Background: Septins assemble into oligomers by interactions at “NC” and “G” interfaces by unknown mechanisms.
Results: NC interface mutations dominantly interfered with polymerization of wild type septins, but G mutations did not.
Conclusion: Septin oligomers likely assemble into dimers with high affinity partners, permitting NC interactions to occur.
Significance: G interactions would dictate the dimeric pairs and contribute to the organization of the oligomer.
Keywords: Actin, Cytokinesis, Cytoskeleton, GTPase, Microtubules, Septin
Abstract
Septins comprise a conserved family of GTPases important in cytokinesis. These proteins polymerize into filaments from rod-shaped heteromeric septin complexes. Septins interact with one another at two interfaces (NC and G) that alternate within the complex. Here, we show that small mutations at the N terminus greatly enhance the formation of SEPT2 homopolymers. Taking advantage of this mutation to examine polymer formation using SEPT2 alone, we show that both NC and G interfaces are required for filament formation. However, co-expression of wild type SEPT2 with SEPT2 containing mutations at either NC or G interfaces revealed that only the NC mutant suppressed filament formation. NC mutants are able to interact with one another at putative G interfaces, whereas G mutants fail to interact at NC interfaces. In addition, all promiscuous septin pairwise interactions occur at the G interface. These findings suggest that G interface interactions must occur before NC interactions during polymer formation.
Introduction
Septins are filamentous GTPases emerging as important cytoskeletal polymers that have been implicated in a diverse array of cellular functions, including cell division, secretion, migration, polarity, dendritogenesis, sperm development, and ciliogenesis (1–19). Four yeast septins (CDC10, CDC11, CDC12, and CDC3) were originally identified in a temperature-sensitive screen for cell division cycle defects (20). Since then, septins have been found in fungi, animals, and protists but are absent from plants (21, 22). In mammals, there are 14 septins (SEPT1–14) that fall into four groups with sequence similarity to the four yeast septins (23). Some of these, including SEPT2, SEPT11, SEPT7, and SEPT9, have been shown to be involved in cytokinesis (24–26). Similar to actin and tubulin, septins are able to polymerize into filaments. Septin filaments are thought to act as scaffolds, recruiting proteins to particular cellular locations. For instance, mammalian septins have been shown to localize to the ingressing furrow in dividing cells where SEPT2 binds to non-muscle myosin II to promote its activation (25). In addition, septin filaments also have a tendency to reorganize into ring structures that have been shown to act as diffusion barriers, preventing the lateral movement of membrane and peripheral proteins (18, 19, 27). The building blocks for septin filaments are heterotypic rod-shaped complexes. Recombinant and immunoaffinity-purified septin complexes from yeast, worm, flies, and human cells have been shown to polymerize into filaments in vitro (28–32). Cryo-electron microscopy of septin protomers and the addition of bulky affinity tags or antibody tagging has provided insight into the apolar nature of septin complexes (28). In budding yeast, the organizational arrangement is 11-12-3-10-10-3-12-11. Thus, in contrast to the unidirectional assembly of actin and microtubule filaments, polymerization of septin filaments is likely bidirectional. Crystal structure of a bacterially expressed preselected mammalian septin complex consisting of SEPT2, SEPT6, and SEPT7 follow a similar apolar organizational arrangement with 7-6-2-2-6-7 (33). We recently confirmed this order in HeLa cells and showed that, as in yeast, septins form octamers with SEPT9 at the ends and organized in the sequence 9-7-6-2-2-6-7-9 (34). Recently, Sellin et al. (35) have shown that immunoaffinity purification of mammalian septins in high salt breaks them into promoteric octamers and that depletion of SEPT9 results in hexameric structures, consistent with its location at the ends of octamers (see Fig. 1B).
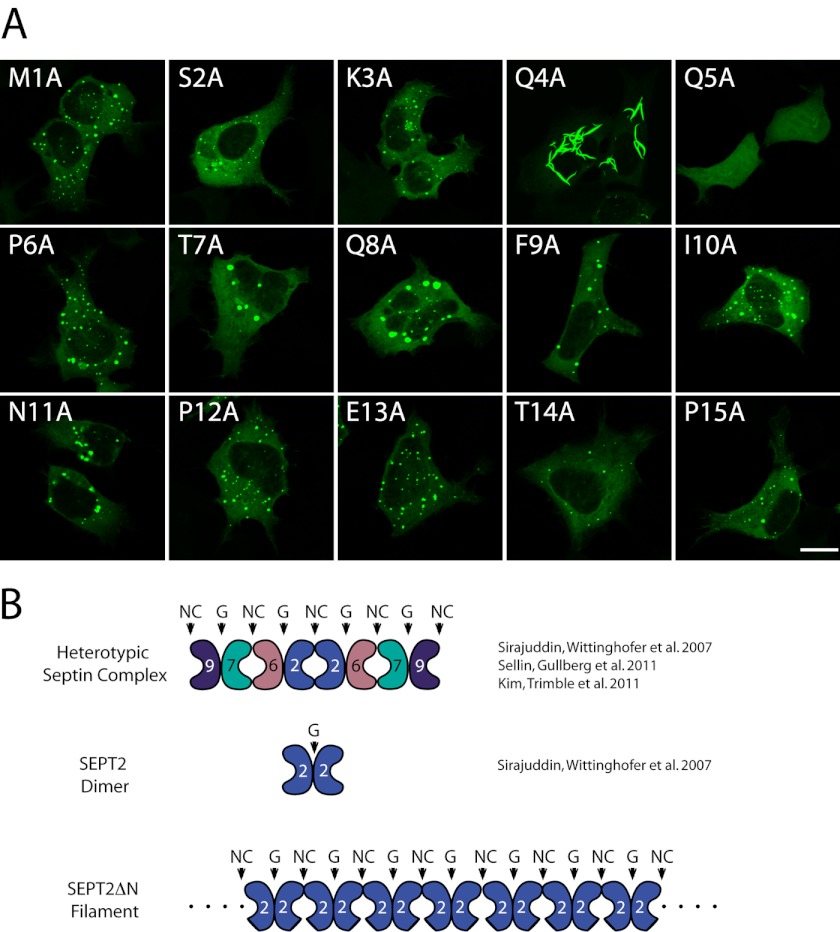
FIGURE 1.
