Background: The interaction between the transactivation domain of p53 with the N-terminal domains of MDM2 and MDMX is critical for many cell functions.
Results: The binding properties of the two subdomains TAD1 and TAD2 of p53 were studied at atomic detail by NMR, mutagenesis, and molecular dynamics simulations.
Conclusion: The binding between subdomains TAD1 and TAD2 is competitive.
Significance: Regulation of the MDM2/MDMX interactions can be modulated by both TAD1 and TAD2.
Keywords: Biophysics, Intrinsically Disordered Proteins, Molecular Dynamics, Nuclear Magnetic Resonance, p53, MDM2, MDMX
Abstract
The interaction between the transactivation domain of p53 (p53TAD) and the N-terminal domain of MDM2 and MDMX plays an essential role for cell function. Mutations in these proteins have been implicated in many forms of cancer. The intrinsically disordered p53TAD contains two subdomains, TAD1 and TAD2. Using NMR spectroscopy, site-directed mutagenesis, and molecular dynamics simulations, we demonstrate that TAD2 directly interacts with MDM2, adopting transient structures that bind to the same hydrophobic pocket of MDM2 as TAD1. Our data show that binding of TAD1 and TAD2 to MDM2 is competitive, which is further supported by the observation that the interaction of TAD2 with MDM2 can be blocked by the small molecule inhibitor nutlin-3. Our data further indicate that TAD2 interacts with MDMX in a fashion very similar to MDM2. Because TAD2 is known to have transcriptional activity, the interaction of TAD2 with MDM2/MDMX may play a direct role in the inhibition of p53 transactivation.
Introduction
The tumor suppressor protein p53 is a key transcription factor that plays an essential role in regulating cell fates (1–3). Under normal cell cycle progression, p53 is maintained at very low levels by negative regulatory proteins including MDM2, MDMX, and various kinases. In response to cellular stresses, p53 is phosphorylated and up-regulated (4–7). Activated p53 induces the transcription of a variety of target genes leading to cell growth arrest, senescence, or apoptosis (8, 9). p53 inactivation is found in almost 50% of human cancers, often caused by elevated levels of MDM2 or MDMX (10–14). In several independent studies it was found that the recovery of p53 antitumor activity leads to tumor growth inhibition in a broad spectrum of human cancers (15–17). Understanding the molecular mechanism of complex formation of p53 with MDM2/MDMX is, therefore, of importance, and it could accelerate the discovery of antagonistic drugs that prevent inactivation of the p53 tumor suppression caused by MDM2/MDMX overexpression.
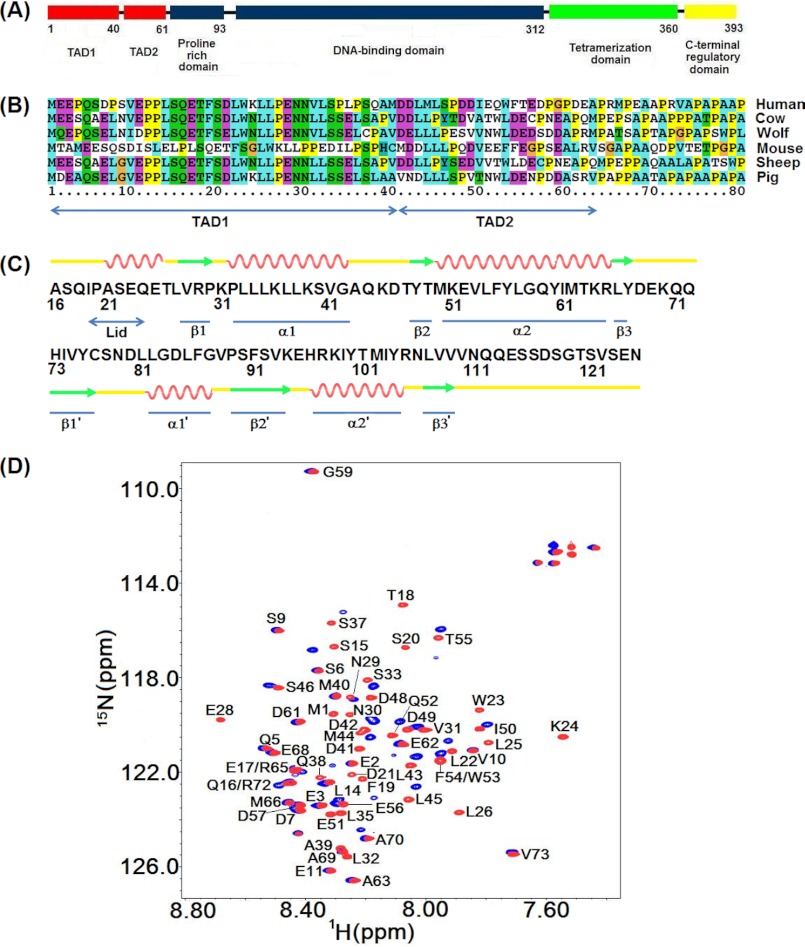
p53 is active as a homo-tetramer formed by four identical chains of 393 residues each. Each p53 subunit consists of an N-terminal transactivation domain or TAD3 (residues 1–61) and a proline-rich region (residues 64–92) followed by a central DNA binding domain, a short tetramerization domain, and a C-terminal regulatory domain (Fig. 1A). TAD can be further divided into two subdomains, TAD1 (residues 1–40) and TAD2 (residues 41–60). The presence of extended segments in TAD that are disordered (18, 19) enables the domain to bind to a range of target proteins including MDM2 (20), MDMX (21, 22), human replication protein A (hRPA70) (23), and the transcriptional coactivator p300/CBP (CREB-binding protein) (24). MDM2 inhibits p53 activity through direct protein-protein interaction. In the cytosol MDM2 mediates polyubiquitination of p53 and subsequent degradation in the proteasome, whereas in the nucleus, MDM2 prevents p53 transactivation by blocking the TAD domain from recruiting transcriptional coactivators (25–27). It is well established that MDM2 interacts with TAD1 through its N-terminal domain (Fig. 1B). The crystal structure of the MDM2 N-terminal domain complexed with a short peptide of TAD1 revealed that the TAD1 peptide binds to a deep hydrophobic cleft of MDM2 through a hydrophobic triad consisting of the residues Phe-19, Trp-23, and Leu-26, whereby the intrinsically partially disordered TAD1 peptide folds into an amphipathic α-helix in the complex (20).
FIGURE 1.
