Background: HBXIP is a novel oncoprotein.
Results: HBXIP was able to up-regulate S100A4 though activating STAT4 and inducing DNA methylation of PTEN.
Conclusion: HBXIP up-regulates S100A4 via two pathways to promote growth and migration of breast cancer cells.
Significance: Our finding provides new insight into the mechanism of HBXIP in promotion of growth and migration of breast cancer.
Keywords: Breast Cancer, Gene Regulation, Migration, Proliferation, Signaling, PTEN, S100A4, STAT4, Hepatitis B X-interacting Protein
Abstract
We have reported that hepatitis B X-interacting protein (HBXIP) promotes the proliferation and migration of breast cancer cells. However, the underlying mechanism is poorly understood. In this study, we report that HBXIP works in the event through up-regulating S100A4. We observed that HBXIP expression was positively correlated to that of S100A4 in 87 clinical breast cancer tissue samples. Then, we identified that HBXIP was able to up-regulate S100A4 expression in breast cancer cells. Notably, we observed the HBXIP nuclear localization, implying that HBXIP may be associated with the promoter of S100A4. Chromatin immunoprecipitation assay (ChIP) and electrophoretic mobility shift assay (EMSA) showed that HBXIP was able to bind to the nucleotides +200∼+239 region of S100A4 promoter, containing two putative recognition motif of transcription factor STAT4 and GRβ. It suggests that HBXIP is able to activate S100A4 promoter via interacting with STAT4 in breast cancer cells, leading to the up-regulation of S100A4. In addition, we identified another pathway of S100A4 up-regulation mediated by HBXIP. We found that HBXIP activated the PTEN/PI3K/AKT signaling by inducing DNA methylation of PTEN, which subsequently boosted S100A4 expression. In function, we demonstrated that HBXIP enhanced the growth or migration of breast cancer cells through S100A4 in vivo and in vitro. Collectively, we conclude that HBXIP up-regulates S100A4 through activating S100A4 promoter involving STAT4 and inducing PTEN/PI3K/AKT signaling to promote growth and migration of breast cancer cells. Our finding provides new insight into the mechanism of HBXIP in promotion of the development of breast cancer.
Introduction
Hepatitis B X-interacting protein (HBXIP),5 a cellular 18 kDa protein, was originally identified by its interaction with the C terminus of hepatitis B virus (HBV) X protein (HBx) (1). It has been reported that HBXIP may form a complex with survivin, an antiapoptotic protein, resulting in the suppression of cell apoptosis through the mitochondrial/cytochrome pathway. HBXIP can also bind to hSuv3p (2). In addition, as a regulator of centrosome dynamics and cytokinesis, HBXIP is necessary for bipolar spindle formation in human HeLa carcinoma cells (3). Our previous studies reported that HBXIP could promote cell proliferation via nucleus factor κB (NF-κB) (4). Recently we have reported that miR-520b plays an impotent role in the migration of breast cancer cells through HBXIP (5). However, the mechanism by which HBXIP enhances proliferation and migration of breast cancer cells is poorly understood.
S100A4 is a member of the S100 family of small, homodimeric, EF-hand Ca2+-binding proteins (6). S100 proteins are able to interact with a variety of target proteins, leading to the regulation of specific cellular processes, such as cell cycle regulation, protein phosphorylation, cell growth, motility, differentiation, and survival (6–9). Growing evidence indicated that the elevated S100A4 protein is associated with the progression and metastasis of several malignant tumors, including esophageal, non-small cell lung, pancreatic, thyroid, colorectal, bladder, gastric, prostate, and breast cancer (11–14). It has been reported that the high levels of S100A4 can be induced by the activation of PI3K/AKT signaling pathway which promotes the proliferation and growth of cancer cells (15). The tumor suppressor PTEN is a physiological inhibitor of the PI3K/Akt signal transduction pathway. The PTEN down- regulation is related to the methylation of PTEN promoter, and this has frequently been found in diverse cancer types, including cervical, adenoid cystic, gastric, and breast cancer (16–18). Loss of PTEN expression results in hyper-activation of the PI3K/AKT signaling pathway, a cascade that plays a vital role in cell proliferation and survival when dysregulated in tumorigenesis (19, 20).
Signal transducers and activators of transcription (STAT) proteins are components of JAK/STAT signal transduction pathways, involving immune response, cell proliferation, migration and programmed cell death in mammalian organisms (21). STAT4, as a member of STAT family, plays a crucial role in biological functions of IL-12, such as the differentiation of T helper type I (Th1) cells and optimal IFN-γ production (22). It has been reported that STAT4 is highly expressed in hematopoietic cells (23, 24). However, the role of STAT4 in carcinogenesis remains unclear.
In the present study, we investigated the effect of HBXIP on regulation of S100A4. Our data show that HBXIP promotes growth and migration of breast cancer cells through up-regulating S100A4 involving activating S100A4 promoter and inducing PTEN/PI3K/AKT signaling. Our finding provides new insight into the mechanism of HBXIP in promotion of the development of breast cancer.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatment
Breast cancer cell lines, MCF-7, T47D, LM-MCF-7 (a metastatic subclone from the MCF-7 breast cancer cell line), were cultured in RPMI Medium 1640 (Invitrogen, Grand Island, NY) with 10% fetal calf serum (FCS). MDA-MB-231 was cultured in DMEM (Invitrogen) supplemented with 10% fetal calf serum (FCS) (Invitrogen), 100 units/ml penicillin, 100 units/ml streptomycin, and 1% glutamine. Stable cell lines were generated by transfected plasmids (pCMV-Tag-2B, pCMV-HBXIP, pSilencer-random and pSilencer-HBXIP vectors) into breast cancer cells with Lipofectamine 2000 and selected with G418 (Invitrogen). The engineered cell lines were named as follows: MCF-7-pCMV (or T47D-pCMV) (stably transfected pCMV-Tag-2B empty vector), MCF-7-HBXIP (or T47D-HBXIP) (stably transfected pCMV-HBXIP plasmid), LM-MCF-7-psi-control (stably transfected pSilencer vector containing a random fragment), LM-MCF-7-psi-HBXIP (stably transfected pSilencer vector containing the HBXIP RNAi fragment). These cell lines were cultured at 37 °C in a humidified atmosphere with 5% CO2. Cells were collected and seeded in 6-well, 24-well or 96-well plates for 24 h and then were transfected with plasmid or siRNA. All transfections were performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol.
Immunohistochemistry (IHC)
Breast cancer tissue array (No.08C14), comprising duplicates of 49 cases of infiltrating primary carcinoma and 38 metastatic tumors, was purchased from Xi'an Aomei Biotechnology (Xi'an, China). Immunohistochemistry assay was performed as described previously (5). The slides were incubated with rabbit anti-HBXIP (or rabbit anti-S100A4) antibody at 4 °C for overnight. After incubation at room temperature for 30 min with biotinylated secondary antibody, the slides were incubated with streptavidin-peroxidase complex at room temperature for 30 min. Immunostaining was developed by using chromogen, 3, 3′-diaminobenzidine (DAB), and counterstained with Mayer's hematoxylin. The staining level of HBXIP and S100A4 was classified into three groups using a modified scoring method based on the intensity of staining (0 = negative; 1 = low; 2 = high) and the percentage of stained cells (0 = 0% stained; 1 = 1–49% stained; 2 = 50–100% stained). A multiplied score (intensity score × percentage score) lower than 1 was considered to be negative staining (−), 1 and 2 were considered to be moderate staining (+), and 4 was considered to be intense staining (++).
Plasmid Construction and Small Interference RNA (siRNA)
PCMV-tag2B, pSilencer, pGL3-Basic, pGL3-Control vectors (Promega, Madison, WI), pCMV-HBXIP, pSilencer-HBXIP were kept in our laboratory. The complete human S100A4 (GenBankTM accession No.NM_002961.2) cDNA or PTEN (GenBankTM accession No.NG_007466.1) cDNA was subcloned into pCMV-tag2B or pcDNA3.0 vector to generate the pCMV-S100A4 or pcDNA3-PTEN construct.
The 5′-flanking region (from −921 to +1001 nt) of S100A4 gene was amplified by PCR from the genomic DNA of MCF-7 cells using specific primers and was inserted into the KpnI/HindIII site in the upstream of the luciferase gene in the pGL3-Basic vector. The resulting plasmid was sequenced and named pGL-P1. Different regions (−51/+1001, +210/+1001, +334/+1001, +487/+1001) of S100A4 promoter were amplified by PCR from the pGL-P1 using specific primers and were inserted into the pGL3-Basic vector, named pGL-P2, pGL-P3, pGL-P4, pGL-P5, respectively. Mutant construction of +210/+1001 region of S100A4 promoter, named as pGL-P3-STAT4mut, carried a substitution of three nucleotides within the binding sites of STAT4. pGL-P3-wt presents the pGL-Basic plasmid containing the wild type of +210/+1001 fragment. All primers are listed in supplemental Table S1.
siRNAs duplexes targeting human S100A4 gene, STAT4 gene (GenBankTM accession No.NM_001243835.1) and AKT gene and siRNA duplexes with nonspecific sequences using as negative control (NC) were designed and synthesized by RiboBio (Guangzhou, China). All oligonucleotide sequences are listed in supplemental Table S1.
