Background: Mdm2 oncogene expression is regulated by p53-dependent and -independent mechanisms.
Results: The NFAT1 transcription factor binds to the mdm2 P2 promoter and transactivates mdm2 independent of p53.
Conclusion: NFAT1 is a transcriptional activator of the mdm2 oncogene.
Significance: The identification of p53-independent control of mdm2 expression by NFAT1 will increase the understanding of the roles of NFAT1 in cancer.
Keywords: Apoptosis, Gene Regulation, NFAT Transcription Factor, p53, Transcription
Abstract
Although the MDM2-p53 interaction has been well documented, MDM2 overexpression is observed in human cancers with little or no functional p53, suggesting that mdm2 expression is regulated by mechanisms independent of p53. Dysregulation of NFAT signaling is associated with malignant transformation and cancer development and progression. In this study, we demonstrate that the human mdm2 P2 promoter contains a consensus binding site for the NFAT1 transcription factor. NFAT1 directly binds the mdm2 P2 promoter in vitro and in vivo, resulting in the up-regulation of mdm2 transcription. Enforced expression of NFAT1 results in an elevated MDM2 protein level and reduces p53 activation and function in response to DNA damage. Both NFAT1 and MDM2 are highly expressed in human hepatocellular carcinoma tissues, compared with adjacent normal liver tissues. There is a positive correlation between the NFAT1 and MDM2 levels in tumor tissues. The novel function of NFAT1 in the control of MDM2 expression provides a basis for future investigations of the role of NFAT1 in cancer development, progression, and therapy.
Introduction
The mdm2 (murine double minute 2) oncogene was initially cloned as a spontaneously amplified gene leading to cellular transformation (1). MDM2 binds to and targets p53 for proteasomal degradation and inhibits its transactivational activity (2–4). However, the role of MDM2 in transformation is not restricted to its regulation of p53 (5, 6). There is growing evidence that MDM2 may contribute to malignant transformation and cancer development and progression through its involvement in other cellular pathways. Genetic data has suggested that there is a higher incidence of sarcomas in p53-null transgenic mice overexpressing mdm2 than in the p53-null mice (7). In addition, tumors bearing both p53 mutations and mdm2 gene amplifications are more aggressive than those with alterations of only one of the genes (8). Moreover, MDM2 is involved in the regulation of cell proliferation, cell cycle control, and apoptosis through interaction with several alternative cellular targets other than p53 (6). High levels of the MDM2 oncoprotein are found in a wide variety of human hematological cancers and solid tumors. The expression of mdm2 is known to be induced by p53 (9), but increasing evidence has supported that MDM2 gene expression is also regulated by mechanisms that are independent of p53 (10–15).
The human nuclear factor of activated T cells (NFAT)3 family comprises five distinct gene products, from NFAT1 to NFAT5 (16, 17). The NFAT proteins are phosphorylated and reside in the cytoplasm in resting cells; upon stimulation, they are dephosphorylated by calcineurin, translocate into the nucleus, and become transcriptionally active, thus providing a direct link between intracellular calcium signaling and gene expression. NFAT functions are not restricted to the immune system, as various NFAT isoforms have been detected in various tissues such as smooth muscle, blood vessels, pancreas, heart, and skin. Furthermore, dysregulation of NFAT signaling is associated with malignant transformation and the development of cancer (18, 19). NFAT isoforms are overexpressed in human solid tumors and hematological malignancies and have roles in invasive migration and cell survival and in regulating the tumor microenvironment (20, 21) and tumor angiogenesis (22, 23).
NFAT1, the first identified member of the NFAT family, has well documented roles in the immune system, but its functions in cancer development and progression remain unclear. It has been demonstrated that the promoter of cyclin-dependent kinase 4 presents a consensus-binding site for NFAT proteins and is negatively regulated by NFAT1 (24). In a mouse model, NFAT1 knock-out causes hyperproliferative cellular responses, altered cell cycle control, and increased stage-specific cyclin expression in lymphocytes, suggesting that NFAT1 plays a major role in regulating cell cycle progression (25). NFAT1 promotes breast cancer cell invasion through the induction of cyclooxygenase-2 and the synthesis of prostaglandins (26, 27). NFAT1 also induces human telomerase reverse transcriptase mRNA expression in activated peripheral blood lymphocytes (28). In addition, the ectopic activation of NFAT1 activates c-Myc, which is a critical mechanism for pancreatic cancer cell growth in vitro and in vivo (29, 30). In our previous work, we have demonstrated that the mdm2 transcription is down-regulated by a dietary isoflavone, genistein, in multiple cancer cell lines (31). Although the underlying mechanism is not fully understood, our preliminary results suggest that the NFAT transcription factors may be involved in the regulation of mdm2 mRNA expression. In the current study, we determined how the NFAT transcription factor might regulate mdm2 and further explored the biological consequence of this NFAT1-mdm2 regulation.
EXPERIMENTAL PROCEDURES
Cells, Plasmids, and Reagents
The NFAT1-inducible cell line was generated as described previously (27). HCT116 cells with or without p53 were provided by Dr. B. Vogelstein (The Johns Hopkins University). MCF7, PC3, and Jurkat cells were purchased from the ATCC (Manassas, VA). Antibodies were purchased from BD Biosciences (NFAT1), Santa Cruz Biotechnology (p53), Covance (HA), Calbiochem (MDM2), and Sigma (FLAG, β-actin, and mouse and rabbit IgG). Vectors expressing HA-NFAT1, CA-NFAT1, and dominant-negative NFAT (DN-NFAT) were kindly provided by Dr. Chi-Wing Chow (Yeshiva University). His-NFAT1-DBD plasmid was provided by Dr. A. Rao (Harvard Medical School). The mdm2 P2 promoter luciferase vectors (Luc01 and Luc03) were provided by Dr. J. Blaydes (University of Southampton). The mdm2 P2 promoter luciferase vector lacking the NFAT1 binding site was generated by site-directed mutagenesis.
Immunoblotting
Plasmids were transfected into cells using Lipofectin (Invitrogen) as indicated. The cells were collected at the indicated times post-transfection and lysed in Nonidet P-40 lysis buffer containing a protease inhibitor mixture from Sigma. Cell lysates were used for immunoblotting as described elsewhere (32).
