Background: Spf1 belongs to the least characterized group of P5-ATPases.
Results: GFP-Spf1 hydrolyzes ATP and forms a phosphoenzyme that rapidly decays in the presence of ADP.
Conclusion: The Spf1 performs well the E1 steps of the reaction cycle, but progression to the E2 forms is slow.
Significance: The study extends the understanding of the catalytic mechanism of P5-ATPases.
Keywords: ATPases, Membrane Enzymes, Membrane Transport, Transporters, Yeast, P5-ATPases
Abstract
The P5-ATPases are important components of eukaryotic cells. They have been shown to influence protein biogenesis, folding, and transport. The knowledge of their biochemical properties is, however, limited, and the transported ions are still unknown. We expressed in Saccharomyces cerevisiae the yeast Spf1 P5A-ATPase containing the GFP fused at the N-terminal end. The GFP-Spf1 protein was localized in the yeast endoplasmic reticulum. Purified preparations of GFP-Spf1 hydrolyzed ATP at a rate of ∼0.3–1 μmol of Pi/mg/min and formed a phosphoenzyme in a simple reaction medium containing no added metal ions except Mg2+. No significant differences were found between the ATPase activity of GFP-Spf1 and recombinant Spf1. Omission of protease inhibitors from the purification buffers resulted in a high level of endogenous proteolysis at the C-terminal portion of the GFP-Spf1 molecule that abolished phosphoenzyme formation. The Mg2+ dependence of the GFP-Spf1 ATPase was similar to that of other P-ATPases where Mg2+ acts as a cofactor. The addition of Mn2+ to the reaction medium decreased the ATPase activity. The enzyme manifested optimal activity at a near neutral pH. When chased by the addition of cold ATP, 90% of the phosphoenzyme remained stable after 5 s. In contrast, the phosphoenzyme rapidly decayed to less than 20% when chased for 3 s by the addition of ADP. The greater effect of ADP accelerating the disappearance of EP suggests that GFP-Spf1 accumulated the E1∼P phosphoenzyme. This behavior may reflect a limiting countertransported substrate needed to promote turnover or a missing regulatory factor.
Introduction
The family of ion transporters known as P-type ATPases comprises a large group of enzymes that couple active ion transport with the hydrolysis of ATP (1). The best known P-ATPases include the Ca2+-ATPase from sarcoendoplasmic reticulum and the Na+-K+-ATPase (2, 3). During ion transport, these enzymes cycle between two major conformations E1-E2 and become transiently phosphorylated at a catalytic aspartate residue. A similar transport mechanism is believed to apply essentially to all P-ATPases. Binding of the transported ion to the E1 form leads to the assembly of the phosphorylation site between the ATP-bound N domain and the P domain, whereas the A domain directs the occlusion of the bound ion. Phosphorylation produces the E1∼P phosphoenzyme and liberates ADP. In the transition from the E1∼P to the E2P form, the A domain associates with the N-P complex and opens the exit site of the occluded ion. The P domain is then dephosphorylated by the A domain, and the cycle completes when the phosphate is released from the enzyme. During this last stage, some ATPases bind a second ion that is countertransported and released when upon returning to the E1 form.
According to the conservation of the amino acid sequence, the P-ATPases can be subdivided in five subfamilies (4). The subfamily P5 includes a group of related enzymes that have been found only in eukaryotes and whose ion specificity is not yet known. Further refinement of the sequence analysis had led to the division of the P5-ATPases in subgroups A and B (5).
In humans, five genes code for P5-ATPases (6). Mutations in P5B-ATPases ATP13A2 and ATP13A4 have been linked to neurological diseases like hereditary parkinsonism and autism spectrum disorder (7, 8). The yeast Saccharomyces cerevisiae contains two genes coding for P5-ATPases, YEL031W coding for P5A-ATPase called Spf12 or Cod1, and YOR291W coding for P5B-ATPase called Ypk9.
The biochemical properties of the P5-ATPases are much less known than those of other members of the P-type ATPase family (9). Here we have expressed and characterized a GFP-tagged yeast Spf1 P5A-ATPase in S. cerevisiae. We found that the purified fusion protein was able to hydrolyze ATP and to form a phosphoenzyme. These activities required only Mg2+ as metallic cofactor. The decomposition of the intermediate phosphoenzyme was highly sensitive to ADP, indicating that most of it was E1∼P.
EXPERIMENTAL PROCEDURES
Chemicals
Polyoxyethylene-10-laurylether (C12E10), l-α-phosphatidylcholine type XVI-E Sigma from fresh egg yolk, brain extract type I Folch fraction I from bovine brain containing ∼10% phosphatidylinositol, 50% phosphatidylserine, and other lipids, ATP (disodium salt, vanadium-free), SDS, yeast synthetic drop-out medium supplement without leucine, yeast nitrogen base without amino acids, dextrose, enzymes, and cofactors for the synthesis of [γ-32P]ATP, and all other chemicals were obtained from Sigma. Tryptone and yeast extract were from Difco. [γ-32P]ATP was provided by PerkinElmer Life Sciences. Salts and reagents were of analytical reagent grade.
Yeast Strains, Transformation, and Growth Media
S. cerevisiae strain DBY 2062 (MATα his4-619 leu2-3,112) (10) was used for expression. Yeast cells were transformed with the pMP625 vector containing a Leu+ marker and the promoter of plasma membrane H+-ATPase from S. cerevisiae. For transformation with the plasmid construct, a lithium acetate/polyethylene glycol method was utilized (11). The cells were grown in complete medium (0.75% yeast extract, 1.13% tryptone, 2.2% dextrose), and transformants were selected for their ability to grow in the absence of leucine on plates containing 6.7% yeast-nitrogen base without amino acids (YNB), 0.67% complete supplemented medium minus Leu (Leu−), 2.2% dextrose, and 1.5% agar.
Membrane Isolation and Purification of GFP-Spf1
Total membranes from four liters of yeasts expressing the GFP-Spf1 protein were obtained as previously described (12, 13). The microsomal membranes (∼200 mg of microsomal protein) were suspended in a purification buffer containing 20 mm MOPS-K (pH 7.4 at 4 °C), 20% glycerol, 130 mm KCl, 1 mm MgCl2, 2 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride; homogenized in a glass homogenizer; and solubilized at 4 °C for 15 min by adding 2 g of C12E10/g of total membrane protein. The final volume of solubilization was adjusted to keep constant the concentration of detergent at 0.5% and 10 mm imidazole. The supernatant was loaded onto a 2-ml nickel-nitrilotriacetic acid-agarose column (Qiagen) and washed with 30 column volumes of purification buffer containing 0.05% C12E10 and 50 mm imidazole. Finally the protein was eluted in purification buffer containing 0.005% C12E10 and 75 mm imidazole. The eluate fractions of higher protein content were pooled, aliquoted, and kept in liquid N2.
Protein Assay
The protein concentration was initially estimated by the method of Bradford (14) and then corrected according to the intensity of the bands after SDS-PAGE on an 8% acrylamide gel according to Laemmli (15) and Coomassie Blue staining, using bovine serum albumin as a standard.