A, shown is the domain structure of p53. B, shown is sequence alignment of the two transactivation subdomains of p53. C, shown are the primary sequence and a schematic representation of the secondary structural elements of the N-terminal domain of MDM2. D, shown are 1H,15N HSQC spectra of p53TAD in the absence (red) and presence of MDM2 (blue). The concentration of apop53TAD was 300 μm, and the MDM2 bound state of p53TAD contained 250 μm p53TAD and 300 μm MDM2.
The crystal structure (20) highlights the hydrophobic nature of the p53·MDM2 (the · symbol signifies an intermolecular complex) interaction, which has promoted the development of a number of antagonistic drug leads such as nutlin-3 (15). The N-terminal lid of MDM2 (Fig. 1C), which is unobservable in the MDM2·p53 crystal structure, self-associates with the p53 binding cleft in the apo state but is disposed away from the hydrophobic pocket in the MDM2·p53 complex (28, 29). By contrast, binding to nutlin-3 still allows the lid to associate with the pocket with direct implications for drug design and lead screening (29).
The TAD2 domain of p53, whose amino acid sequence succeeds the one of TAD1 (Fig. 1B), has been shown to activate various p53 response genes and induce apoptosis (30). Hence, transcriptional activity of TAD2 in the nucleus must be subjected to a regulatory mechanism. The potential interaction between TAD2 and MDM2 has been previously investigated by NMR titration experiments, and an extended binding region of p53TAD that includes both TAD1 and TAD2 has been implicated (24). However, the disappearance of the resonances of both the TAD1 and TAD2 subdomains in the bound state prevented any detailed characterization of the p53TAD·MDM2 complex (24). In addition, the disordered TAD domain so far has rendered it inaccessible to structure determination by x-ray crystallography. A structural model has been proposed on the basis of transferred NOE data in combination of molecular docking, which suggested that the isolated TAD2 peptide comprising residues 49–54 of p53 binds to MDM2 and forms a well-folded helical turn structure (31). However, it remains unknown how the TAD2 region in full-length p53TAD interacts with MDM2 and how this interaction relates to TAD1 binding. Therefore, despite of the functional importance of both p53 transactivation subdomains TAD1 and TAD2, the mechanism for the formation and stabilization of the complex between p53TAD and MDM2 remains unclear.
Here, we report the characterization of the structural dynamics of full-length p53TAD and its interaction with MDM2 at atomistic detail. Our NMR and molecular dynamics simulation data in combination with mutational analysis show that TAD2 preferentially interacts with the hydrophobic cleft of MDM2 in a manner that is competitive with TAD1 binding without undergoing a significant change in its structural propensity. Our results provide a comprehensive structural-dynamic view of the full-length p53TAD in its interaction with MDM2 as well as MDMX with important implications for the function of this intrinsically disordered transactivation domain of p53.
EXPERIMENTAL PROCEDURES
Sample Preparation
The DNA fragment corresponding to residues 1–73 of the p53 transactivation domain was PCR-amplified from an EST clone (access number BU157354) and cloned into the cloning vector pTBGST, which encodes a N-terminal His6 tag with a tobacco etch virus recognition site upstream of the cloning site. The primers for generating the TAD1 mutations were purchased from Integrated DNA Technologies (IDT). Wild-type p53TAD and its mutants as well as MDM2 were expressed and purified as previously described (29). The TAD1 peptide consisting of p53 residues 17–29 was prepared as previously described (29). The purified TAD2 peptide corresponding to residues 40–57 of p53TAD was purchased from Neo Group, Inc. The DNA fragment corresponding to residues 14–111 of MDMX was PCR-amplified from an EST clone (access number BC067299.1) and cloned into the cloning vector pTBGST. The C17S mutation was introduced to avoid protein purification problems due to low protein stability as this mutation does not affect the ligand binding behavior of MDMX (32, 33).
NMR Experiments
Purified proteins were concentrated using an Amicon Ultra centrifugal filter device (Millipore) and buffer exchanged to 50 mm pH 7.0 HEPES that contains 100 mm NaCl, 5 mm DTT, and 0.05% (w/v) NaN3. The concentration of the apop53TAD and its mutants were 300 μm, and the bound samples contained 250 μm p53TAD and 300 μm MDM2. NMR experiments were conducted at 25 °C on a Bruker Avance II 800 MHz spectrometer equipped with a cryogenic TCI probe. Standard triple resonance experiments were performed to obtain the resonance assignments. The backbone T1, T1ρ, and heteronuclear NOE experiments were performed as previously described (29). All spectra were processed using NMRPipe (34) and analyzed by NMRViewJ (35). Secondary structural propensity (SSP) scores were calculated as described by Marsh et al. (36). The dissociation constants between p53TAD F19A/W23A and MDM2/MDMX were obtained by fitting of 1H and 15N chemical shift changes as a function of ligand concentration to a two-site binding model.
Molecular Dynamics Simulations
MD simulations of the TAD2 domain (residues Met-40 to Asp-57) were performed at 330 K using the GROMACS software package Version 4.5.3 (37) with the TIP3P explicit water model and the Amber ff99SB force field (38). As the initial state, the TAD2 peptide in an extended conformation was placed near the hydrophobic pocket of MDM2 followed by standard equilibration procedures (39). Two independent 1-μs simulations were performed with different starting peptide orientations.
RESULTS
MDM2 Binding Modulates NMR Resonances of Both p53 Transactivation Subdomains
To characterize the interaction between full-length p53TAD and MDM2, a construct consisting of the N-terminal 73 residues of p53 was used here, which is known to be sufficient to activate transcription (40). The 1H,15N HSQC spectrum of apop53TAD (Fig. 1D) shows limited dispersion of the resonances in both the 1HN and the 15N dimensions, characteristic for an intrinsically disordered protein. Binding to MDM2 led to the disappearance of the cross-peaks of 15 consecutive non-proline residues between Glu-17 to Leu-32, which constitute the binding interface of TAD1, consistent with previous studies (24, 31). Importantly, for the first time NMR signals belonging to TAD2 in the MDM2-bound state could be observed as weak cross-peaks (Fig. 1D). As shown in Fig. 2F, the average cross-peak intensities for these TAD2 residues in the bound state are ∼20–30% of those in the apo state at the same p53TAD concentration.