Colony Formation, Apoptosis, Quantitative PCR, and Luciferase Reporter Assays
The colony formation assay, cell apoptosis assay, quantitative PCR assay, and luciferase reporter assay were performed as described previously (31).
Chromatin Immunoprecipitation Assay
The chromatin immunoprecipitation assay was performed in HCT116 and PC3 cells as indicated. The cells were cross-linked with 1% formaldehyde for 10 min at 37 °C and harvested in sodium dodecyl sulfate lysis buffer (Millipore), and then the DNA was shredded to fragments of 200 base pairs by sonication. Antibodies against NFAT1, HA, or nonspecific IgG were added, and the precleared chromatin was incubated overnight. Protein G-agarose beads were added and incubated for 1 h at 4 °C. After reversing the cross-links, the DNA was isolated and used for polymerase chain reaction (PCR). Specific primer pairs were designed as follows: 5′-cccccgtgacctttaccctg-3′ and 5′-agcctttgtgcggttcgtg-3′ for qualitative or quantitative PCR amplification of the responsive element of the mdm2 promoter.
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extracts of Jurkat cells were prepared as described (10). The expression and purification of the His-NFAT1-DBD recombinant protein was performed as described previously (33). For the protein-DNA binding reaction, prepared nuclear extracts or recombinant proteins were preincubated with 1 μg of poly(dI:dC) (Pierce) and then reacted with a biotin-labeled NFAT1 probe at room temperature for 30 min. For the competition assay, a 100-fold molar excess of cold probes was preincubated with the samples for 10 min. The incubated samples were separated by 5% non-denaturing PAGE and transferred to Biodyne B nylon membranes. The signals were observed using a Light Super-shifted module kit (Pierce). The sequences for the biotin-labeled probe and unlabeled wild-type competitor were as follows: NFAT1 WT (forward, 5′-gcaggttgactcagcttttcctcttgagctggtcaagttca-3′; reverse, 5′-tgaacttgaccagctcaagaggaaaagctgagtcaacctgc-3′); unlabeled mutant competitor (forward, 5′-gcaggttgactcagctttagatcttgagctggtcaagttca-3′; reverse, 5′-tgaacttgaccagctcaagatctaaagctgagtcaacctgc-3′). The mutated site in the probe is underlined.
Tissue Microarrays and Immunohistochemistry
This study was approved by the Institutional Review Board of Zhong Shan Hospital (Fudan University). The clinicopathologic characteristics of the patients were summarized previously (34). The tissue microarray was constructed as described previously (34).
Statistical Analysis
The statistical analyses were done using the SPSS 17.0 software program. The χ2 test, Fisher's exact probability, and Student's t test were used for comparisons between groups. p < 0.05 was considered to be statistically significant.
RESULTS
NFAT1 Up-regulates MDM2 Expression
To demonstrate that NFAT1 regulates MDM2 protein expression in a p53-independent manner, we transiently transfected HCT116 (p53+/+ and p53−/−) cells with increasing amounts of a NFAT1 plasmid (HA-NFAT1). As shown in Fig. 1A, NFAT1 overexpression up-regulated the MDM2 protein levels in both HCT116 p53+/+ and HCT116 p53−/− cells. The expression of p53 protein was down-regulated by NFAT1 transfection in HCT116 p53+/+ cells (Fig. 1A). We confirmed the up-regulation of MDM2 protein levels by NFAT1 using a constitutively activated NFAT1 vector (CA-NFAT1) in HCT116 p53−/− cells (Fig. 1B). We then demonstrated that DN-NFAT, the competitive form of wild-type NFAT, inhibited the MDM2 protein levels in HCT116 p53−/− cells (Fig. 1C). Knockdown of NFAT1 in PC3 (p53 null) cells with a specific interfering RNA decreased the MDM2 protein levels in a dose-dependent manner (Fig. 1D). Because NFAT is primarily regulated by calcineurin in the cells, we next wanted to know whether modification of the calcineurin activity would affect the MDM2 protein levels. HCT116 (p53−/−) and PC3 (p53 null) cells were treated with increasing concentrations of cyclosporine A (CsA), a calcineurin activity inhibitor, and the result showed that CsA decreased the MDM2 protein levels in a dose-dependent manner (Fig. 1E). To demonstrate that NFAT1 may mediate the increase in the MDM2 protein levels induced by calcineurin activation, we knocked down NFAT1 in HCT116 p53−/− cells and then treated the cells with ionomycin, a calcineurin activator. The result showed that NFAT1 knockdown remarkably suppressed the ionomycin-mediated up-regulation of MDM2 protein level compared with cells treated with the control siRNA (Fig. 1F). Consistent with the results of the knockdown experiment, CsA treatment attenuated the up-regulation of the MDM2 protein level by ectopic expression of NFAT1 (Fig. 1G). Taken together, these results indicate that NFAT1 positively regulates the expression of the MDM2 protein.
FIGURE 1.
NFAT1 induces MDM2 expression. A, HCT116 (p53+/+and p53−/−) cells were transiently transfected with increasing amounts of an HA-tagged NFAT1 plasmid (0, 3, and 5 μg) for 24 h. The transfected NFAT1, MDM2, and p53 protein levels were detected by Western blot using antibodies against HA, MDM2, and p53. B, HCT116 (p53−/−) cells were transfected with increasing amounts of a HA-tagged constitutively activated NFAT1 (CA-NFAT1) plasmid (0, 3 and 5 μg) for 24 h. The transfected CA-NFAT1 and MDM2 protein levels were detected by Western blot using antibodies against HA and MDM2. C, HCT116 (p53−/−) cells were transfected with the FLAG-tagged dominant-negative NFAT (DN-NFAT) plasmid for 24 h. The transfected DN-NFAT and MDM2 protein levels were detected by Western blot using antibodies against FLAG and MDM2. D, PC3 (p53 null) cells were transfected with increasing amounts of NFAT1 siRNA (si-NFAT1) (0, 25, 50 nm) for 36 h. The NFAT1 and MDM2 protein levels were detected by Western blot using antibodies against NFAT1 and MDM2. E, HCT116 (p53−/−) and PC3 (p53 null) cells were treated with CsA (0, 1, and 2 μm) for 24 h. The cells were then collected, and the NFAT1 and MDM2 protein levels were detected by Western blot using antibodies against NFAT1 and MDM2. F, HCT116 (p53−/−) cells were transfected with NFAT siRNA (si-NFAT1) (50 nm) for 36 h, and then the cells were treated with ionomycin (ION; 4 μm) for another 24 h. The NFAT1 and MDM2 protein levels were detected by Western blot using antibodies against NFAT1 and MDM2. G, HCT116 (p53−/−) cells were transfected with the HA-tagged NFAT1 plasmid (5 μg) for 24 h. The cells were treated with CsA (2 μm) for another 24 h. The NFAT1 and MDM2 protein levels were detected by Western blot using antibodies against HA and MDM2.