Reverse-transcription PCR (RT-PCR) and Quantitative Real-time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from the cells using Trizol reagent (Invitrogen, Carlsbad, CA). First-strand cDNA was synthesized by PrimeScript reverse transcriptase (TaKaRa Bio, Dalian, China) and oligo(dT) following the manufacturer's instructions. To examine the expression, real-time PCR was performed by a Bio-Rad sequence detection system according to the manufacturer's instructions using double-stranded DNA-specific SYBR Premix Ex TaqTM II Kit (TaKaRa Bio). Double-stranded DNA specific expression was tested by the comparative Ct method using 2−ΔΔCt. Primer sets for specific genes are showed in supplemental Table S1.
Western Blot Analysis
Western blotting was carried out with standard protocols. Primary antibodies used were rabbit anti-S100A4 (Proteintech Group), rabbit anti-PTEN (Proteintech Group), rabbit anti-HBXIP (Proteintech Group), mouse anti-pAKT (Abcam, Cambridge, UK), rabbit anti-AKT (BOSTER, Wuhan, China), rabbit anti-STAT4 (BOSTER), rabbit anti-DNMT1 (BOSTER), rabbit anti-pSTAT4 (SAB), rabbit anti-HistoneH3 (Proteintech Group), and mouse anti-β-actin (Sigma, Aldrich). Protein bands were quantified using Quantity One software (Bio-Rad).
Luciferase Reporter Gene Assay
Adherent cells were seeded into 24-well plates and co-transfected with the constructs containing different length fragments of S100A4 promoter or pGL3-Basic, pGL-P3-STAT4mut, and the pRL-TK plasmid (Promega, Madison, WI) which is used as internal normalization. Cell extracts were harvested after 36 h and lysed using lysis buffer (Promega). Luciferase reporter gene assay was implemented using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. All experiments were performed at least three times.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed using EpiQuikTM Chromatin Immunoprecipitation Kit from Epigentek Group Inc (Brooklyn, NY). Protein-DNA complexes were immunoprecipatited with HBXIP or STAT4 antibody, with mouse IgG as a control, respectively. DNA from these samples was then subjected to PCR analysis. Primers sets for S100A4 promoter were 5′-GAG ATC CAG ATG TGA GAT TC-3′ and 5′-GGG TTG GAA GAG AAG CTG CA-3′. Primers sets for a distance region of MEKK2 gene promoter using as the negative control were 5′-TCC ACC TGT TCA TCC CTG-3′ and 5′-GAG CCA AGA TTC CAC CAC-3′, followed by sequencing. Sonicated DNA fragment prior to immunoprecipitation was used as an input control.
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear protein extracts were prepared from MCF-7 cells. Probes were generated by annealing single strand oligonucleotides containing the S100A4 promoter and labeling the ends with [γ-32P]ATP using T4 polynucleotide kinase (TaKaRa Bio). Binding reactions were performed at 4 °C for 1 h, in 10 μl mixtures containing 20 mm Hepes, 50 mm KCl, 0.05 mm DTT, 0.05 mm EDTA, 1 mm MgCl2, 5% glycerol, 0.05 μg/μl of poly (dI·dC), 0.05% Nonidet P-40, 50 ng probe, and 1.5 μg nuclear extract. Specificity of HBXIP-DNA or STAT4-DNA interaction was confirmed by competition or supershift with HBXIP or STAT4 antibody, respectively. For the antibody competitive or supershift experiment, 1 μg of HBXIP or STAT4 antibody were added into the reaction mixture and incubated at 4 °C for 30 min before the DNA probe was added. For the competitive binding experiment of cold competitor, 500 ng unlabeled DNA was added after the initial incubation for additional 30 min. The binding mixtures were then resolved on a native 6% polyacrylamide gel in 0.5× TBE at 4 °C. The gel was dried and exposed to x-ray film for autoradiography. Primers for each DNA fragment and oligonucleotide sequences are listed in supplemental Table S1.
Preparation of Recombinant Proteins
The plasmid pET-28a-HBXIP was used to express recombinant His-HBXIP in Escherichia coli strain BL21. The recombinant proteins were expressed by induction with 0.5 mm isopropyl β-d-1-thiogalactopyranoside at 16 °C for 20 h and purified by Ni2+-NTA affinity chromatography according to standard procedures.
Co-immunoprecipitation(Co-IP) Assay
MCF-7 cells(2 × 106) were harvested and lysed in a lysis buffer (50 mm Tris-HCl pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.3% Triton X-100, 1 mm protease inhibitor PMSF). The lysates were incubated with antibodies and protein G-conjugated agarose beads at 4 °C for 3 h. The precipitates were washed six times with ice-cold lysis buffer, resuspended in the same buffer, and resolved by SDS-PAGE followed by immunoblotting.
Methylation-specific PCR and Bisulfite Sequencing
Promoter methylation of PTEN gene was analyzed by methylation-specific PCR (MSP), as described previously (17). Genomic DNA from MCF-7 stable cell lines was subjected to bisulfite modification using Epitect Bisulfite Kit (Qiagen) according to the manufacturer's protocol. MSP experiments were performed at least in duplicate. Bisulfite sequencing was performed to further investigate the methylation frequency of CpG site in PTEN promoter, as described previously (26). We randomly selected three clones from MCF-7-PCMV or MCF-7-HBXIP stable cell lines to examine methylation status. The primer sequences are showed in supplemental Table S1.
Analysis of Cell Proliferation and Migration
Cell proliferation was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma) assay as described previously (27) and 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay, which was carried out using the Cell-Light TM EdU imaging detection kit (RiboBio) according to the manufacturer's instructions. Wound healing assay for migration analysis was performed as described previously (5).
Animal Transplantation
Three groups of 4-week-old female BALB/c athymic nude mice (Experiment Animal Center of Peking, China) (each group, n = 6) were housed and treated according to guidelines established by the National Institutes of Health Guide for the Care and Use of Laboratory Animals. According to the report (28), MCF-7-pCMV, MCF-7-HBXIP, and MCF-7-HBXIP-siS100A4 (MCF-7-HBXIP cells were transfected with 100 nm siS100A4) cells were harvested and suspended at 5 × 107 cells/ml with phosphate saline and then subcutaneously injected at the shoulder with 0.2 ml of the cell suspensions. After injected 10 days, the tumor growth was measured every 3 days. Tumor volume (V) was monitored by measuring the length (L) and width (W) with calipers and calculated with the formula (L ×W2) × 0.5. After 30 days, three groups of the mice were sacrificed, and the tumors were excised and measured.
Statistical Analysis
Each experiment was repeated at least three times. Statistical significance was assessed by comparing mean values (± S.D.) using a Student's t test for independent groups and was assumed for p < 0.05 (*) and p < 0.01 (**).