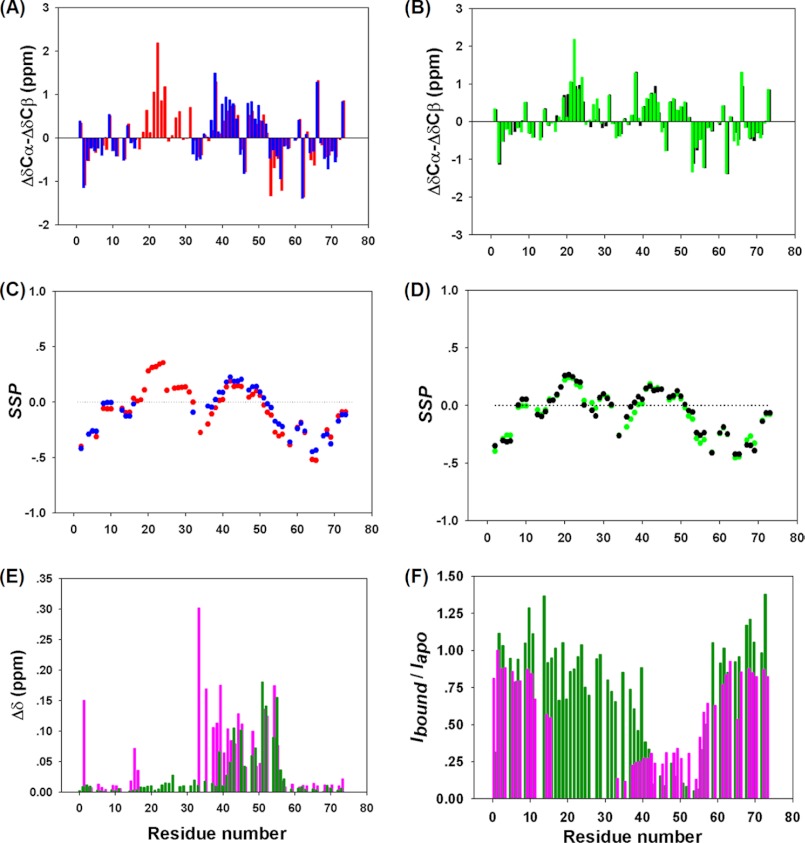
FIGURE 2.
A, 13C ΔδCα-ΔδCβ secondary shifts for the apop53TAD (red) and p53TAD·MDM2 (blue) are shown. Positive numbers indicate α-helical structure, and negative values suggest β-strand structural propensity. B, shown are 13C ΔδCα-ΔδCβ secondary shifts for the apop53TAD F19A/W23A (green) and p53TAD F19A/W23A·MDM2 (black). C, shown is an SSP (Marsh et al. (36) analysis of apop53TAD (red) and p53TAD·MDM2 (blue). Positive SSP indicates propensity to form α-helical structure, and negative SSP suggests β-strand structure. D, shown is a SSP analysis of apop53TAD F19A/W23A (green) and p53TAD F19A/W23A·MDM2 (black). E, shown are chemical shift perturbations induced by MDM2 binding Δδ = √(ΔδHN2 + (0.15ΔδN)2). p53TAD WT is in magenta; p53TAD F19A/W23A is in dark green). F, experimental peak intensity ratios (Ibound/Iapo) for p53TAD WT (magenta) and p53TAD F19A/W23A (green) after and before the addition of MDM2.
To investigate this binding process in more detail, an HSQC titration experiment was performed by incrementally adding non-labeled MDM2 to 15N-labeled p53TAD. The results show that p53TAD residues located in the two termini, i.e. residues Glu-2 to Glu-11 and residues Glu-56 to Val-73, experienced very little change both in terms of peak positions and peak intensities. By contrast, resonances of residues Leu-14 to Thr-55 in the free state gradually diminished with increasing MDM2 concentrations without changing their positions. In addition, a new set of peaks corresponding to the MDM2-bound state became resolved for residues Ser-33 to Thr-55 that encompasses most of the TAD2 subdomain as well as residues Leu-14 to Gln-16 (Fig. 1D). These results suggest the presence of a slow conformational exchange between the free state of p53TAD and p53TAD·MDM2 with lifetimes of the order of hundreds of milliseconds or slower.
Interestingly, bound-state resonances Glu-17 to Leu-32 of TAD1 residues, which dominate the p53TAD·MDM2 interaction, were too weak to be observed during the entire titration. The severe line-broadening upon MDM2 binding by far exceeds what is expected from a slowdown of overall rotational diffusion due to the volume increase of the p53TAD·MDM2 complex compared with apop53TAD. Moreover, the line widths of the resonances in 15N-MDM2 were only little affected by complex formation with TAD (see supplemental Fig. S1A), and for the TAD1 peptide (i.e. in the absence of TAD2), the TAD1 cross-peaks only underwent moderate broadening upon binding to MDM2 supplemental Fig. S1B). Taken together, these results show that TAD2 directly interferes with TAD1 binding in the p53TAD·MDM2 complex, inducing conformational exchange that leads to the broadening of the TAD1 resonances beyond detection. Although it is quite possible that in the bound state the TAD1 region adopts predominantly the type of helical structure exhibited by TAD1 peptide·MDM2 in the x-ray crystal structure (20), TAD1 in p53TAD·MDM2 must experience significant conformational exchange between two or more alternative structures.
TAD2 of p53TAD Interacts Directly with MDM2
To address whether the TAD2 resonance changes are caused merely by a tethering effect due to their vicinity to TAD1, a mutational study was conducted. For this purpose the mutant p53TAD F19A/W23A was made to disrupt binding of TAD1 to MDM2. Although wild-type TAD1 binds to the deep hydrophobic cleft of MDM2 with its hydrophobic amino acid triad Phe-19, Trp-23, and Leu-26, which form the primary driving force of the TAD1·MDM2 interaction (20), the double-point mutation of Phe-19 and Trp-23 to alanines completely abolished the interaction between TAD1 and MDM2 as was manifested by large peak intensities of F19A, W23A, and other TAD1 residues in the MDM2-bound form, which is in sharp contrast to wild-type p53TAD·MDM2 (Fig. 2F). In fact, the TAD1 cross-peak intensities were comparable to those of apop53TAD, and their positions are almost unchanged upon the addition of MDM2 (Fig. 2, E and F). At the same time, TAD2 residues experienced peak intensity attenuations and chemical shift perturbations, providing clear evidence that TAD2 directly interacts with MDM2 (Fig. 2, E and F). In addition, TAD2 in the double-point mutant showed similar chemical shift perturbations upon MDM2 binding as in wild-type p53TAD (Fig. 2E), suggesting that TAD1 binding only weakly affects the average TAD2 conformational ensemble in the MDM2-bound state. This finding is further corroborated by secondary chemical shift and SSP analysis (36) (see supplemental Fig. S2, A and B). Titration of non-labeled MDM2 to 15N-p53TAD F19A/W23A causes the TAD2 residues to gradually move from their positions in the free state to their positions in the bound state, suggesting that the free and the bound forms undergo exchange that is fast on the chemical shift timescale, a behavior that is often accompanied by relatively low binding affinity.