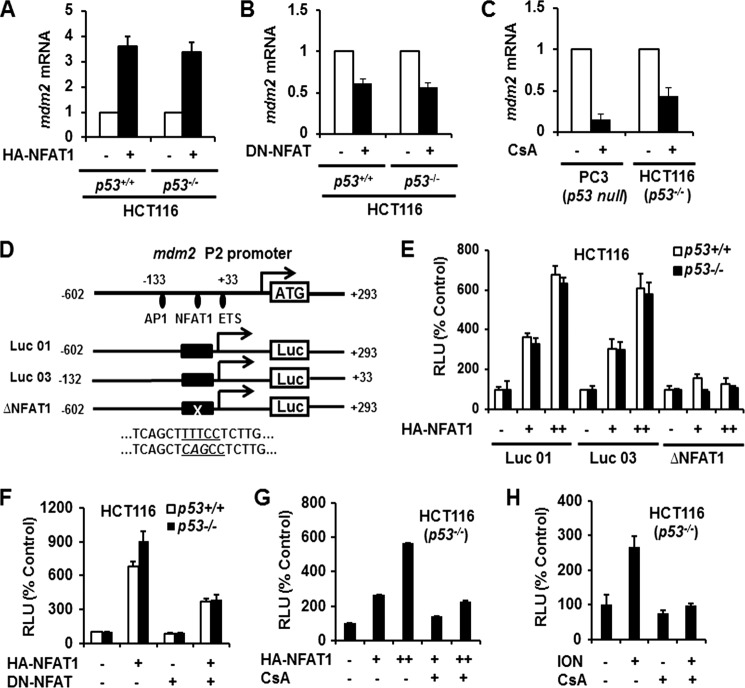
NFAT1 Induces mdm2 Transcription
To demonstrate that NFAT1 regulates MDM2 protein expression at the transcriptional level, we transfected HCT116 p53+/+ and p53−/− cells with HA-NFAT1 and DN-NFAT plasmids and determined the mdm2 mRNA levels in transfected cells. The results showed that NFAT1 overexpression increased, whereas NFAT1 inhibition decreased the mdm2 mRNA levels (Figs. 2, A and B). There was no significant difference of mdm2 mRNA expression increased by NFAT1 between p53+/+ and p53−/− cells. To further confirm that endogenous NFAT1 regulates MDM2 transcription independent of p53, we treated PC3 and HCT116 p53−/− cells with CsA and then determined the mdm2 mRNA levels. As shown in Fig. 2C, CsA treatment decreased the mdm2 mRNA levels in both PC3 and HCT116 p53−/− cells. To demonstrate that NFAT1 is a potential transcriptional activator of mdm2, we performed a transcription factor binding site analysis for the mdm2 promoter using the TFSEARCH online software program. As shown in Fig. 2D, a putative NFAT1 binding site was identified within the region from −109 to −105 bases of the mdm2 P2 promoter. To confirm that NFAT1 binding to this site regulates mdm2 transcription, HCT116 p53+/+ and p53−/− cells were transfected with NFAT1 and the full-length (Luc 01), deleted (Luc 03), or mutant (Luc01-ΔNFAT1) mdm2 P2 promoter luciferase vectors. We observed that NFAT1 increased the MDM2 luciferase activity in those transfected with the full-length mdm2 P2 promoter luciferase vector (Luc 01) in a dose-dependent manner (Fig. 2E). NFAT1 increased MDM2 luciferase activity almost equally in HCT116 p53+/+ and p53−/− cells. The mdm2 P2 promoter deletion (Luc 03) appeared to be as efficient as Luc 01, likely because the putative NFAT1 binding site resides in this region. However, the mutant luciferase vector lacking NFAT1 binding site was unable to increase the MDM2 luciferase activity (Fig. 2E).
FIGURE 2.
NFAT1 up-regulates mdm2 transcription. A, HCT116 (p53+/+ and p53−/−) cells were transfected with the HA-NFAT1 plasmid for 24 h. The relative mRNA levels of mdm2 were determined by real-time quantitative PCR. B, HCT116 (p53+/+ and p53−/−) cells were transfected with the DN-NFAT plasmid for 24 h. The relative mRNA levels of mdm2 were determined by real-time quantitative PCR. C, PC3 and HCT116 (p53−/−) cells were treated with CsA (2 μm) for 24 h. The relative mRNA levels of mdm2 were determined by real-time quantitative PCR. D, the structure of the mdm2 P2 promoter and the luciferase reporter vectors. E, HCT116 (p53+/+and p53−/−) cells were transfected with the NFAT1 plasmid (0, 3, and 5 μg) combined with a luciferase (Luc) reporter vector as indicated. At 24 h after transfection, the cells were harvested, and the relative luciferase levels were determined using a dual-reporter gene detection system. F, HCT116 (p53+/+and p53−/−) cells were co-transfected with a combination of NFAT1, DN-NFAT, and a luciferase reporter plasmid for 24 h. The relative luciferase levels were then determined using a dual-reporter gene detection system. G, HCT116 (p53−/−) cells were transfected with the NFAT1 plasmid (0, 3, and 5 μg) with a luciferase reporter vector for 24 h. The cells were treated with CsA (2 μm) for an additional 24 h, and then the cells were harvested, and the relative luminescence units (RLU) were determined using a dual-reporter gene detection system. H, HCT116 (p53−/−) cells were transfected with a luciferase reporter plasmid for 24 h, followed by treatment with ionomycin (4 μm) with or without CsA (2 μm) for 24 h. The cells were harvested, and the relative luciferase levels were determined using a dual-reporter gene detection system.
