Background: Ca2+-calmodulin (CaM) and apo-CaM regulate ryanodine receptors (RyRs) differently.
Results: Mutant and wild-type CaM-binding locations on RyRs have been determined.
Conclusion: The distinct binding locations of apo- and Ca2+-CaM on RyR1 have been identified and are likely related to the different regulation effects.
Significance: A CaM-binding location on RyR2 has been determined for the first time.
Keywords: Calcium Intracellular Release, Calmodulin, Electron Microscopy (EM), Ryanodine Receptor, Single-particle Analysis
Abstract
Ryanodine receptor types 1 (RyR1) and 2 (RyR2) are calcium release channels that are highly enriched in skeletal and cardiac muscle, respectively, where they play an essential role in excitation-contraction coupling. Apocalmodulin (apo-CaM) weakly activates RyR1 but inhibits RyR2, whereas Ca2+-calmodulin inhibits both isoforms. Previous cryo-EM studies showed distinctly different binding locations on RyR1 for the two states of CaM. However, recent studies employing FRET appear to challenge these findings. Here, using cryo-EM, we have determined that a CaM mutant that is incapable of binding calcium binds to RyR1 at the apo site, regardless of the calcium concentration. We have also re-determined the location of RyR1-bound Ca2+-CaM using uniform experimental conditions. Our results show the existence of the two overlapping but distinct binding sites for CaM in RyR1 and imply that the binding location switch is due to Ca2+ binding to CaM, as opposed to direct effects of Ca2+ on RyR1. We also discuss explanations that could resolve the apparent conflict between the cryo-EM and FRET results. Interestingly, apo-CaM binds to RyR2 at a similar binding location to that of Ca2+-CaM on RyR1, in seeming agreement with the inhibitory effects of these two forms of CaM on their respective receptors.
Introduction
Ryanodine receptors (RyRs)3 are calcium release channels that are located in the sarcoplasmic or endoplasmic reticulum of muscle and non-muscle cells, where they play a major role in controlling cytoplasmic levels of free calcium (1, 2). RyRs are the largest known ion channels, existing as homotetramers with a net mass in excess of 2 MDa. More than 80% of the receptors' mass exists as a multidomain cytoplasmic assembly, which serves as a platform for interactions with numerous regulatory enzymes and binding proteins. RyR1 and RyR2 are mammalian RyR isoforms and play an essential role in excitation-contraction coupling in skeletal and cardiac muscle, respectively.
One of the regulatory binding proteins for RyRs is calmodulin (CaM), which binds tightly to the receptors in both its Ca2+-bound (Ca2+-CaM) and Ca2+-free (apo-CaM) states (3–5). The stoichiometry of CaM binding is 1 mol of CaM/mol RyR subunit, regardless of whether Ca2+ is bound (6, 7), and a region of the RyR1 amino acid sequence involving residues 3614–3643 (residues 3581–3612 for RyR2) is involved in binding both apo- and Ca2+-CaM (6, 8). Apo-CaM weakly activates RyR1, whereas Ca2+-CaM inhibits RyR1 (9–12). In contrast, for RyR2, CaM inhibits receptor activity at both high and low [Ca2+] (7, 13). Previous cryo-EM and single-particle three-dimensional reconstruction studies demonstrated that apo- and Ca2+-CaM bind to overlapping but spatially distinct locations on the cytoplasmic assembly of RyR1, in which the centers of mass of CaM differ by 3–4 nm (14, 15). The binding positions of apo- and Ca2+-CaM on RyR2 have not yet been reported.
Recently, the technique of FRET was applied to monitor changes in the modes of apo- and Ca2+-CaM binding to RyR1 and RyR2 (16, 17). In these studies, fluorescently labeled FKBP12.6 served as a reference point from which distances were determined to CaM that was fluorescently labeled within its N-terminal domain, its C-terminal domain, or the central region that connects the N- and C-terminal domains. The results from FRET for RyR1 indicated no significant difference in the distances between apo-CaM and FKBP12.6 and between Ca2+-CaM and FKBP12.6. Identical results were obtained by FRET for CaM-RyR2 complexes (17). This result was unexpected because the cryo-EM studies had shown a several-nanometer shift in the location of apo-CaM relative to Ca2+-CaM.
Here, in an effort to better understand the mode of CaM binding to RyR1 and RyR2, we investigate in further detail the mode of CaM binding to RyR1 and RyR2 by cryo-EM and three-dimensional reconstruction. Mutant CaM1234, in which all four of the Ca2+-binding domains in CaM have been rendered incapable of binding Ca2+ (18) so that it constantly mimics the apo-CaM conformation, was used to form CaM1234-RyR1 complexes in both low and high calcium buffers. Our results show that CaM1234 binds at the location (between domains 3 and 4) previously identified by Samsó and Wagenknecht (14) for apo-CaM. We found no difference in the binding location of CaM1234 at micromolar and nanomolar Ca2+, from which we infer that the binding location of CaM does not depend on Ca2+ acting directly on regulator sites present on RyR1 itself; rather, a reconfiguration of the CaM mode of binding to RyR1 occurs as a result of Ca2+ associating with CaM. Finally, complexes of apo-CaM and RyR2 were obtained, and cryo-EM analysis revealed that apo-CaM, an inhibitor of RyR2, localizes to the region (between domains 3 and 7) where Ca2+-CaM, an inhibitor of RyR1, binds RyR1, rather than the apo-CaM site found for RyR1.
EXPERIMENTAL PROCEDURES
Materials and Chemicals
The soybean phospholipid phosphatidylcholine, the detergent CHAPS, the redox reagent DTT, and the protease inhibitor leupeptin were purchased from Calbiochem. All other chemicals were purchased from Sigma-Aldrich. RyR1 was purified from skeletal terminal cisternal vesicles obtained from New Zealand White rabbits (Pel-Freez Biologicals, Rogers, AZ) as described previously (19). Mouse RyR2 was cloned, expressed in HEK293 cells, and purified as described (20). Wild-type CaM and mutant CaM were expressed and purified as described previously (21).
Cryo-grid Preparation
The previously published localizations of apo- and Ca2+-CaM on RyR1 were obtained under different buffer conditions (14, 15). Notably, the concentration of the detergent CHAPS used for apo-CaM localization (14) was ∼20 times higher than that used for Ca2+-CaM localization (15), and exogenous lipid (phosphatidylcholine) was introduced into the buffer system used for apo-CaM localization, but not for Ca2+-CaM localization. A uniform buffer system was used for the studies described here. Low calcium buffer contained 20 mm NaMOPS (pH 7.4), 0.108 m NaCl, 0.278 m KCl, 2 mm EGTA, 1% CHAPS, 0.5% phosphatidylcholine, 2 mm DTT, and 2 μg/ml leupeptin in the incubation condition and 20 mm NaMOPS (pH 7.4), 0.0216 m NaCl, 0.176 m KCl, 2 mm EGTA, 0.2% CHAPS, 0.1% phosphatidylcholine, 2 mm DTT, and 2 μg/ml leupeptin in the final on-grid condition. High calcium buffer was the same as low calcium buffer except that 0.1 mm CaCl2 was substituted for 2 mm EGTA.