To further probe the interaction of TAD2 with MDM2, we studied the interaction between MDM2 and the isolated TAD2 peptide that corresponds to TAD2 residues Met-40 to Asp-57. The addition of the TAD2 peptide induced very similar chemical shift perturbations in MDM2 as the p53TAD F19A/W23A double-point mutant (data not shown). Therefore, the chemical shift perturbations observed in the TAD2 subdomain in wild-type p53TAD are not secondary effects of residual TAD1 binding. They provide unambiguous evidence for the presence of direct interactions between TAD2 and MDM2 as an alternative binding mode to TAD1·MDM2.
TAD2 and the Two Termini Remain Significantly Disordered in the MDM2-bound State
Binding of p53TAD to other proteins often accompanies considerable structural change of those residues that directly participate in the interaction (20, 23, 41, 42). To gain insight into the global and local structural changes of p53TAD in response to MDM2 binding, 13Cα, 13Cβ, and 13CO chemical shifts were determined for p53TAD in both the apo state and the MDM2-bound state. ΔδCα-ΔδCβ secondary chemical shifts and SSP analysis (36) both indicated that apop53TAD is largely unfolded, whereas the TAD1 subdomain adopts a nascent helical structure (Fig. 2, A and C), in agreement with a previous NMR residual dipolar coupling study of apop53TAD (43). In addition, two small segments in TAD2, corresponding to residues 40–44 and residues 48–52, showed residual propensity for helical structure. Upon binding to MDM2, the observable residues of TAD, including those residues corresponding to TAD2 and the two termini, displayed very little change in their secondary structure (Fig. 2, A and C).
Although the secondary structure remained unaffected, TAD2 residues 41–55 exhibited moderately strong amide 1HN and 15N chemical shift perturbations upon MDM2 binding, with an average change of 0.08 ppm (Fig. 2E). Moreover, the average intensity for TAD2 residues was only about 20–30% that in the bound state compared with the apo state (Fig. 2F). When taking into consideration that under the present experimental conditions p53TAD predominantly exists in the bound state according to its submicromolar dissociation constant Kd (19, 24), these findings are indicative of the presence of a heterogeneous distribution of bound TAD2 conformations that are in medium-fast exchange. This picture was further corroborated by the microsecond MD simulation during which TAD2 sampled a broad distribution of poses (see Fig. 4). Although the microsecond MD trajectory is too short for full equilibration, it qualitatively illustrates the dynamic nature of TAD2 while interacting with MDM2. Finally, 13Cα, 13Cβ, and 13CO chemical shifts for the p53TAD F19A/W23A double-point mutant in both the free and the bound forms were measured. The double mutations slightly increase the helicity of the neighboring residues of the mutation sites, but the secondary structural propensity of TAD2 remained essentially unaffected; the average observed ΔδCα-ΔδCβ secondary chemical shift was 0.342 ppm for TAD2 residues 41–52 in the bound form, whereas the average value for the same segment was 0.336 ppm in the apo state. According to a Kd of 119 μm determined by NMR titration experiments (see “Experimental Procedures”), ∼50% of the p53TAD double-point mutant populated the MDM2-bound state under the experimental conditions of Fig. 2E (because of limited solubility of MDM2 at concentrations of >300 μm (44, 45), this percentage cannot be easily increased). Due to fast exchange, the observed secondary chemical shifts (Fig. 2B) were population-weighted averages of the secondary chemical shifts between the bound and the free states. Therefore, the secondary chemical shifts of the fully bound state were estimated by scaling up the secondary shifts of the MDM2-bound state of p53TAD F19A/W23A by a factor of 2 (supplemental Fig. S2C). The resulting shifts are very similar to those in the free state, indicating minimal secondary structural changes upon binding.
FIGURE 4.
A, evidence for competitive binding between TAD2 and TAD1 is shown. Superimposed 1H,15N HSQC spectra of 15N-p53TAD F19AW23A (150 μm): apostate (green) in the presence of 160 μm unlabeled MDM2 (black) and in the presence of 160 μm unlabeled MDM2 and 200 μm unlabeled TAD1-peptide (residues 17–29; orange). B, shown is a model of competitive binding dynamics between the two transactivation subdomains TAD1 and TAD2 of p53 to MDM2. When interacting with the hydrophobic cleft of MDM2, the TAD2 subdomain adopts a wide range of transient structures, including helical conformers, which undergo fast exchange on the NMR chemical shift time scale. The MDM2·TAD2 structures shown represent snapshots selected from two 1-μs MD trajectories in explicit solvent.
TAD2 Binds to the Hydrophobic Cleft of MDM2 and Nutlin-3 Antagonizes the Interaction
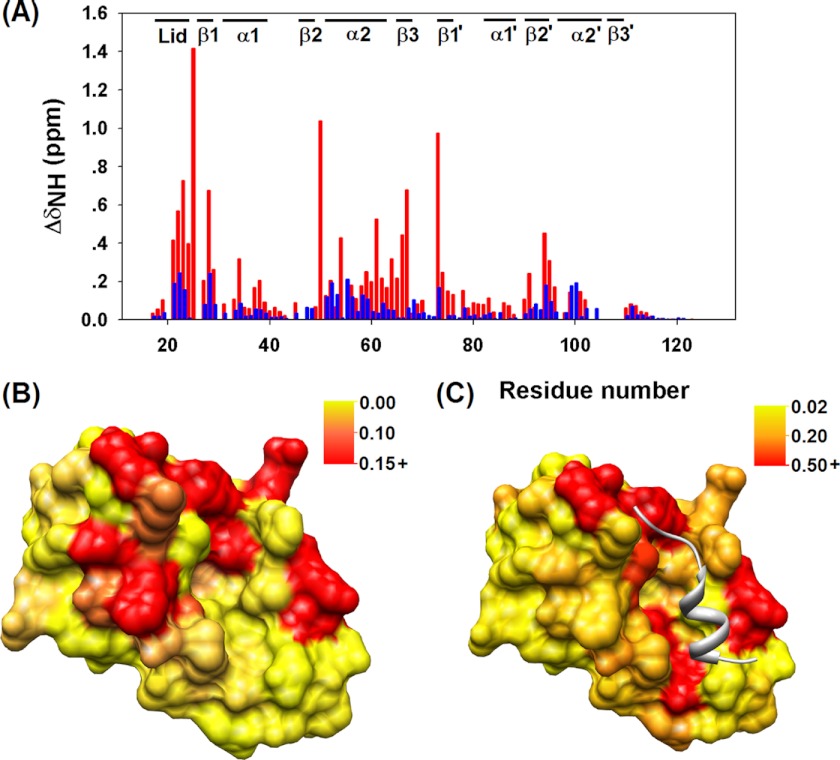
An HSQC titration experiment was performed by adding unlabeled p53TAD F19A/W23A in a stepwise fashion to 15N-labeled MDM2 to determine the TAD2 binding interface of MDM2. A cluster of significant changes in the amide chemical shifts was found in the N-terminal lid of MDM2 as well as helices α2 and α2′ and strand β2′ that formed the large hydrophobic cleft (Fig. 3, A–C). Interestingly, this is the same cleft that binds the isolated TAD1 peptide (20). Because nutlin-3 antagonistically blocks the interaction between MDM2 and the isolated TAD1 peptide (15), we tested whether nutlin-3 can also inhibit the TAD2·MDM2 interaction by recording an HSQC spectrum with 1 mm of nutlin-3 added to the p53TAD F19A/W23A·MDM2 complex. Remarkably, in the presence of nutlin-3 the spectrum closely resembles that of free p53TAD F19A/W23A (see supplemental Fig. S3, A and B), indicating that nutlin-3 effectively blocks the interaction between TAD2 and MDM2. Because x-ray crystal structural data showed that nutlin-3 specifically binds to the hydrophobic cleft of MDM2 (15), its disruption of the binding of TAD2 suggests that the hydrophobic cleft is also the binding site for TAD2.