A single point mutant, Q4A, phenocopies SEPT2 Δ15 filaments. A, alanine-scanning mutagenesis of first 15 amino-terminal residues in SEPT2. Mutations were made in full-length GFP-SEPT2 and transfected into HeLa cells. Scale bar indicates 8 μm. B, heterotypic mammalian septin complex (SEPT2-SEPT6-SEPT7-SEPT9) is an octamer with alternating NC and G interfaces. Each colored jellybean represents a different septin. The concave surface of the jellybean represent the NC interface, whereas the convex surface represents the G interface. SEPT2 interacts with itself at the NC interface within the octamer; however, it is a dimer at the G-interface in the absence of the other septins (top and middle row). SEPT2ΔN mutant forms filaments (bottom row).
The precise and consistent order of septins within the octameric complexes implies some fundamental principles of assembly. There are two septin-septin interfaces (NC and G) that alternate within the complex (33). However, the relative affinities of the NC and G interface are not known, and how the septin heteromeric complex assembles remains elusive. For example, it is possible that the tetrameric halves assemble first, followed by their assembly into an octamer. Another mode of assembly could involve the formation of a SEPT2-SEPT2 seed on which monomers then assemble. Alternatively, it is possible that dimers preassemble first and that these then assemble into tetramers and finally octamers. However, in this latter case, it is not clear whether the dimers form at the NC interface or the G interface. Understanding the principles of septin assembly will shed light on how these important filaments are turned over in cells.
One challenge in determining how septin assembly occurs is the fact that it involves the assembly of at least four different proteins. However, we have found a mutation that promotes the self-assembly of SEPT2 into homotypic filaments, thereby simplifying the analysis of how septin assembly occurs. In this unique scenario, we have probed the relative affinities of the NC and G interfaces. We find that interaction at the G interface is much stronger than that of the NC interface and must occur before NC interaction takes place. Expanding the analysis to SEPT6, SEPT7, and SEPT9 suggests that septins associate with one another in preferred pairs through dimers at the G interface. Thus, septin heterotypic complexes appear to begin with an initial G interaction followed by an NC interaction.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HeLa Tet-ON (Clontech), COS, B35 neuroblastoma, and HEK293 cells (ATCC) were subcultured in DMEM + 10% FBS in 37 °C incubator with 5% CO2. CHO cells were grown in Alpha Minimal Essential Medium (AMEM) + 10% FBS. Cells were transfected with either Lipofectamine 2000 (Invitrogen) or JetPrime Van Waters and Rogers, Inc. (VWR) following manufacturer's protocol.
Plasmid Construction
The multiple cloning site of pEGFP-C1 (Clontech) was modified so that EcoRI and XhoI are in-frame with GFP creating a fusion protein. FLAG oligo nucleotide was inserted into pCDNA3.0+ (Invitrogen), and the multiple cloning site was changed similarly. Thus, we generated two N-terminal fusion vectors with similar multiple cloning sites. Human SEPT2 (GenbankTM accession no. NP_001008491.1) was subcloned from pBlueScript-SK(−) into modified GFP and FLAG vectors using EcoRI and XhoI. SEPT2 truncation mutants (N- and C-terminal truncations) were PCR-cloned into modified GFP or FLAG vectors using custom primers (Sigma-Aldrich Genosys) containing EcoRI and XhoI overhangs. Point mutants were introduced into SEPT2 pBlueScript-SK(−) using a site-directed mutagenesis kit (Stratagene) and subsequently subcloned into modified FLAG or GFP vectors.
Immunoprecipitation
Eighteen hours post transfection, HeLa Tet-ON cells were washed with ice-cold phosphate-buffered saline (PBS). Cells were immediately lysed using 1% Triton X-100 in PBS with protease inhibitor tablet (Roche Diagnostics). Cells were clarified in a microcentrifuge, 13,000 × g at 4 °C. Clarified supernatant were incubated with 2 μg of rabbit polyclonal GFP antibody (Invitrogen) for 1 h. Cell lysate-antibody mixture was incubated with protein A-agarose (Bioshop, Inc.) for an additional hour. Beads were washed three times in 0.1% Triton X-100 in PBS and resuspended in 1× SDS-loading buffer.
Immunoblotting
Samples were run on 10% SDS-PAGE and transferred to PVDF membrane using standard procedures. Blots were blocked in 5% nonfat milk in Tris-buffered saline with 0.05% Tween 20 (TBST) and incubated with primary and then secondary antibodies for one hour each. Blots were extensively washed in TBST between incubations. Mouse FLAG (Sigma), rabbit GFP (Invitrogen), and mouse GAPDH antibodies were used at 1:7,500, 1:20,000, and 1:50,000 respectively. Horseradish peroxidase secondary antibodies were used at 1:5,000 and developed on film using ECL reagent.
Immunostaining and Microscopy
HeLa Tet-On cells were seeded on glass coverslips. Eighteen hours post-transfection, cells were washed briefly in PBS and fixed using 1% paraformaldehyde for 20 min at room temperature. Paraformaldehyde was inactivated and permeabilized using 25 mm glycine, 25 mm ammonium chloride, and 0.1% Triton X-100 in PBS for 20 min. Cells were blocked in 5% horse serum in phosphate-buffered saline with 0.05% Tween 20 (PBST), incubated with primary, then secondary antibodies for 1 h each. Blots were washed three times in PBST between incubations. Antibodies to FLAG (Sigma), SEPT2, SEPT11, SEPT7 (Santa Cruz Biotechnology), and SEPT9 were used at 1:1,000, 1:100, 1:500, 1:10, and 1:100, respectively. Secondary antibodies conjugated to Cy3 were used at 1:500 (Jackson Immunologicals). Cells were imaged using Zeiss LSM 510 Meta confocal microscope (Figs. 1, 2, 4, and 5, or Leica DMIRE2 epifluorescence microscope (Fig. 3).