To demonstrate that manipulating NFAT1 activity would affect mdm2 transcription, we transfected HCT116 p53+/+ and p53−/− cells with a combination of NFAT1 and DN-NFAT plasmids. As shown in Fig. 2F, enforced expression of NFAT1 increased the MDM2 luciferase activity, whereas simultaneous expression of DN-NFAT significantly antagonized this change. We also observed that CsA treatment antagonized the NFAT1-mediated increase in theMDM2 luciferase activity (Fig. 2G). To demonstrate that the activation of physiological NFAT1 promotes MDM2 transcription, we treated HCT116 p53−/− cells with ionomycin in the presence or absence of CsA. We found that ionomycin treatment up-regulated the MDM2 luciferase activity, whereas the simultaneous treatment with CsA down-regulated the MDM2 luciferase activity (Fig. 2H). Together, these results indicate that NFAT1 transcriptionally activates mdm2 independent of p53.
NFAT1 Binds to mdm2 P2 Promoter
We next performed a ChIP assay to determine the binding of NFAT1 to the mdm2 P2 promoter. As shown in Fig. 3A, endogenous NFAT1 from PC3 cells specifically bound to the P2 promoter of MDM2. We further confirmed this binding using exogenous NFAT1. Transfection of HCT116 p53+/+ and p53−/− cells with an HA-NFAT1 plasmid, followed by immunoprecipitation (IP) with an HA antibody showed that there was specific binding between ectopic NFAT1 and the mdm2 P2 promoter, whereas IP with the IgG control showed no binding (Fig. 3B). To determine the interaction between NFAT1 and the mdm2 P2 promoter in response to NFAT1 activation, we treated PC3 cells with ionomycin, alone or together with CsA, and then analyzed the relative enrichment of NFAT1 on the mdm2 P2 promoter using a quantitative PCR analysis combined with a ChIP assay. The results showed that ionomycin treatment significantly recruited NFAT1 to the mdm2 P2 promoter, which was abolished by CsA treatment (Fig. 3C).
FIGURE 3.
NFAT1 directly binds to mdm2 P2 promoter. A, PC3 cell lysates were immunoprecipitated with NFAT1 or IgG antibodies. The DNA bound to the endogenous NFAT1 was eluted and detected with primers specific for the mdm2 P2 promoter using PCR. NFAT1 RE, NFAT1 responsive element. B, HCT116 (p53+/+and p53−/−) cells were transfected with the HA-NFAT1 plasmid for 24 h. The transfected cell lysates were immunoprecipitated with HA or IgG antibodies. The DNA bound to the exogenous NFAT1 was eluted and then detected using primers specific for the mdm2 P2 promoter by PCR. C, PC3 cells were pre-treated with CsA (2 μm) for 30 min and then followed by treatment with ionomycin (4 μm) for 6 h. The cell lysates were immunoprecipitated with NFAT1 or IgG antibodies. The DNA bound to the endogenous NFAT1 was eluted and quantified using primers specific for the mdm2 P2 promoter by real-time PCR. D, increasing amounts of NFAT1-DBD proteins (0, 15, 30, and 60 ng) were incubated with an mdm2 probe in the presence or absence of a competitor probe. An EMSA assay was performed to detect the binding between the NFAT1-DBD protein and the mdm2 probe. E, His-NFAT1-DBD protein (60 ng) was incubated with the mdm2 probe in the presence or absence of increasing levels of wild-type or mutant competitor probes (100- and 200-fold molar excess of biotin-labeled probes). An EMSA assay was performed to detect the binding between the NFAT1-DBD protein and the mdm2 probe. F, the nuclear extract (NE) from Jurkat cells was incubated with the mdm2 probe in the presence or absence of the competitor probe. An EMSA assay was performed to detect the binding between the endogenous NFAT1 protein and the mdm2 probe. G, nuclear extracts from Jurkat cells treated with ionomycin (ION) in the presence or absence of CsA were incubated with the mdm2 probe. An EMSA assay was performed to detect the binding between the endogenous NFAT1 protein and the mdm2 probe. H, nuclear extracts from Jurkat cells were incubated with the mdm2 probe in the presence or absence of a NFAT antibody (Ab) with the wild-type or mutant competitor probe. An EMSA assay was performed to detect the binding between the endogenous NFAT1 protein and the mdm2 probe. Mut, mutant.
We further performed an EMSA assay to confirm the binding between NFAT1 and the mdm2 P2 promoter. The purified NFAT1 DNA binding domain (NFAT1-DBD) protein was incubated with a biotin-labeled mdm2 probe in vitro. The results showed that NFAT1-DBD bound to the mdm2 probe in a dose-dependent manner and that the binding was obviously antagonized by the presence of excessive unlabeled competitor probe (Fig. 3D). We demonstrated that the unlabeled wild-type competitor probe could antagonize the binding of NFAT1-DBD to the probe in a dose-dependent manner, whereas the unlabeled mutant competitor probe could not, indicating that the interaction is dependent on the NFAT1 binding site on the probe (Fig. 3E).
We also determined the binding between endogenous NFAT1 and the mdm2 probe using the EMSA assay. Consistent with the results obtained using in vitro-purified NFAT1-DBD, the nuclear extracts from Jurkat cells showed obvious binding to the mdm2 probe, and the unlabeled wild-type competitor probe could antagonize this binding (Fig. 3F). In addition, the nuclear extracts from ionomycin-treated Jurkat cells showed enhanced binding to the mdm2 probe, whereas simultaneous treatment with CsA inhibited the binding (Fig. 3G). We also demonstrated that the unlabeled mutant competitor probe could not antagonize the binding between endogenous NFAT1 and the mdm2 probe. However, preincubation with the anti-NFAT1 antibody efficiently prevented the binding between endogenous NFAT1 and the mdm2 probe (Fig. 3H). These results demonstrate that NFAT1 binds to the mdm2 P2 promoter both in vitro and in vivo.
