Background: RPE65 retinol isomerohydrolase is essential for vision, but its catalytic mechanism is poorly understood.
Results: Mutating aromatic residues in the substrate cleft abolishes or modifies RPE65 activity to make 13-cis- instead of 11-cis-retinol.
Conclusion: The RPE65 substrate-binding cleft is molded by aromatic residues to promote specific isomerization of retinol.
Significance: This further defines the vitamin A isomerization step central to the vertebrate visual cycle.
Keywords: Enzyme Catalysis, Enzyme Mechanisms, Molecular Modeling, Retinal Metabolism, Retinoid, Vitamin A, 11-cis-Retinol, RPE65 Isomerase, Retinyl Ester, Visual Cycle
Abstract
Previously, we showed that mutating RPE65 residue Phe-103 preferentially produces 13-cis-retinol instead of 11-cis-retinol, supporting a carbocation/radical cation mechanism of retinol isomerization. We asked whether this modulation of specificity can occur with residues other than Phe-103 and what role it plays in substrate binding and isomerization. We modeled the substrate-binding cleft of RPE65 to identify residues lining its surface. Many are phenylalanines and tyrosines, including three Phe residues (Phe-61, Phe-312, and Phe-526) forming an arch-like arrangement astride the cleft and Tyr-338. Also, Phe-418 sits at the neck of the cleft, lending a bend to the volume enclosed by the cleft. All mutations of Phe-61, Phe-312, and Phe-418 result in severely impaired or inactive enzyme. However, mutation of Phe-526 and Tyr-338, like Phe-103, decreases 11-cis-retinol formation, whereas increasing the 13-cis isomer. Significantly, 2 of these 3 residues, Phe-103 and Tyr-338, are located on putatively mobile interstrand loops. We propose that residual densities located in the binding cleft of the RPE65 structure represents a post-cleavage snapshot consistent not only with a fatty acid product, as originally modeled, but also an 11-cis-retinol product. Substrate docking simulations permit 11-cis- or 13-cis-retinyl ester binding in this relatively closed cleft, with the latter favored in F103L, F526A, and Y338A mutant structures, but prohibit binding of all-trans-retinyl ester, suggesting that isomerization occurs early in the temporal sequence, with O-alkyl ester cleavage occurring later. These findings provide insight into the mechanism of isomerization central to the visual cycle.
Introduction
The process of vision is initiated by photoisomerization of 11-cis-retinylidene bound to photoreceptor visual pigments, such as rhodopsin and cone opsins, to the all-trans conformation. Rhodopsin is thereby activated to metarhodopsin, initiating the phototransduction cascade. All-trans-retinal is ultimately released following the decay of metarhodopsin. To keep the visual pigments in a state capable of responding to light, they must be regenerated with 11-cis-retinal. This is achieved in the vertebrate retina by a process termed the visual cycle (1, 2). The visual cycle is localized in the retinal pigment epithelium (RPE)2 and retina and involves an assemblage of enzymes and isomer-specific binding proteins, some specific to the retina/RPE and some not. In the retinal visual cycle, RPE65 (3, 4) is the enzyme that isomerizes and cleaves all-trans-retinyl esters (all-trans-RE) into 11-cis-retinol (11-cis-ROL) (5–7), which is then converted to the 11-cis-retinal chromophore of opsins. Recently, our laboratory has shown that RPE65 acts as a leaky isomerase that can isomerize all-trans-RE to both 11-cis-ROL and 13-cis-ROL (8). Although there is no known physiological role for 13-cis-ROL, and 13-cis-retinal cannot regenerate opsin, it has been shown to accumulate as esters in Rdh5 knock-out mouse RPE (9). RDH5 is the primary short-chain retinol dehydrogenase in RPE that oxidizes 11-cis-ROL to 11-cis-retinal (10). Its loss may disrupt the process of mass action crucial for 11-cis-retinoid production in the visual cycle, thereby allowing accumulation of 13-cis-retinoids (9). In our previous studies, we could change the ratio of 11-cis- to 13-cis-ROL with specific conservative mutations of RPE65; for example, F103L increased the proportion of 13-cis-ROL, whereas T147S increased the proportion of 11-cis-ROL (8). We concluded that the mechanism underlying the leaky isomerization implicates a carbocation or a radical cation intermediate with loss of polyene bond order allowing for production of either 11-cis-ROL or 13-cis-ROL by RPE65 and that ultimate specificity for 11-cis isomers occurs downstream by a process of mass action applied by 11-cis-specific binding proteins such as CRALBP and apoprotein opsins (8), as suggested previously (9, 11). We hypothesize that the aromatic side chain-rich environment of the RPE65 substrate-binding cleft is primarily responsible for favoring an 11-cis-specific outcome.