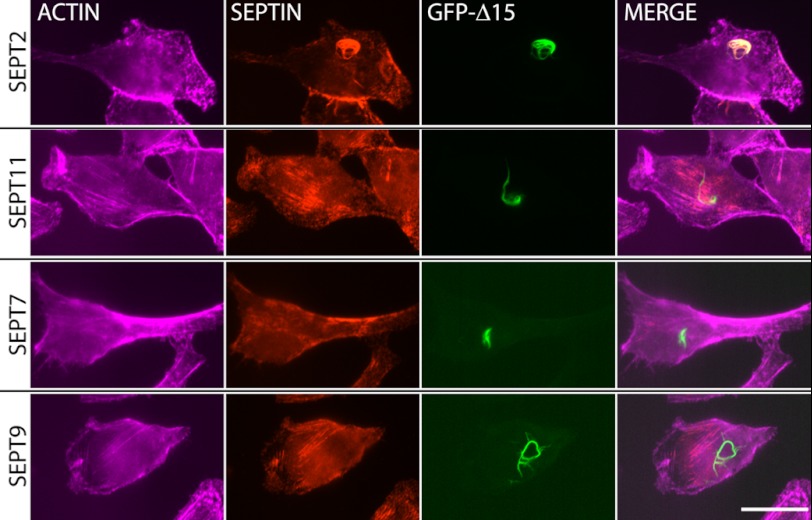
FIGURE 2.
SEPT2 Δ15 filaments are homotypic filaments. HeLa cells were transfected with GFP-SEPT2 Δ15 and stained with antibodies to endogenous SEPT2 (top row), SEPT11 (second row), SEPT7 (third row), and SEPT9 (bottom row). SEPT2 Δ15 filaments colocalize with endogenous SEPT2 (top row) but not with SEPT11, SEPT7, and SEPT9. SEPT11, -7, and -9 show unaffected actin-templated septin fibers upon SEPT2 Δ15 overexpression. In contrast, endogenous SEPT2 protein has largely been incorporated into SEPT2 Δ15 filaments, although at this level of overexpression, not all endogenous SEPT2 proteins were incorporated. The SEPT2 antibody was raised against a peptide containing the first 15 amino acids of SEPT2. Scale bar indicates 10 μm.
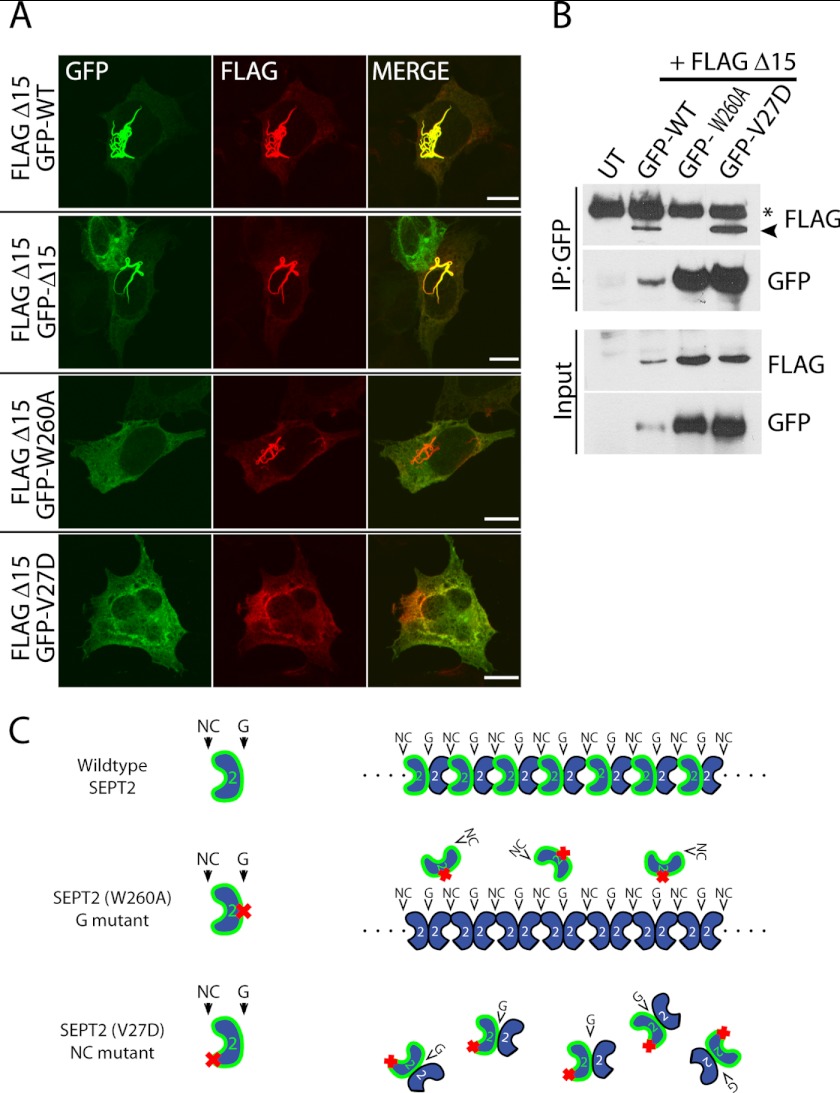
FIGURE 4.
NC mutant interferes with SEPT2 Δ15 filament formation. A, FLAG- SEPT2 Δ15 was cotransfected with full-length GFP-SEPT2 (first row), GFP-SEPT2 Δ15 (second row), GFP-G mutant (third row), and GFP-NC mutant (last row) in HeLa cells. Scale bar indicates 10 μm. B, GFP-WT, NC mutant, or G mutant were immunoprecipitated (IP) with GFP antibody and immunoblotted with FLAG antibody to detect FLAG- SEPT2 ΔN15. An asterisk denotes nonspecific bands. The nonspecific band is present in the immunoprecipitated lanes but not in the input lanes. Because this band runs at the same molecular weight as IgG, cross-reactivity of secondary antibodies is likely. UT denotes untransfected control lane. C, schematic depicting the effects of the SEPT2 mutants when coexpressed with SEPT2 Δ15 filaments. Wild type SEPT2 incorporates into the filament (top), G mutant does not incorporate into the filament (middle), and the NC mutant blocks SEPT2 Δ15 filaments by interacting with SEPT2 Δ15 (bottom).