Cryo-grids prepared for two-dimensional localization of RyR1-bound CaMWT and CaM1234 in low and high calcium buffers were prepared as follows. Complexes of CaM and RyR1 were prepared by mixing equal volumes of CaMWT or CaM1234 (in CaM buffer containing 20 mm NaMOPS (pH 7.4), 2 mm DTT, and 2 μg/ml leupeptin) and RyR1 (in 20 mm NaMOPS (pH 7.4), 150 mm NaCl, 1 m KCl, 0.4% CHAPS, 2 mm DTT, and 2 μg/ml leupeptin). The CaM/RyR1 molar ratio was 18:1. CaM-free RyR1 control reactions contained the relevant buffer lacking CaM. CaM and RyR1 mixtures were used to generate different CaM-RyR1 complexes by incubating CaM and RyR1 mixtures with either low or high calcium buffer for 20 min at room temperature to achieve the incubation conditions mentioned above. Just before cryo-plunging, the incubated mixtures were diluted to the final on-grid condition listed above. The concentration of RyR1 in the final on-grid condition was ∼0.05–0.1 mg/ml. 4-μl diluted mixtures were applied to 300-mesh grids coated with a thin continuous carbon film (∼5 nm) suspended over a thick holey carbon film (>10 nm) for 30 s. The grids were blotted with either Whatman No. 540 (low calcium buffer) or No. 1 (high calcium buffer) filter paper for 4–4.5 s before being plunged into liquid ethane.
Cryo-grids for three-dimensional localization of RyR1-bound CaM1234 in low and high calcium buffers were prepared as described above. Cryo-grids for three-dimensional localization of RyR1-bound CaMWT in high calcium buffer were separated into two parts. The cryo-grids for low tilt (0–30°) image collection were prepared in the same way as described above, whereas grids for high tilt (30–50°) image collection were prepared by the carbon film sandwich method (22). The buffer conditions and Ca2+-CaMWT-RyR1 complex concentration were the same. After applying 4-μl diluted mixtures to the grids, an additional thin continuous carbon film was placed on top of the diluted mixture. Side blotting followed the second carbon film introduction before cryo-plunging.
Cryo-grids prepared for two- and three-dimensional localization of RyR2-bound CaMWT in low and high calcium buffers were prepared as follows. Purified RyR2 (7 μl at a concentration of 0.2 mg/ml in 20 mm NaMOPS (pH 7.4), 1 m NaCl, 1% CHAPS, 2 mm DTT, and 2 μg/ml leupeptin) was mixed with 1.4 μl of 0.5 mg/ml CaM (in 20 mm NaMOPS (pH 7.4) and 400 mm KCl) and 28 μl of low calcium buffer containing 20 mm NaMOPS (pH 7.4), 400 mm KCl, 3.0 mm EGTA, 0.5% CHAPS, 2 mm DTT, and 2 μg/ml leupeptin. Control RyR2 was prepared identically except that CaM was omitted. The calcium-containing buffer contained 0.15 mm CaCl2 in place of 3.0 mm EGTA. The mixtures were incubated on ice for 30–40 min, and 4 μl was applied to grids and frozen as described above.
Cryo-EM
Cryo-EM was done using an FEI Tecnai F20 transmission electron microscope equipped with a field emission gun and operated at 200 kV. An Oxford CT3500 cryo-transfer holder (Gatan, Inc., Warrendale, PA) was used with the temperature maintained within a few degrees of −175 °C. Micrographs of CaMWT-RyR1 and CaM1234-RyR1 complexes were recorded using a Tietz 415 4k×4k CCD camera (Tietz Video and Image Processing Systems GmbH, Gauting, Germany) using low dose protocols; CaMWT-RyR2 complexes were recorded on films and digitized using a Zeiss imaging scanner (Z/I Imaging Corp., Huntsville, AL) with a step size of 14 μm. The objective lens defocus range was from 1.5 to 4.5 μm. Magnification was ×50,760 (±2%). For three-dimensional reconstruction, images were collected with the grids tilted by 0°, 10°, 20°, 30°, 40°, and 50°, whereas for two-dimensional analyses, only 0° data were used. Each exposure corresponded to an electron dose of ∼10 e−/Å2.
For the Ca2+-CaMWT-RyR1 complex, we modified the grid preparation by applying a second carbon film, which resulted in the specimen being sandwiched between two layers of carbon (22). This was done to reduce the specimen charging or beam-induced specimen movement that usually occurs when the specimen grid is tilted, especially to tilt angles above 20°. This method helped to reduce the charging and image blurring in micrographs of tilted specimens and improved the ratio of usable images in the 40° and 50° tilted images from <5 to ∼20%. However, at the same time, introduction of the second carbon film decreased the signal-to-noise ratio of the images and might have caused some minor structural alterations in the Ca2+-CaMWT-RyR1 complexes. The use of this technique for the Ca2+-CaMWT-RyR1 specimen might have been responsible for the less robust differences attributable to CaM than were observed in the previous study of the same complex (15) (see “Results” for further discussion).
Image Processing
Micrographs were processed using the SPIDER/WEB software package (23). A phase-flipping method (24) was used to correct the contrast transfer function effect on each particle. Two-dimensional analyses were done using 400 particles (defocus range of 3.0 ± 0.1 μm) for each experimental group. A projection-matching procedure (25) was used for three-dimensional reconstructions with 4-fold symmetry enforced. The final resolutions were estimated by Fourier shell correlation with a cutoff value of 0.5 (26), and the final three-dimensional reconstructions were filtered to the estimated resolutions. The final three-dimensional reconstructions of CaM1234-RyR1 in low (resolution of 27 Å) and high (resolution of 28 Å) calcium buffers were computed from 5561 and 5033 particles, respectively. The three-dimensional reconstruction of CaM-free RyR1 (control) in low calcium buffer was reported previously (27). The three-dimensional reconstructions of Ca2+-CaMWT-RyR1 (resolution of 27 Å) and the CaM-free RyR1 control (resolution of 30 Å) in high calcium buffer were computed from 7000 and 4932 particles, respectively. The three-dimensional reconstruction of Ca2+-CaMWT-RyR1 following supervised classification (see below) attained a resolution of 28 Å and was computed from 6969 particles. A three-dimensional reconstruction of apo-CaM-RyR2 was computed from 14,568 particles and attained a resolution of 30 Å. The CaM-free RyR2 control was computed from 7102 particles at a resolution of 30 Å.
An initial three-dimensional reconstruction of Ca2+-CaMWT-RyR1 indicated less than stoichiometric binding of CaM and other unexpected structural differences from the control reconstruction. The unexpected differences could be artifactual (e.g. induced by the carbon film sandwich technique that was used for Ca2+-CaMWT-RyR1; see “Results”) or reflect genuine small structural alterations in RyR1. To enhance the signal from Ca2+-CaMWT, supervised classification (28) was applied to the images of the Ca2+-CaMWT-RyR1 complexes; which we applied masks to the region between domains 3 and 7, that were used to screen for the fully bound Ca2+-CaMWT-RyR1 complexes to be included in the computation of the three-dimensional reconstruction that is shown in Fig. 3 (E and F).