RESULTS
HBXIP Up-regulates S100A4 in Breast Cancer Cells
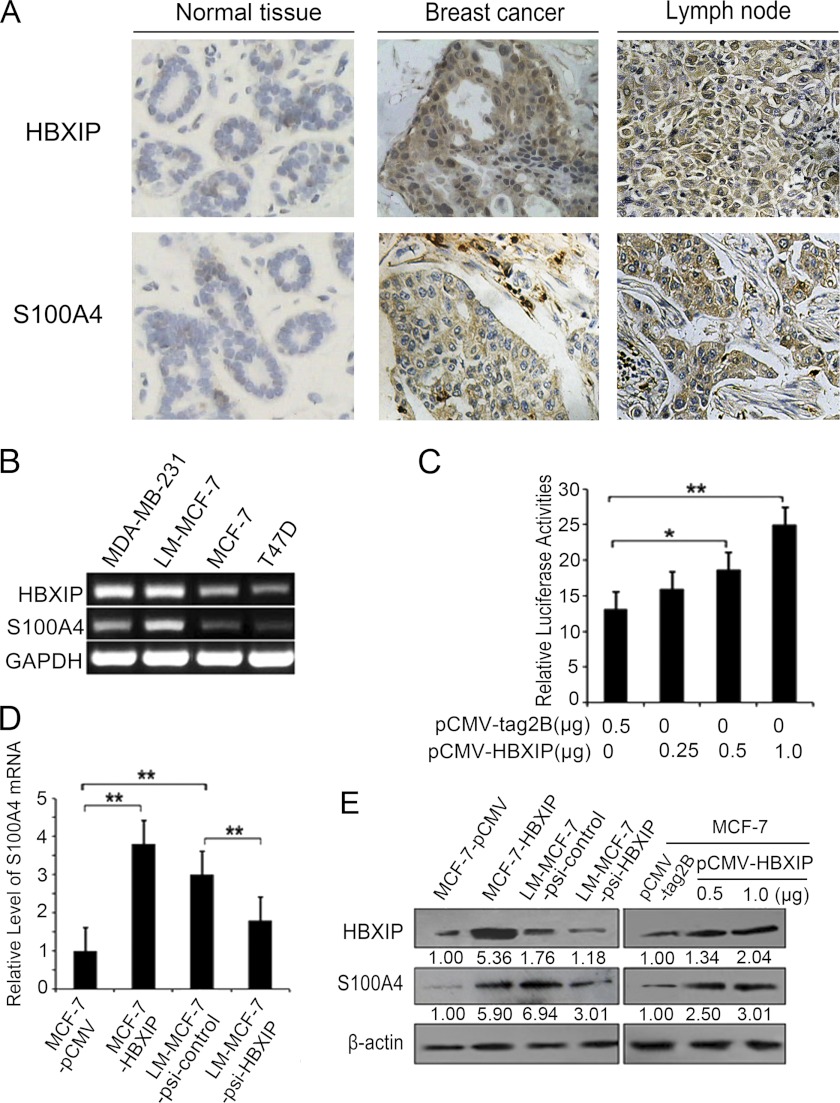
Previously, we found that HBXIP expression was positively associated with the tumor carcinogenesis and metastasis. It may be a novel oncoprotein (5). Accordingly, S100A4 is an important regulator and closely associated with the progression of breast cancer (12, 13). Therefore, we supposed that HBXIP may be associated with S100A4 in promotion of cell proliferation and migration. Then, we try to investigate the expression relationship between HBXIP and S100A4 by IHC using tissue arrays which were from the same tissue paraffin block. We found that the positive rate of HBXIP was 77.6% (38/49) in clinical primary breast cancer tissue samples, in which the positive rate of S100A4 was 81.8% (31/38) in the HBXIP-positive specimens, while the positive rate of HBXIP was 94.7% (36/38) in clinical metastasis breast cancer tissue samples, in which the positive rate of S100A4 was 88.9% (32/36) in the HBXIP-positive specimens (Fig. 1A, supplemental Table S2). The data suggest that the expression of S100A4 is relevant to that of HBXIP in the same tissues. Then, we further observed the expression relationship between HBXIP and S100A4 in breast cancer cell lines including LM-MCF-7, MDA-MB-231, MCF-7, and T47D by RT-PCR (Fig. 1B). The data supported the above conclusion. Therefore, our data imply that HBXIP may regulate S100A4. To examine the effect of HBXIP on S100A4, we cloned the promoter region of S100A4 (−921/+1001) into pGL3-Basic plasmid. Luciferase reporter gene assay showed that HBXIP could increase the promoter activity of S100A4 in a dose-dependent manner in MCF-7 cells (*, p < 0.05, **, p < 0.01, Student's t test, Fig. 1C). Furthermore, we demonstrated that S100A4 was up-regulated in stably HBXIP-transfected cell line (MCF-7-HBXIP) and down-regulated in stably HBXIP knockdown cell line (LM-MCF-7-psi-HBXIP) at the levels of mRNA and protein (Fig. 1, D and E) in a dose-dependent manner in MCF-7 cells, suggesting that HBXIP is capable of up-regulating S100A4 in breast cancer cells.
FIGURE 1.
HBXIP up-regulates S100A4 in breast cancer cells. A, expression of HBXIP and S100A4 was examined by immunohistochemistry staining in normal breast tissues, breast cancer tissues, and metastatic lymph node tissues. B, expression levels of HBXIP and S100A4 mRNA were examined by RT-PCR in breast cancer cell lines. C, promoter activity of S100A4 was examined by luciferase reporter gene assays in MCF-7 cells transfected with different amounts of the pCMV-HBXIP plasmids. Luciferase activities were measured 24 h after transfection (*, p < 0.05, **, p < 0.01, versus control, Student's t test). D and E, expression levels of S100A4 were examined using qRT-PCR and Western blot analysis in the cell lines of MCF-7-pCMV, pCMV-HBXIP, LM-MCF-7-psi-control, and LM-MCF-7-psi-HBXIP, respectively. The expression levels of HBXIP and S100A4 were analyzed by Western blot assay in MCF-7 cells transiently transfected with pCMV-HBXIP in a dose-dependent manner (**, p < 0.01, versus control, Student's t test). The data presented are from three independent experiments; error bars represent standard error. The intensity for each band was densitometrically quantified. The value under each lane indicates the relative amounts of protein relative to control group. The value is obtained by the intensity ratio between the target protein and β-actin band in each lane. Protein bands were quantified using Quantity One software (Bio-Rad).
HBXIP Activates S100A4 via Binding to +200/+239 Region in S100A4 Promoter
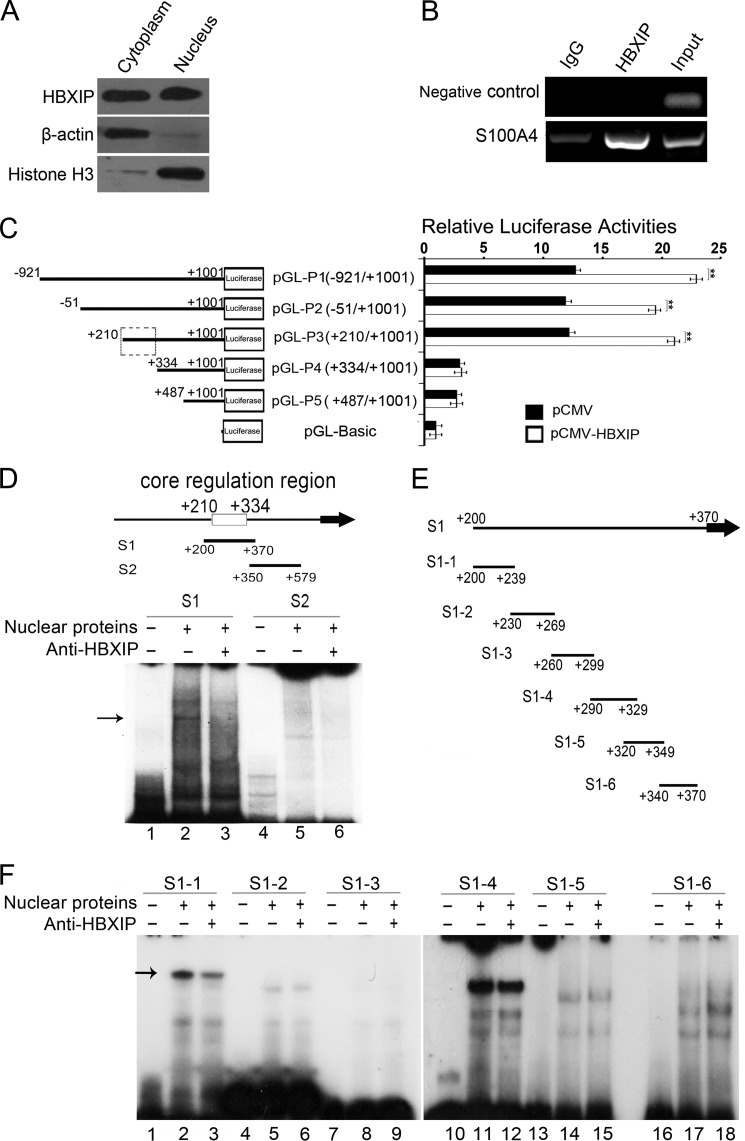
Next, we try to investigate the underlying mechanism by which HBXIP up-regulates S100A4. IHC staining showed that HBXIP expression could be observed in both cytoplasm and nucleus in breast cancer tissues (Fig. 1A). Western blot assay validated the distribution of HBXIP in both cytoplasm and nucleus of MCF-7 cells (Fig. 2A). Thus, we speculated that HBXIP might be involved in the transcriptional regulation of S100A4 in the nucleus. Interestingly, ChIP assays showed HBXIP was available to occupy the S100A4 promoter (Fig. 2B). To map the HBXIP binding site in S100A4 promoter, we cloned a series of fragments of S100A4 promoter 5′-flanking region, including nucleotides −921/+1001 (pGL-P1), −51/+1001 (pGL-P2), +210/+1001 (pGL-P3), +334/+1001 (pGL-P4), and +487/+1001 (pGL-P5). We found that the overexpression of HBXIP was able to obviously enhance the luciferase activities of the plasmids, such as pGL-P1, pGL-P2, and pGL-P3, rather than pGL-P4 and pGL-P5 (**, p < 0.01, Student's t test. Fig. 2C), suggesting that the +210/+334 region in S100A4 promoter is the regulatory target sequence of HBXIP. We further examined whether HBXIP was able to bind to the +210/+334 region in S100A4 promoter by EMSA. The DNA segment S1 (+200/+370) and S2 (+350/+579) were used as probes. We observed an obvious interaction between segment S1 and proteins of nuclear extract (Fig. 2D, lane 2), which could be blocked by anti-HBXIP antibody (Fig. 2D, lane 3), suggesting that HBXIP is able to bind to the segment S1. However, the segment S2 failed to work (Fig. 2D, lanes 4–6). Furthermore, the segment S1 was divided into a series of six overlapping DNA fragments (Fig. 2E). EMSA indicated that HBXIP could interact with segment S1-1 (+200/+239) (Fig. 2F). Thus, we conclude that HBXIP activates S100A4 promoter activity via binding to the +200/+239 region in S100A4 promoter.