FIGURE 3.
A, chemical shift perturbations of MDM2 induced by p53TAD WT binding (blue) and p53TAD F19A/W23A binding (red) are shown. ΔδNH = √(ΔδH2 + (0.15ΔδN)2). Mapping of chemical shift changes (ppm) of MDM2 upon p53TAD F19A/W23A binding (B) and p53TAD WT binding (C) on the surface of MDM2 (using PDB 1YCR as template) is shown. The largest chemical shift changes are clustered around the hydrophobic cleft.
In apoMDM2 the N-terminal lid self-associates with the hydrophobic cleft and thereby plays a regulatory role in ligand binding (29). To probe the lid structure in the TAD2·MDM2 complex, the amide chemical shifts of the lid residues were examined. Several MDM2 lid residues, including Ile-19, Ala-21, and Ser-22, display substantial changes in peak position in the TAD2-bound form, trending clearly toward the TAD1-bound state resonances where the lid moves freely and is fully exposed to the water solvent (supplemental Fig. S3, C–E). The shift of the lid resonances toward the open state suggests that TAD2 significantly destabilizes self-association between the lid and the cleft.
Competitive Binding
When bound to MDM2, the TAD2 motif in full-length p53 has a very weak helical propensity, which only slightly exceeds that of apoTAD2. Nonetheless, this subdomain exhibited substantially slower overall backbone dynamics in the bound state than in the apo state (supplemental Fig. S2, D and E), indicating that TAD2 is an active participant in p53·MDM2 interaction. Abolishment of the TAD1·MDM2 interaction in the p53TAD double-point mutant led to a further decrease of the resonance intensities of TAD2 residues (supplemental Fig. S2, D and E), suggesting tighter coupling between TAD2 and MDM2 in the absence of TAD1 binding. Taken together our data support a competitive MDM2 binding mechanism between the TAD1 and TAD2 subdomains to the hydrophobic cleft of MDM2: (i) interaction of both TAD1 and TAD2 regions with MDM2 can be inhibited by nutlin-3; (ii) the presence of the TAD1-peptide directly competes with p53TAD F19A/W23A binding to MDM2, leading to the complete disruption of the p53TAD F19A/W23A·MDM2 interaction shown in Fig. 4; (iii) competitive binding also works the other way around (supplemental Fig. S4), where the addition of TAD2 peptide disrupts the association of MDM2 with the TAD1 peptide (17–29). Isothermal calorimetry measurements yielded for MDM2·p53TAD and MDM2·TAD1 Kd values of 0.077 ± 0.006 and 0.44 ± 0.1 μm, respectively, which are consistent with results reported previously (24). Although the strengthening of binding is consistent with a competitive binding mechanism (see the supplemental binding polynomial analysis), the observed quantitative increase of binding affinity suggests, in addition, the presence of some degree of binding cooperativity between TAD1 and TAD2, i.e. when either one of the TAD domains is bound, the other domain interacts favorably with MDM2.