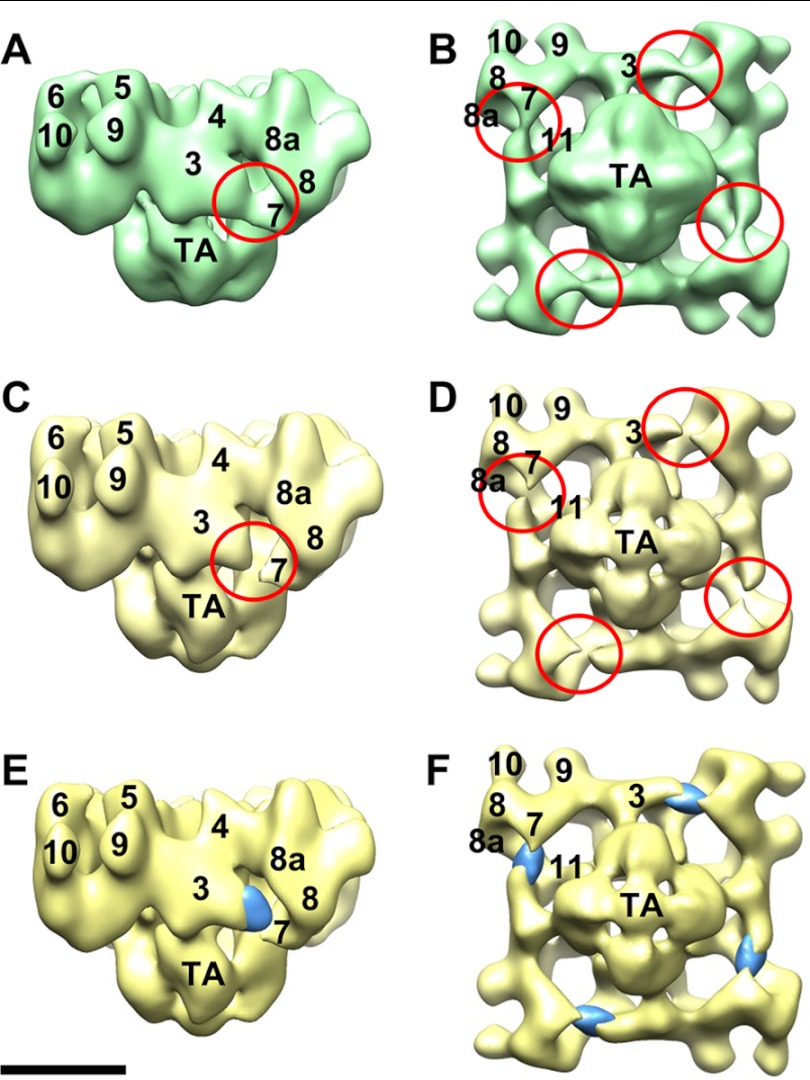
FIGURE 3.
Three-dimensional localization of RyR1-bound Ca2+-CaMWT. A and B, three-dimensional reconstruction of Ca2+-CaMWT-RyR1. A shows a side view, and B shows a 4-fold view of the bottom (membrane-facing) surface. C and D, side view and bottom 4-fold view, respectively, of the CaM-free RyR1 control. Red circled regions in A–D highlight the major differences between Ca2+-CaMWT-RyR1 and the CaM-free RyR1 control. E and F, densities likely due to RyR1-bound Ca2+-CaMWT that were generated by volume subtraction of the RyR1 control from Ca2+-CaMWT-RyR1. Scale bar = 10 nm.
RESULTS
Two-dimensional Localization of RyR1-bound CaMWT and Mutant CaM1234 at Low and High Calcium Concentrations
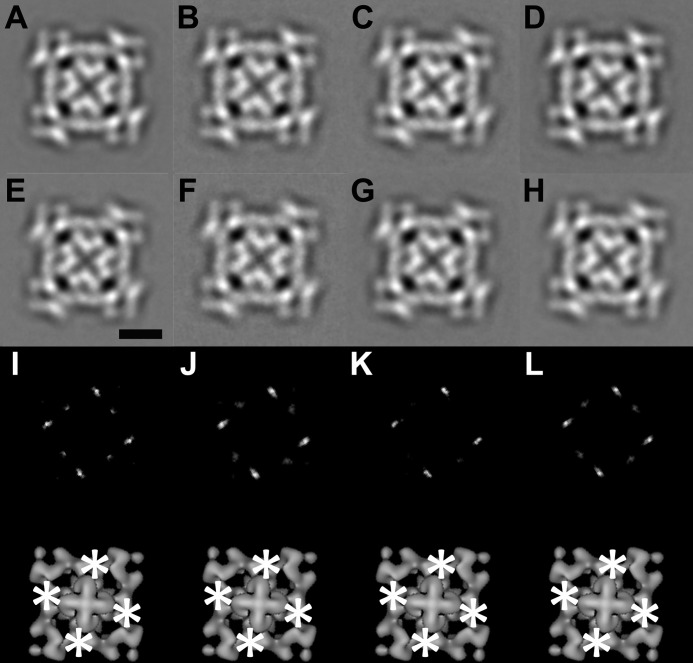
Frozen hydrated complexes of CaMWT-RyR1 and CaM1234-RyR1 in buffers containing either high (100 μm) or low (<10 nm) [Ca2+], as well as the CaM-free RyR1 control in the same buffers, were imaged by cryo-EM. Under all of the conditions, the images displayed the standard tetrameric RyR shape, a square with four protruding corners (“clamps”) and a central cross, which is characteristic of RyRs adsorbed to carbon-coated EM grids. Because of the high noise level of individual images and the small mass of CaM (16.5 kDa) relative to RyR1 (565 kDa/subunit), RyR-bound CaM is not discernible in images of CaMWT-RyR1 or CaM1234-RyR1, which appear to be identical to their respective control specimens lacking bound CaM. After averaging the images, small symmetrically distributed regions of increased density are apparent in CaMWT-RyR1 and CaM1234-RyR1 (Fig. 1, A–D) compared with the respective RyR1 controls (Fig. 1, E–H) under both low and high calcium conditions. To make these subtle differences clearer, we subtracted the averaged RyR1 controls from the averaged complexes and generated difference maps (Fig. 1, I–L), each presenting four strong positive regions of density (white) having the expected 4-fold symmetry. Application of t tests showed these positive differences to be statistically significant at >99.9% confidence levels. We attribute these differences to RyR-bound CaMWT or CaM1234 present at four copies/tetrameric RyR1. These localization results are consistent with published apo- and Ca2+-CaM localizations (14, 15). However, two-dimensional analysis did not distinguish between the apo- and Ca2+-CaM positions, as both appear on one side of and at the base of the clamp structures. Some weaker positive differences also appeared in difference maps (Fig. 1, I–L), and they might have been the result of subtle conformational changes in RyR1 triggered by CaM binding.
FIGURE 1.