FIGURE 2.
HBXIP activates S100A4 via binding to +200/+239 region in S100A4 promoter. A, distribution of HBXIP in the cytoplasm or nucleus of MCF-7 cells was detected by Western blot assay, respectively. B, interaction of HBXIP with promoter region of S100A4 was determined by ChIP assay. The +208/+584 region of S100A4 promoter was amplified. The data presented are from three independent experiments. C, activities of various length fragments of S100A4 promoter constructs and the effect of HBXIP on different S100A4 promoter regions were determined by luciferase reporter gene assays in MCF-7 cells, respectively (**, p < 0.01, versus control, Student's t test). The data presented are from three independent experiments; error bars represent standard error. D, binding of endogenous HBXIP to S100A4 promoter was showed by EMSA. 32P-labeled S1 and S2 probes were incubated with nuclear extracts containing endogenous HBXIP in the absence (lanes 2 and 5) or presence (lanes 3 and 6) of anti-HBXIP antibody as indicated. Lanes 1 and 4 show the control reactions without nuclear extracts. The arrow denoted that anti-HBXIP antibody blocked the HBXIP binding to fragment S1. E, scheme shows that the 6 probes (S1-1 to S1-6) were from S1. F, interaction of HBXIP with probe S1-1 to S1-6 was examined by EMSA. The arrow denoted that anti-HBXIP antibody blocked the HBXIP binding to fragment S1-1.
HBXIP Activates S100A4 via Binding STAT4 Involving the STAT4 Element in the +200/+239 Region of S100A4 Promoter
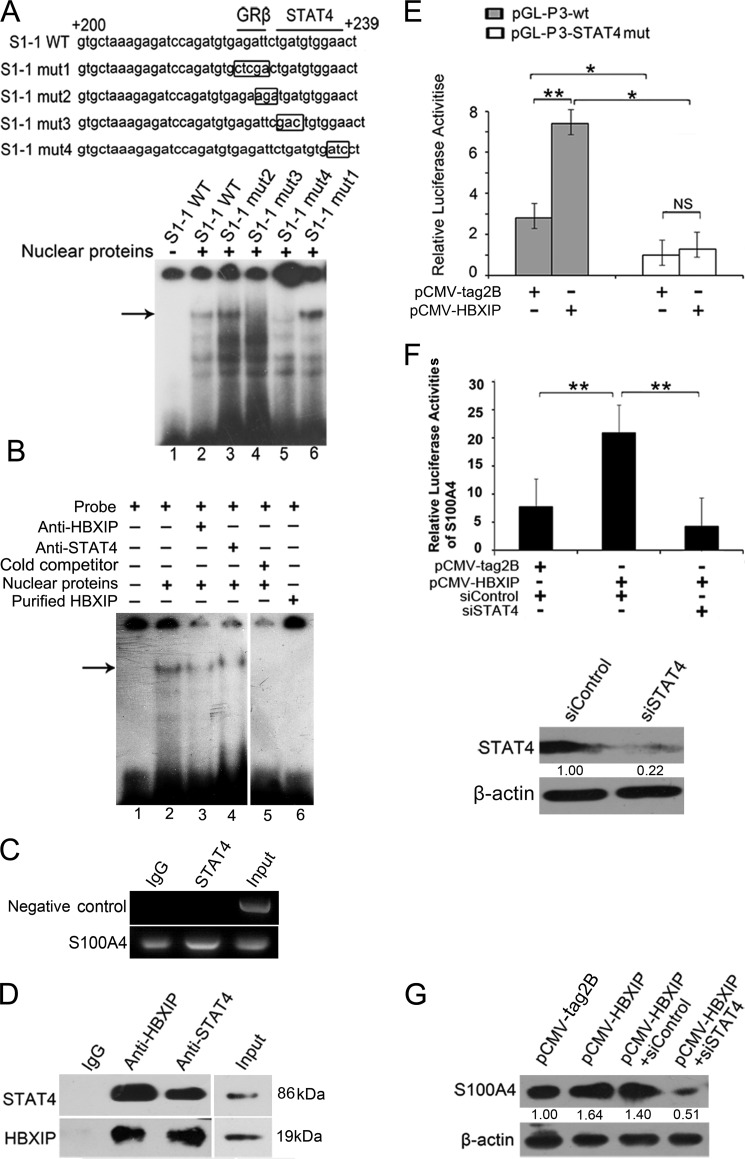
Using online promoter analysis tool Search Promoter Site, we predicted two putative transcription factor binding sites in the +200 ∼+239 promoter region of S100A4, such as GRβ and STAT4. Accordingly, we further determined whether the interaction of HBXIP with S100A4 promoter was related to them. EMSA showed that both of the two STAT4 binding site mutants (S1-1 mut3 and S1-1 mut4) in STAT4 recognition motif were able to disrupt the protein-DNA interaction, rather than GRβ binding site mutant (S1-1 mut1) or the mutant in 5′-flanking nucleotides of STAT4 binding sequence (S1-1 mut2) (Fig. 3A). We further validated the finding that HBXIP was able to interact with STAT4 binding site by using anti-STAT4 antibody or anti-HBXIP antibody (Fig. 3B), suggesting that the transcription factor STAT4 is responsible for the HBXIP-DNA interaction. In addition, we further observed that STAT4 was able to interact with S100A4 promoter by ChIP assay (Fig. 3C), supporting the above conclusion.
FIGURE 3.
HBXIP activates S100A4 via binding STAT4 involving the STAT4 element in +200/+239 region of S100A4 promote. A, interaction of nuclear proteins with mutants of +200/+239 region in S100A4 promoter was determined by EMSA. The mutated nucleotides were showed in box. B, binding of nuclear proteins to S1-1 fragment with the addition of anti-HBXIP or anti-STAT4 antibody was examined by EMSA. C, interaction of STAT4 with promoter region of S100A4 was examined by ChIP assay. The +208/+584 region of S100A4 promoter was amplified by PCR. The data presented are from three independent experiments. D, interaction between endogenous HBXIP and STAT4 was examined in MCF-7 cells by Co-IP assay. HBXIP and STAT4 in cell lysate were examined by Western blot, which was used as an input control. E, MCF-7 cells were transiently co-transfected with plasmids of pCMV-HBXIP (or pCMV-tag2B) and pGL-P3 wt (or pGL-P3-STAT4mut) constructs (*, p < 0.05, **, p < 0.01, versus control, Student's t test). The data presented are from three independent experiments; error bars represent standard error. F, MCF-7 cells were transiently co-transfected with plasmid pCMV-HBXIP (or pCMV-tag2B), 50 nm STAT4 siRNA (or control siRNA) and pGL-P3 constructs. Western blot showed the knockdown efficiency of 50 nm siRNAs for STAT4 in MCF-7 cells. (**, p < 0.01, versus control, Student's t test). The data presented are from three independent experiments; error bars represent standard error. G, MCF-7 cells were transiently transfected with pCMV-HBXIP (or pCMV-tag2B), pCMV-HBXIP, and 50 nm siSTAT4 (or siControl), respectively. The expression levels of S100A4 were examined by Western blot analysis.
Next, we try to demonstrate the interaction between HBXIP and transcription factor STAT4 by co-immunoprecipitations. Our data revealed that HBXIP was able to interact with STAT4 in MCF-7 cells (Fig. 3D). However, the purified recombinant HBXIP protein alone failed to bind to S1-1 oligos by EMSA (Fig. 3B, lane 6). Thus, we conclude that HBXIP indirectly occupies S100A4 promoter through interaction with STAT4.
Then, we tested whether STAT4 was involved in the activation of S100A4 mediated by HBXIP. Luciferase reporter gene assays indicated that HBXIP failed to increase the activity of S100A4 promoter (+210/+1001) when STAT4 binding site was mutated (S1-1 mut4) (*, p < 0.05, **, p < 0.01, Student's t test, Fig. 3E). Moreover, the silence of STAT4 by siRNA abolished HBXIP-enhanced activity of S100A4 promoter. Meanwhile, Western blot analysis showed the silenced efficiency of STAT4 in MCF-7 cells (**, p < 0.01, Student's t test, Fig. 3F). Western blot analysis further confirmed that the silence of STAT4 by siRNA was able to down-regulate the expression of S100A4 at the protein level (Fig. 3G). Thus, we conclude that HBXIP is able to activate S100A4 promoter through interacting with transcription factor STAT4.