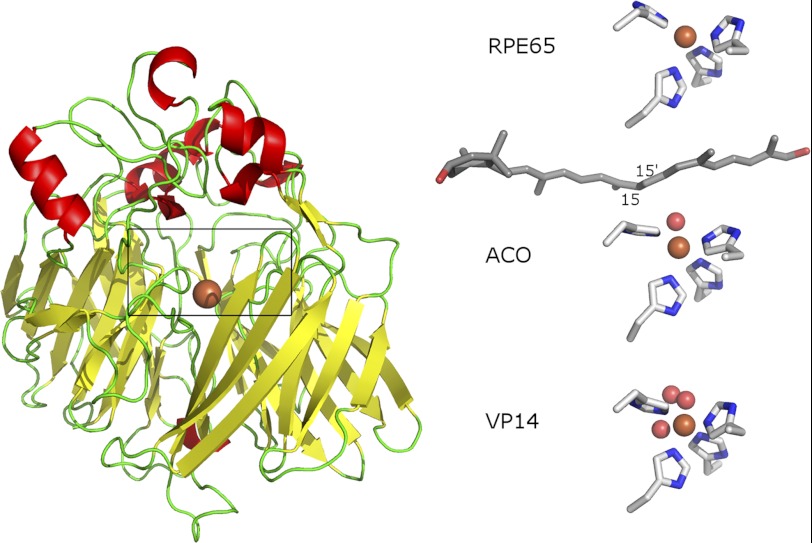
At present, there are crystal structures for three members of the carotenoid oxygenase family: a cyanobacterial apocarotenoid oxygenase (ACO (12, 13)), the plant 9-cis-epoxycarotenoid dioxygenase VP14 (14), and bovine RPE65 (15). All three show a core seven-bladed β propeller structure centered on an iron atom coordinated by 4 conserved histidines. However, the substrate-binding cleft in each differs with respect to what one might term a snapshot of the cleft occupancy (Fig. 1). The ACO has a closed, compact binding cleft in which, in the presence of substrate, a density was seen that could be modeled into the cleft as the 13,14-13′,14′-di-cis-apocarotenol with the 15-15′ bond centered over the iron, although the β-ionone ring was not visible (12). In contrast, the VP14 crystal has a dioxygen coordinated in the sixth coordination position of the iron, but no carotenoid substrate appears evident, and the binding cleft is open (14). The RPE65 crystal structure has the most restricted binding cleft of all, but no modeled substrate (15).
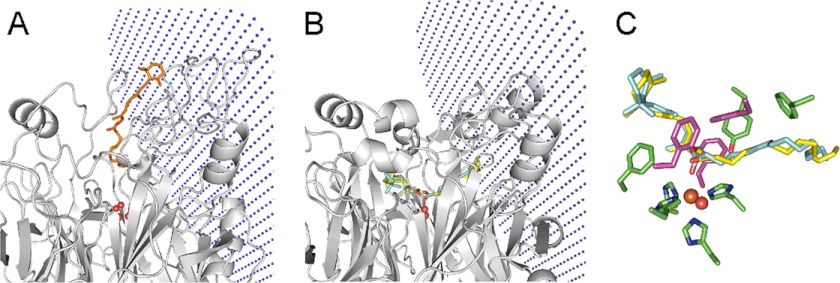
FIGURE 1.
Carotenoid oxygenase crystal structures. The structure of RPE65 includes a seven-bladed β propeller fold showing iron at the center. Above the propeller, many loops are arrayed to complete the structure. The boxed region is magnified on the right to show the contents of the three crystal structures, respectively. Iron is shown as orange sphere, iron-coordinating histidines are shown as sticks in elemental colors, ACO apocarotenol is in gray, and oxygen is shown as red spheres.