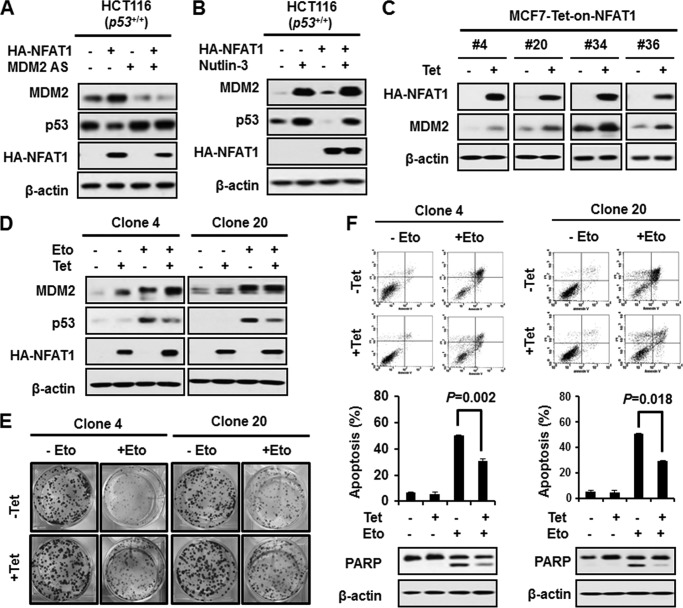
NFAT1-MDM2 Pathway Inhibits p53 Activation and Function
To demonstrate the biological consequence of the NFAT1-mediated up-regulation of MDM2, we transfected HCT116 p53+/+ cells with an NFAT1 plasmid and detected the p53 protein levels by immunoblotting. As shown in Fig. 4A, overexpression of NFAT1 increased the MDM2 but decreased the p53 protein levels. We confirmed that the decrease in p53 induced by NFAT1 overexpression was due to MDM2 up-regulation because the knockdown of MDM2 by a specific antisense oligonucleotide reversed the NFAT1-mediated decrease in the p53 protein level. We further demonstrated that the MDM2-p53 interaction was responsible for the down-regulation of p53 by NFAT1. Nutlin-3, a specific MDM2 inhibitor that disrupts the MDM2-p53 interaction, restored the p53 protein expression in the presence of NFAT1 (Fig. 4B).
FIGURE 4.
NFAT1 destabilizes p53 through its up-regulation of MDM2. A, HCT116 (p53+/+) cells were transfected with the NFAT1 plasmid (5 μg) in combination with an antisense oligonucleotide targeting mdm2 (50 nm) for 24 h. The levels of the MDM2 and p53 proteins were detected by Western blotting. B, HCT116 (p53+/+) cells were transfected with the NFAT1 plasmid (5 μg), and then 24 h after transfection, the cells were treated with Nutlin-3 (10 nm) for another 12 h. The levels of the MDM2 and p53 proteins were detected by Western blot. C, various NFAT1 inducible MCF7 clones were induced with tetracycline (Tet; 1.0 μg/ml) for 24 h. The levels of MDM2 proteins were detected by Western blot. D, NFAT1-inducible MCF7 clones 4 and 20 were treated with etoposide (Eto; 100 μm) for 24 h in the presence or absence of tetracycline (1.0 μg/ml). The expression levels of MDM2 and p53 proteins were detected by Western blotting. E, NFAT1 inducible MCF7 clones 4 and 20 were seeded at 1000 cells/well and continuously cultured for 10 days and then treated with etoposide (100 μm) for 6 h. The medium was then changed, and the cells were cultured for another 6 days. The colonies in each well were fixed and stained with crystal violet. F, NFAT1-inducible MCF7 clones 4 and 20 were treated with etoposide (100 μm) for 48 h in the presence or absence of tetracycline (1.0 μg/ml). The cells were stained with annexin V/propidium iodide to evaluate their rate of apoptosis, and the stained cells were detected by flow cytometry. PARP, poly(ADP-ribose) polymerase.
We then generated MCF7 cells that had inducible expression of NFAT1 to determine the effects of the NFAT1-MDM2 pathway on p53 activation and function under stress. As shown in Fig. 4C, various NFAT1-inducible clones were established, and two of these were used for the further investigations. The induced expression of NFAT1 increased MDM2 level and attenuated etoposide-induced p53 activation (Fig. 4D). Etoposide did not affect NFAT1 level in HCT116 (p53+/+ and p53−/−) and PC3 (p53 null) cells (data not shown). The results of a colony formation assay showed that the inducible expression of NFAT1 promoted colony formation and protected the cells from etoposide-induced cell death (Fig. 4E). We further demonstrated that the effects of the induction of NFAT1 on cell growth were due to decreased apoptosis of the cells. As shown in Fig. 4F, treatment with etoposide for 48 h induced a high percentage of apoptotic cells, whereas the apoptotic rate decreased in the presence of induced NFAT1. The inhibitory role of the inducible NFAT1 on etoposide-induced cell apoptosis was also confirmed by the decreased level of cleaved poly(ADP-ribose) polymerase, which is a critical effector for apoptosis.