ATPase Activity
The ATPase activity was estimated from the release of 32P from [γ-32P]ATP at 37 °C (16) in a final volume of 0.3 ml of ATPase medium containing, 20 mm Hepes-K (pH 7.4 at 37 °C), 100 mm KCl, 5 mm N3Na, 5 mm MgCl2, 3 mm ATP, and 2 μg of GFP-Spf1 in 15–100 μl of elution buffer. In some experiments, the ATPase medium also contained 0.5 mm EGTA and no KCl. Where the indicated lipids were added to the purified GFP-Spf1 protein, the suspension was thoroughly mixed and preincubated for at least 5 min on ice before being added to the ATPase reaction medium. ATP and ADP solutions were prepared from the disodium salts and neutralized at pH 7.1. The reaction was initiated by the addition of GFP-Spf1 and terminated by acid denaturation. To assure that the total ATP hydrolysis was lower than 20%, the reaction time was varied between 5 and 10 min, depending on the activity of the sample and the ATP concentration in the medium.
Phosphorylation of GFP-Spf1
Approximately 2.5 μg of purified GFP-Spf1 was phosphorylated at 4 or 37 °C in 0.25 ml of reaction buffer containing 50 mm Tris-HCl (pH 7.6) and MgCl2 to give a concentration of 2 mm free Mg2+. The reaction was started by the addition of 30 μm [γ32P]ATP and was stopped after 60 s with 10% ice-cold trichloroacetic acid. After adding 10 μg of bovine serum albumin, the denatured proteins were collected by centrifugation at 20,000 × g for 10 min, washed once with 5% trichloroacetic acid and 150 mm NaH2PO4, and washed once more with distilled water. The precipitated protein was suspended in sample buffer and separated by acidic SDS-PAGE. The gels were dried, and the radioactivity was detected using a Storm Molecular Image System. The dephosphorylation was initiated by the addition of 23 mm of cold ATP-Mg or ADP-Mg and continued for 3 or 5 s on ice before the addition of 10% ice-cold trichloroacetic acid.
RESULTS
Expression and Purification of GFP-Spf1
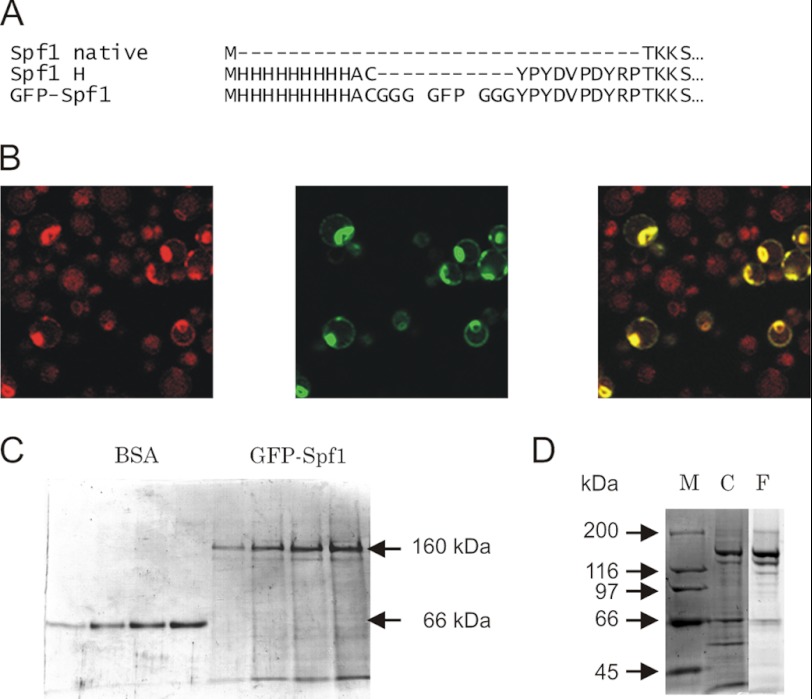
The cDNA coding for either N-terminal poly-His- or poly His-GFP-tagged Spf1 were cloned into the pMP625 vector for expression in S. cerevisiae cells (Fig. 1). We determined the localization of the GFP-Spf1 by confocal fluorescence microscopy. The observed fluorescence of GFP-Spf1 was typical for ER localization with a ring around the nucleous and additional peripheral staining. The distribution of GFP-Spf1 overlapped with that of Cherry fatty acid elongase, ELO3, a protein of the ER membrane. The localization of GFP-Spf1 is in agreement with the ER localization of Spf1 reported previously (9).
FIGURE 1.
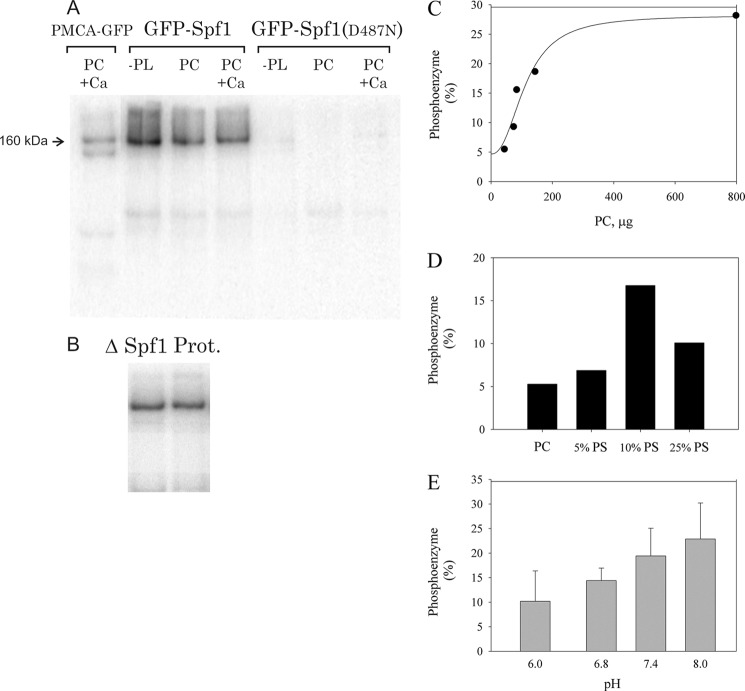
A, alignment of the N-terminal amino acid sequence of the Spf1 protein (Spf1 native), the recombinant Spf1 9×His-HA-tagged as used in Ref. 9 (Spf1 H), and recombinant Spf1 fused to GFP (GFP-Spf1). B, confocal microscopy of yeasts expressing Cherry-ELO3 and GFP-Spf1. Yeast cells expressing a chromosome-integrated Cherry-ELO3 were transfected with pMP625-GFP-Spf1. Fluorescence images of Cherry (left panel), GFP (center panel), and merged (right panel) are shown. C, purification of GFP-Spf1. Different amounts of GFP-Spf1 protein purified by Ni2+-NTA chromatography as described under “Experimental Procedures” were loaded onto an 8% SDS-PAGE and stained with Coomassie Blue. On the left part of the gel, increasing amounts of BSA corresponding to 0.12, 0.24, 0.36, and 0.48 μg of protein were loaded for estimating the concentration of GFP-Spf1 protein. On the right, increasing volumes of purified GFP-Spf1 were loaded. The estimated amounts of GFP-Spf1 protein corresponded to 0.8, 0.15, 0.24, and 0.30 μg of protein. D, endogenous proteolysis of GFP-Spf1; SDS-PAGE of purified as in C, except for the lack of protease inhibitor during the purification protocol. Lane M, marker; lane C, Coomassie Blue staining; lane F, green fluorescence.