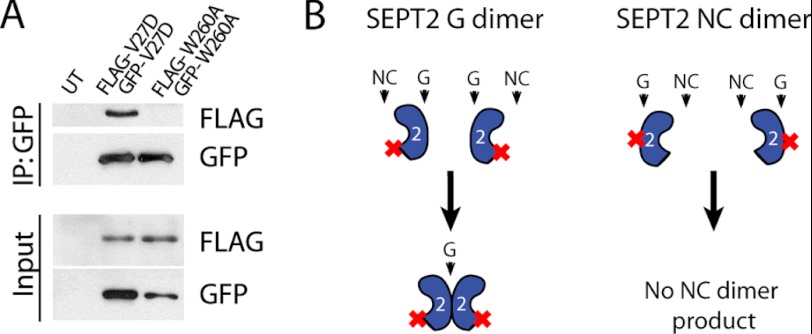
FIGURE 5.
Preferred binding of septin interface mutants at the G interface. A, HeLa cells were transfected with GFP- and FLAG-NC mutant or GFP-and FLAG-G mutant. Cell lysates were immunoprecipitated (IP) with GFP antibody and immunoblotted with FLAG antibody. UT denotes untransfected control lane. B, schematic of dimer formation by the NC or G interface. G dimers interact strongly, whereas NC dimers do not.
FIGURE 3.
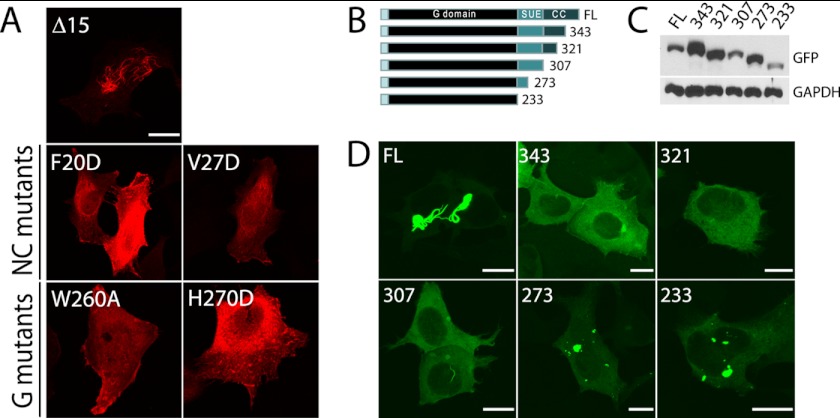
SEPT2 Δ15 filaments interact at NC and G interfaces and require the C terminus. A, FLAG-SEPT2 Δ15 filaments are disrupted when NC and G interface point mutants are introduced. B, schematic of serial C-terminal truncations in Q4A mutant. Full-length (FL) SEPT2 is 361 amino acids. G domain represents GTPase domain, SUE represents the septin unique element (as defined in Ref. 23), and CC represents a putative coiled-coil domain. C, immunoblot with GFP to confirm the expression of each C-terminal truncation in transfected HeLa cell lysates. D, fluorescent images of transfected C-terminal truncations to Q4A.
RESULTS
N-terminal Truncation Mutant of SEPT2 Is Filamentous
To gain insights into how septin oligomers assemble, we attempted to produce homotypic filaments in cells by overexpression of a single septin. Despite the heterogeneity in septin filaments, most individually overexpressed septins do not have the innate ability to polymerize into exogenous filaments. Typically, overexpressed septins localize diffusely in the cytosol or form perinuclear punctae, as is seen for N-terminal GFP-fusions to SEPT2 (supplemental Fig. 1, top panel). In our efforts to characterize septin-septin interactions, we generated a series of N- and C-terminal truncations in many human septins. One such truncation in SEPT2, lacking the first 15 amino acids, was originally constructed to be an siRNA-resistant rescue construct that lacked the epitope to our peptide-derived SEPT2 antibody. Surprisingly, this mutant (referred from now on as SEPT2 Δ15) when fused to either N-terminal FLAG or GFP, tended to form exogenous filaments in multiple cell lines including HeLa, human embryonic kidney 293 (HEK293), African monkey kidney (COS), CHO, and B35 neuroblastoma cells (supplemental Fig. 1, middle and bottom). The overexpression of this truncated mutant did not have any obvious cytokinesis defects at 18-h post-transfection (data not shown). These results suggest that the first 15 amino acids of SEPT2 inhibit exogenous filament assembly and that its removal relieves this inhibition.
A Single Point Mutant, Q4A, Phenocopies SEPT2 Δ15 Filaments
Next, we asked whether a protein motif or phosphorylation site located within the first 15 amino acids was responsible for inhibiting exogenous SEPT2 filaments. Using alanine-scanning mutagenesis, we systematically mutated each of the first 15 amino acids to alanine and asked whether any mutants phenocopied the ability of SEPT2 Δ15 to form filaments (Fig. 1). Unexpectedly, a single amino acid substitution, glutamine at position 4, mutated to alanine (Q4A), phenocopied the ability of SEPT2 Δ15 to form filaments. Because of the singular nature of this point mutant, we could not reliably map this mutant on any of the other human septins.
SEPT2 Δ15 Filaments Are Homomeric
To determine whether these SEPT2 Δ15 exogenous filaments were sequestering endogenous septins, we co-immunostained with endogenous septin antibodies to SEPT2, SEPT11, SEPT7, and SEPT9. Endogenous SEPT2 co-localized strongly with SEPT2 Δ15 filaments (Fig. 2, top panel), but no co-localization with the other septin antibodies was detected (Fig. 2, middle and bottom panels). Thus, these filaments were homomeric, consisting of a mix of full-length endogenous SEPT2 and exogenous truncated SEPT2 Δ15. The crystal structure of the recombinant SEPT2-SEPT6-SEPT7 complex revealed that septins interact with each other at two interfaces, the NC and G interface (33). Because mammals encode 14 septins, studying the combinatorial possibilities of each NC and G interface is challenging, even though the inherent assembly of the SEPT2-SEPT6-SEPT7 complex implies preferred NC and G interactions. To simplify the complexities of heterotypic interactions, we use the SEPT2 Δ15 filaments as a model to study NC and G interactions in a homomeric septin filament.