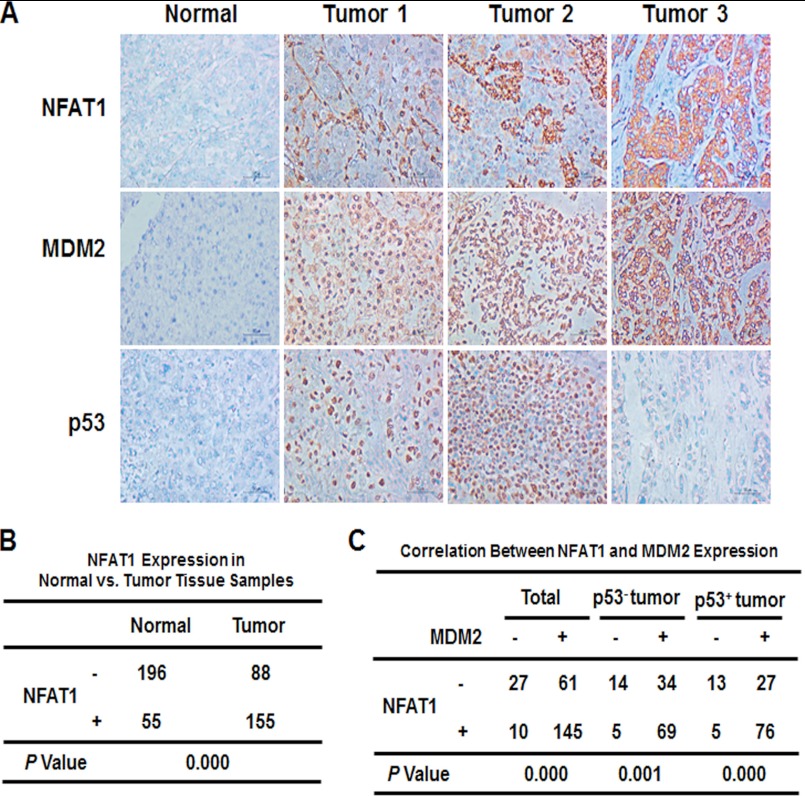
Correlation between NFAT1 and MDM2 Expression in Clinical Hepatoma Tissues
We further performed a tissue microarray and immunohistochemical staining to examine the expression of NFAT1 in human hepatocellular carcinoma tissue samples. NFAT1 was stained in the cytoplasm or nucleus of tumor cells and exhibited a variety of staining patterns with respect to the staining intensity and percentage of positive cells (Fig. 5A). We observed that 63.8% (155 of 243) of the hepatocellular carcinoma patient samples had positive scores for NFAT1. Most of the peritumoral tissues showed no or low NFAT1 expression, with only 55 of 251 samples showing positive expression. There were significant differences in NFAT1 expression between the tumor and peritumoral tissues (Fig. 5B; p = 0.000). The staining for NFAT1 and MDM2 was simultaneously positive in 145 of the 243 tumor tissue samples, of which 69 cases were p53 negative (69 of 122) and 76 cases were p53 positive (76 of 121). The MDM2 expression showed a significant correlation with the NFAT1 expression, regardless of the p53 status (Fig. 5C; p = 0.000).
FIGURE 5.
The correlation between NFAT1 and MDM2 expression in human hepatocellular carcinoma. A, representative images of the NFAT1, MDM2, and p53 staining in tissue microarrays as determined by an immunohistochemical analysis. B, the rate of positive NFAT1 staining in the tumor and adjacent normal tissue samples. C, the clinical correlation of the NFAT1 and MDM2 expression levels in hepatocellular carcinoma.
DISCUSSION
In this study, we identified and validated NFAT1 as a novel transcription factor for the mdm2 oncogene. The activation or overexpression of NFAT1 resulted in elevated mdm2 transcription. This study describes at least five new discoveries. First, an NFAT consensus binding site was identified within the mdm2 P2 promoter. Second, NFAT1 directly bound to the mdm2 P2 promoter and transactivated mdm2. Third, the activation of NFAT1 up-regulated whereas inactivation of NFAT1 down-regulated the MDM2 protein level. Fourth, the NFAT1-MDM2 pathway inhibited p53 activation and function in response to DNA damage. Finally, the NFAT1-MDM2 pathway was demonstrated to be active in human hepatocellular carcinoma. In general, these results help provide a better understanding of the p53-independent regulation of the mdm2 oncogene. In addition to their well documented role in T cell activation, NFAT proteins are now being increasingly recognized for their implications in tumorigenesis. Considering that high levels of NFAT1 and MDM2 proteins have been observed in various human cancers (18, 19), the identification of the NFAT1-MDM2 pathway provides evidence of a novel function for NFAT1 and a mechanism for the p53-independent regulation of MDM2.
A role for the calcineurin-NFAT signaling pathway in cancer has been suggested (18). In this study, we have extended the knowledge on the role of the calcineurin-NFAT pathway in the regulation of cancer development and progression and identified that it modulates the transcription of the mdm2 oncogene. Despite the previously described roles of this pathway in regulating COX-2 (26, 27), c-Myc (29, 30), and more recently, p15INK4b (35), to the best of our knowledge, this is the first report to show the direct transcriptional activation of mdm2 by NFAT1. The calcineurin-NFAT-MDM2 axis may constitute an intrinsic pathway that links NFAT1 to cancer. The ability of mdm2 to be up-regulated by a variety of mechanisms and to mediate p53-independent oncogenic effects could be a reflection of its more pervasive role in tumorigenesis than has previously been believed.
The overexpression of mdm2 in cancer is not always dependent on p53 because p53 is mutated in more than half of human tumors (36). Our findings that NFAT1 can directly bind to mdm2 P2 promoter, up-regulate the basal level of MDM2, and inhibit p53-mediated apoptosis suggest an important pathogenic role for MDM2 in cancer development driven by aberrant NFAT1 activation. Previously, a single nucleotide polymorphism within the mdm2 promoter (SNP309) was demonstrated to have enhanced affinity for the transcription factor Sp1, which increases mdm2 expression and was shown to be associated with accelerated tumor formation (37). The stabilization of p53 after DNA damage is impaired in cells homozygous for SNP309. Although our data demonstrated the positive regulation of MDM2 by NFAT1, and a high correlation of NFAT1 with MDM2 expression in human hepatocellular carcinoma, whether MDM2 overexpression is caused by aberrant NFAT1 activation (high affinity for the mdm2 P2 promoter) in cancer patients still needs to be investigated in further studies.
An elevated MDM2 level has been suggested be a prognostic factor for cancer; however, the mechanism(s) responsible for this effect have remained unclear. The current study, together with the previous work by other investigators, provides a basis for understating the increased level of MDM2 expression in cancer, particularly late-stage and metastatic cancer (13), in which mdm2 is regulated independent of the p53 status. Moreover, MDM2 has been suggested to be an ideal target for cancer therapy because of its diverse oncogenic roles (38). The previous anti-cancer strategies that were based on disturbing the MDM2-p53 interaction may be beneficial to those cancer patients who harbor functional p53, however, would not benefit the population expressing little or no functional p53. Alternative approaches that target the p53-independent regulation of MDM2 represent a new direction for MDM2 inhibitor design and development.