Yeast microsomes were solubilized with detergent C12E10, and GFP-Spf1 was purified by standard metal affinity chromatography. With the aim of preserving the functional properties of GFP-Spf1, the amount of detergent used for solubilizing the membranes was kept at minimum and in the same range of concentration as that used for purification of other recombinant P-ATPases (12, 13). Analysis by SDS-PAGE of the purified sample revealed the presence of a fluorescent polypeptide with the expected migration according to the predicted size of GFP-Spf1 (∼160 kDa) (Fig. 1B). Interestingly, the omission of protease inhibitors during the purification procedure led to the appearance of several smaller fragments that presumably arose by the action of the yeast proteases (Fig. 1D). Because the major proteolytic fragments were still recovered after purification and exhibited fluorescence, they still retained the poly-His tag-GFP portion of the molecule, and hence proteolysis must have shortened the molecule from the C-terminal end.
ATP Hydrolysis by the Purified GFP-Spf1
The functional state of the purified GFP-Spf1 was tested by measuring its ability to catalyze hydrolysis of ATP (Fig. 2A). The GFP-Spf1 exhibited an ATPase activity that, depending on the purification experiment, was between 0.3 and 1.2 μmol of Pi/mg/min. This value of ATPase activity is somewhat higher than that reported previously for Spf1 (9). Although the fusion of GFP to the N-terminal end of other P-ATPases has been reported to have minimal functional consequences (12, 17), its effects on the Spf1 enzyme are not known. To this end, Spf1 was expressed and purified, and its ATPase activity was compared with that of GFP-Spf1. We found that both proteins had similar ATPase activity, suggesting that the presence of GFP at N-terminal end did not impair the function of Spf1 (not shown).
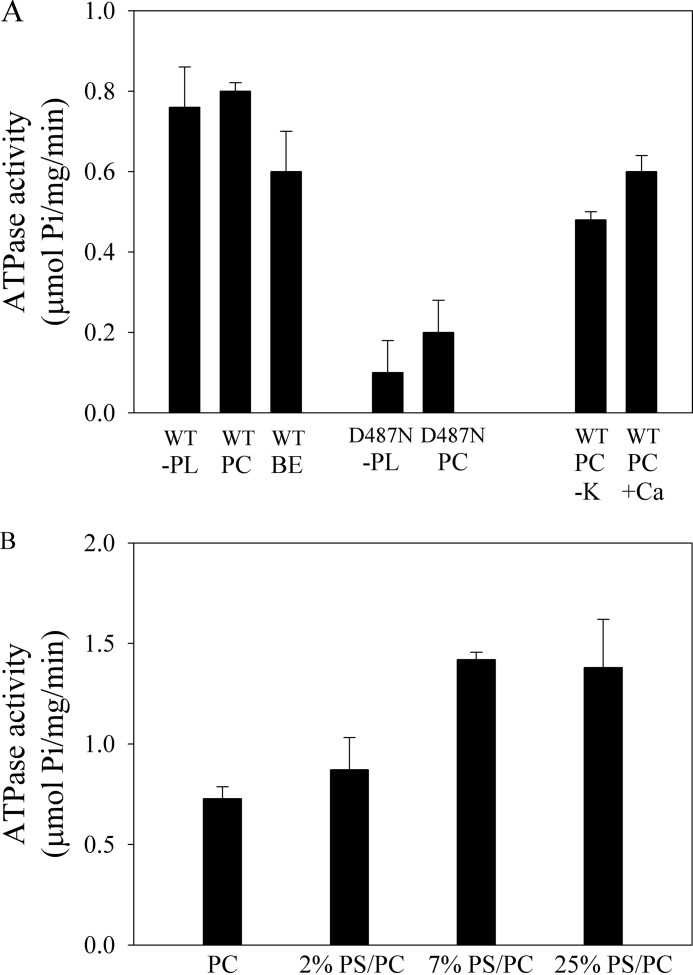
FIGURE 2.
A, ATPase activity of purified GFP-Spf1 (WT) and GFP-Spf1 containing the mutation D487N. The purified proteins (∼2 μg) in elution buffer were incubated with 3 mm ATP for 10 min at 37 °C. The purified proteins were either not supplemented (-PL) or supplemented with 85 μg of C12E10 and 43 μg of phosphatidylcholine (PC) or acidic lipids extracted from bovine brain extract (BE). The activity was also measured in the absence of added KCl (-K) and in the presence of 0.5 mm EGTA and 10 μm free Ca2+. B, ATPase activity of purified GFP-Spf1 reconstituted in PC or mixtures of PS/PC at increasing concentrations of PS. 1 μg of GFP-Spf1 (10 μl in elution buffer) was supplemented with 85 μg of C12E10 and 43 μg of PC or a mixture of PC/PS. The detergent/protein/phospholipid mixture was kept for 5 min under gentle stirring, and then the detergent was removed by adding 0.25 g of prewashed wet Bio-Beads SM-2 (Bio-Rad). The mixture was stirred at room temperature for 30 min, and the Bio-Beads were separated by filtration. The activity was measured as indicated under “Experimental Procedures.”
The ATPase activity of GFP-Spf1 was approximately the same when the purified protein was either in micelles of C12E10, C12E10 plus PC, or C12E10 plus a mixture of brain acidic phospholipids (brain extract), which has been shown to be a potent activator of the plasma membrane Ca2+ pump (13).
Previous studies of other recombinant P-ATPases purified from yeasts have reported the presence of contaminating ATPase activities (18). To establish that ATP was specifically hydrolyzed by GFP-Spf1, a mutant GFP-Spf1 in which the catalytic residue Asp487 was substituted by Asn was prepared and purified following the same purification protocol as for the wild-type GFP-Spf1. The GFP-Spf1 D487N mutant had only marginal ATPase, indicating that most of the ATP hydrolytic activity of the purified sample was due to GFP-Spf1. The standard medium used for the purification of the protein and the ATPase measurements contained KCl. In the absence of added KCl, the ATPase activity of GFP-Spf1 decreased ∼40%. This effect is similar to the activating effect of alkali metal ions on other P-ATPases (19) and probably is not due to the transport specificity of Spf1.
It was previously suggested that Spf1 could be a Ca2+ transporter involved in the regulation of Ca+ concentration in the yeast endoplasmic reticulum (9). However, we found that the addition to the reaction medium of either CaCl2 to obtain 10 μm free Ca2+ or 0.5 mm EGTA did not change the ATPase activity of GFP-SPf1.
The effect of lipids was further investigated following a different protocol in which the purified GFP-SPf1 enzyme was supplemented with a mixture of PC/PS/C12E10, and then the detergent was removed by treatment with biobeads. The results in Fig. 2B show that as the relative abundance of PS increased, the ATPase activity of GFP-Spf1 slightly increased ∼1.7-fold.