SEPT2 Δ15 Filaments Require NC and G Interfaces
Previously, SEPT2 has been shown to interact with itself at both the NC interface and the G interface (33). Based on the crystal structure of the SEPT2 dimer, we introduced the same mutations in SEPT2 Δ15 that Sirajuddin et al. (33) used to determine whether SEPT2 was dimerizing at the NC or G interface. These mutants were predicted to block interactions at the NC interface (F20D, V27D) or G interface (W260A, H270D). Mutations in either the NC interface or the G interface prevented SEPT2 Δ15 filaments, suggesting that both interfaces are required for filament assembly (Fig. 3). Most septins, including SEPT2, contain a C-terminal coiled-coil that projects perpendicular to the axis of the filament (33). We generated serial truncations at the C terminus of the SEPT2 (Q4A) mutant. Removal of just 18 amino acids from the C terminus prevented SEPT2 (Q4A) from forming filaments. Thus, exogenous SEPT2 filaments are disrupted when septin-septin interactions are inhibited by mutations at the interfaces (NC or G mutant) or truncation of the coiled-coil domain (C-terminal mutant).
NC Mutant Acts to Interfere in a Dominant-negative Manner with SEPT2 Δ15 Filaments
Given that both NC and G interfaces are required for SEPT2 Δ15 filaments, we reasoned that SEPT2 Δ15 filaments could be disrupted equally well by coexpression of either the NC or G mutant; both mutants would be predicted to act in a dominant-negative manner. In one case, the NC mutant would bind to the wild type SEPT2 Δ15 by its G interface to inhibit filaments. Similarly, the G mutant would bind to wild type SEPT2 Δ15 by its NC interface to inhibit filaments. As a control, coexpression with full-length wild type SEPT2 would be expected to efficiently incorporate into SEPT2 Δ15 filaments, as endogenous SEPT2 was enriched in these structures (Fig. 4A, top panel). As expected, full-length GFP-SEPT2 efficiently incorporated into SEPT2 Δ15 filaments (Fig. 4A, top row, and Fig. 4C, top row). Surprisingly, only the NC mutant was capable of dominant-negatively interfering with SEPT2 Δ15 filaments, whereas the G mutant had no effect (Fig. 4A, third and fourth rows, and Fig. 4C, bottom two rows). Furthermore, immunoprecipitation revealed that the GFP-NC mutant bound to FLAG-SEPT2 Δ15, similar to wild type, whereas there was no appreciable binding with the G mutant (Fig. 4B). These results show that the NC mutant interferes with filament formation by interacting with SEPT2 Δ15, presumably by its functional G interface, whereas the G mutant cannot bind to wild type SEPT2 Δ15 by its functional NC interface. This would suggest that interaction between septin monomers at the G interface predominates, perhaps due to a stronger binding affinity than that of the NC interface (Fig. 4C).
Isolation of SEPT2 Δ15 NC and G Interfaces and Preferred Binding at G Interface
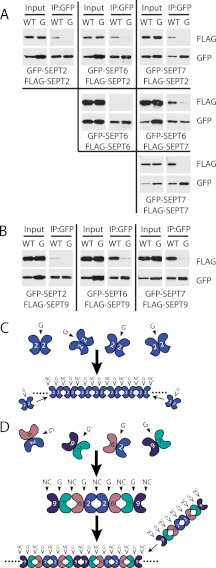
To test the possibility of stronger binding at one interface relative to the other, we tried to “force” the interaction of SEPT2 interfaces. To do this, we coexpressed two different epitope-tagged NC mutants and asked whether both mutants could interact at their G interfaces. Conversely, we coexpressed two different epitope-tagged G mutants and asked whether both mutants could interact at their NC interface (Fig. 5B). Immunoprecipitation of GFP-NC mutant resulted in binding to FLAG-NC mutant, but the G mutants failed to co-immunoprecipitate each other (Fig. 5A). Consistent with the results in Fig. 4, these results indicate that NC mutants bind to each other through their G interfaces, but G mutants do not bind to each other through their NC interfaces. Because we could not detect any NC interactions in the G mutants, this raises the intriguing idea that septins may require an interaction at the G interface before an NC interaction is permitted.
All Potential Septin-Septin Pairwise Interactions Are G Interface Interactions
Because septins are in a heterotypic complex, we asked whether other septins interact preferentially at their G interface. In the prototypical 7-6-2-2-6-7 septin hexameric complex, NC interactions are found at the 2-2 and 6-7 interfaces, whereas G interactions are at the 2-6 and 7-7 interfaces (33). We examined all septin-septin pairwise interactions involving SEPT2, SEPT6, and SEPT7. To do so, we co-expressed two different epitope-tagged septins (one with GFP and one with FLAG) and asked which septin pairs could interact by immunoprecipitating the GFP-septin and blotting for the FLAG-septin. Surprisingly, all septin-septin pairwise combinations were detected except the SEPT6-SEPT6 pair, despite the fact that some of these septins do not interact within the protomer. (Fig. 6A, WT lanes). Interestingly, introducing G interface mutations into the GFP-septin in each of the septin pairs resulted in severely compromised binding, suggesting that all potential septin-septin pairwise interactions are G interface interactions. This was unexpected since the 2-2 and 6-7 septin pairs are expected to interact at their NC interface and were therefore predicted to be insensitive to G mutations. Finally, we examined the pairwise interaction of SEPT9 with SEPT2, SEPT6, or SEPT7. SEPT9 displays complex alternative splicing, and we chose SEPT9_i3 as a representative form of the protein because we know that this form is sufficient to rescue cytokinesis defects caused by siRNA-mediated SEPT9 depletion (24). We have previously found that it coimmunoprecipitates with the 2-6-7 complex and co-localizes with actin stress fiber-associated septins and localizes to the ends of septin octamers (24–26, 34). Similar to the different combinatorial septin pairs for 2-6-7, SEPT9_i3 interacted with the other septins in a G interface-dependent manner (Fig. 6B). Endogenous septins did not copurify with the overexpressed septin pair (supplemental Fig. 2), suggesting that the septins are likely dimers interacting at the G interface. However, we cannot rule out the possibility that non-septin protein interactors may mediate these interactions. A more rigorous analysis of each septin pair in vitro with associated binding constants are needed to corroborate our immunoprecipitation and pulldown data. It is important to note that in these experiments, we have exposed the blots for different amounts of time ranging from 1 s to 10 min to normalize the intensity such that the WT and G mutant phenotypes were clearly discernable. Although we cannot make accurate affinity estimates because the overexpressed septin pairs were expressed at different levels in the lysates, by far the strongest interactions (1-s exposure times) were seen between the SEPT2-SEPT6 and SEPT7-SEPT9 pairs, similar to results we have obtained in yeast two-hybrid experiments (data not shown). Notably, these are the two septin pairs that would be expected to interact at their G interface in an octameric septin complex. Thus, septin heterotypic complex assembly may first involve interactions of the strongest G interface septin dimers (2-6 and 7-9), which would then permit secondary interactions at the available NC interfaces (Fig. 6, C and D).
