actin. A possible link between mRNA and microfilaments. J. Cell Sci. 20, 3487–3496 [DOI] [PubMed] [Google Scholar]
- 40. Uchiumi T., Fotovati A., Sasaguri T., Shibahara K., Shimada T., Fukuda T., Nakamura T., Izumi H., Tsuzuki T., Kuwano M., Kohno K. (2006) Yb-1 is important for an early stage embryonic development. Neural tube formation and cell proliferation. J. Biol. Chem. 281, 40440–40449 [DOI] [PubMed] [Google Scholar]
- 41. Chernov K. G., Mechulam A., Popova N. V., Pastre D., Nadezhdina E. S., Skabkina O. V., Shanina N. A., Vasiliev V. D., Tarrade A., Melki J., Joshi V., Baconnais S., Toma F., Ovchinnikov L. P., Curmi P. A. (2008) Yb-1 promotes microtubules assembly in vitro through interaction with tubulin and microtubules. BMC Biochem. 9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]