HBXIP Up-regulates S100A4 through PTEN/PI3K/AKT Signaling Pathway Independent of STAT4
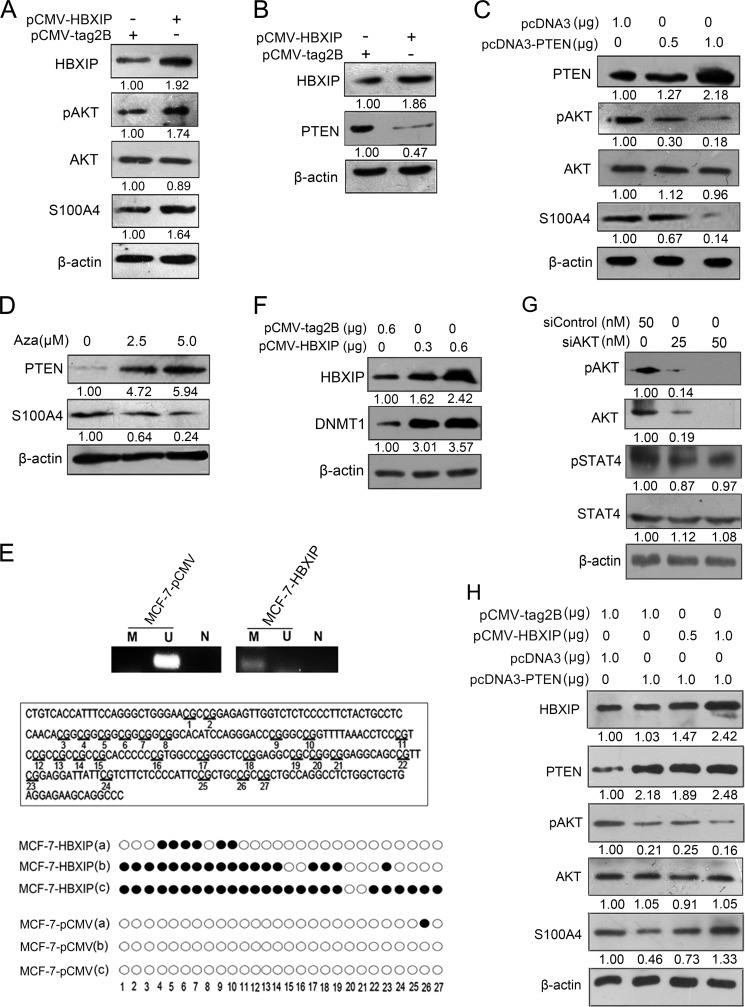
We try to identify other mechanisms by which HBXIP up-regulates S100A4. It has been reported that S100A4 expression can be enhanced by AKT kinase in breast cancer cell lines (29) and HBXIP is able to active AKT signaling in hepatoma HepG2 cells (30). Thus, we supposed that PI3K/AKT pathway may be involved in the regulation of S100A4 meditated by HBXIP in breast cancer cells. Our data showed that the over-expression of HBXIP resulted in greatly increase of the pAKT levels and up-regulation of S100A4 in MCF-7 cells (Fig. 4A), suggesting that PTEN/PI3K/AKT pathway may be involved in the up-regulation of S100A4 mediated by HBXIP.
FIGURE 4.
HBXIP up-regulates S100A4 through PTEN/PI3K/AKT signaling pathway independently of STAT4. A, Western blot analysis showed the expression levels of HBXIP, pAKT, total AKT, and S100A4 levels in MCF-7 cells transfected with pCMV-tag2B or pCMV-HBXIP constructs. B, Western blot analysis showed the expression levels of HBXIP, PTEN in MCF-7 cells transfected with pCMV-tag2B or pCMV-HBXIP constructs. C, pcDNA3 empty vector or pcDNA3-PTEN constructs were transfected into MCF-7 cells in a dose-dependent manner. The expression levels of PTEN, pAKT, total AKT, and S100A4 were determined by Western blot. D, MCF-7 cells treated with 2.5 or 5.0 μm 5-aza-2′-deoxycytidine (Aza) (Sigma) for 72 h. Western bolt showed the expression levels of PTEN and S100A4 in the cells. E, CpG island methylation in PTEN promoter was examined by methylation-specific PCR (MSP) and bisulfite sequencing PCR (randomly selected three clones, such as a, b, c) in MCF-7-pCMV, or MCF-7-HBXIP stable cell lines, respectively. N, negative control using distilled water as the template. Open circles, unmethylated CpG site. Closed circles, methylated CpG site. F, aberrant expression of DNMT1 was examined by Western blot after transfection with pCMV-HBXIP plasmids into MCF-7 cells. G, levels of pAKT, total AKT, pSTAT4, and total STAT4 were examined by Western blot analysis in MCF-7 cells transfected by siControl or siAKT. H, expression levels of HBXIP, PTEN, pAKT, total AKT, and S100A4 were examined by Western blot assay in MCF-7 cells cotransfected with pCMV-tag2B or pCMV-HBXIP constructs and pcDNA3 empty vector or pcDNA3-PTEN constructs.
It has been reported that the tumor suppressor PTEN is a physiological inhibitor of the PI3K/Akt signal transduction pathway (31). Therefore, we next investigated whether HBXIP was able to down-regulate PTEN expression and lead to consequent activation of PTEN/PI3K/AKT pathway in regulation of S100A4. As shown in Fig. 4B, the over-expression of HBXIP dramatically decreased the PTEN expression levels in MCF-7 cells, indicating that PTEN/PI3K/AKT signaling pathway may be involved in the up-regulation of S100A4 mediated by HBXIP. Conversely, the overexpression of PTEN resulted in the decrease of pAKT and down-regulation of S100A4 in a dose-dependent manner in MCF-7 cells (Fig. 4C), suggesting that PTEN/PI3K/AKT signaling pathway is involved in the regulation of S100A4 mediated by HBXIP.
Next, we try to identify the mechanism by which HBXIP inhibits PTEN. It has been reported that PTEN inactivation is associated with DNA methylation (16–18, 32). In our study, we validated the finding in MCF-7 cells using DNA methylation inhibitor 5-aza-2′-deoxycytidine (Aza) (Fig. 4D). We speculated that HBXIP may down-regulate PTEN through inducing methylation of PTEN promoter. The CpG island in PTEN promoter located at ∼216 bp upstream to the start codon is the most frequent site of CpG methylation. DNMT1 was found to play an essential role in aberrant methylation of PTEN promoter in cancer cells (33). Then, we observed the CpG methylation of PTEN gene in stably pCMV- HBXIP-transfected MCF-7 (MCF-7-HBXIP) cells by methylation- specific PCR (MSP) analysis. Meanwhile, the bisulfate sequencing of this CpG island showed the highly frequent methylation of 27 CpG sites in stably HBXIP-transfected MCF-7 cells (Fig. 4E). We further found that the over-expression of HBXIP significantly elevated the DNMT1 protein levels in MCF-7 cells (Fig. 4F). Thus, we conclude that HBXIP down-regulates PTEN through inducing methylation of PTEN promoter involving DNMT1 in breast cancer cells.
So far, we identified two pathways which up-regulate S100A4 mediated by HBXIP. Certainly, we are interested in the relationship between them. We wonder whether STAT4 is one of downstream effectors of PTEN/PI3K/AKT signaling pathway. It is reported that the activity of STATs family is latent until phosphorylation by receptor-associated kinases (34). However, we observed that the inactivation of AKT signaling by siAKT led to no appreciable changes of levels of pSTAT4 in MCF-7 cells (Fig. 4G). In addition, we observed that HBXIP up-regulated S100A4 as well in a dose-dependent manner when activation of AKT signaling was suppressed by over-expressing PTEN in the cells (Fig. 4H). Thus, we conclude that HBXIP up-regulates S100A4 through PTEN/PI3K/AKT signaling pathway independently of STAT4.