In the present study, we have analyzed the RPE65-binding cleft to identify additional residues that may contribute to the process of substrate binding and isomerization, extending the mutational analyses with attention to the 11-cis-ROL/13-cis-ROL ratio. The phenylalanines surrounding the iron center (Phe-61, Phe-312, and Phe-526) were considered likely to be of importance in the substrate binding, comparable with the paralogous residues in VP14 (14). We have constructed homology models of RPE65 based on the more open conformations of ACO and VP14 and carried out docking simulations of substrate (all-trans-RE), potential intermediates (11-cis-RE and 13-cis-RE), and products (11-cis-ROL, 13-cis-ROL, and palmitate) on all three structures. By comparison of these simulations in the three structures, contributions of the relevant residues to the reaction mechanism of RPE65 are proposed. Docking was also investigated in mutant RPE65 models to clarify possible mechanisms for the changes in activities seen in these mutants.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis of RPE65
QuikChange XL site-directed mutagenesis kit (Agilent, La Jolla, CA) was used for mutagenesis of the RPE65 open reading frame cloned in pVitro2 (InvivoGen, San Diego, CA). Primer sequences for mutagenesis are given in supplemental Table S1. Mutants were verified by sequence analysis of DNA minipreps (Northwoods DNA, Solway, MN). Validated mutant and wild-type plasmids were purified by Qiagen purification kits (Qiagen, Valencia, CA).
Transient Transfection and Cell Culture
Cell culture methods and transient transfection protocols were previously published (7, 8). In a typical experiment, 3 × 107 HEK293-F (Invitrogen) cells were transfected with 30 μg of pVitro2 plasmid (containing RPE65 (wild type or mutant) and CRALBP open reading frames) and 30 μg of pVitro3 (InvivoGen) plasmid (containing lecithin-retinol acyltransferase (LRAT)) and retinol dehydrogenase 5 (RDH5)) open reading frames in the presence of 40 μl of 293Fectin transfection reagent (Invitrogen), all in a total volume of 30 ml. 24 h after transfection, all-trans-ROL was added to a final concentration of 2.5 μm, and the cells were cultured for a further 7 h and then harvested for analysis. Immunoblot analysis was carried out as described previously (8).
Retinoid Extractions and HPLC
Culture fractions of 29-ml volumes of transfected HEK293-F cells were centrifuged, cells were harvested, and retinoids were extracted and saponified as described previously (7). Isomeric retinols were analyzed on 5-μm particle LiChrospher (Alltech, Deerfield, IL) normal phase columns (2 × 250 mm) on an isocratic HPLC system equipped with a diode array UV-visible detector (Agilent 1100/1200 series, Agilent Technologies, New Castle, DE), following Landers and Olson (16) as modified by us (7). Data were analyzed using ChemStation32 software (Agilent).
Construction of Models and Ligand Docking Simulations
A structural alignment template was made from the crystal structures of ACO (Protein Data Bank (PDB) ID: 2BIW), VP14 (PDB ID: 3NPE), and bovine RPE65 (PDB ID: 3KVC). The murine RPE65 sequence was then aligned to this template and submitted to the Swiss Model server (17, 18) to obtain a generalized model that covered loops missing in the crystal structures (residues 109–126 are missing in the bovine RPE65 structure). The substrate-binding cleft was visualized using the MSMS reduced surface package in Chimera (19). It was noted that the three crystal structures appear to represent differing degrees of openness at the presumed substrate-binding cleft, VP14 being the most open and the bovine RPE65 being the most closed. Therefore, an additional open murine RPE65 model based on the VP14 structure was also constructed with the water and dioxygen coordinated to the iron, as seen in VP14 structure. Three-dimensional models of ligands were either obtained from the PubChem database when available or made using the PRODRG program (20, 21). Ligand dockings of substrate (all-trans-retinyl palmitate), intermediates (11-cis- and 13-cis-retinyl palmitate), and products (11-cis-ROL and palmitate) were carried out using Autodock Vina (22). In general, the ligand constraints chosen allowed for all possible torsions. Docking of all-trans, 11-cis, and 13-cis isomers of both esters and retinols were carried out on both the bovine RPE65 structure and the murine models. The top hits from the docking simulations were chosen based on two criteria. The first was that they were in the correct orientation, and the second was that the retinyl O-alkyl bond was positioned over the iron center. Except where noted, models were visualized using the PyMOL Molecular Graphics System (Version 1.4, Schrödinger, LLC). The σ-A-weighted difference density for 3KVC, calculated using structure factors deposited by Golczak et al. (31) on the PDB, was obtained from the EDS Uppsala Electron Density Server (EDS) (23).