FIGURE 6.
Septin pairwise interactions are G interface-dependent. A, combinatorial septin pairwise interactions using SEPT2, SEPT6, and SEPT7. Cell lysates were immunoprecipitated (IP) with GFP antibody and immunoblotted with FLAG antibody. B, septin pairwise interaction of SEPT9_i3 with SEPT2, SEPT6, or SEPT7. Lanes marked G represent G mutants and correspond to W260A for SEPT2, W258A for SEPT6, and W249A for SEPT7. C, proposed model for septin complex assembly and filament formation. SEPT2 Δ15 filaments are made from initial G interface dimers that then interact at their NC interfaces. D, heterotypic septin filaments are composed of septin octamers that likely assemble through the strongest G dimer pairs.
DISCUSSION
N Terminus of SEPT2 Inhibits Homotypic Polymer Formation
Crystallography of septins revealed that in the heterotypic 7-6-2-2-6-7 complex, SEPT2 interacts with itself at the NC interface when SEPT6 and SEPT7 are present, yet SEPT2 alone formed a dimer through interaction at the G interface (33). Thus, SEPT2 has the potential to interact with itself at both G and NC interfaces depending on the presence of SEPT6. In the SEPT2 dimer, a filamentous lattice crystal was not observed, implying that interactions at the NC interface did not occur. Unfortunately, the crystal structures did not reveal the organization of the N termini. However, the N terminus of SEPT2 appears to project toward the NC interface (33), making it possible that non-ideal SEPT2-SEPT2 G dimers are inhibited from forming NC interactions by the N terminus. In our overexpression model of SEPT2 in HeLa cells, full-length SEPT2 overexpression likely results in mostly G interface dimeric SEPT2 with non-productive NC interactions. Our lab and Mendoza et al. (37) have previously shown that SEPT2 alone can form filaments in vitro (36). This polymerization required high concentrations of SEPT2 (millimolar amounts). Such concentrations are probably not achieved in our overexpression system, and we speculate that the N-terminally mutated SEPT2 would polymerize at a much lower concentration in vitro. Removal of the N-terminal 15 amino acids of SEPT2, or the point mutation Q4A, would likely result in stabilization of the NC interaction (a gain-of-function mutation), with the end result leading to filament formation. However, this interaction only occurs after the formation of the G-G dimer because G mutants of SEPT2 Δ15 did not interact via their NC interface (Fig. 6). We cannot rule out the possibility that the N-terminal mutation might artificially emphasize the importance of the G-G interaction on assembly. However, we do not believe this is likely as (i) the crystal structure of non-mutated wild type SEPT2 is a dimer at the G interface, and (ii) we show here that different combinatorial septin pairs (i.e. ones that do not contain the N-terminal mutation) are G interface-dependent.
We reasoned that there might be cellular factors that could bind to the first 15 amino acids to inhibit SEPT2 filaments but not the Q4A mutant peptide. However, passing HeLa cell lysates over WT and Q4A peptide-conjugated beads did not enrich for any particular protein (data not shown). In addition, we expressed these peptides conjugated to TAP tags and did not see enrichment of any bands when compared with the tag alone. Finally, we attempted overexpression of the WT and Q4A N-terminal peptides in cells and did not see any changes in endogenous SEPT2 staining patterns (data not shown). Hence, further work is needed to understand how the glutamine at position 4 of the N terminus is able to stabilize the NC of SEPT2.
SEPT2 Δ15 Forms Filamentous Structures That Require the Coiled-coil
SEPT4 is found enriched in Tau-based filamentous deposits and cytoplasmic inclusions in Parkinson and Alzheimer disease. Garratt et al. (38, 39) have shown previously that SEPT4 can be partially thermally denatured, generating a nucleotide-free intermediate that stains positive with Congo Red and thioflavin-T, a marker for structures rich in β-sheets such as those found in amyloid plaques. However, the filaments formed by N-terminal mutations in SEPT2 do not appear to be amyloid in nature. First, heterologous expression of SEPT2 Q4A in Sf21 insect cells showed a similar level of GDP binding compared with WT SEPT2 (data not shown). Furthermore, SEPT2 Q4A filaments in overexpressed HeLa cells did not stain positive with thioflavin-S (data not shown). These filaments also relied on NC and G interfaces that are characteristic of typical septin-septin interactions associated with septin protomer complex assembly. Taken together, this suggests that the SEPT2 Δ15 higher-order structures are distinctly different from amyloid-based filaments that septins can form.
Interestingly, removal of just 18 amino acids from the C terminus of SEPT2 Δ15 resulted in complete disruption of filaments. Septin coiled-coils are thought to promote filament pairing by lateral association (28). This strong phenotype suggests that the C terminus of SEPT2 also plays a prominent role in septin complex assembly in addition to the lateral pairing of septin filaments. However, it remains possible that uncoupling bundles of septin filaments presumably into single thin filaments could make them difficult to detect by fluorescence microscopy, or that the bundling of septin filaments provides stability to the oligomers and in the absence of this lateral association the NC interface cannot remain associated.