Dependence of the GFP-Spf1 ATPase Activity with ATP
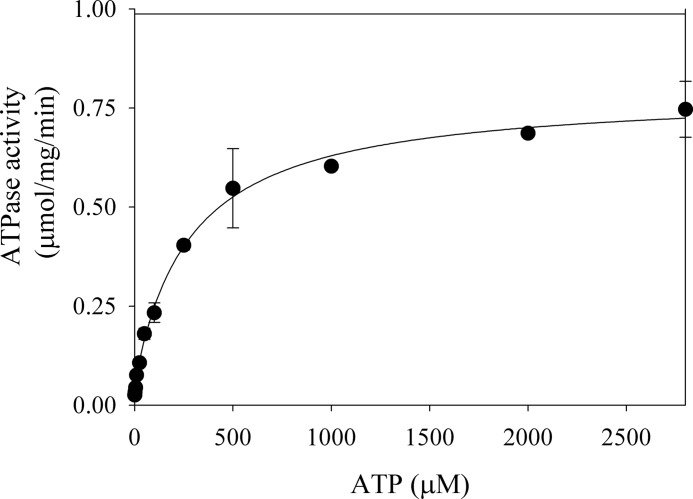
P-ATPases usually display a complex dependence with ATP, having a high affinity catalytic site and a lower affinity regulatory site (20, 21). However, in a previous study the ATPase activity of Spf1 was shown to increase with ATP along a hyperbolic curve that reached saturation at ∼50 μm ATP (9). We examined the effect of an extended range of ATP concentrations on the ATPase activity of GFP-Spf1 (Fig. 3). The maximal activity was reached at mm concentrations of ATP, and the best fit to the experimental points was obtained by fitting a double reciprocal equation indicating the presence of a high affinity catalytic site of Km1 ∼3 μm and a modulatory site of Km2 ∼280 μm for ATP.
FIGURE 3.
ATP dependence of the ATPase activity of GFP-Spf1. The measurement was performed as indicated under “Experimental Procedures” in a reaction medium containing 1 μg of GFP-Spf1, 43 μg of PC, 1.2 mm free Mg2+, and increasing concentrations of ATP. The points shown are duplicates from two experiments performed in duplicate. The error bars are the standard errors of the mean. The line represents the best fit to the data given by the following equation: v = v1·Km1/(ATP + Km1) + v2·Km2/(ATP + Km2) with the following parameters v1 = 0.06 ± 0.02 μmol/mg/min, v2 = 0.73 ± 0.02 μmol/mg/min, Km1 = 3 ± 2 μm, and Km2 = 285 ± 41 μm.
Dependence of GFP-Spf1 ATPase with the Concentration of Mg2+, Mn2+, and H+
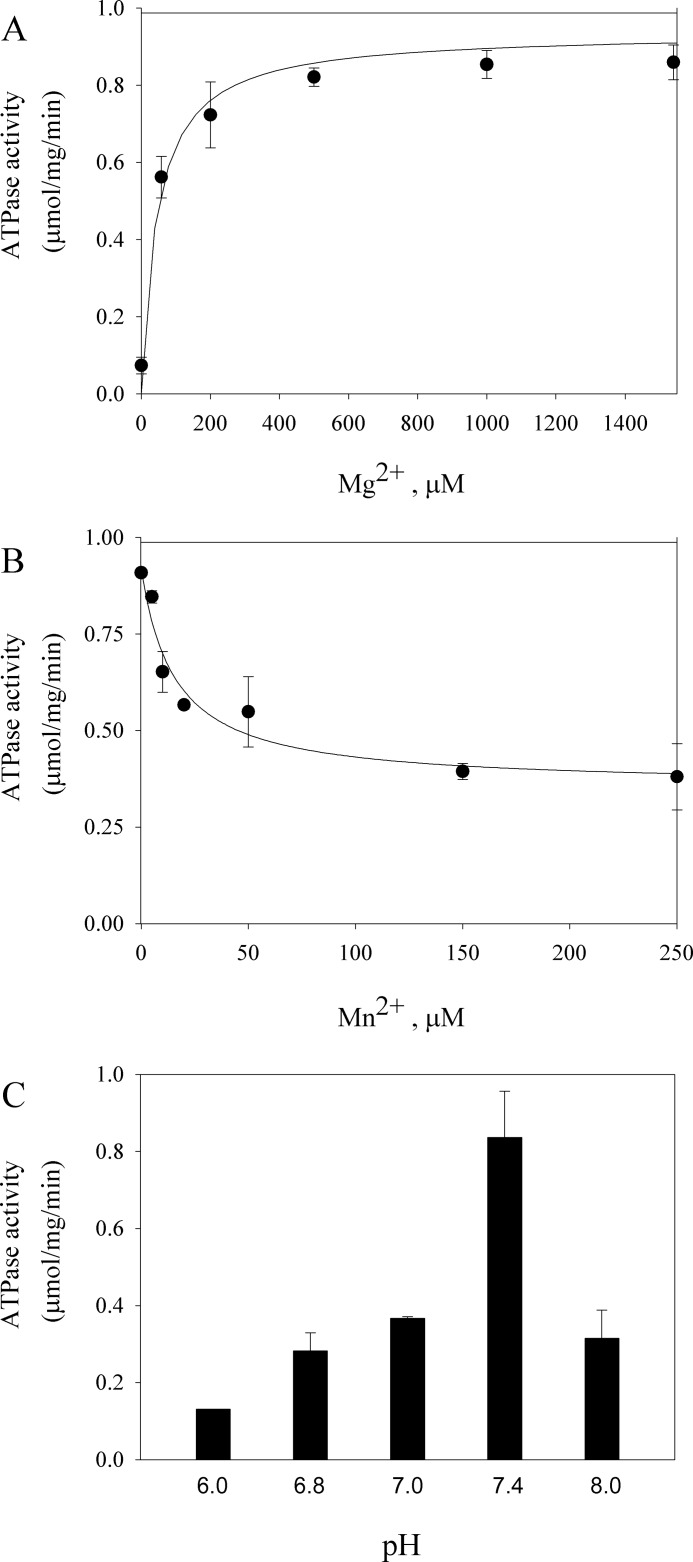
A characteristic of all ion transporting P-type ATPases is the stimulation of their ATPase activity by the presence of the transported ionic substrates. Because it has been previously observed that Mg2+ stimulates the ATPase activity of Spf1, it was suggested that Mg2+ might be transported by Spf1 (9). Fig. 4A shows the ATPase activity of GFP-Spf1 measured at 3 mm ATP-Mg and increasing concentrations of free Mg2+. In these conditions, the GFP-Spf1 activity increased with the concentration of Mg2+ along a hyperbolic curve that reached half-maximal activity at 45 μm Mg2+. This dependence of the GFP-Spf1 ATPase with Mg2+ is similar to that of other P-ATPases where Mg2+ acts as cofactor. A recent study has shown that the human P5B-ATPase ATP13A2 plays a role in regulating the intracellular manganese concentration (22). Fig. 4B shows that increasing the concentration of manganese in the reaction medium from 0 to 250 μm decreased ∼40% in the ATPase activity of GFP-Spf1. The dependence of the GFP-Spf1 ATPase with the H+ concentration was also examined. As shown in Fig. 4C, the maximal activity was reached at near neutral pH.