The roles of NFAT1 in carcinogenesis, cancer development, and progression, and clinical relevance remain elusive. In our study, we found a higher positive rate of NFAT1 in primary hepatoma tissues than adjacent non-tumor tissues, which is correlated with increased MDM2 level in tumor. Baumgart et al. (35) report that the NFAT1 protein levels are up-regulated in human advanced pancreatic cancer and positively related to disease progression. Chen et al. (39) report that the positive rate of NFAT1 is significantly higher in non-small cell lung cancer tissues than adjacent normal lung tissues and also linked to disease progression and patient survival. However, Maxeiner et al. (40) report that NFAT1 mRNA level is decreased in bronchial adenocarcinoma issues compared with control tissues and pneumonia tissues. In the same report, transgenic mice lacking NFAT1 were shown to have more and larger bronchioalveolar adenocarcinoma than wild-type littermates, which may be associated with the immunomodulatory function of NFAT1. In contrary, Gerlach et al. (41) suggest that NFAT1-deficient mice are protected from colitis-associated colon cancer development compared with wild-type animals. In brief, NFAT1 expression and activation may be tumor specific and stage-dependent. Future studies need to clarify the role of NFAT1-MDM2 pathway in cancer onset, development, and progression.
In conclusion, in this study, we have demonstrated that the calcineurin-NFAT1 pathway regulates MDM2 transcription by a p53-independent mechanism. The identification of this calcineurin-NFAT1-MDM2 axis provides additional information that will increase the understanding of the p53-independent control of mdm2 expression in cancer and the roles of NFAT1 in cancer development and progression.
Acknowledgments
We thank Drs. C. W. Chow, A. Rao, and A. Toker for the NFAT plasmids and Drs. D. Chen, L. Ao, and J. Qin and S. Voruganti for helpful discussions and excellent technical support.
This work was supported in part by National Institutes of Health Grants R01 CA112029 and R01 CA121211. This work was also supported by Susan G. Komen Foundation Grant BCTR0707731 (to R. Z.).
- NFAT
- nuclear factor of activated T cells
- CA-NFAT1
- constitutively activated NFAT1
- DN-NFAT
- dominant-negative NFAT
- DBD
- DNA binding domain
- CsA
- cyclosporine A.
REFERENCES
- 1. Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J. (1992) The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 2. Haupt Y., Maya R., Kazaz A., Oren M. (1997) Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- 3. Oliner J. D., Pietenpol J. A., Thiagalingam S., Gyuris J., Kinzler K. W., Vogelstein B. (1993) Oncoprotein MDM2 conceals the activation domain of tumor suppressor p53. Nature 362, 857–860 [DOI] [PubMed] [Google Scholar]
- 4. Kubbutat M. H., Jones S. N., Vousden K. H. (1997) Regulation of p53 stability by Mdm2. Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- 5. Daujat S., Neel H., Piette J. (2001) MDM2: Life without p53. Trends Genet. 17, 459–464 [DOI] [PubMed] [Google Scholar]
- 6. Ganguli G., Wasylyk B. (2003) p53-independent functions of MDM2. Mol. Cancer Res. 1, 1027–1035 [PubMed] [Google Scholar]
- 7. Jones S. N., Hancock A. R., Vogel H., Donehower L. A., Bradley A. (1998) Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 95, 15608–15612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cordon-Cardo C., Latres E., Drobnjak M., Oliva M. R., Pollack D., Woodruff J. M., Marechal V., Chen J., Brennan M. F., Levine A. J. (1994) Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer research 54, 794–799 [PubMed] [Google Scholar]
- 9. Wu X., Bayle J. H., Olson D., Levine A. J. (1993) The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 7, 1126–1132 [DOI] [PubMed] [Google Scholar]
- 10. Ries S., Biederer C., Woods D., Shifman O., Shirasawa S., Sasazuki T., McMahon M., Oren M., McCormick F. (2000) Opposing effects of Ras on p53: Transcriptional activation of mdm2 and induction of p19ARF. Cell 103, 321–330 [DOI] [PubMed] [Google Scholar]
- 11. Truong A. H., Cervi D., Lee J., Ben-David Y. (2005) Direct transcriptional regulation of MDM2 by Fli-1. Oncogene 24, 962–969 [DOI] [PubMed] [Google Scholar]
- 12. Slack A., Chen Z., Tonelli R., Pule M., Hunt L., Pession A., Shohet J. M. (2005) The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc. Natl. Acad. Sci. U.S.A. 102, 731–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Araki S., Eitel J. A., Batuello C. N., Bijangi-Vishehsaraei K., Xie X. J., Danielpour D., Pollok K. E., Boothman D. A., Mayo L. D. (2010) TGF-β1-induced expression of human Mdm2 correlates with late-stage metastatic breast cancer. J. Clin. Invest. 120, 290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Busuttil V., Droin N., McCormick L., Bernassola F., Candi E., Melino G., Green D. R. (2010) NF-κB inhibits T cell activation-induced, p73-dependent cell death by induction of MDM2. Proc. Natl. Acad. Sci. U.S.A. 107, 18061–18066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tergaonkar V., Pando M., Vafa O., Wahl G., Verma I. (2002) p53 stabilization is decreased upon NFκB activation: A role for NFκB in acquisition of resistance to chemotherapy. Cancer cell 1, 493–503 [DOI] [PubMed] [Google Scholar]
- 16. Hogan P. G., Chen L., Nardone J., Rao A. (2003) Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17, 2205–2232 [DOI] [PubMed] [Google Scholar]
- 17. Macian F. (2005) NFAT proteins: Key regulators of T cell development and function. Nat. Rev. Immunol. 5, 472–484 [DOI] [PubMed] [Google Scholar]
- 18. Müller M. R., Rao A. (2010) NFAT, immunity, and cancer: A transcription factor comes of age. Nat. Rev. Immunol. 10, 645–656 [DOI] [PubMed] [Google Scholar]