Two-dimensional analysis of CaMWT-RyR1 and CaM1234-RyR1 in 4-fold view. A and B, 4-fold symmetrized two-dimensional averages of apo-CaMWT-RyR1 and CaM1234-RyR1, respectively, in low calcium buffer (2 mm EGTA). C and D, 4-fold symmetrized two-dimensional averages of Ca2+-CaMWT-RyR1 and CaM1234-RyR1, respectively, in high calcium buffer (0.1 mm CaCl2). E and F, 4-fold symmetrized two-dimensional averages of the RyR1 control in low calcium buffer. G and H, 4-fold symmetrized two-dimensional averages of the RyR1 control in high calcium buffer. Scale bar = 10 nm (E). I–L, upper panels, difference maps generated by subtracting images in E–H from A–D, respectively. The white regions in I–L are the positions of positive differences. The biggest differences are in bright white. Lower panels, the positive differences (asterisks) in the upper panels superimposed on the bottom view of a three-dimensional reconstruction of RyR1.
Three-dimensional Localization of CaM1234 on RyR1 at Low and High Calcium Concentrations
The three-dimensional reconstructions of CaM1234-RyR1 in both low and high calcium buffers display the typical RyR1 appearance (29, 30), which is a square prism shape (28-nm edge length) comprising 14 distinguishable domains connecting to a smaller transmembrane region (Fig. 2, A and D). Compared with the CaM-free RyR1 control (Fig. 2, B and E), in both low and high calcium buffers, CaM1234-RyR1 shows a major difference in domain 3 close to domain 4 (yellow boxed regions in Fig. 2). Subtractions of the CaM-free RyR1 controls from the CaM1234-RyR1 complexes in the same buffer were performed to more clearly display the excess densities that are present in CaM1234-RyR1 but absent in the RyR1 controls. For both the high and low Ca2+ conditions, the strongest positive mass differences appear in domain 3 (Fig. 2, C and F), as expected from visual comparisons of the reconstructions of RyR1 with and without CaM. Based upon the sizes of the major differences and the absence of other substantial differences, they are interpreted as RyR1-bound CaM1234. The locations of these strongest masses are consistent with the 4-fold view positions of CaM1234 in the two-dimensional analyses (Fig. 1). The locations, as well as the overall ellipsoidal shapes of the major differences, closely match the results of the published localization of apo-CaMWT on RyR1 (14).
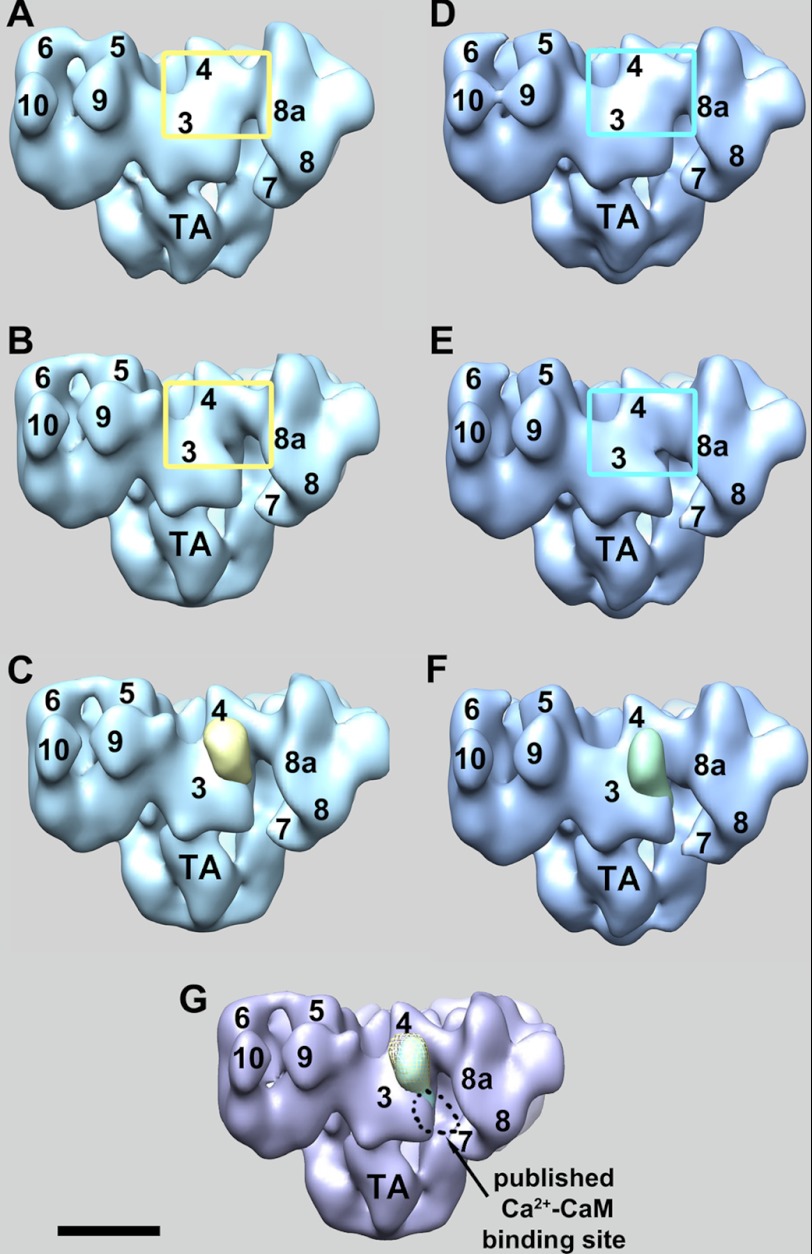
FIGURE 2.
Three-dimensional localization of RyR1-bound CaM1234 at low and high calcium. A and B, solid-body representations of the three-dimensional reconstructions of CaM1234-RyR1 complexes and the CaM-free RyR1 control, respectively, in low calcium buffer. The yellow boxed regions highlight the areas with the biggest difference. C, subtraction of B from A yields four symmetrically related positive differences, one of which is shown superimposed on the RyR1 control (yellow mass) and which is attributed to RyR1-bound CaM1234. D and E, solid-body representations of the three-dimensional reconstructions of CaM1234-RyR1 complexes and the CaM-free RyR1 control, respectively, in high calcium buffer. The cyan boxed regions highlight the areas with the biggest difference. F, the green mass is the result of subtraction of E from D and represents RyR1-bound CaM1234 in high calcium buffer. G, locations of RyR1-bound CaM1234 in low (yellow mesh) and high (green solid) calcium buffers. Both were localized on domain 3 close to domain 4, which closely matches the previously published apo-CaMWT site. The numerals on the cytoplasmic assembly indicate the distinguish domains as numbered previously (30). TA, transmembrane assembly. Scale bar = 10 nm.
Fig. 2 shows only the differences due to additional mass present in the CaM1234-RyR1 complexes. Analysis of the negative differences indicated that none were significant enough to compensate for the positive masses attributed to CaM1234. Therefore, the positive CaM1234-like masses are not the result of RyR1 conformational changes, and we conclude that each of these positive masses corresponds to RyR1-bound CaM1234.