FIGURE 4.
A, Mg2+ dependence of the ATPase activity of the GFP-Spf1 protein. The ATPase activity was measured as described under “Experimental Procedures,” at 37 °C for 10 min in a medium containing 3 mm ATP and increasing concentrations of MgCl2. The points shown are the averages from two experiments performed in duplicate. The error bars are the standard errors of the mean. The line represents the best fit to the data given by a Michaelis-Menten equation with Km = 46 ± 9 μm and Vmax = 0.94 ± 0.02 μmol/mg/min. B, effect of Mn2+ on the ATPase activity of the GFP-Spf1 protein. The ATPase activity was measured as described under “Experimental Procedures,” at 37 °C for 10 min in a medium containing 3 mm ATP and increasing concentrations of MnCl2. The line represents the best fit to the data given by a hyperbolic decay equation: v = v0 + v1·Mn/(Ki + Mn) with the following parameters v0 = 0.35 ± 0.04 μmol/mg/min, v1 = 0.56 ± 0.06 μmol/mg/min, and Ki = 15 ± 6 μm. C, pH dependence of the GFP-Spf1 ATPase. The ATPase activity was measured as described under “Experimental Procedures” in the standard medium containing 50 mm MOPS-imidazole adjusted at the indicated pH.
Expression, Purification, and ATPase Activity of GFP-Spf1 Purified from Yeast Cells Lacking the Endogenous Spf1
It has been shown that yeast cells lacking the endogenous Spf1 are hypersensitive to lovastatin (9). However, transforming the cells with a plasmid expressing Spf1 sufficed for restoring wild-type sensitivity. This result indicates that in ΔSpf1 cells, either the Spf1 by itself or by interacting with other endogenous components is able to complement the ΔSpf1 phenotype. To investigate whether the lack of the endogenous Spf1 changes the functional properties of the recombinant GFP-Spf1 protein, GFP-Spf1 was purified from ΔSpf1 yeasts. No differences were detected in the ATPase activity of the GFP-Spf1 purified either from ΔSpf1 or wild-type yeasts (not shown).
Formation and Decay of the Phosphoenzyme Intermediate
P-type ATPases react with ATP to form a phosphoenzyme in a reaction that is greatly stimulated by the presence of the transported ion. Purified GFP-Spf1 in elution buffer was incubated at 4 °C for 1 min with 30 μΜ [γ-32P]ATP and 2 mm MgCl2. As shown in Fig. 5A, a strong radioactive band corresponding to the GFP-Spf1 phosphoenzyme was detected. When the phosphorylation medium was supplemented with PC as mixed micelles of lipid-C12E10, the amount of phosphoenzyme formed was the same or slightly lower. Similarly, the addition of 10 μm CaCl2 did not have a significant effect on the amount of GFP-Spf1 phosphoenzyme. However, the removal of Mg2+, by preincubating the enzyme with 1 mm EDTA, eliminated the formation of phosphoenzyme (not shown). Moreover, no phosphoenzyme was detected in the GFP-Spf1 mutant D487N in any of the conditions tested. Results in Fig. 5B show that the amount of phosphoenzyme formed did not change when the GFP-Spf1 protein was purified from yeasts lacking the endogenous Spf1. It is also worth noticing that endogenous proteolysis seemed to eliminate the ability to form the phosphoenzyme because only one band corresponding to the intact protein was observed in the proteolysed sample. By comparing the intensity of the phosphoenzyme bands, it was estimated that in these conditions ∼5–10% of the GFP-Spf1 protein was phosphorylated. Although this value may seem rather low, it represents a lower limit for the amount of EP formed, because part of the phosphoenzyme decomposes during electrophoresis. In any case, the amount of GFP-Spf1 phosphoenzyme is in the range of the maximal levels obtained for other P-ATPases like the plasma membrane Ca2+ pump in the presence of La3+ (23).
FIGURE 5.
Formation of the GFP-Spf1 phosphoenzyme. A, 5 μg of purified wild-type or D487N GFP-Spf1 protein or 5 μg of purified human plasma membrane Ca2+ pump (PMCA) were incubated with 30 μm ATP for 1 min at 4 °C as described under “Experimental Procedures.” The purified GFP-Spf1 proteins in elution buffer containing detergent C12E10 were either not supplemented with lipids or supplemented with PC or PC plus 10 μm Ca2+. B, phosphoenzyme formation of GFP-Spf1 purified from ΔSpf1 cells (ΔSpf1) and GFP-Spf1 sample proteolysed as shown in Fig. 1D (Prot.). C, effect of increasing amounts of PC on the amount of EP formed by GFP-Spf1. 1 μg of GFP-Spf1was supplemented with 85 μg of C12E10 and increasing amounts of PC. The suspension was preincubated for at least 5 min on ice before being added to the phosphorylation medium. The phosphorylation reaction was started by the addition of 30 μm ATP and was performed for 1 min at 4 °C. The results are expressed as percentages. 100% is the theoretical value if all the protein was phosphorylated at a stoichiometric ratio of 1. The line represents the best fit to the data given by a Hill equation EP = EP0 + EPmax*(PC)n/(Kn + (PC)n) with EP0 = 4 ± 2%, EPmax = 24 ± 1%, n = 2 ± 1, and K = 110 ± 24 μm. D, effect of increasing amounts of PS on the amount of EP formed by GFP-Spf1. 1 μg of GFP-Spf1was supplemented with 85 μg of C12E10 and 43 μg of PC or a mixture of PC/PS. The suspension was preincubated for at least 5 min on ice before being added to the phosphorylation medium. The phosphorylation reaction was started by the addition of 30 μm ATP and was performed for 1 min at 4 °C. The results are expressed as percentages. 100% is the theoretical value if all the protein was phosphorylated at a stoichiometric ratio of 1. E, effect of the pH on the EP formation. Phosphorylation was performed as indicated under “Experimental Procedures” in a reaction medium containing 50 mm of buffer MOPS-Tris adjusted at different pH. 1 μg of GFP-Spf1 (15 μl) in elution buffer containing 0.75 μg of C12E10 was supplemented with 25 μg of PC as described under “Experimental Procedures.” The results are expressed as percentages. 100% is the theoretical value if all the protein was phosphorylated at a stoichiometric ratio of 1.
The addition of increasing amounts of PC while keeping constant the concentration of C12E10 produced slightly higher levels of EP (Fig. 5C). On the other hand, if the total amount of lipids was kept constant, and part of PC was replaced by PS, the EP increased reaching a maximal level when PS was ∼10% of the total lipid and decreased at higher contents of PS (Fig. 5D). Fig. 5E shows the effect of pH on the level of EP. Maximal levels of EP were obtained at pH 7.4–8.0, consistent with the higher levels of ATPase activity detected at this range of pH.