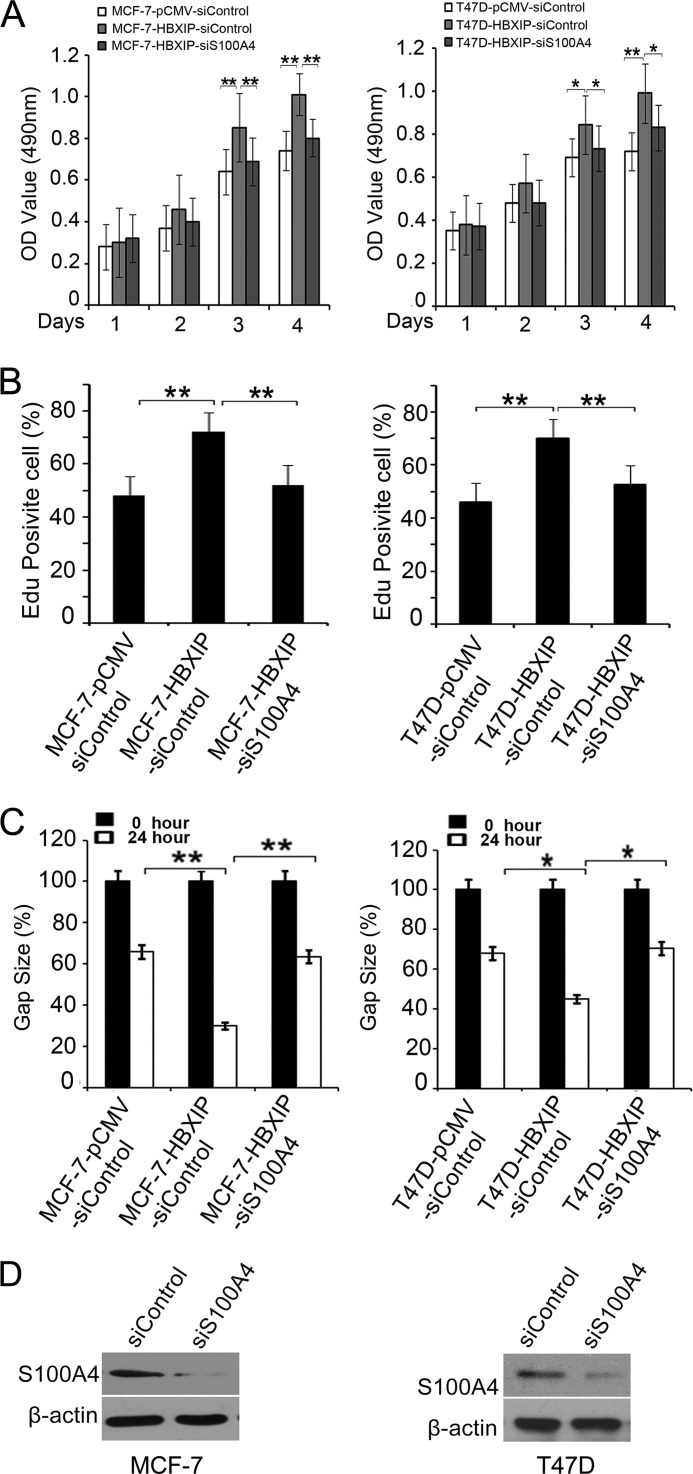
S100A4 Is Responsible for HBXIP-enhanced Proliferation and Migration of Breast Cancer Cells
MTT assays showed that HBXIP was able to promote the growth of MCF-7 and T47D cells in a time course manner, while the treatment with S100A4 siRNAs abolished the enhancement (*, p < 0.05, **, p < 0.01, Student's t test, Fig. 5A). EdU incorporation assays showed the similar results (*, p < 0.05, **, p < 0.01, Student's t test, Fig. 5B). In addition, wound healing assays revealed that HBXIP enhanced the migration capability of MCF-7 and T47D cells, while the treatment with S100A4 siRNAs abolished the enhancement in the cells (*, p < 0.05, **, p < 0.01, Student's t test, Fig. 5C). The knockdown efficiency of S100A4 by siRNA in MCF-7 cells was showed by Western blot (Fig. 5D). These data suggest that HBXIP promotes the proliferation and migration of breast cancer cells through S100A4.
FIGURE 5.
S100A4 is responsible for HBXIP-enhanced proliferation and migration of breast cancer cells. A and B, effect of S100A4 siRNA on the HBXIP enhanced cell proliferation was determined by MTT and EdU incorporation assays, respectively. C, effect of S100A4 siRNA on HBXIP-enhanced cell migration was determined by wound healing assay. The images are representative of at least three independent experiments. Statistically significant differences are indicated: (*, p < 0.05, **, p < 0.01, versus control, Student's t test). The data presented are from three independent experiments; error bars represent standard error. D, knockdown efficiency of S100A4 by siRNA in MCF-7 cells and T47D cells. The cells were treated with 50 nm siS100A4 (or siRNA control) for 48 h, and the cell lysate was subjected to Western blot analysis.
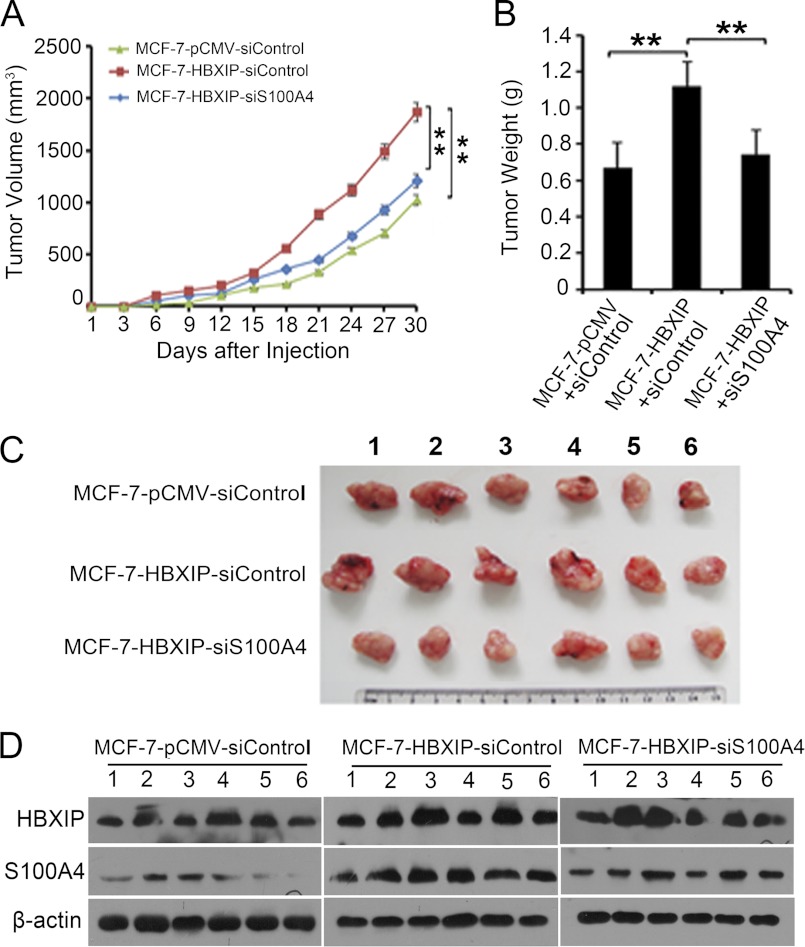
HBXIP Promotes the Tumor Growth through S100A4 in Mice
To further validate the effect of HBXIP on tumor growth through S100A4 in vivo, we performed tumor formation assay. We observed that the volume and weight of tumors were highly enhanced in MCF-7-HBXIP group compared with MCF-7-pCMV control group, whereas the S100A4 knockdown group displayed a significant decreased tumor growth (**, p < 0.01, Student's t test, Fig. 6, A–C). Meanwhile, Western blot analysis showed the HBXIP protein levels and silence efficiency of S100A4 in the tumor tissues from mice (Fig. 6D). Thus, we conclude that HBXIP is capable of promoting growth of breast cancer cells in mice through S100A4.
FIGURE 6.
HBXIP promotes tumor growth through S100A4 in mice. A, growth curve of tumors in nude mice. B, average weight of tumors. C, image of tumors. D, relative expression levels of HBXIP and S100A4 in the tumor tissues from the mice were detected by Western blot analysis, respectively. Statistically significant differences are indicated: (**, p < 0.01, versus control, Student's t test). The data presented are from three independent experiments; error bars represent standard error.
DISCUSSION
Growing evidence reveals that HBXIP is a novel oncoprotein (1–5). Our previous studies showed that HBXIP was highly expressed in breast cancer tissues and metastatic lymph node tissues and significantly associated with the growth and metastasis of breast cancer cells (4, 5). However, the underling mechanism is poorly understood. S100A4 has an established role in tumor progression and metastasis of several malignant tumors (12–14). Therefore, we are interested in whether HBXIP regulates S100A4 in promotion of growth and migration of breast cancer cells.
In this study, we observed that the expression of HBXIP was significantly positive correlation to that of S100A4 in breast cancer tissues. Interestingly, we identified that HBXIP was able to up-regulate S100A4 in breast cancer cells. Next, we sought to elucidate the underlying mechanism by which HBXIP up-regulates S100A4. We observed the HBXIP nuclear localization in breast cancer tissues and MCF-7 cells, implying that HBXIP may be involved in the transcriptional regulation of S100A4. Moreover, we identified that HBXIP was able to bind to the promoter of S100A4 and activated S100A4 promoter through binding to the nucleotides +200/+239 region containing a STAT4 regulating element of S100A4 promoter. We further demonstrated that HBXIP activated S100A4 promoter through binding to the transcription factor STAT4. Previous studies reported that the functions of STAT transcription factor family are mostly dedicated to hematopoiesis and immunity. STAT4, as a member of STAT family, has well documented roles in hematopoiesis and immunity (35). However, the role of STAT4 in carcinogenesis is poorly understood. Interestingly, in our study we identified a new STAT4 recognition sequence TGAT (N3) GAA in S100A4 promoter, which is different from the STAT4 consensus TTC (N3) GAA sequence (36, 37). Moreover, EMSA and ChIP assays confirmed that the three-nucleotide mutants of TGA or GAA within STAT4 binding site (S1-1 mut3 or S1-1 mut4) were able to abolish the protein-DNA interaction, and STAT4 was involved in the interaction between HBXIP and S100A4 promoter. Thus, we identify that TGAT (N3) GAA is a novel STAT4 binding motif in S100A4 promoter. Therefore, here we first report that the transcription factor STAT4 plays a role in regulating S100A4 mediated by HBXIP in breast cancer.