In summary, RyR1-bound CaM1234 in low calcium buffer is localized on domain 3 close to domain 4 on RyR1, which is the same position as the published apo-CaMWT site. RyR1-bound CaM1234 in high calcium buffer is also localized to this region; the CaM1234 masses in both the low and high calcium buffer experimental groups almost completely overlap (Fig. 2G) with the distance between the masses of <0.1 nm. Most significantly, this result shows that the previous localization of RyR-bound CaM at high [Ca2+], in which Ca2+-CaM was localized to the cleft formed by domains 3 and 4 and the clamp (arrow in Fig. 2G), was a consequence of Ca2+ binding to CaM and was not due to a direct effect of Ca2+ on the structure of RyR1.
Three-dimensional Localization of Ca2+-CaMWT on RyR1
To check that the Ca2+-CaM-binding site that we identified previously (15) is present under the conditions used in this study, we determined a three-dimensional reconstruction of the Ca2+-CaM-RyR1 complex. The three-dimensional reconstruction of Ca2+-CaMWT-RyR1 shows the typical RyR1 appearance (Fig. 3). The major differences between Ca2+-CaMWT-RyR1 (Fig. 3, A and B) and the CaM-free RyR1 control (Fig. 3, C and D) involve domains 3 and 7 and the space between them. The strongest differences are located in domain 3. In Ca2+-CaMWT-RyR1, domains 3 and 7 are connected, whereas in the CaM-free RyR1 control, they are separated. This result is consistent with previous results (15), which placed Ca2+-CaM in the gap between domains 3 and 7.
However, volume subtraction of the CaM-free RyR1 control from Ca2+-CaMWT-RyR1 showed a more complex distribution of positive and negative differences (mainly in domain 3) than seen previously. Several differences in experimental parameters are evidently responsible for these discrepancies, which caused less than stoichiometric binding (or increased mobility) of Ca2+-CaM to RyR1 and possibly some structural alterations involving domain 3 not seen previously. Therefore, to clarify the CaM-binding position, we applied a supervised classification step to the images that were used to generate the final differences shown in Fig. 3 (E and F) (see “Experimental Procedures”). However, even without this additional image-processing step, it is evident that Ca2+-CaM binding to RyR1 occurs within the cleft between domains 3 and 7 (red circled regions in Fig. 3, A and B), in agreement with previous work, and not in the upper regions of domain 3 where apo-CaM and CaM1234 bind (Fig. 2).
Localization of Apo-CaMWT on RyR2
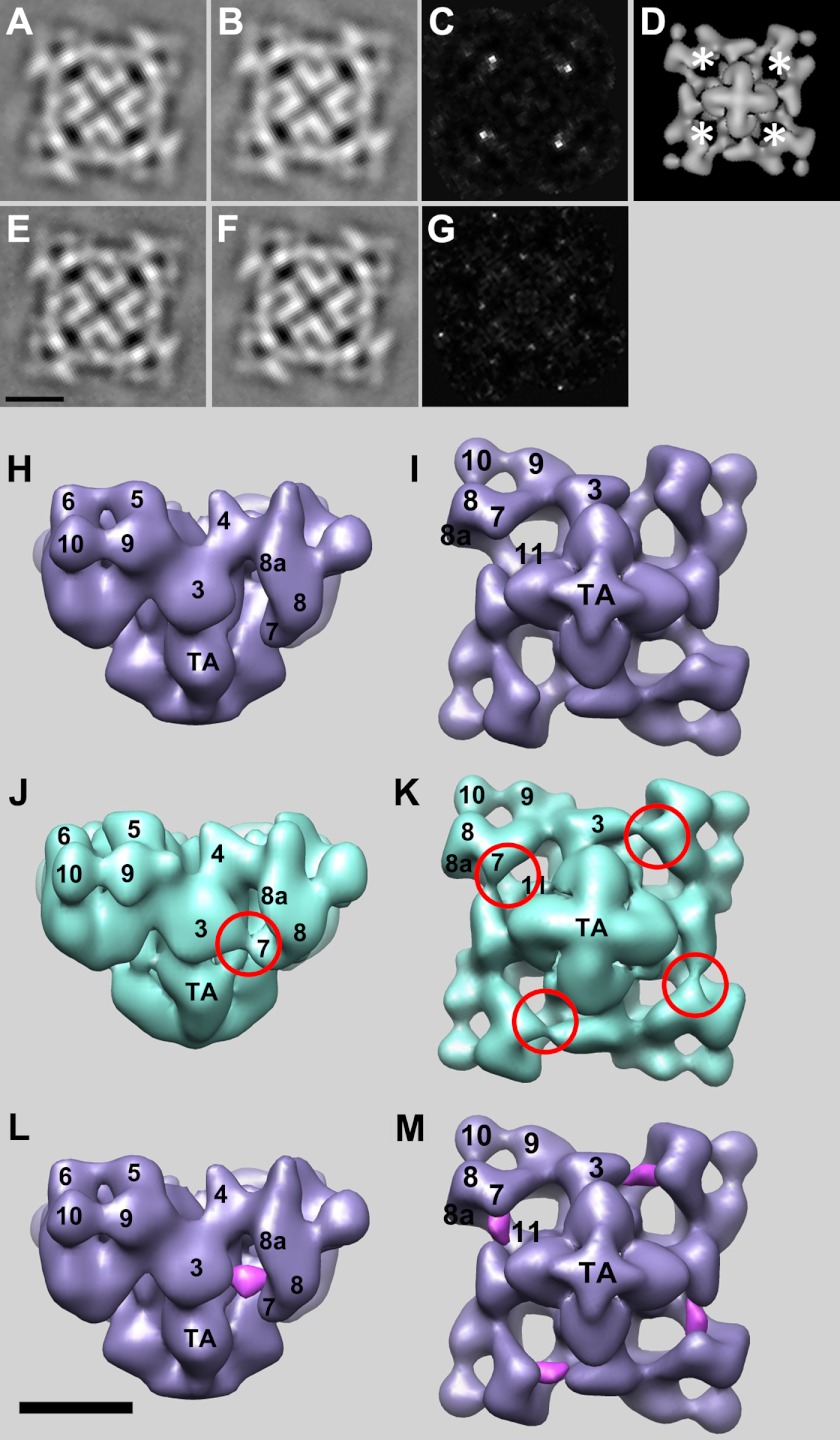
The effects of apo-CaM on RyR2 calcium channel activity differ from its effects on RyR1, but CaM interactions with RyR2 have not been investigated previously by cryo-EM. Two-dimensional averages of apo-CaMWT-RyR2 and Ca2+-CaMWT-RyR2 complexes showed typical RyR shapes, but when difference images were formed with control (CaM-free) RyR2, only apo-CaM-RyR2 showed significant excess mass density compared with control RyR2 (Fig. 4, A–D). Several experimental conditions were tested for Ca2+-CaMWT-RyR2, but for unknown reasons, none resulted in significant differences compared with control RyR2. Therefore, data for three-dimensional reconstruction were collected for apo-CaM-RyR2, but not for Ca2+-CaMWT-RyR2. The three-dimensional reconstruction of apo-CaMWT-RyR2 localizes apo-CaMWT to the gap between domains 3 and 7 (Fig. 4, H–M). The strongest differences appear to be associated with domain 7, perhaps indicating that the mode of binding is not identical to that of Ca2+-CaM binding to RyR1, in which Ca2+-CaM appears to be more closely associated with domain 3. Nevertheless, the binding of apo-CaM on RyR2 to the cleft formed by domains 3 and 7 is much more similar to that of Ca2+-CaM compared with apo-CaM on RyR1.