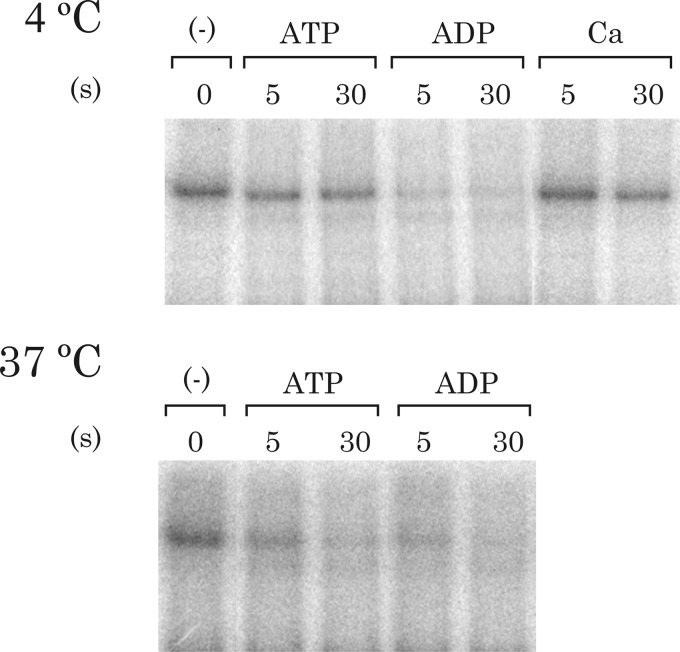
The decay of the phosphoenzyme formed by the GFP-Spf1 protein was investigated (Fig. 6). When the phosphoenzyme was chased with 23 mm of cold ATP, the level of phosphoenzyme after 5 s showed a slight decrease. However, if ADP was used instead of ATP, the EP decayed much faster, and only ∼20% of the original amount of phosphoenzyme remained after 5 s. Interestingly, the addition of 5 mm CaCl2 to the phosphorylation medium after 30 s of phosphorylation slightly reduced the level of phosphoenzyme, suggesting that Ca2+ may inhibit the formation or accelerate the decomposition of EP.
FIGURE 6.
Phosphoenzyme decay. GFP-Spf1 protein supplemented with PC was phosphorylated for 1 min with 30 μm ATP, after which phosphorylation was either stopped by acid quenching (time 0 of dephosphorylation) or continued for 3 or 5 s after the addition of either 23 mm cold ATP, 23 mm ADP, or 5 mm CaCl2 (Ca) and then stopped. The experiment was conducted at either 4 or 37 °C as indicated.
To compare the results with the measurements of ATPase activity that were done at 37 °C, the formation and decomposition of the EP were also studied at 37 °C. At 37 °C the reactions were considerably faster than at 4 °C, but still ADP was more effective than ATP in promoting a faster dephosphorylation.
DISCUSSION
The number of studies showing the importance of the P5-ATPases in eukaryotic cells is rapidly growing. The P5A-ATPAse from S. cerevisiae Spf1 is located in the ER, and several studies have shown that it is crucial for ER-associated processes as proper ER folding of proteins and N-linked glycosylation (9, 24–27). Several observations indicate that Spf1 is involved in calcium homeostasis (9): (a) disruption of the SPF1 gene increases the transcription of calcium regulated genes; (b) the ΔSpf1 mutant yeast is hypersensitive to growth inhibition by extracellular calcium, similar to the mutants defective in the Golgi-localized Pmr1 Ca2+/Mn2+ pump; and (c) deletion of Spf1 has a synergistic effect on the increase of cellular calcium content that results from deletion of Pmr1. Alterations in calcium regulation have also been associated with mutations of mammalian P5B-ATPase ATP13A4 (28). However, the results presented here and those from previous studies do not support the idea that the P5-ATPases are directly involved in the active transport of Ca2+ (9, 29).
The P5-ATPases are assumed to share the biochemical properties of other P-ATPases but had not been examined in detail. We overexpressed in S. cerevisiae its P5A-ATPase Spf1 fused to a His-GFP tag at the N-terminal end. Because the ionic substrate of Spf1 is not known and yeasts possess many P-type ATPases, we chose to perform this study using a detergent-solubilized and affinity-purified preparation of the recombinant enzyme to ascribe the measured enzymatic activities specifically to Spf1. The protocol followed for purifying GFP-Spf1 was similar to that employed previously for purification of other P-ATPases in a functional form (13). The preparation of purified GFP-Spf1 exhibited an ATPase activity that was specifically associated with the function of the Spf1 protein because it was nearly absent in the mutant of the catalytic residue D487N (Fig. 2A). The presence of the His-GFP tag at the N terminus of the Spf1 protein did not alter the ability of Spf1 to hydrolyze ATP. In contrast, it became apparent that the C-terminal portion of the molecule is functionally relevant because its removal by the endogenous yeast proteases abolished the ability of Spf1 to react with ATP to form the phosphoenzyme (Fig. 5B).
Under physiological conditions, ATP hydrolysis by P-ATPases is coupled to ion transport, and ATP hydrolysis is stimulated by the transported ions. We found that the ATPase activity of C12E10 solubilized-GFP-Spf1 required only Mg2+ as cofactor, a requirement indeed shared by all P-ATPases. Three recent studies indicate that P5-ATPases may transport Fe2+, Ca2+, or Mn2+ (30–33). Our results do not support this view. If the ATPase activity of GFP-Spf1 would be related to traces of these ions present in the reaction medium, the addition of EGTA should nearly eliminate the activity, contrary to what was observed. Indeed, increasing concentrations of Mn2+ resulted in a decrease of the activity, probably by competition with Mg2+ (Fig. 4B).
We found that in the standard phosphorylation medium containing Tris-HCl, MgCl2, and the components from the elution solution brought by the purified protein, GFP-Spf1 was capable of phosphoenzyme formation (Fig. 5A). The turnover number based on the level of phosphorylated intermediate gives a value of 3 s−1. This turnover number is in the lower limit for P-ATPases, which have values in the range of 100 s−1 (34). Our estimation of the turnover number may not be accurate because the conditions used for ATPase and EP determination were not exactly the same, i.e., ATP concentration, and also because part of the EP may decompose during electrophoresis. Despite these precautions, it is interesting to note that a molecular activity of 3–10 s−1 can be calculated from the value of Spf1 ATPase activity, assuming a 100% pure protein, a fact that is in agreement with our proposal that the decay of EP limits the ATPase activity.
A well known property of the P-ATPases is their use of ATP both as a catalytic substrate and as a allosteric ligand. ATP at physiological concentrations exhibits a modulatory effect on the rate of ion transport that has been attributed to the acceleration of several partial reactions including the transition between E1P to E2P and the dephosphorylation of the later form. However, in the absence of the countertransported ion, high concentrations of ATP may inhibit phosphoenzyme decay as has been shown for the Na+,K+-ATPase in the absence of K+ (35, 36).
During ion translocation, P-ATPases undergo large conformational changes between two major conformations, E1 and E2. The transported ion binds to E1 and is occluded in the phosphoenzyme E1P. Although in the absence of the transported ion a marginal amount of phosphoenzyme can be produced from ATP, the formation of E1P requires the presence of the transported substrate (37). Indeed it is usually assumed that in the P-ATPases the γ-phosphate of bound ATP is optimally positioned for phosphoryl transfer in the E1 conformation attained by the binding of the transported ion (20). In this conformation (XE1-ATP), the N- and P-domains come together to form the catalytic site. Our results may indicate that a transported ion, or an analog, was already present in the phosphorylation medium. If that were the case, traces of the ion needed for E1P formation may be tightly bound and copurified with the Spf1 protein or MOPS, Tris, imidazole, or glycerol, or the detergent C12E10 may itself activate the GFP-Spf1 acting as a surrogate of the true transported substrate. Moreover, H+ could be the ion required for phosphorylation. However, our measurements of ATPase and phosphorylation do not support this idea because the optimal activity was observed at neutral pH.