TAD2 of p53TAD Interacts with MDMX in a Similar Manner to MDM2
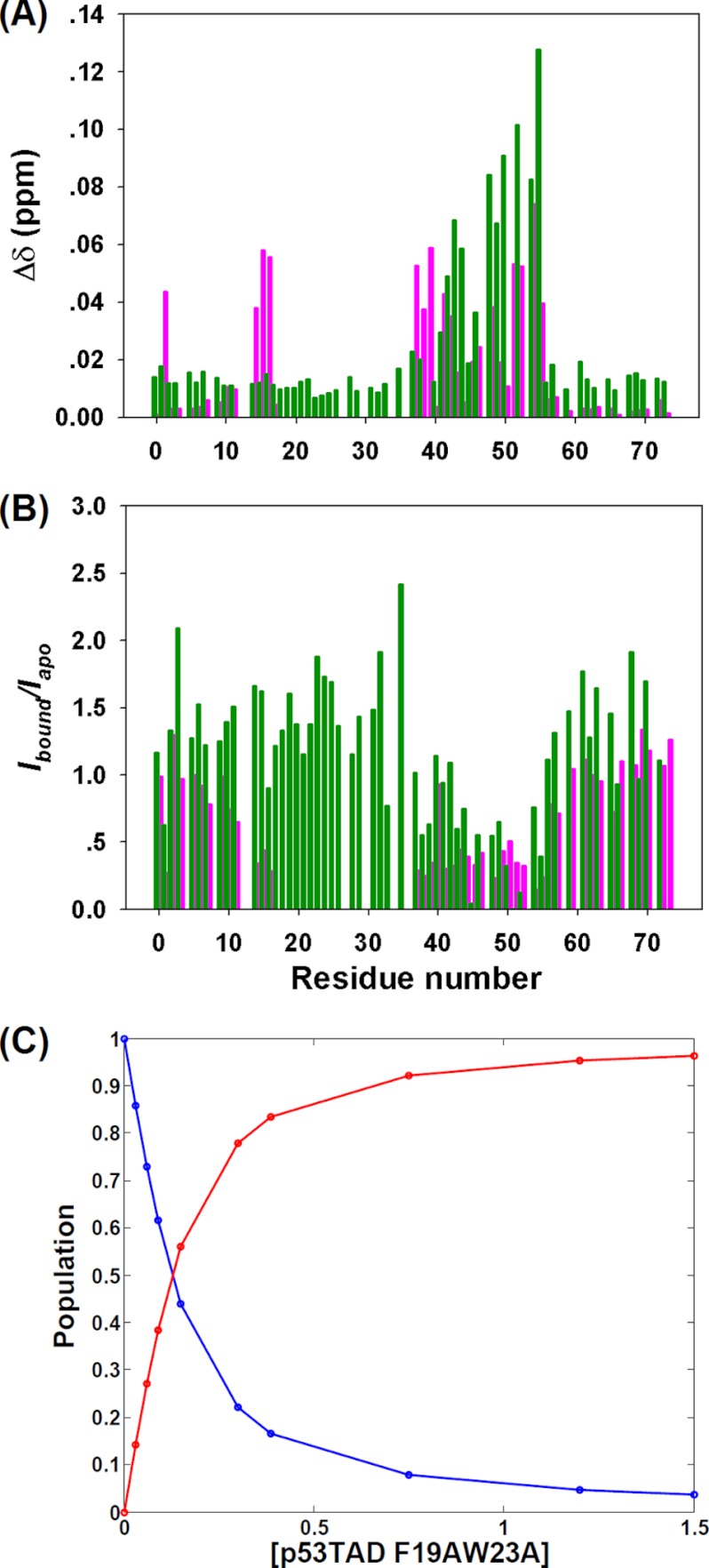
The MDM2 homologue MDMX cooperatively interacts with MDM2 and impedes p53-induced tumor growth inhibition (46, 47). Concomitant targeting of both MDM2 and MDMX interactions with p53 is necessary for the full activation of p53 activity (48–51). NMR titration experiments of 15N-labeled p53TAD with non-labeled MDMX showed a similar behavior as in the case of MDM2. Although the bound state resonances Glu-17–Leu-32 of TAD1, which dominate the interaction with MDMX, were too weak to be observed, bound-state resonances of TAD2 residues gradually emerged at new peak positions with increasing MDMX concentration, characteristic of slow exchange. In comparison, titration of MDMX to the double-point mutant p53TAD-F19A/W23A induced continuous changes of peak positions, characteristic of fast exchange confirming the weak but direct interaction between the TAD2 motif and MDMX. The binding affinity between TAD2 and MDMX was determined at Kd = 52 μm by NMR titration experiments, which corresponds to more than a 2-fold increase in binding affinity over MDM2 (Fig. 5).
FIGURE 5.
TAD2·MDMX interaction. A, amide chemical shift perturbations of p53TAD WT (magenta) and p53TAD-F19AW23A (green) induced by MDMX binding; Δδ = √(ΔδHN2 + (0.15ΔδN)2). B, the calculated peak intensity ratios (Ibound/Iapo) for p53TAD WT (magenta) and p53TAD F19A/W23A (green) after the addition of MDMX are shown. C, fitting of the titration data using 15N-MDMX and non-labeled TAD mutant is shown. The red curve indicates the population of the bound state of MDMX, and blue curve represents the free state of MDMX. The fitted Kd is 52 μm.
DISCUSSION
Intrinsic disorder is a common structural motif in proteins that play central roles in signal transduction networks (52). The transactivation domains of transcriptional activators such as p53TAD are capable of binding to diverse target proteins with high specificity. As a typical molecular recognition motif (53), binding of p53TAD to partner proteins often accompanies disorder-to-order transitions in partially populated helical structural segments. Regions in the TAD2 subdomain fold into an α-helical structure upon binding to the replication protein A (hRPA70) (23) and to the Tfb1 subunit of the yeast TFIIH (41). Synergistic binding between both p53 transactivation subdomains and partner proteins has been found in the interaction between p53TAD and four individual domains of transcription factor p300 (24). A recent NMR structure revealed that TAD1 and TAD2 simultaneously bind to the nuclear coactivator binding domain (NCBD) of CREB-binding protein (CBP) and form a pair of α-helices in which TAD2 dominates the interaction with NCBD with a Kd of 13.6 μm (42). Our data for p53TAD·MDM2 binding suggest that both TAD1 and TAD2 subdomains actively participate in the interaction with MDM2, but TAD2 does not show a pronounced binding-induced folding behavior. This differs from the previously reported formation of a helical turn by isolated TAD2 peptide when bound to MDM2 (31). Our results highlight the relevance of studying the intact full-length p53 domain for elucidating the binding behavior of its subdomains with the MDM2 binding partner.
It has been recognized that for intrinsically disordered proteins involved in regulatory functions in the cell, binding to their receptors is often accompanied by disorder-to-order transitions of various degrees (54–57). In an alternative mechanism, sequence motifs may bind to a modular binding domain via extended conformations (58). Multiple binding modes associated with disordered proteins allow for highly dynamic complex structures, including the interaction between CFTR regulatory region and nucleotide binding domain (NBD1) as well as the interaction between the yeast cyclin-dependent kinase inhibitor Sic1 and its cognate receptor Cdc4 (59, 60). The quantitative atomic-detail data presented here further adds to the understanding of the rich structural-dynamic behavior displayed by ensembles of intrinsically disordered proteins involved in protein-protein interactions.
Previously reported apoptotic and transcriptional activities of both p53(Δ1–42) and p53 L22Q/W23S mutants further underlined the importance of the TAD2 transactivation subdomain in mediating the cell cycle (30). TAD mutations associated with transactivation deficiency, including L22Q/W23S and W53Q/F54S double-point mutants, were found to significantly weaken the binding of p53TAD to MDM2 or transcription coactivator p300 (24), respectively, thus demonstrating the determinant role of protein-protein interactions in p53 transcription activity. Despite the fact that the transcriptional activity of TAD2 has been established, this subdomain has evaded structural investigation by crystallographic methods, presumably due to its disordered nature. The poor solution stability of MDM2 as well as the reduction of TAD2 resonance intensities also limited direct characterization of TAD2·MDM2 interaction by NMR. Our optimized conditions for MDM2 stability at 300 μm enabled the discovery of a competitive MDM2 binding mode between the TAD1 and TAD2 subdomains, reported here for the first time with a similar mode observed for interactions between p53 and MDMX. This result has direct implications for the observed linkage between TAD2 mutations and p53 transcription deficiency. In particular, the effect of TAD2·MDM2 association may be quite pronounced in the case of MDM2 overexpression in cancer cells, considering that the high level of MDM2 will prevent the TAD subdomain from recruiting transcriptional activators in the nucleus and hence prohibit p53 transactivation. Our studies underscore the significance of the simultaneous inhibition of the MDM2/MDMX interactions with both TAD1 and TAD2 subdomains of p53.