Gene regulatory process in cells is a complex dynamic network that involving numerous regulators that interact with each other and crosstalks between signaling pathways (38, 39). Therefore, we tried to identify other mechanisms by which HBXIP regulates S100A4. It has been reported that the up-regulation of S100A4 involves PI3K/AKT signaling pathway which promotes the proliferation and growth of cancer cells (29). HBXIP was able to active AKT signaling in HepG2 cells (30). Accordingly, we proposed the possibility that HBXIP might up-regulate S100A4 through PI3K/AKT signaling pathway as well. Indeed, we observed that HBXIP could induce PI3K/AKT pathway and consequent S100A4 expression. It has been reported that PTEN mediates inhibition of PI3K/AKT pathway (25). Our data indicated that HBXIP was able to down-regulate PTEN, resulting in the activation of PI3K/AKT pathway and consequent S100A4 expression. Then, we identified the mechanism by which HBXIP suppressed PTEN expression. It is reported that one of the mechanisms of PTEN inactivation is epigenetic silencing of the PTEN gene, such as DNA methylation, which has been observed in a lot of cancers, including brain tumors, hematologic malignancies, malignant melanoma, and carcinomas of the cervical, adenoid cystic, ovarian, colorectal, lung, gastric, and breast cancers (16–18, 32). DNMT1 has been recently suggested to play an essential role in aberrant de novo methylation in various carcinomas and its abnormal expression is responsible for the loss of PTEN function in breast cancer cells (10, 33). In our study, we found that HBXIP induced the hypermethylation of PTEN promoter by dramatically stimulating DNMT1 expression in MCF-7 cells. The mechanism by which AKT signaling regulates expression of S100A4 in cancer cells has few reports. Tae Hyong Kim et al. reported that integrin (α6β4) signals stimulated S100A4 through PI3K/AKT pathway, which activated transcription factor NFAT5 in breast cancer cells (29). Therefore, we speculate that NFAT5 may be involved in the process of S100A4 up-regulation mediated by HBXIP. So far, we identified two pathways which up-regulate S100A4 mediated by HBXIP. We further identified that HBXIP up-regulated S100A4 through activating PTEN/PI3K/AKT signaling pathway independently of STAT4 in breast cancer cells.
It has been reported that elevated S100A4 protein level associates with the progression and metastasis of breast cancer (12–14). In function, we provided evidence that HBXIP promoted the growth or migration of breast cancer cells via up-regulating of S100A4 in vivo and in vitro.
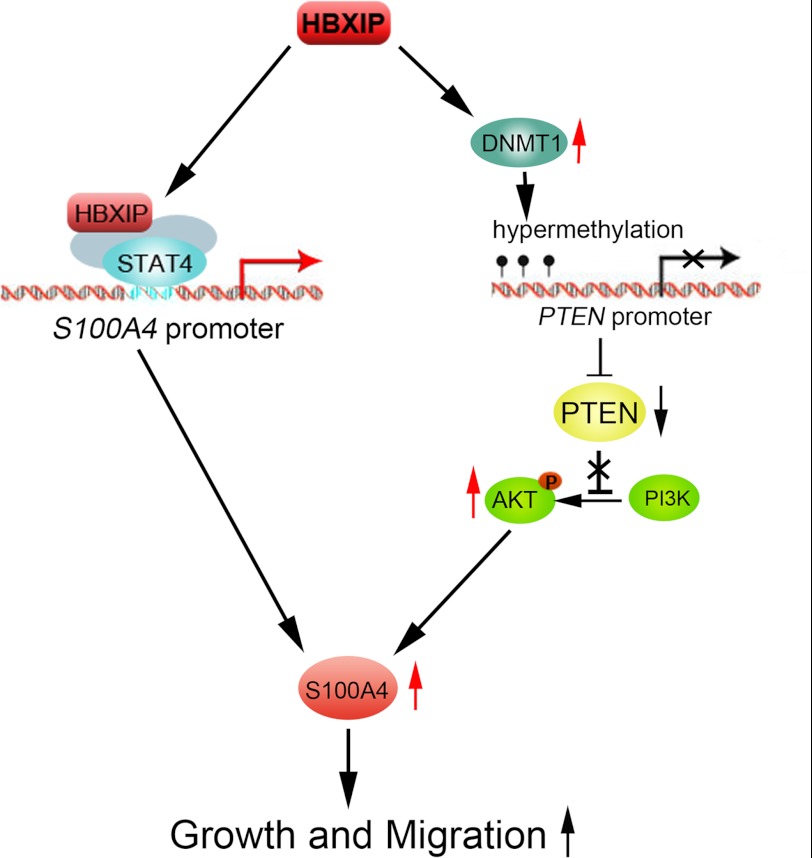
In summary, our finding indicates that HBXIP promotes the proliferation and migration of breast cancer cells through up-regulating S100A4 in two pathways, such as: 1) HBXIP activates the transcription of S100A4 through interaction with transcription factor STAT4; 2) HBXIP up-regulates S100A4 through inducing the CpG island methylation in PTEN promoter via up-regulation of DNMT1, resulting in the activation of PTEN/PI3K/AKT signaling pathway, which is summarized in Fig. 7. Therefore, our finding provides new insight into the mechanism of HBXIP in promotion of growth and migration of breast cancer cells. HBXIP may serve as a target for the therapy of breast cancer.
FIGURE 7.
The model illustrates proposed two pathways of regulation of S100A4 mediated by HBXIP. The model shows that HBXIP up-regulates S100A4 through not only binding to STAT4, but also activating PTEN/PI3K/AKT signaling pathway independently of STAT4.
Supplementary Material
This work was supported in part by National Basic Research Program of China (973 Program, No. 2011CB512113, No. 2009CB521702) and the Natural Scientific Foundation of China (No. 81071623, No. 81071624, No. 30960120).

This article contains supplemental Tables S1 and S2.
- HBXIP
- hepatitis B X-interacting protein
- STAT
- signal transducers and activators of transcription protein
- DNMT
- DNA methyltransferase
- NFAT
- nuclear factor of activated T-cells
- MSP
- methylation-specific PCR
- BSP
- bisulfite sequencing PCR
- siRNA
- small interfering RNA.