Septin Interactions at G Interface Are Stronger than NC Interface
Three key observations into the nature of G and NC interfaces suggest that the G interface has a stronger affinity than the NC interface. First, SEPT2 NC mutants are capable of disrupting SEPT2 Δ15 filaments likely by binding at the G interface. Second, overexpression of mutants capable of interaction only at NC or G interfaces revealed preferred binding at the G interface. Last, septin-septin pairwise interactions between cognate or non-cognate interaction partners are all predominantly G interface-dependent. The strong tendency of septins to interact at the G interface and the complete absence of an interaction at the NC interface when the G interface is mutated suggest that septins associate first through G interfaces as dimers, and this interaction causes a conformational change that then permits NC interactions. These conformational changes may be nucleotide-dependent, as we have observed a strong preference for GTP binding in monomeric SEPT2 G mutants, compared with predominantly GDP-bound dimeric wild type SEPT2 (data not shown). This is consistent with the observations of Sirajuddin et al. (40), who showed that the nucleotide bound to the G domain affected the conformation of both the G and NC interfaces of SEPT2. Because septins have low intrinsic GTPase activities (36, 41) but are usually found bound to GDP, septin oligomerization may occur when newly formed septin monomers bound to GTP dimerize first at the G interface. Subsequent GTP hydrolysis would result in conformational changes at the NC interface that would then allow dimers to interact into higher-order structures. This would indicate that the first step in septin oligomer formation would be interaction at the G interface, a step for which the second interaction at the NC interface is dependent. This conformational change may not depend on GTP hydrolysis in all septins because SEPT6 lacks the critical threonine responsible for GTP hydrolysis (40).
Our data also suggest that interaction at the G interface is somewhat promiscuous, and this could explain why there is some plasticity in septin organization in yeast (42). Yeast lacking the septin normally found at the ends (Cdc11) or middle (Cdc10) of the octamer are still viable, and the remaining septins form hexamers that are capable of forming filaments in vitro. Correct assembly likely results because each septin pair has different affinities relative to one another, and the strongest G dimers likely set the stage for septin complex assembly. In the case of HeLa cells, SEPT2-SEPT6 and SEPT7-SEPT9 pairs interacted with each other via their G interfaces with the highest relative affinity. Presumably the same is true at the NC interfaces with SEPT2 and SEPT9 preferring to bind homotypically, whereas SEPT6 prefers to bind to SEPT7. In this context, septin depletion would likely change the binding partners at the G interface in such a way that the next strongest G interface septin pair would predominate. This would allow septin complex plasticity in cases where septins are not able to functionally swap within the same septin family group. Septin pairs (such as 6-7 and 2-2) that interact at NC interfaces in the 7-6-2-2-6-7 heterotypic complex will interact at the G interface when coexpressed in pairs. Given the propensity of septins to interact at the G interface, we suggest careful examination and interpretation of septin-septin interaction data particularly when septins are expressed in pairs, such as the yeast two-hybrid.
Supplementary Material
Acknowledgments
We thank members of the Trimble laboratory for many helpful discussions.
Footnotes
This work was supported by a Canadian Cancer Society grant (to W. S. T.).

This article contains supplemental “Experimental Procedures” and Figs. 1 and 2.
REFERENCES
- 1. Estey M. P., Kim M. S., Trimble W. S. (2011) Septins. Curr. Biol. 21, R384–387 [DOI] [PubMed] [Google Scholar]
- 2. Joo E., Tsang C. W., Trimble W. S. (2005) Septins: Traffic control at the cytokinesis intersection. Traffic 6, 626–634 [DOI] [PubMed] [Google Scholar]
- 3. Spiliotis E. T., Kinoshita M., Nelson W. J. (2005) A mitotic septin scaffold required for mammalian chromosome congression and segregation. Science 307, 1781–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oh Y., Bi E. (2011) Septin structure and function in yeast and beyond. Trends Cell Biol. 21, 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barral Y., Kinoshita M. (2008) Structural insights shed light onto septin assemblies and function. Curr. Opin. Cell Biol. 20, 12–18 [DOI] [PubMed] [Google Scholar]
- 6. Faty M., Fink M., Barral Y. (2002) Septins: A ring to part mother and daughter. Curr. Genet. 41, 123–131 [DOI] [PubMed] [Google Scholar]
- 7. Beites C. L., Xie H., Bowser R., Trimble W. S. (1999) The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat. Neurosci. 2, 434–439 [DOI] [PubMed] [Google Scholar]
- 8. Dent J., Kato K., Peng X. R., Martinez C., Cattaneo M., Poujol C., Nurden P., Nurden A., Trimble W. S., Ware J. (2002) A prototypic platelet septin and its participation in secretion. Proc. Natl. Acad. Sci. U.S.A. 99, 3064–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tooley A. J., Gilden J., Jacobelli J., Beemiller P., Trimble W. S., Kinoshita M., Krummel M. F. (2009) Amoeboid T lymphocytes require the septin cytoskeleton for cortical integrity and persistent motility. Nat. Cell Biol. 11, 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chacko A. D., Hyland P. L., McDade S. S., Hamilton P. W., Russell S. H., Hall P. A. (2005) SEPT9_v4 expression induces morphological change, increased motility, and disturbed polarity. J. Pathol. 206, 458–465 [DOI] [PubMed] [Google Scholar]
- 11. Gladfelter A. S., Pringle J. R., Lew D. J. (2001) The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 4, 681–689 [DOI] [PubMed] [Google Scholar]
- 12. Spiliotis E. T., Hunt S. J., Hu Q., Kinoshita M., Nelson W. J. (2008) Epithelial polarity requires septin coupling of vesicle transport to polyglutamylated microtubules. J. Cell Biol. 180, 295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xie Y., Vessey J. P., Konecna A., Dahm R., Macchi P., Kiebler M. A. (2007) The GTP-binding protein Septin 7 is critical for dendrite branching and dendritic-spine morphology. Curr. Biol. 17, 1746–1751 [DOI] [PubMed] [Google Scholar]