Supplementary Material
This work was supported by National Science Foundation Grant MCB-0918362 and the National High Magnetic Field Laboratory at Florida State University.

This article contains supplemental Figs. S1–S4 and a binding polynomial analysis.
- TAD
- transactivation domain
- SSP
- secondary structural propensity
- HSQC
- heteronuclear single quantum correlation.
REFERENCES
- 1. Vogelstein B., Lane D., Levine A. J. (2000) Surfing the p53 network. Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 2. Levine A. J., Hu W., Feng Z. (2006) The P53 pathway. What questions remain to be explored? Cell Death Differ. 13, 1027–1036 [DOI] [PubMed] [Google Scholar]
- 3. Joerger A. C., Fersht A. R. (2008) Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 77, 557–582 [DOI] [PubMed] [Google Scholar]
- 4. Lambert P. F., Kashanchi F., Radonovich M. F., Shiekhattar R., Brady J. N. (1998) Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 273, 33048–33053 [DOI] [PubMed] [Google Scholar]
- 5. Dumaz N., Meek D. W. (1999) Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 18, 7002–7010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sakaguchi K., Saito S., Higashimoto Y., Roy S., Anderson C. W., Appella E. (2000) Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J. Biol. Chem. 275, 9278–9283 [DOI] [PubMed] [Google Scholar]
- 7. Schon O., Friedler A., Bycroft M., Freund S. M., Fersht A. R. (2002) Molecular mechanism of the interaction between MDM2 and p53. J. Mol. Biol. 323, 491–501 [DOI] [PubMed] [Google Scholar]
- 8. Levine A. J. (1997) p53, the cellular gatekeeper for growth and division. Cell 88, 323–331 [DOI] [PubMed] [Google Scholar]
- 9. Römer L., Klein C., Dehner A., Kessler H., Buchner J. (2006) p53. A natural cancer killer. Structural insights and therapeutic concepts. Angew. Chem. Int. Ed. Engl. 45, 6440–6460 [DOI] [PubMed] [Google Scholar]
- 10. Hollstein M., Rice K., Greenblatt M. S., Soussi T., Fuchs R., Sørlie T., Hovig E., Smith-Sørensen B., Montesano R., Harris C. C. (1994) Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 22, 3551–3555 [PMC free article] [PubMed] [Google Scholar]
- 11. Oliner J. D., Kinzler K. W., Meltzer P. S., George D. L., Vogelstein B. (1992) Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358, 80–83 [DOI] [PubMed] [Google Scholar]
- 12. Momand J., Jung D., Wilczynski S., Niland J. (1998) The MDM2 gene amplification database. Nucleic Acids Res. 26, 3453–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marine J. C., Dyer M. A., Jochemsen A. G. (2007) MDMX. From bench to bedside. J. Cell Sci. 120, 371–378 [DOI] [PubMed] [Google Scholar]
- 14. Wade M., Wahl G. M. (2009) Targeting Mdm2 and Mdmx in cancer therapy. Better living through medicinal chemistry? Mol. Cancer Res. 7, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E. A. (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 [DOI] [PubMed] [Google Scholar]
- 16. Ringshausen I., O'Shea C. C., Finch A. J., Swigart L. B., Evan G. I. (2006) Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell 10, 501–514 [DOI] [PubMed] [Google Scholar]
- 17. Marx J. (2007) Oncology. Recruiting the cell's own guardian for cancer therapy. Science 315, 1211–1213 [DOI] [PubMed] [Google Scholar]
- 18. Lee H., Mok K. H., Muhandiram R., Park K. H., Suk J. E., Kim D. H., Chang J., Sung Y. C., Choi K. Y., Han K. H. (2000) Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J. Biol. Chem. 275, 29426–29432 [DOI] [PubMed] [Google Scholar]
- 19. Dawson R., Müller L., Dehner A., Klein C., Kessler H., Buchner J. (2003) The N-terminal domain of p53 is natively unfolded. J. Mol. Biol. 332, 1131–1141 [DOI] [PubMed] [Google Scholar]
- 20. Kussie P. H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A. J., Pavletich N. P. (1996) Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274, 948–953 [DOI] [PubMed] [Google Scholar]
- 21. Popowicz G. M., Czarna A., Rothweiler U., Szwagierczak A., Krajewski M., Weber L., Holak T. A. (2007) Molecular basis for the inhibition of p53 by Mdmx. Cell Cycle 6, 2386–2392 [DOI] [PubMed] [Google Scholar]
- 22. Popowicz G. M., Czarna A., Holak T. A. (2008) Structure of the human Mdmx protein bound to the p53 tumor suppressor transactivation domain. Cell Cycle 7, 2441–2443 [DOI] [PubMed] [Google Scholar]
- 23. Bochkareva E., Kaustov L., Ayed A., Yi G. S., Lu Y., Pineda-Lucena A., Liao J. C., Okorokov A. L., Milner J., Arrowsmith C. H., Bochkarev A. (2005) Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc. Natl. Acad. Sci. U.S.A. 102, 15412–15417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Teufel D. P., Freund S. M., Bycroft M., Fersht A. R. (2007) Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc. Natl. Acad. Sci. U.S.A. 104, 7009–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J. (1992) The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 26. Roth J., Dobbelstein M., Freedman D. A., Shenk T., Levine A. J. (1998) Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 17, 554–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chène P. (2004) Inhibition of the p53-MDM2 interaction. Targeting a protein-protein interface. Mol. Cancer Res. 2, 20–28 [PubMed] [Google Scholar]
- 28. McCoy M. A., Gesell J. J., Senior M. M., Wyss D. F. (2003) Flexible lid to the p53-binding domain of human Mdm2. Implications for p53 regulation. Proc. Natl. Acad. Sci. U.S.A. 100, 1645–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Showalter S. A., Bruschweiler-Li L., Johnson E., Zhang F., Brüschweiler R. (2008) Quantitative lid dynamics of MDM2 reveals differential ligand binding modes of the p53-binding cleft. J. Am. Chem. Soc. 130, 6472–6478 [DOI] [PubMed] [Google Scholar]
- 30. Zhu J., Zhou W., Jiang J., Chen X. (1998) Identification of a novel p53 functional domain that is necessary for mediating apoptosis. J. Biol. Chem. 273, 13030–13036 [DOI] [PubMed] [Google Scholar]
- 31. Chi S. W., Lee S. H., Kim D. H., Ahn M. J., Kim J. S., Woo J. Y., Torizawa T., Kainosho M., Han K. H. (2005) Structural details on mdm2-p53 interaction. J. Biol. Chem. 280, 38795–38802 [DOI] [PubMed] [Google Scholar]