An alternative explanation for the results presented here is that the catalytic phosphoenzyme intermediate of Spf1 can be formed spontaneously. This idea is mechanistically interesting because it would imply that Spf1 is naturally “E1-stabilized” and would not require the binding of the transported ion for phosphorylation. This hypothesis would also explain the higher abundance of E1P.
Because ADP acts on the E1 phosphoforms by reversing the phosphorylation with reformation of ATP, the high rate of ADP-induced phosphoenzyme decay (Fig. 6) is indicative of the accumulation of GFP-Spf1 phosphoenzyme as E1P. This is in contrast with the fact that under usual conditions the E2 phosphoform is predominant in most P-ATPases (35, 36). The high ADP sensitivity of the GFP-Spf1 phosphoenzyme suggests that the transition from E1P to E2P occurs at a limiting rate, and thus Spf1 may be functioning at suboptimal conditions. This behavior may result from the need for an as yet unidentified component of the system that is missing in the recombinant protein.
Although most P-ATPases have a catalytic subunit that mediates ATP hydrolysis and ion transport, it is possible that the ion transport specificity results from the interaction with a subunit different from that endowed with the ATPase activity. Indeed in the Escherichia coli KdpFABC K+ transporter, the KdpB subunit mediates ATP hydrolysis and has the P-type ATPase motifs, whereas the KdpA subunit has homology to K+ channels and is required for K+ binding and transport (39). Another possibility is that the behavior of Spf1 described here reflects the absence of the countertransported ion. Ion countertransport has been suggested to be a characteristic of all P-ATPases (40). Because ion countertransport would be coupled to the dephosphorylation reaction, the absence of the countertransported ion may lead to the accumulation of phosphoenzyme, as experimentally observed. As in the case of other P-ATPases, the ion countertransported by the P5-ATPases may be physiologically as relevant as that transported one.
It has been pointed out that, by sequence homology, the P5-ATPases are closest to the P4 subfamily, and thus it is tempting to think that they may share transport specificity for lipids (41). Noticeably, the P4-ATPases Drs2p and ATP8A2 require the presence of Cdc50 proteins to yield EP levels similar to those reported here for Spf1 (18, 38). In contrast we found that the purified GFP-Spf1 was capable of forming the phosphoenzyme. The addition of PC to the purified GFP-Spf1 protein increased the yield of EP (Fig. 5C), an effect apparently not accompanied by an increase in the ATPase activity (Fig. 2A). On the other hand, only a modest increment in the ATPase activity was observed when the amount of C12E10 detergent was decreased, and the protein was reconstituted in lipid mixtures containing increasing amounts of PS (Fig. 2B). Although these results suggest that Spf1 has functional properties that differ from those of the P4-ATPases, future work will be needed to examine in more detail the involvement of specific lipids as substrates or regulators of P5-ATPases.
Acknowledgments
We thank Dr. R. Y. Hampton for the generous gift of the cDNA sequence coding for the Spf1 protein. We thank Marien Gautier and Javier Calvo for collaboration during the first stage of this work and Dr. Pablo Aguilar for the generous gift of the Cherry-ELO3 expressing yeast.
This work was supported by Grant BID PICT 2134 from Agencia Nacional de Promoción Científica y Tecnológica, Grant 2122 from Consejo Nacional de Investigaciones Científicas y Tecnológicas, and Universidad de Buenos Aires Grant UBACyT B009.
- Spf1
- sensitivity to Pichia farinosa killer toxin
- C12E10
- polyoxyethylene 10-lauryl ether
- EP
- phosphorylated enzyme
- PC
- phosphatidylcholine
- PS
- phosphatidylserine
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Palmgren M. G., Nissen P. (2011) P-ATPases. Annu. Rev. Biophys. 40, 243–266 [DOI] [PubMed] [Google Scholar]
- 2. Toyoshima C., Inesi G. (2004) Structural basis of ion pumping by Ca2+-ATPase of the sarcoplasmic reticulum. Annu. Rev. Biochem. 73, 269–292 [DOI] [PubMed] [Google Scholar]
- 3. Morth J. P., Pedersen B. P., Toustrup-Jensen M. S., Sørensen T. L., Petersen J., Andersen J. P., Vilsen B., Nissen P. (2007) Crystal structure of the sodium-potassium pump. Nature 450, 1043–1049 [DOI] [PubMed] [Google Scholar]
- 4. Palmgren M. G., Axelsen K. B. (1998) Evolution of P-type ATPases. Biochim. Biophys. Acta 1365, 37–45 [DOI] [PubMed] [Google Scholar]
- 5. Sørensen D. M., Buch-Pedersen M. J., Palmgren M. G. (2010) Structural divergence between the two subgroups of P5 ATPases. Biochim. Biophys. Acta 1797, 846–855 [DOI] [PubMed] [Google Scholar]
- 6. Schultheis P. J., Hagen T. T., O'Toole K. K., Tachibana A., Burke C. R., McGill D. L., Okunade G. W., Shull G. E. (2004) Characterization of the P5 subfamily of P-type transport ATPases in mice. Biochem. Biophys. Res. Commun. 323, 731–738 [DOI] [PubMed] [Google Scholar]
- 7. Ramirez A., Heimbach A., Gründemann J., Stiller B., Hampshire D., Cid L. P., Goebel I., Mubaidin A. F., Wriekat A. L., Roeper J., Al-Din A., Hillmer A. M., Karsak M., Liss B., Woods C. G., Behrens M. I., Kubisch C. (2006) Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet. 38, 1184–1191 [DOI] [PubMed] [Google Scholar]
- 8. Kwasnicka-Crawford D. A., Carson A. R., Roberts W., Summers A. M., Rehnström K., Järvelä I., Scherer S. W. (2005) Characterization of a novel cation transporter ATPase gene (ATP13A4) interrupted by 3q25-q29 inversion in an individual with language delay. Genomics 86, 182–194 [DOI] [PubMed] [Google Scholar]
- 9. Cronin S. R., Rao R., Hampton R. Y. (2002) Cod1p/Spf1p is a P-type ATPase involved in ER function and Ca2+ homeostasis. J. Cell Biol. 157, 1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bauer A., Kölling R. (1996) Characterization of the SAC3 gene of Saccharomyces cerevisiae. Yeast 12, 965–975 [DOI] [PubMed] [Google Scholar]
- 11. Elble R. (1992) A simple and efficient procedure for transformation of yeasts. BioTechniques 13, 18–20 [PubMed] [Google Scholar]
- 12. Corradi G. R., Adamo H. P. (2007) Intramolecular fluorescence resonance energy transfer between fused autofluorescent proteins reveals rearrangements of the N- and C-terminal segments of the plasma membrane Ca2+ pump involved in the activation. J. Biol. Chem. 282, 35440–35448 [DOI] [PubMed] [Google Scholar]