RESULTS
Structural Modeling
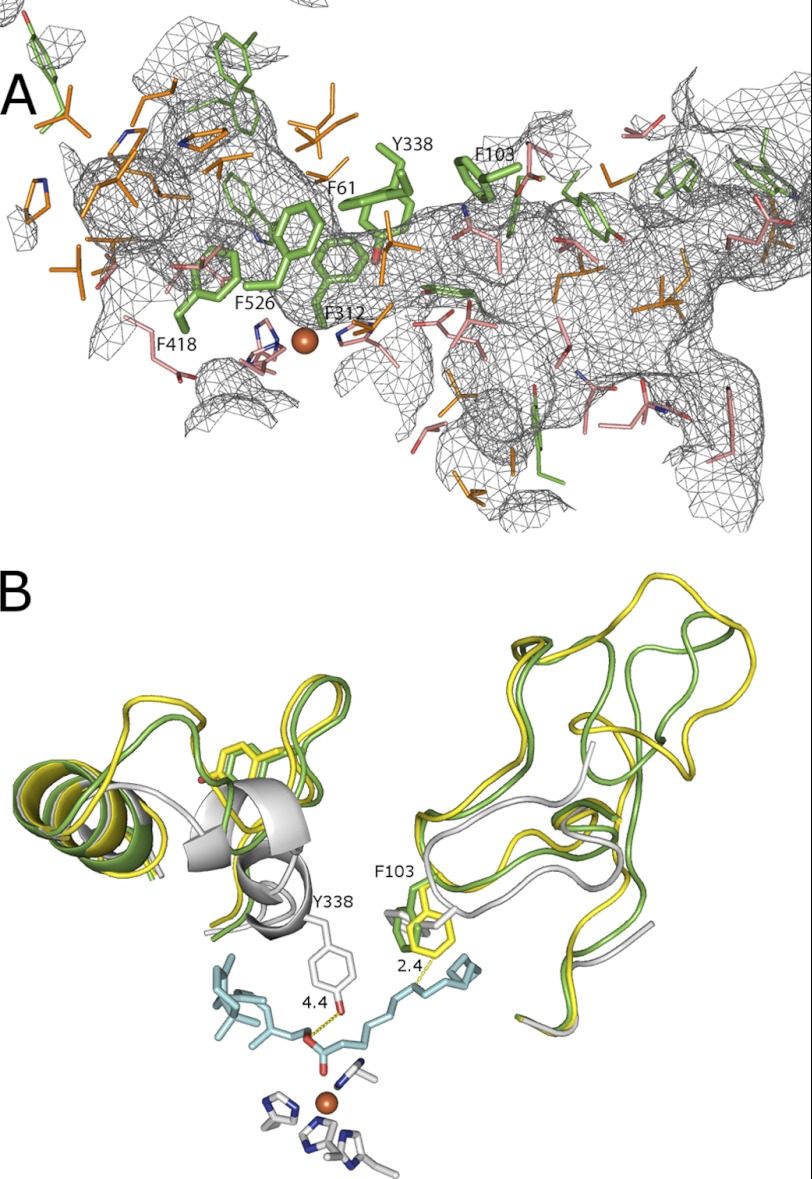
Examination of the three crystal structures of the carotenoid oxygenase family showed that they share a common seven-bladed β-propeller fold, although the three proteins are less than 30% identical in sequence. For the propeller region, the root mean square deviation between RPE65 and ACO is 1.1 Å (for 209 Cα atoms) and between RPE65 and VP14 is 1.4 Å (for 206 Cα atoms). A cleft was observed above the propeller region, over the iron center that was assumed to be the substrate-binding site. Comparison of these clefts showed the RPE65 cleft to be more compact than the ACO cleft. VP14, which contains a dioxygen on the sixth iron coordination position, showed the most extended conformation (Fig. 2). The differences in cleft volumes can be explained by contributions from the dynamic loops in the upper half of the proteins that likely undergo order-disorder transition. In RPE65, these include residues 108–127 (missing in both crystal structures), 258–281 (missing and high B-factor in 3KVC and 3FSN, respectively), and 332–361. This would be modified by the state of the protein during crystallization, i.e. with or without substrate or product. The residues comprising the surface of the cavity in RPE65 were identified using the MarkUs Cavity Server (24). The cavity overlaps with the residual density, as seen in the RPE65 crystal structure, and the surrounding accessible area (supplemental Fig. S1). A total of 62 residues were identified as contributing to this surface (Fig. 3A; supplemental Table S2), with 6 of these contributing to the iron-coordinating complex (3 histidine (out of 4 coordinating) and 3 glutamic acid residues). Of the remaining, 10 were phenylalanine, 5 were tyrosine, 2 were tryptophan, 9 were leucine, and 3 were isoleucine residues. Thus, aromatic residues contribute about 30% of the cavity internal surface with the majority being due to phenylalanines or tyrosines. In the three structures, an arch of phenylalanines (with one leucine replacement in ACO) surmounts the iron center: in RPE65, Phe-61, Phe-312, and Phe-526; in ACO, Phe-69, Phe-311, and Leu-483; and in VP14, Phe-171, Phe-411, and Phe-589. The RPE65 arch residues include Phe-61 that is part of the FDG conserved motif and 2 residues immediately adjacent to two of the iron-coordinating histidines (Phe-312 and