- 32. Kallen J., Goepfert A., Blechschmidt A., Izaac A., Geiser M., Tavares G., Ramage P., Furet P., Masuya K., Lisztwan J. (2009) Crystal structures of human MdmX (HdmX) in complex with p53 peptide analogues reveal surprising conformational changes. J. Biol. Chem. 284, 8812–8821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sanchez M. C., Renshaw J. G., Davies G., Barlow P. N., Vogtherr M. (2010) MDM4 binds ligands via a mechanism in which disordered regions become structured. FEBS Lett. 584, 3035–3041 [DOI] [PubMed] [Google Scholar]
- 34. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe. A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 35. Johnson B. A. (2004) Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- 36. Marsh J. A., Singh V. K., Jia Z., Forman-Kay J. D. (2006) Sensitivity of secondary structure propensities to sequence differences between α- and γ-synuclein. Implications for fibrillation. Protein Sci. 15, 2795–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hess B., Kutzner C., van der Spoel D., Lindahl E. (2008) GROMACS 4. Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 38. Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. (2006) Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li D. W., Brüschweiler R. (2011) Iterative Optimization of molecular mechanics force fields from NMR data of full-length proteins. J. Chem. Theory Comput. 7, 1773–1782 [DOI] [PubMed] [Google Scholar]
- 40. Fields S., Jang S. K. (1990) Presence of a potent transcription activating sequence in the p53 protein. Science 249, 1046–1049 [DOI] [PubMed] [Google Scholar]
- 41. Di Lello P., Jenkins L. M., Jones T. N., Nguyen B. D., Hara T., Yamaguchi H., Dikeakos J. D., Appella E., Legault P., Omichinski J. G. (2006) Structure of the Tfb1/p53 complex. Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol. Cell 22, 731–740 [DOI] [PubMed] [Google Scholar]
- 42. Lee C. W., Martinez-Yamout M. A., Dyson H. J., Wright P. E. (2010) Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein. Biochemistry 49, 9964–9971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wells M., Tidow H., Rutherford T. J., Markwick P., Jensen M. R., Mylonas E., Svergun D. I., Blackledge M., Fersht A. R. (2008) Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc. Natl. Acad. Sci. U.S.A. 105, 5762–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Uhrinova S., Uhrin D., Powers H., Watt K., Zheleva D., Fischer P., McInnes C., Barlow P. N. (2005) Structure of free MDM2 N-terminal domain reveals conformational adjustments that accompany p53 binding. J. Mol. Biol. 350, 587–598 [DOI] [PubMed] [Google Scholar]
- 45. Tang C., Iwahara J., Clore G. M. (2006) Visualization of transient encounter complexes in protein-protein association. Nature 444, 383–386 [DOI] [PubMed] [Google Scholar]
- 46. Francoz S., Froment P., Bogaerts S., De Clercq S., Maetens M., Doumont G., Bellefroid E., Marine J. C. (2006) Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc. Natl. Acad. Sci. U.S.A. 103, 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xiong S., Van Pelt C. S., Elizondo-Fraire A. C., Liu G., Lozano G. (2006) Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc. Natl. Acad. Sci. U.S.A. 103, 3226–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Khoury K., Grzegorz M. Popowicz, Holak T. A., Domling A. (2011) The p53-MDM2/MDMX axis. A chemotype perspective. Medchemcomm 2, 246–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hu B., Gilkes D. M., Chen J. (2007) Efficient p53 activation and apoptosis by simultaneous disruption of binding to MDM2 and MDMX. Cancer Res. 67, 8810–8817 [DOI] [PubMed] [Google Scholar]
- 50. Pazgier M., Liu M., Zou G., Yuan W., Li C., Li C., Li J., Monbo J., Zella D., Tarasov S. G., Lu W. (2009) Structural basis for high affinity peptide inhibition of p53 interactions with MDM2 and MDMX. Proc. Natl. Acad. Sci. U.S.A. 106, 4665–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li C., Pazgier M., Li C., Yuan W., Liu M., Wei G., Lu W. Y., Lu W. (2010) Systematic mutational analysis of peptide inhibition of the p53-MDM2/MDMX interactions. J. Mol. Biol. 398, 200–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dyson H. J., Wright P. E. (2002) Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 12, 54–60 [DOI] [PubMed] [Google Scholar]
- 53. Mohan A., Oldfield C. J., Radivojac P., Vacic V., Cortese M. S., Dunker A. K., Uversky V. N. (2006) Analysis of molecular recognition features (MoRFs). J. Mol. Biol. 362, 1043–1059 [DOI] [PubMed] [Google Scholar]
- 54. Dyson H. J., Wright P. E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 55. Demarest S. J., Deechongkit S., Dyson H. J., Evans R. M., Wright P. E. (2004) Packing, specificity, and mutability at the binding interface between the p160 coactivator and CREB-binding protein. Protein Sci. 13, 203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. De Guzman R. N., Martinez-Yamout M. A., Dyson H. J., Wright P. E. (2004) Interaction of the TAZ1 domain of the CREB-binding protein with the activation domain of CITED2. Regulation by competition between intrinsically unstructured ligands for non-identical binding sites. J. Biol. Chem. 279, 3042–3049 [DOI] [PubMed] [Google Scholar]
- 57. Sugase K., Dyson H. J., Wright P. E. (2007) Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 447, 1021–1025 [DOI] [PubMed] [Google Scholar]
- 58. Pawson T. (1995) Protein modules and signaling networks. Nature 373, 573–580 [DOI] [PubMed] [Google Scholar]
- 59. Baker J. M., Hudson R. P., Kanelis V., Choy W. Y., Thibodeau P. H., Thomas P. J., Forman-Kay J. D. (2007) CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat. Struct. Mol. Biol. 14, 738–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mittag T., Orlicky S., Choy W. Y., Tang X., Lin H., Sicheri F., Kay L. E., Tyers M., Forman-Kay J. D. (2008) Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc. Natl. Acad. Sci. U.S.A. 105, 17772–17777 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.