FIGURE 4.
Two- and three-dimensional localizations of RyR2-bound apo-CaM. A and B, 4-fold symmetrized two-dimensional averages of apo-CaMWT-RyR2 and the CaM-free control, respectively, in low calcium buffer (3 mm EGTA). C, difference map generated by subtracting B from A. The white regions are the positions of positive differences. The biggest differences are in bright white. D, locations of the positive differences (asterisks) in C superimposed on the bottom view of a three-dimensional reconstruction of RyR1. E and F, 4-fold symmetrized two-dimensional averages of Ca2+-CaMWT-RyR2 and the CaM-free control, respectively, in high calcium buffer (0.1 mm CaCl2). G, difference map generated by subtracting F from E, showing no significant positive differences. H and I, three-dimensional reconstruction of the CaM-free RyR2 control. H shows a side view, and I shows a 4-fold view of the bottom (membrane-facing) surface. J and K, side view and bottom 4-fold view, respectively, of the apo-CaMWT-RyR2 complex. Red circled regions highlight the major differences between the apo-CaMWT-RyR2 complex and the CaM-free RyR2 control. L and M, positive densities (purple) attributed to RyR2-bound apo-CaMWT that were generated by volume subtraction of the CaM-free RyR2 control from apo-CaMWT-RyR2, which suppresses the positive differences due to FKBP12.6. Scale bar = 10 nm.
DISCUSSION
The motivation for this study was to clarify the modes of apo- and Ca2+-CaM binding to RyR1 and to test the hypothesis that Ca2+ binding to CaM promotes large-scale structural rearrangements in RyR1. Previous results obtained by cryo-EM indicated that apo- and Ca2+-CaM bind to clearly distinct but partially overlapping locations on the lateral surface of the cytoplasmic region of the receptor (Fig. 2G), ∼10 nm from the Ca2+-conducting channel in the transmembrane region (14, 15). The straightforward interpretation of this finding is that Ca2+- and apo-CaM bind differently to RyR1 such that their centers of mass are displaced from one another by several nanometers. Analysis of CaM binding to a CaM-binding peptide corresponding to RyR1 residues 3614–3643 indicated that CaM moves toward the N terminus of the peptide upon binding Ca2+ (31), an observation that is not inconsistent with the movement detected by cryo-EM, but additional structural changes would seem to be required to account for the large (∼3 nm) shift in the CaM location found by cryo-EM. Furthermore, because RyR1 is also regulated by Ca2+ independently of CaM, it has been suggested that Ca2+ itself could induce a structural change in RyR1 that reconfigures the binding site for CaM (2); this interpretation is consistent with results from multiple studies showing involvement of a 30-residue amino acid sequence within RyR1 (residues 3614–3643) in binding both apo- and Ca2+-CaM (6, 8). To test for this latter possibility, we have determined the three-dimensional reconstructions of mutant CaM (CaM1234) in which all four Ca2+-binding motifs have been rendered incapable of binding Ca2+, and so CaM1234 should behave as apo-CaM even in the presence of micromolar [Ca2+].
The three-dimensional reconstructions of CaM1234-RyR1 complexes determined at either nanomolar or micromolar [Ca2+] show the CaM1234 mutant to be bound at the location previously determined for apo-CaM (14). Two aspects of this result are significant. First, it provides confirmatory evidence of the apo-CaM-binding location, which has been reported only once before. The apo-CaM site has also been called into question based upon estimations by FRET of the distance between fluorescently tagged CaM and FKBP12.6 that were more consistent with apo-CaM being bound nearer to the location found by cryo-EM for Ca2+-CaM (16). Second, the result rules out the hypothesis that direct binding of Ca2+ to RyR1 causes a restructuring of the CaM-binding site that accounts for the difference in apo- and Ca2+-CaM binding observed by cryo-EM. On this point, our results agree with FRET results, which also showed no significant Ca2+-dependent changes in FRET for RyR1-bound CaM1234 (16, 17).
We have also re-determined the location of RyR1-bound Ca2+-CaM and have confirmed that Ca2+-CaM binds in the deep cleft formed between the “handle” and clamp regions of the cytoplasmic region of RyR1. For unknown reasons, either the binding occupancy of Ca2+-CaM was <100%, or Ca2+-CaM was somewhat mobile because the density attributed to Ca2+-CaM was weaker than expected, but nevertheless highly significant. Thus, increasing evidence from cryo-EM supports a model in which apo- and Ca2+-CaM bind to distinct sites on RyR1. Meanwhile, more recent studies by FRET (17), employing fluorophore-labeled CaM and FKBP12.6, continue to report small changes in FRET that appear to be inconsistent with the nanometer-scale movements seen by cryo-EM of CaM relative to FKBP12.6 when CaM switches between the Ca2+-free and Ca2+-bound states for both RyR1 and RyR2.
Are the results from FRET and cryo-EM necessarily discordant? In the FRET study, distances between fluorophore-labeled CaM and FKBP12.6 were determined for CaM that was labeled in its N-terminal domain (N-terminal lobe), central connecting peptide, or C-terminal lobe. In all three instances, no significant Ca2+-dependent changes in FRET were found, and the N-terminal lobe consistently yielded higher FRET than the C-terminal lobe (16). The FRET experiments were conducted on unpurified RyR1 that was present in vesicles derived from skeletal sarcoplasmic reticulum, as well as on purified detergent-solubilized RyR1, the latter conditions being similar to those used for cryo-EM. No instances were found of Ca2+-dependent changes in FRET of a magnitude that might be expected for nanometer-scale shifts in the CaM-binding location as have been inferred from cryo-EM (14).
Cornea et al. (16) discussed possible ways in which their FRET results could be interpreted so as to be consistent with the cryo-EM results. One possibility is that FKBP12.6, like CaM, switches between two binding sites such that there is little net change in the distance between the two bound ligands for the two states (high and low [Ca2+]). However, this possibility was considered to be unlikely given the lack of evidence that FKBP12 undergoes any significant changes in its mode of binding as a function of Ca2+. It was also suggested that if the transition of apo-CaM to Ca2+-CaM approximates CaM moving along a circular arc relative to FKBP12.6, then no change in the distance from CaM to FKBP12.6 would be observed using FRET. However, the cryo-EM results do not appear to lend support to this interpretation either.