- 13. Cura C. I., Corradi G. R., Rinaldi D. E., Adamo H. P. (2008) High sensibility to reactivation by acidic lipids of the recombinant human plasma membrane Ca2+-ATPase isoform 4xb purified from Saccharomyces cerevisiae. Biochim. Biophys. Acta 1778, 2757–2764 [DOI] [PubMed] [Google Scholar]
- 14. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 15. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 16. Richards D. E., Rega A. F., Garrahan P. J. (1978) Two classes of site for ATP in the Ca2+-ATPase from human red cell membranes. Biochim. Biophys. Acta 511, 194–201 [DOI] [PubMed] [Google Scholar]
- 17. Winters D. L., Autry J. M., Svensson B., Thomas D. D. (2008) Interdomain fluorescen resonance energy transfer in SERCA probed by cyan-fluorescent protein fused to the actuator domain. Biochemistry 47, 4246–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lenoir G., Williamson P., Puts C. F., Holthuis J. C. (2009) Cdc50p plays a vital role in the ATPase reaction cycle of the putative aminophospholidip transporter Drs2p. J. Biol. Chem. 284, 17956–17967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sørensen T. L., Clausen J. D., Jensen A. M., Vilsen B., Møller J. V., Andersen J. P., Nissen P. (2004) Localization of a K+-binding site involved in dephosphorylation of the sarcoplasmic reticulum Ca2+-ATPase. J. Biol. Chem. 279, 46355–46358 [DOI] [PubMed] [Google Scholar]
- 20. Jensen A. M., Sørensen T. L., Olesen C., Møller J. V., Nissen P. (2006) Modulatory and catalytic modes of ATP binding by the calcium pump. EMBO J. 25, 2305–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rossi J. P., Rega A. F., Garrahan P. J. (1985) Compound 48/80 and calmodulin modify the interaction of ATP with the (Ca2+ + Mg2+)-ATPase of red cell membranes. Biochim. Biophys. Acta 816, 379–386 [DOI] [PubMed] [Google Scholar]
- 22. Tan J., Zhang T., Jiang L., Chi J., Hu D., Pan Q., Wang D., Zhang Z. (2011) Regulation of intracellular manganese homeostasis by Kufor-Rakeb syndrome associated ATP13A2. J. Biol. Chem. 286, 29654–29662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Echarte M. M., Rossi R. C., Rossi J. P. (2007) Phosphorylation of the plasma membrane calcium pump at high ATP concentration. On the mechanis of ATP hydrolysis. Biochemistry 46, 1034–1041 [DOI] [PubMed] [Google Scholar]
- 24. Suzuki C., Shimma Y. I. (1999) P-type ATPase Spf1 mutants show a novel resistance mechanism for the killer toxin SMKT. Mol. Microbiol. 32, 813–823 [DOI] [PubMed] [Google Scholar]
- 25. Cronin S. R., Khoury A., Ferry D. K., Hampton R. Y. (2000) Regulation of HMG-CoA reductase degradation requires the P-type ATPase Cod1p/Spf1p. J. Cell Biol. 148, 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tipper D. J., Harley C. A. (2002) Yeast genes controlling responses to topogenic signals in a model transmembrane protein. Mol. Biol. Cell 13, 1158–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vashist S., Frank C. G., Jakob C. A., Ng D. T. (2002) Two distinctly localized P-type ATPases collaborate to maintain organelle homeostasis required for glycoprotein processing and quality control. Mol. Biol. Cell 13, 3955–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vallipuram J., Grenville J., Crawford D. A. (2010) The E646D-ATP13A4 mutation associated with autism reveals a defect in calcium regulation. Cell Mol. Neurobiol. 30, 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Tezanos Pinto F., Corradi G. R., Adamo H. P. (2011) The human P5B-ATPase ATP13A2 is not a Ca2+ transporting pump. J. Life Sci. 5, 1–6 [Google Scholar]
- 30. Schmidt K., Wolfe D. M., Stiller B., Pearce D. A. (2009) Cd2+, Mn2+, Ni2+ and Se2+ toxicity to Saccharomyces cerevisiae lacking YPK9p the orthologue of human ATP13A2. Biochem. Biophys. Res. Commun. 383, 198–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gitler A. D., Chesi A., Geddie M. L., Strathearn K. E., Hamamichi S., Hill K. J., Caldwell K. A., Caldwell G. A., Cooper A. A., Rochet J. C., Lindquist S. (2009) α-Synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nat. Genet. 41, 308–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schneider S. A., Paisan-Ruiz C., Quinn N. P., Lees A. J., Houlden H., Hardy J., Bhatia K. P. (2010) ATP13A2 mutations (PARK9) cause neurodegeneration with brain iron accumulation. Mov. Disord. 25, 979–984 [DOI] [PubMed] [Google Scholar]
- 33. Lustoza A. C., Palma L. M., Facanha A. R., Okorokov L. A., Okorokova-Facanha A. L. (2011) P5A-type ATPase Cta4p is essential for Ca2+ transport in the endoplasmic reticulum of Schizosaccharomyces pombe. PLoS ONE 6, e27843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adamo H. P., Rega A. F., Garrahan P. J. (1988) Pre-steady-state phosphorylation of the human red cell Ca2+-ATPase. J. Biol. Chem. 263, 17548–17554 [PubMed] [Google Scholar]
- 35. Ronald J., Clarke R. J., Apell H. J., Kong B. Y. (2007) Allosteric effect of ATP on Na+, K+-ATPase conformational kinetics. Biochemistry 46, 7034–7044 [DOI] [PubMed] [Google Scholar]
- 36. Froehlich J. P., Heller P. F. (1985) Transient-state kinetics of the ADP-insensitive phosphoenzyme in sarcoplasmic reticulum. Implications for transient-state calcium translocation. Biochemistry 24, 126–136 [DOI] [PubMed] [Google Scholar]
- 37. Mazzitelli L. R., Rinaldi D. E., Corradi G. R., Adamo H. P. (2010) The plasma membrane Ca2+ pump catalyzes the hydrolysis of ATP at low rate in the absence of Ca2+. Arch. Biochem. Biophys. 495, 62–66 [DOI] [PubMed] [Google Scholar]
- 38. Coleman J. A., Vestergaard A. L., Molday R. S., Vilsen B., Peter Andersen J. (2012) Critical role of a transmembrane lysine in aminophospholipid transport by mammalian photoreceptor P4-ATPase ATP8A2. Proc. Natl. Acad. Sci. U.S.A. 109, 1449–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Greie J. C. (2011) The KdpFABC complex from Escherichia coli. A chimeric K+ transporter merging ion pumps with ion channels. Eur. J. Cell Biol. 90, 705–710 [DOI] [PubMed] [Google Scholar]
- 40. Niggli V., Sigel E. (2008) Anticipating antiport in P-type ATPases. Trends Biochem. Sci. 33, 156–160 [DOI] [PubMed] [Google Scholar]
- 41. Møller A. B., Asp T., Holm P. B., Palmgren M. G. (2008) Phylogenetic analysis of P5 P-type ATPases, a eukaryotic lineage of secretory pathway pumps. Mol. Phylogenet. Evol. 46, 619–634 [DOI] [PubMed] [Google Scholar]