Phe-526). Therefore, these 3 residues were selected for mutational studies. (Phe-61 was addressed in our previous study (8), but we provide additional insight into its role herein.) In addition, comparisons of the mouse RPE65 models based on ACO and VP14 indicated a change in the positions of both Phe-103 (mutation of which was previously shown to favor 13-cis-ROL production, (8)) and Tyr-338 in the different conformations. This suggests that the loops on which these residues are located, amino acids 84–133 for Phe-103 and amino acids 333–361 for Tyr-338, are relatively dynamic or mobile (Fig. 3B). This is known to be the case for the amino acids 82–133 loop as the center residues of this loop (amino acids 109–126) were poorly ordered in the RPE65 crystal (15). Loop residues Thr-101, Glu-102, Phe-103, Glu-127, Val-128, and Thr-129 contribute to the internal surface of the cleft (supplemental Table S2). Secondary structure topology of the amino acids 333–361 loop reveals three predicted α helices (15), and residues in two of these helices (Val-337, Tyr-338, Leu-341, and Leu-346) contribute to the internal surface of the cleft (supplemental Table S2). Another residue of interest was Phe-418. Phe-418 is a paralog of residues Val-478 and Phe-409 in VP14 and ZmCCD1, respectively, that appear to regulate substrate specificity in these enzymes (14). Because of these additional considerations, we analyzed Tyr-338 and Phe-418.
FIGURE 2.
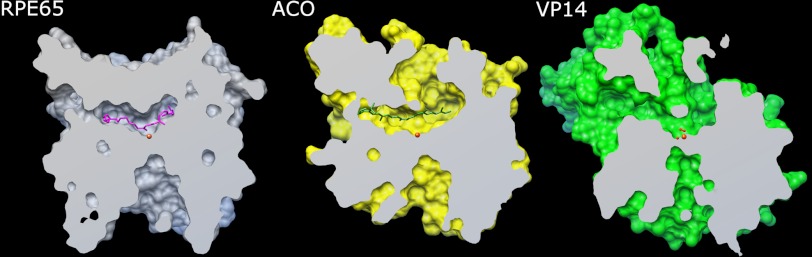
Binding clefts of the three crystal structures. Sliced views vertically bisecting the binding clefts of bovine RPE65, Synechocystis ACO, and maize VP14 at the plane of the iron atom were made using the MSMS package in Chimera. Ligands are shown as sticks (RPE65 with docked retinyl palmitate in magenta, ACO apocarotenol in green), iron is shown as orange sphere, and oxygen is shown as red spheres.
FIGURE 3.
Residues enclosing the RPE65 cavity. A, the gray mesh represents the cavity; residues that enclose the cavity are shown as lines (aromatic residues are in green; aliphatic residues are in orange; and hydrophilic residues are in peach). Residues examined in this study are represented as thick sticks. B, positions of Phe-103 and Tyr-338 in the bovine RPE65 crystal structure (gray) and in the mouse RPE65 models built on ACO (yellow) and VP14 (green) are shown. Models were visualized using PyMOL.