In light of the results presented here, we argue that it is worthwhile to consider additional explanations. We contend that there are other (probably many) ways in which CaM binding to RyR1 could occur so as not to contradict either the FRET or cryo-EM results. For example, we note that in the FRET studies (16), the N- and C-terminal lobe-labeled versions of CaM had the fluorophores on the same side of the CaM, assuming that Ca2+-CaM binds to the RyR1 CaM-binding domain as determined by x-ray crystallography (32). One could envision scenarios in which the center of mass of the CaM undergoes a 3–4-nm translational shift (in agreement with the cryo-EM) while, at the same time, the N- and C-terminal lobes change their orientations, perhaps independently of each other (e.g. by rotations about the long axis of CaM), such that the attached fluorophores undergo movements that compensate for the translational shift (Fig. 5). The central region of CaM almost certainly differs substantially in conformation in apo-CaM compared with Ca2+-CaM, and therefore, in the absence of structural information, FRET changes for probes placed in this region are unpredictable. This speculation illustrates how small changes in FRET could still allow for large structural rearrangements, but the actual explanation for the apparent disagreement of the FRET and cryo-EM results may well be more complex than this. For instance, whereas the C-terminal lobe of CaM is thought to bind tightly and constitutively to the well documented CaM-binding region of RyR1 (residues 3614–3643), some studies suggest of independent movements of the N-terminal lobe relative to the C-terminal lobe of CaM, perhaps with the N-terminal lobe undergoing major changes in its mode of interaction with RyR1 (32–34). Further investigation is needed to resolve the nature of potentially complex dynamical interactions of the N- and C-terminal lobes of CaM with multiple domains within RyR1.
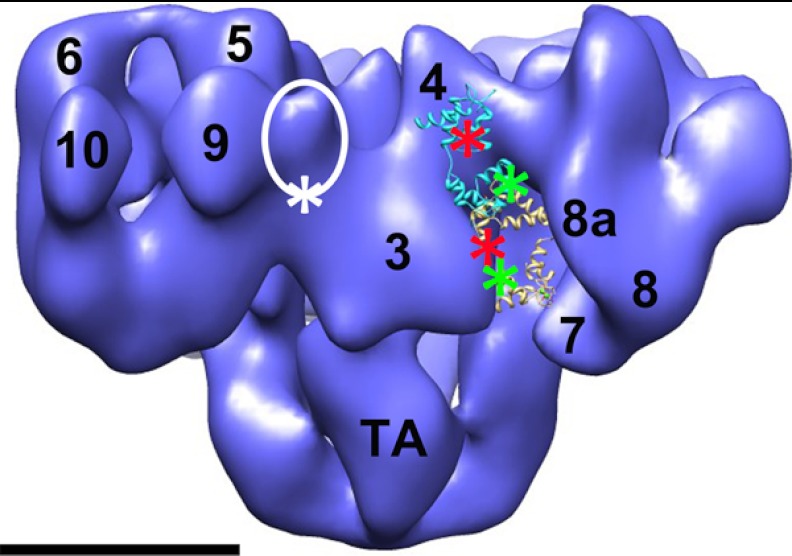
FIGURE 5.
Hypothetical model of FRET pair locations on RyR1-bound apo-CaM, Ca2+-CaM, and FKBP12.6. The white circled region indicates the location of FKBP12.6 on the RyR1 surface, which is between domains 3 and 9. The white asterisk shows one possible donor fluorophore location on the FK506-binding protein. The red asterisks are the potential acceptor fluorophore locations on the N-terminal lobe of apo-CaM (backbone in cyan) and Ca2+-CaM (backbone in yellow). The green asterisks are the acceptor locations on the C-terminal lobe. CaMs were positioned and oriented so as to be consistent with both the cryo-EM localizations and the FRET data that indicate small differences between donor/acceptor fluorophore pairs in apo-CaM versus Ca2+-CaM. Scale bar = 10 nm. The Protein Data Bank code for apo-CaM is 2IX7 (35), and that for Ca2+-CaM is 2BCX (32).
Our finding that the apo-CaM-binding site on RyR2 is in the deep cleft between domains 3 and 7 (Fig. 4), similar to the Ca2+-CaM-binding position on RyR1, is perhaps the structural correlate of functional studies showing that both apo- and Ca2+-CaM inhibit the calcium channel activity of RyR2, whereas Ca2+-CaM inhibits but apo-CaM activates the channel activity of RyR1 (7, 9–13). A structural model that posits two CaM-binding sites on the RyR, one in the cleft between domains 3 and 7, which has an inactivating effect on the RyR, and the second near the juncture of domains 3 and 4, which has an activating effect, is consistent with existing structural and functional data.
Acknowledgments
We thank Xing Meng for technical support in cryo-EM data collection and Dr. S. R. Wayne Chen and laboratory members for RyR2 expression and purification. We gratefully acknowledge use of the 3D-EM Facilities at the Wadsworth Center.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL095541 (to Z. L.), R01 AR040615 (to T. W.), and R01 HL076433 (to B. F.).
- RyR
- ryanodine receptor
- CaM
- calmodulin.
REFERENCES
- 1. Wagenknecht T., Liu Z. (2010) Electron microscopy of ryanodine receptors. Curr. Top. Membr. 66, 27–47 [DOI] [PubMed] [Google Scholar]
- 2. Lanner J. T., Georgiou D. K., Joshi A. D., Hamilton S. L. (2010) Ryanodine receptors: structure, expression, molecular details, and function in calcium release. Cold Spring Harbor Perspect. Biol. 2, a003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prosser B. L., Hernández-Ochoa E. O., Schneider M. F. (2011) S100A1 and calmodulin regulation of ryanodine receptor in striated muscle. Cell Calcium 50, 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aracena P., Hidalgo C., Hamilton S. L. (2005) in Ryanodine Receptors: Structure, Function and Dysfunction in Clinical Diseases (Wehrens X. H. T., Marks A. R., eds) pp. 163–168, Springer, New York [Google Scholar]
- 5. Meissner G. (2004) Molecular regulation of cardiac ryanodine receptor ion channel. Cell Calcium 35, 621–628 [DOI] [PubMed] [Google Scholar]
- 6. Moore C. P., Rodney G., Zhang J. Z., Santacruz-Toloza L., Strasburg G., Hamilton S. L. (1999) Apocalmodulin and Ca2+-calmodulin bind to the same region on the skeletal muscle Ca2+ release channel. Biochemistry 38, 8532–8537 [DOI] [PubMed] [Google Scholar]
- 7. Balshaw D. M., Xu L., Yamaguchi N., Pasek D. A., Meissner G. (2001) Calmodulin binding and inhibition of cardiac muscle calcium release channel (ryanodine receptor). J. Biol. Chem. 276, 20144–20153 [DOI] [PubMed] [Google Scholar]
- 8. Yamaguchi N., Xin C., Meissner G. (2001) Identification of apocalmodulin and Ca2+-calmodulin regulatory domain in skeletal muscle Ca2+ release channel, ryanodine receptor. J. Biol. Chem. 276, 22579–22585 [DOI] [PubMed] [Google Scholar]
- 9. Rodney G. G., Williams B. Y., Strasburg G. M., Beckingham K., Hamilton S. L. (2000) Regulation of RyR1 activity by Ca2+ and calmodulin. Biochemistry 39, 7807–7812 [DOI] [PubMed] [Google Scholar]
- 10. Buratti R., Prestipino G., Menegazzi P., Treves S., Zorzato F. (1995) Calcium-dependent activation of skeletal muscle Ca2+ release channel (ryanodine receptor) by calmodulin. Biochem. Biophys. Res. Commun. 213, 1082–1090 [DOI] [PubMed] [Google Scholar]