- 19. Mancini M., Toker A. (2009) NFAT proteins: Emerging roles in cancer progression. Nat. Rev. Cancer 9, 810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jauliac S., López-Rodriguez C., Shaw L. M., Brown L. F., Rao A., Toker A. (2002) The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat. Cell Biol. 4, 540–544 [DOI] [PubMed] [Google Scholar]
- 21. Medyouf H., Alcalde H., Berthier C., Guillemin M. C., dos Santos N. R., Janin A., Decaudin D., de Thé H., Ghysdael J. (2007) Targeting calcineurin activation as a therapeutic strategy for T cell acute lymphoblastic leukemia. Nat. Med. 13, 736–741 [DOI] [PubMed] [Google Scholar]
- 22. Ryeom S., Baek K. H., Rioth M. J., Lynch R. C., Zaslavsky A., Birsner A., Yoon S. S., McKeon F. (2008) Targeted deletion of the calcineurin inhibitor DSCR1 suppresses tumor growth. Cancer Cell 13, 420–431 [DOI] [PubMed] [Google Scholar]
- 23. Hernández G. L., Volpert O. V., Iñiguez M. A., Lorenzo E., Martínez-Martínez S., Grau R., Fresno M., Redondo J. M. (2001) Selective inhibition of vascular endothelial growth factor-mediated angiogenesis by cyclosporin A: Roles of the nuclear factor of activated T cells and cyclooxygenase 2. J. Exp. Med. 193, 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baksh S., Widlund H. R., Frazer-Abel A. A., Du J., Fosmire S., Fisher D. E., DeCaprio J. A., Modiano J. F., Burakoff S. J. (2002) NFATc2-mediated repression of cyclin-dependent kinase 4 expression. Mol. Cell 10, 1071–1081 [DOI] [PubMed] [Google Scholar]
- 25. Hodge M. R., Ranger A. M., Charles de la Brousse F., Hoey T., Grusby M. J., Glimcher L. H. (1996) Hyperproliferation and dysregulation of IL-4 expression in NF-ATp-deficient mice. Immunity 4, 397–405 [DOI] [PubMed] [Google Scholar]
- 26. Duque J., Fresno M., Iñiguez M. A. (2005) Expression and function of the nuclear factor of activated T cells in colon carcinoma cells: Involvement in the regulation of cyclooxygenase-2. J. Biol. Chem. 280, 8686–8693 [DOI] [PubMed] [Google Scholar]
- 27. Yiu G. K., Toker A. (2006) NFAT induces breast cancer cell invasion by promoting the induction of cyclooxygenase-2. J. Biol. Chem. 281, 12210–12217 [DOI] [PubMed] [Google Scholar]
- 28. Chebel A., Rouault J. P., Urbanowicz I., Baseggio L., Chien W. W., Salles G., Ffrench M. (2009) Transcriptional activation of hTERT, the human telomerase reverse transcriptase, by nuclear factor of activated T cells. J. Biol. Chem. 284, 35725–35734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buchholz M., Schatz A., Wagner M., Michl P., Linhart T., Adler G., Gress T. M., Ellenrieder V. (2006) Overexpression of c-Myc in pancreatic cancer caused by ectopic activation of NFATc1 and the Ca2+/calcineurin signaling pathway. EMBO J. 25, 3714–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Köenig A., Linhart T., Schlengemann K., Reutlinger K., Wegele J., Adler G., Singh G., Hofmann L., Kunsch S., Büch T., Schäfer E., Gress T. M., Fernandez-Zapico M. E., Ellenrieder V. (2010) NFAT-induced histone acetylation relay switch promotes c-Myc-dependent growth in pancreatic cancer cells. Gastroenterology 138, 1189–1199.e1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li M., Zhang Z., Hill D. L., Chen X., Wang H., Zhang R. (2005) Genistein, a dietary isoflavone, down-regulates the MDM2 oncogene at both transcriptional and posttranslational levels. Cancer Res. 65, 8200–8208 [DOI] [PubMed] [Google Scholar]
- 32. Li M., Zhang Z., Hill D. L., Wang H., Zhang R. (2007) Curcumin, a dietary component, has anticancer, chemosensitization, and radiosensitization effects by down-regulating the MDM2 oncogene through the PI3K/mTOR/ETS2 pathway. Cancer Res. 67, 1988–1996 [DOI] [PubMed] [Google Scholar]
- 33. Jain J., Burgeon E., Badalian T. M., Hogan P. G., Rao A. (1995) A similar DNA-binding motif in NFAT family proteins and the Rel homology region. J. Biol. Chem. 270, 4138–4145 [PubMed] [Google Scholar]
- 34. Yang G. H., Fan J., Xu Y., Qiu S. J., Yang X. R., Shi G. M., Wu B., Dai Z., Liu Y. K., Tang Z. Y., Zhou J. (2008) Osteopontin combined with CD44, a novel prognostic biomarker for patients with hepatocellular carcinoma undergoing curative resection. Oncologist 13, 1155–1165 [DOI] [PubMed] [Google Scholar]
- 35. Baumgart S., Glesel E., Singh G., Chen N. M., Reutlinger K., Zhang J., Billadeau D. D., Fernandez-Zapico M. E., Gress T. M., Singh S. K., Ellenrieder V. (2012) Restricted heterochromatin formation links NFATc2 repressor activity with growth promotion in pancreatic cancer. Gastroenterology 142, 388–398.e1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hollstein M., Sidransky D., Vogelstein B., Harris C. C. (1991) p53 mutations in human cancers. Science 253, 49–53 [DOI] [PubMed] [Google Scholar]
- 37. Bond G. L., Hu W., Bond E. E., Robins H., Lutzker S. G., Arva N. C., Bargonetti J., Bartel F., Taubert H., Wuerl P., Onel K., Yip L., Hwang S. J., Strong L. C., Lozano G., Levine A. J. (2004) A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119, 591–602 [DOI] [PubMed] [Google Scholar]
- 38. Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E. A. (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 [DOI] [PubMed] [Google Scholar]
- 39. Chen Z. L., Zhao S. H., Wang Z., Qiu B., Li B. Z., Zhou F., Tan X. G., He J. (2011) Expression and unique functions of four nuclear factor of activated T cells isoforms in non-small cell lung cancer. Chin. J. Cancer 30, 62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maxeiner J. H., Karwot R., Sauer K., Scholtes P., Boross I., Koslowski M., Türeci O., Wiewrodt R., Neurath M. F., Lehr H. A., Finotto S. (2009) A key regulatory role of the transcription factor NFATc2 in bronchial adenocarcinoma via CD8+ T lymphocytes. Cancer Res. 69, 3069–3076 [DOI] [PubMed] [Google Scholar]
- 41. Gerlach K., Daniel C., Lehr H. A., Nikolaev A., Gerlach T., Atreya R., Rose-John S., Neurath M. F., Weigmann B. (2012) Cancer Res. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]