- 14. Tada T., Simonetta A., Batterton M., Kinoshita M., Edbauer D., Sheng M. (2007) Role of Septin cytoskeleton in spine morphogenesis and dendrite development in neurons. Curr. Biol. 17, 1752–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ihara M., Kinoshita A., Yamada S., Tanaka H., Tanigaki A., Kitano A., Goto M., Okubo K., Nishiyama H., Ogawa O., Takahashi C., Itohara S., Nishimune Y., Noda M., Kinoshita M. (2005) Cortical organization by the septin cytoskeleton is essential for structural and mechanical integrity of mammalian spermatozoa. Dev. Cell 8, 343–352 [DOI] [PubMed] [Google Scholar]
- 16. Kissel H., Georgescu M. M., Larisch S., Manova K., Hunnicutt G. R., Steller H. (2005) The Sept4 septin locus is required for sperm terminal differentiation in mice. Dev. Cell 8, 353–364 [DOI] [PubMed] [Google Scholar]
- 17. Kim S. K., Shindo A., Park T. J., Oh E. C., Ghosh S., Gray R. S., Lewis R. A., Johnson C. A., Attie-Bittach T., Katsanis N., Wallingford J. B. (2010) Planar cell polarity acts through septins to control collective cell movement and ciliogenesis. Science 329, 1337–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu Q., Milenkovic L., Jin H., Scott M. P., Nachury M. V., Spiliotis E. T., Nelson W. J. (2010) A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science 329, 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kwitny S., Klaus A. V., Hunnicutt G. R. (2010) The annulus of the mouse sperm tail is required to establish a membrane diffusion barrier that is engaged during the late steps of spermiogenesis. Biol. Reprod. 82, 669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hartwell L. H., Culotti J., Reid B. (1970) Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc. Natl. Acad. Sci. U.S.A. 66, 352–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nishihama R., Onishi M., Pringle J. R. (2011) New insights into the phylogenetic distribution and evolutionary origins of the septins. Biol. Chem. 392, 681–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pan F., Malmberg R. L., Momany M. (2007) Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol. Biol. 7, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Versele M., Thorner J. (2005) Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol. 15, 414–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Estey M. P., Di Ciano-Oliveira C., Froese C. D., Bejide M. T., Trimble W. S. (2010) Distinct roles of septins in cytokinesis: SEPT9 mediates midbody abscission. J. Cell Biol. 191, 741–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Joo E., Surka M. C., Trimble W. S. (2007) Mammalian SEPT2 is required for scaffolding nonmuscle myosin II and its kinases. Dev. Cell 13, 677–690 [DOI] [PubMed] [Google Scholar]
- 26. Surka M. C., Tsang C. W., Trimble W. S. (2002) The mammalian septin MSF localizes with microtubules and is required for completion of cytokinesis. Mol. Biol. Cell 13, 3532–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luedeke C., Frei S. B., Sbalzarini I., Schwarz H., Spang A., Barral Y. (2005) Septin-dependent compartmentalization of the endoplasmic reticulum during yeast polarized growth. J. Cell Biol. 169, 897–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bertin A., McMurray M. A., Grob P., Park S. S., Garcia G., 3rd, Patanwala I., Ng H. L., Alber T., Thorner J., Nogales E. (2008) Saccharomyces cerevisiae septins: Supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc. Natl. Acad. Sci. U.S.A. 105, 8274–8279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. John C. M., Hite R. K., Weirich C. S., Fitzgerald D. J., Jawhari H., Faty M., Schläpfer D., Kroschewski R., Winkler F. K., Walz T., Barral Y., Steinmetz M. O. (2007) The Caenorhabditis elegans septin complex is nonpolar. EMBO J. 26, 3296–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kinoshita M., Field C. M., Coughlin M. L., Straight A. F., Mitchison T. J. (2002) Self- and actin-templated assembly of mammalian septins. Dev. Cell 3, 791–802 [DOI] [PubMed] [Google Scholar]
- 31. Frazier J. A., Wong M. L., Longtine M. S., Pringle J. R., Mann M., Mitchison T. J., Field C. (1998) Polymerization of purified yeast septins: Evidence that organized filament arrays may not be required for septin function. J. Cell Biol. 143, 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Field C. M., al-Awar O., Rosenblatt J., Wong M. L., Alberts B., Mitchison T. J. (1996) A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J. Cell Biol. 133, 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sirajuddin M., Farkasovsky M., Hauer F., Kühlmann D., Macara I. G., Weyand M., Stark H., Wittinghofer A. (2007) Structural insight into filament formation by mammalian septins. Nature 449, 311–315 [DOI] [PubMed] [Google Scholar]
- 34. Kim M. S., Froese C. D., Estey M. P., Trimble W. S. (2011) SEPT9 occupies the terminal positions in septin octamers and mediates polymerization-dependent functions in abscission. J. Cell Biol. 195, 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sellin M. E., Sandblad L., Stenmark S., Gullberg M. (2011) Deciphering the rules governing assembly order of mammalian septin complexes. Mol. Biol. Cell 22, 3152–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang Y. W., Surka M. C., Reynaud D., Pace-Asciak C., Trimble W. S. (2006) GTP binding and hydrolysis kinetics of human septin 2. FEBS J. 273, 3248–3260 [DOI] [PubMed] [Google Scholar]
- 37. Mendoza M., Hyman A. A., Glotzer M. (2002) GTP binding induces filament assembly of a recombinant septin. Curr. Biol. 12, 1858–1863 [DOI] [PubMed] [Google Scholar]
- 38. Garcia W., Rodrigues N. C., de Oliveira Neto M., de Araújo A. P., Polikarpov I., Tanaka M., Tanaka T., Garratt R. C. (2008) The stability and aggregation properties of the GTPase domain from human SEPT4. Biochim. Biophys. Acta 1784, 1720–1727 [DOI] [PubMed] [Google Scholar]
- 39. Garcia W., de Araújo A. P., Lara F., Foguel D., Tanaka M., Tanaka T., Garratt R. C. (2007) An intermediate structure in the thermal unfolding of the GTPase domain of human septin 4 (SEPT4/Bradeion-β) forms amyloid-like filaments in vitro. Biochemistry 46, 11101–11109 [DOI] [PubMed] [Google Scholar]
- 40. Sirajuddin M., Farkasovsky M., Zent E., Wittinghofer A. (2009) GTP-induced conformational changes in septins and implications for function. Proc. Natl. Acad. Sci. U.S.A. 106, 16592–16597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Farkasovsky M., Herter P., Voss B., Wittinghofer A. (2005) Nucleotide binding and filament assembly of recombinant yeast septin complexes. Biol. Chem. 386, 643–656 [DOI] [PubMed] [Google Scholar]
- 42. McMurray M. A., Bertin A., Garcia G., 3rd, Lam L., Nogales E., Thorner J. (2011) Septin filament formation is essential in budding yeast. Dev. Cell 20, 540–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.