- 11. Damiani E., Margreth A. (2000) Pharmacological clues to calmodulin-mediated activation of skeletal ryanodine receptor using [3H]ryanodine binding. J. Muscle Res. Cell Motil. 21, 1–8 [DOI] [PubMed] [Google Scholar]
- 12. Tripathy A., Xu L., Mann G., Meissner G. (1995) Calmodulin activation and inhibition of skeletal muscle Ca2+ release channel (ryanodine receptor). Biophys. J. 69, 106–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamaguchi N., Xu L., Pasek D. A., Evans K. E., Meissner G. (2003) Molecular basis of calmodulin binding to cardiac muscle Ca2+ release channel (ryanodine receptor). J. Biol. Chem. 278, 23480–23486 [DOI] [PubMed] [Google Scholar]
- 14. Samsó M., Wagenknecht T. (2002) Apocalmodulin and Ca2+-calmodulin bind to neighboring locations on the ryanodine receptor. J. Biol. Chem. 277, 1349–1353 [DOI] [PubMed] [Google Scholar]
- 15. Wagenknecht T., Radermacher M., Grassucci R., Berkowitz J., Xin H. B., Fleischer S. (1997) Locations of calmodulin and FK506-binding protein on the three-dimensional architecture of the skeletal muscle ryanodine receptor. J. Biol. Chem. 272, 32463–32471 [DOI] [PubMed] [Google Scholar]
- 16. Cornea R. L., Nitu F., Gruber S., Kohler K., Satzer M., Thomas D. D., Fruen B. R. (2009) FRET-based mapping of calmodulin bound to the RyR1 Ca2+ release channel. Proc. Natl. Acad. Sci. U.S.A. 106, 6128–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guo T., Fruen B. R., Nitu F. R., Nguyen T. D., Yang Y., Cornea R. L., Bers D. M. (2011) FRET detection of calmodulin binding to the cardiac RyR2 calcium release channel. Biophys. J. 101, 2170–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maune J. F., Klee C. B., Beckingham K. (1992) Ca2+ binding and conformational change in two series of point mutations to the individual Ca2+-binding sites of calmodulin. J. Biol. Chem. 267, 5286–5295 [PubMed] [Google Scholar]
- 19. Inui M., Saito A., Fleischer S. (1987) Purification of the ryanodine receptor and identity with feet structures of junctional terminal cisternae of sarcoplasmic reticulum from fast skeletal muscle. J. Biol. Chem. 262, 1740–1747 [PubMed] [Google Scholar]
- 20. Liu Z., Zhang J., Li P., Chen S. R., Wagenknecht T. (2002) Three-dimensional reconstruction of the recombinant type 2 ryanodine receptor and localization of its divergent region 1. J. Biol. Chem. 277, 46712–46719 [DOI] [PubMed] [Google Scholar]
- 21. Fruen B. R., Black D. J., Bloomquist R. A., Bardy J. M., Johnson J. D., Louis C. F., Balog E. M. (2003) Regulation of the RyR1 and RyR2 Ca2+ release channel isoforms by Ca2+-insensitive mutants of calmodulin. Biochemistry 42, 2740–2747 [DOI] [PubMed] [Google Scholar]
- 22. Gyobu N., Tani K., Hiroaki Y., Kamegawa A., Mitsuoka K., Fujiyoshi Y. (2004) Improved specimen preparation for cryo-electron microscopy using a symmetric carbon sandwich technique. J. Struct. Biol. 146, 325–333 [DOI] [PubMed] [Google Scholar]
- 23. Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 [DOI] [PubMed] [Google Scholar]
- 24. Frank J. (2006) Three-dimensional Electron Microscopy of Macromolecular Assemblies, pp. 58–62, Oxford University Press, New York [Google Scholar]
- 25. Liu Z., Zhang J., Sharma M. R., Li P., Chen S. R., Wagenknecht T. (2001) Three-dimensional reconstruction of the recombinant type 3 ryanodine receptor and localization of its amino terminus. Proc. Natl. Acad. Sci. U.S.A. 98, 6104–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Malhotra A., Penczek P., Agrawal R. K., Gabashvili I. S., Grassucci R. A., Jünemann R., Burkhardt N., Nierhaus K. H., Frank J. (1998) Escherichia coli 70 S ribosome at 15 Å resolution by cryo-electron microscopy: localization of fMet-tRNAfMet and fitting of L1 protein. J. Mol. Biol. 280, 103–116 [DOI] [PubMed] [Google Scholar]
- 27. Meng X., Wang G., Viero C., Wang Q., Mi W., Su X. D., Wagenknecht T., Williams A. J., Liu Z., Yin C. C. (2009) CLIC2-RyR1 interaction and structural characterization by cryo-electron microscopy. J. Mol. Biol. 387, 320–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Frank J. (2006) Three-dimensional Electron Microscopy of Macromolecular Assemblies, pp. 145–192, Oxford University Press, New York [Google Scholar]
- 29. Ludtke S. J., Serysheva I. I., Hamilton S. L., Chiu W. (2005) The pore structure of the closed RyR1 channel. Structure 13, 1203–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Samsó M., Wagenknecht T., Allen P. D. (2005) Internal structure and visualization of transmembrane domains of the RyR1 calcium release channel by cryo-EM. Nat. Struct. Mol. Biol. 12, 539–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodney G. G., Moore C. P., Williams B. Y., Zhang J. Z., Krol J., Pedersen S. E., Hamilton S. L. (2001) Calcium binding to calmodulin leads to an N-terminal shift in its binding site on the ryanodine receptor. J. Biol. Chem. 276, 2069–2074 [DOI] [PubMed] [Google Scholar]
- 32. Maximciuc A. A., Putkey J. A., Shamoo Y., Mackenzie K. R. (2006) Complex of calmodulin with a ryanodine receptor target reveals a novel, flexible binding mode. Structure 14, 1547–1556 [DOI] [PubMed] [Google Scholar]
- 33. Xiong L. W., Newman R. A., Rodney G. G., Thomas O., Zhang J. Z., Persechini A., Shea M. A., Hamilton S. L. (2002) Lobe-dependent regulation of ryanodine receptor type 1 by calmodulin. J. Biol. Chem. 277, 40862–40870 [DOI] [PubMed] [Google Scholar]
- 34. Zhang H., Zhang J. Z., Danila C. I., Hamilton S. L. (2003) A noncontiguous, intersubunit binding site for calmodulin on the skeletal muscle Ca2+ release channel. J. Biol. Chem. 278, 8348–8355 [DOI] [PubMed] [Google Scholar]
- 35. Houdusse A., Gaucher J. F., Krementsova E., Mui S., Trybus K. M., Cohen C. (2006) Crystal structure of apocalmodulin bound to the first two IQ motifs of myosin V reveals essential recognition features. Proc. Natl. Acad. Sci. U.S.A. 103, 19326–19331 [DOI] [PMC free article] [PubMed] [Google Scholar]