Aside from changes to residues that do not cause protein folding alterations, we identified two groups of residue changes: (i) where mutation results in total loss of activity and (ii) where mutation results in modulation/alteration of enzyme specificity. The former are associated with mutation of residues associated with catalytic function or with family conserved structural elements, whereas the latter are associated with substrate-binding and/or variable loops. In the inactivating mutations, 13-cis-retinol production was comparable with thermal levels, as described previously (8).
Inactivating Mutations
Mutations of the “Phe arch” residues Phe-61 or Phe-312 to other conservative substitutions severely diminished or eliminated all catalytic activity (Table 1). Phe-61, part of the FDG motif seen in all carotenoid oxygenases, is located on the top of the Phe arch. It is positioned right above the iron, on the other side of the substrate in the cleft. Phe-61 displays negligible activity upon any substitution, suggesting a conserved role for this residue. Tyrosine, due to its hydrophilic nature, may destroy the putative carbocation/cation radical intermediate, and tryptophan may be too bulky a substitution for that location, sterically hindering the substrate. Phe-312 is also irreplaceable, suggesting a high invariance at this position. Due to its location and proximity to the substrate, Phe-312 possibly plays a role in steric shielding to allow proper hydrolysis of the C–O bond. Similarly, mutation of Phe-418 to Ala and other residues, including aromatics Tyr and Trp, resulted in inactivation, whereas the Leu mutant retained only 4% of WT 11-cis-retinol production activity (Table 1). We suggest that Phe-418 may function by forming a bottleneck in the volume of the cleft that constrains the intermediate in precise conformational register to ensure isomerization and/or O-alkyl cleavage (see below).
TABLE 1.
Expression and cis-retinol production of HEK293-F cells with mutants inactivating RPE65 activity
| Expressiona | ± S.D. | 11-cis-ROLb | ± S.D. | 13-cis-ROLb | ± S.D. | |
|---|---|---|---|---|---|---|
| WT | 1 | 1 | 1 | |||
| F61A | 0.11 | 0.04 | 0 | 0 | 0.35 | NAc |
| F61Y | 0.56 | 0.08 | 0.08 | 0.01 | 0.42 | 0.07 |
| F61L | 0.39 | 0.15 | 0.01 | 0.01 | 0.37 | 0.05 |
| F61W | 0.22 | 0.05 | 0.02 | 0.00 | 0.38 | 0.06 |
| F312L | 0.21 | 0.11 | 0.03 | 0.01 | 0.32 | 0.04 |
| F312Y | 0.2 | 0.08 | 0.02 | 0.01 | 0.30 | 0.03 |
| F312W | 0.3 | 0.1 | 0 | 0 | 0.29 | 0.03 |
| F418A | 0.13 | 0 | 0 | 0 | 0.51 | 0.11 |
| F418L | 0.18 | 0 | 0.04 | 0 | 0.42 | 0.1 |
| F418Y | 0.12 | 0.08 | 0 | 0 | 0.4 | 0.08 |
| F418W | 0.15 | 0.04 | 0 | 0 | 0.39 | 0.11 |
a Data are given as -fold change from wild-type expression ± S.D.; n ≥ 3.
b Data are given as -fold wild-type RPE65 isomerase activity ± S.D.; n ≥ 3.
c NA, not applicable.