REFERENCES
- 1. Melegari M., Scaglioni P. P., Wands J. R. (1998) Cloning and characterization of a novel hepatitis B virus x binding protein that inhibits viral replication. J. Virol. 72, 1737–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Minczuk M., Mroczek S., Pawlak S. D., Stepien P. P. (2005) Human ATP-dependent RNA/DNA helicase hSuv3p interacts with the cofactor of survivin HBXIP. FEBS J. 272, 5008–5019 [DOI] [PubMed] [Google Scholar]
- 3. Wen Y., Golubkov V. S., Strongin A. Y., Jiang W., Reed J. C. (2008) Interaction of hepatitis B viral oncoprotein with cellular target HBXIP dysregulates centrosome dynamics and mitotic spindle formation. J. Biol. Chem. 283, 2793–2803 [DOI] [PubMed] [Google Scholar]
- 4. Wang F. Z., Sha L., Ye L. H., Zhang X. D. (2008) Promotion of cell proliferation by HBXIP via upregulation of human telomerase reverse transcriptase in human mesenchymal stem cells. Acta Pharmacol. Sin. 29, 83–89 [DOI] [PubMed] [Google Scholar]
- 5. Hu N., Zhang J., Cui W., Kong G., Zhang S., Yue L., Bai X., Zhang Z., Zhang W., Zhang X., Ye L. (2011) miR-520b regulates migration of breast cancer cells by targeting hepatitis B X-interacting protein and interleukin-8. J. Biol. Chem. 286, 13714–13722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rivard C. J., Brown L. M., Almeida N. E., Maunsbach A. B., Pihakaski-Maunsbach K., Andres-Hernando A., Capasso J. M., Berl T. (2007) Expression of the calcium-binding protein S100A4 is markedly up-regulated by osmotic stress and is involved in the renal osmoadaptive response. J. Biol. Chem. 282, 6644–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garrett S. C., Varney K. M., Weber D. J., Bresnick A. R. (2006) S100A4, a mediator of metastasis. J. Biol. Chem. 281, 677–680 [DOI] [PubMed] [Google Scholar]
- 8. Miranda K. J., Loeser R. F., Yammani R. R. (2010) Sumoylation and nuclear translocation of S100A4 regulate IL-1β-mediated production of matrix metalloproteinase-13. J. Biol. Chem. 285, 31517–31524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang H., Zhao K., Yu Q., Wang X., Song Y., Li R. (2012) Evaluation of plasma and tissue S100A4 protein and mRNA levels as potential markers of metastasis and prognosis in clear cell renal cell carcinoma. J. Int. Med. Res. 40, 475–485 [DOI] [PubMed] [Google Scholar]
- 10. Phuong N. T., Kim S. K., Lim S. C., Kim H. S., Kim T. H., Lee K. Y., Ahn S. G., Yoon J. H., Kang K. W. (2011) Role of PTEN promoter methylation in tamoxifen-resistant breast cancer cells. Breast Cancer Res. Treat. 130, 73–83 [DOI] [PubMed] [Google Scholar]
- 11. Zhang G., Li M., Jin J., Bai Y., Yang C. (2011) Knockdown of S100A4 decreases tumorigenesis and metastasis in osteosarcoma cells by repression of matrix metalloproteinase-9. Asian Pac. J. Cancer Prev. 12, 2075–2080 [PubMed] [Google Scholar]
- 12. Cabezón T., Celis J. E., Skibshøj I., Klingelhöfer J., Grigorian M., Gromov P., Rank F., Myklebust J. H., Maelandsmo G. M., Lukanidin E., Ambartsumian N. (2007) Expression of S100A4 by a variety of cell types present in the tumor microenvironment of human breast cancer. Int. J. Cancer 121, 1433–1444 [DOI] [PubMed] [Google Scholar]
- 13. Malashkevich V. N., Varney K. M., Garrett S. C., Wilder P. T., Knight D., Charpentier T. H., Ramagopal U. A., Almo S. C., Weber D. J., Bresnick A. R. (2008) Structure of Ca2+-bound S100A4 and its interaction with peptides derived from nonmuscle myosin-IIA. Biochemistry 47, 5111–5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ismail N. I., Kaur G., Hashim H., Hassan M. S. (2008) S100A4 overexpression proves to be independent marker for breast cancer progression. Cancer Cell Int. 8, 12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hernan R., Fasheh R., Calabrese C., Frank A. J., Maclean K. H., Allard D., Barraclough R., Gilbertson R. J. (2003) ERBB2 up-regulates S100A4 and several other prometastatic genes in medulloblastoma. Cancer Res. 63, 140–148 [PubMed] [Google Scholar]
- 16. Fan X., Chen B., Xu J., Zhang H., Deng F., Xiang X. (2010) Methylation status of the PTEN gene in adenoid cystic carcinoma cells. Mol. Med. Report 3, 775–779 [DOI] [PubMed] [Google Scholar]
- 17. Rizvi M. M., Alam M. S., Ali A., Mehdi S. J., Batra S., Mandal A. K. (2011) Aberrant promoter methylation and inactivation of PTEN gene in cervical carcinoma from Indian population. J. Cancer Res. Clin. Oncol. 137, 1255–1262 [DOI] [PubMed] [Google Scholar]
- 18. Yang J., Ikezoe T., Nishioka C., Takezaki Y., Hanazaki K., Taguchi T., Yokoyama A. (2012) Long-term exposure of gastrointestinal stromal tumor cells to sunitinib induces epigenetic silencing of the PTEN gene. Int. J. Cancer 130, 959–966 [DOI] [PubMed] [Google Scholar]
- 19. Chow L. M., Endersby R., Zhu X., Rankin S., Qu C., Zhang J., Broniscer A., Ellison D. W., Baker S. J. (2011) Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell 19, 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goltsov A., Faratian D., Langdon S. P., Mullen P., Harrison D. J., Bown J. (2012) Features of the reversible sensitivity-resistance transition in PI3K/PTEN/AKT signaling network after HER2 inhibition. Cell Signal. 24, 493–504 [DOI] [PubMed] [Google Scholar]
- 21. Kawata T. (2011) STAT signaling in Dictyostelium development. Dev Growth Differ. 53, 548–557 [DOI] [PubMed] [Google Scholar]
- 22. Lupov I. P., Voiles L., Han L., Schwartz A., De La Rosa M., Oza K., Pelloso D., Sahu R. P., Travers J. B., Robertson M. J., Chang H. C. (2011) Acquired STAT4 deficiency as a consequence of cancer chemotherapy. Blood 118, 6097–6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Torpey N., Maher S. E., Bothwell A. L., Pober J. S. (2004) Interferon α but not interleukin 12 activates STAT4 signaling in human vascular endothelial cells. J. Biol. Chem. 279, 26789–26796 [DOI] [PubMed] [Google Scholar]
- 24. Kim E. S., Kim S. W., Moon C. M., Park J. J., Kim T. I., Kim W. H., Cheon J. H. (2012) Interactions between IL17A, IL23R, and STAT4 polymorphisms confer susceptibility to intestinal Behcet's disease in Korean population. Life Sci. 90, 740–746 [DOI] [PubMed] [Google Scholar]
- 25. Carnero A., Blanco-Aparicio C., Renner O., Link W., Leal J. F. (2008) The PTEN/PI3K/AKT signaling pathway in cancer, therapeutic implications. Curr. Cancer Drug Targets 8, 187–198 [DOI] [PubMed] [Google Scholar]
- 26. Ushiku T., Chong J. M., Uozaki H., Hino R., Chang M. S., Sudo M., Rani B. R., Sakuma K., Nagai H., Fukayama M. (2007) Int. J. Cancer 120, 60–66 [DOI] [PubMed] [Google Scholar]
- 27. Shan C., Xu F., Zhang S., You J., You X., Qiu L., Zheng J., Ye L., Zhang X. (2010) Hepatitis B virus X protein promotes liver cell proliferation via a positive cascade loop involving arachidonic acid metabolism and p-ERK1/2. Cell Res. 20, 563–575 [DOI] [PubMed] [Google Scholar]
- 28. Shan C., Zhang S., Cui W., You X., Kong G., Du Y., Qiu L., Ye L., Zhang X. (2011) Hepatitis B virus X protein activates CD59 involving DNA binding and let-7i in protection of hepatoma and hepatic cells from complement attack. Carcinogenesis. 32, 1190–1197 [DOI] [PubMed] [Google Scholar]
- 29. Kim T. H., Kim H. I., Soung Y. H., Shaw L. A., Chung J. (2009) Integrin (α6β4) signals through Src to increase expression of S100A4, a metastasis-promoting factor: implications for cancer cell invasion. Mol. Cancer Res. 7, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 30. Wang F. Z., Fei H. R., Lian L. H., Wang J. M., Qiu Y. Y. (2011) Hepatitis B x-interacting protein induces HepG2 cell proliferation through activation of the phosphatidylinositol 3-kinase/Akt pathway. Exp. Biol. Med. 236, 62–69 [DOI] [PubMed] [Google Scholar]
- 31. Martelli A. M., Evangelisti C., Chappell W., Abrams S. L., Bäsecke J., Stivala F., Donia M., Fagone P., Nicoletti F., Libra M., Ruvolo V., Ruvolo P., Kempf C. R., Steelman L. S., McCubrey J. A. (2011) Targeting the translational apparatus to improve leukemia therapy: roles of the PI3K/PTEN/Akt/mTOR pathway. Leukemia 25, 1064–1079 [DOI] [PubMed] [Google Scholar]
- 32. Wiencke J. K., Zheng S., Jelluma N., Tihan T., Vandenberg S., Tamgüney T., Baumber R., Parsons R., Lamborn K. R., Berger M. S., Wrensch M. R., Haas-Kogan D. A., Stokoe D. (2007) Methylation of the PTEN promoter defines low-grade gliomas and secondary glioblastoma. Neuro. Oncol. 9, 271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hino R., Uozaki H., Murakami N., Ushiku T., Shinozaki A., Ishikawa S. (2009) Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 69, 2766–2774 [DOI] [PubMed] [Google Scholar]
- 34. Nicolas C. S., Peineau S., Amici M., Csaba Z., Fafouri A. (2012) The Jak/STAT pathway is involved in synaptic plasticity. Neuron 73, 374–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Korman B. D., Kastner D. L., Gregersen P. K., Remmers E. F. (2008) STAT4: genetics, mechanisms, and implications for autoimmunity. Curr. Allergy Asthma Rep. 8, 398–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ehret G. B., Reichenbach P., Schindler U., Horvath C. M., Fritz S., Nabholz M., Bucher P. (2001) DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J. Biol. Chem. 276, 6675–6688 [DOI] [PubMed] [Google Scholar]
- 37. Yamamoto K., Miura O., Hirosawa S., Miyasaka N. (1997) Binding sequence of STAT4: STAT4 complex recognizes the IFN-gamma activation site (GAS)-like sequence (T/A)TTCC(C/G)GGAA(T/A). Biochem. Biophys. Res. Commun. 233, 126–132 [DOI] [PubMed] [Google Scholar]
- 38. Stork P. J., Schmitt J. M. (2002) Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol. 12, 258–266 [DOI] [PubMed] [Google Scholar]
- 39. Macneil L. T., Walhout A. J. (2011) Gene regulatory networks and the role of robustness and stochasticity in the control of gene expression. Genome Res. 21, 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.