Modulating Mutations
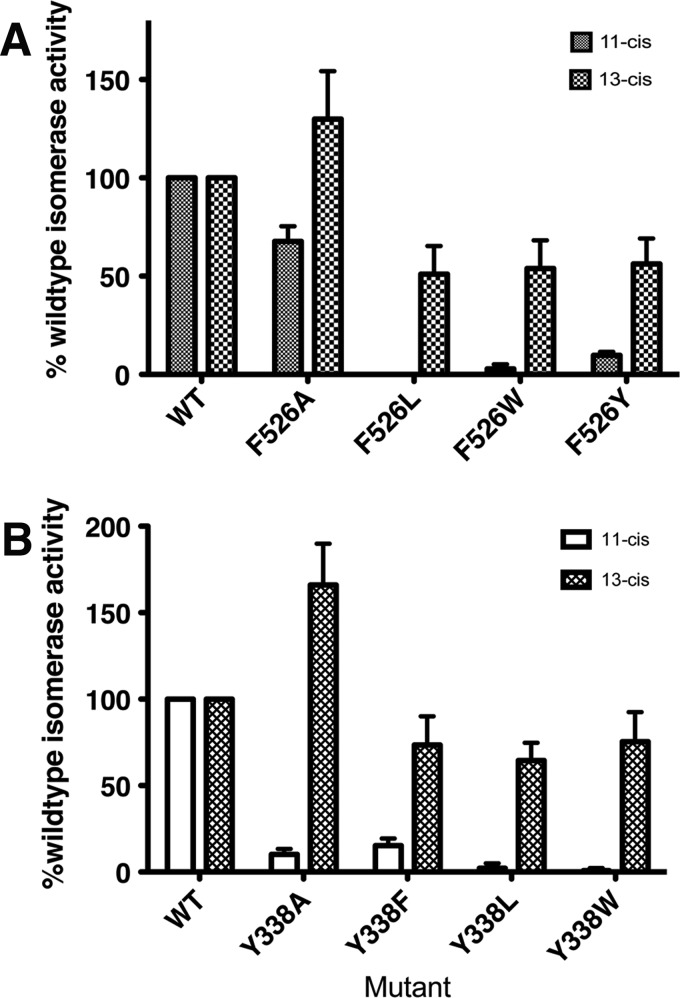
In contrast to the inactivating effects of mutations at Phe-61 and Phe-312, substitution at the 3rd Phe arch residue Phe-526 altered but did not abolish activity. Mutation with any residue other than alanine decreased 11-cis-ROL (but not 13-cis-ROL) production in excess of what would be expected by the lower expression levels of these mutants (Table 2 and Fig. 4A). However, the F526A mutation promoted leaky isomerase activity, selectively increasing 13-cis-ROL production with a concomitant reduction in 11-cis-ROL production. Tyr-338, one of the dynamic residues seen in the crystal structures, selectively reduced 11-cis-ROL production in all its mutants. Y338L, Y338F, and Y338W produced the 13-cis-ROL product in similar amounts to wild type when corrected for expression (Table 2 and Fig. 4B), and Y338A displayed increased 13-cis-ROL (1.63-fold increase) similar to the F103L or F103I mutations previously reported, although somewhat more severe in the reduction of 11-cis production (8). This result suggests that Tyr-338 substitutions also reduce 11-cis-ROL isomerization specifically, but that 13-cis isomerization is essentially unaffected by most substitutions or increased by the Y338A mutation. We observed this increased 13-cis-retinol production by the Y338A mutant to be elevated when compared with thermal production of 13-cis-retinol either in the presence or in the absence of wild-type RPE65 at all time points analyzed (supplemental Fig. S2). We suggest that the major effect of Tyr-338, just as in Phe-103 and Phe-526, is to stabilize the cis isomers as they are being positioned in the cleft prior to cleavage; 13-cis-RE, being lower in its energy of formation, may be more favored to bind in the cleft of these mutants as they are more permissive.
TABLE 2.
Expression and cis-retinol production of HEK293-F cells with mutants modulating RPE65 activity
| Expressiona | ± S.D. | 11-cis-ROLb | ± S.D. | 13-cis-ROLb | ± S.D. | 11-cis expression | 13-cis expression | |
|---|---|---|---|---|---|---|---|---|
| WT | 1 | 1 | 1 | |||||
| F526A | 0.88 | 0.12 | 0.68 | 0.08 | 1.30 | 0.24 | 0.77 | 1.48 |
| F526Y | 0.54 | 0.09 | 0.10 | 0.02 | 0.56 | 0.13 | 0.18 | 1.04 |
| F526L | 0.46 | 0.10 | 0 | 0 | 0.51 | 0.14 | 0.00 | 1.11 |
| F526W | 0.38 | 0.05 | 0.03 | 0.02 | 0.54 | 0.14 | 0.07 | 1.42 |
| Y338A | 0.31 | 0.11 | 0.10 | 0.03 | 1.63 | 0.22 | 0.31 | 5.25 |
| Y338L | 0.29 | 0.10 | 0.03 | 0.03 | 0.62 | 0.10 | 0.09 | 2.14 |
| Y338W | 0.24 | 0.11 | 0.01 | 0.02 | 0.74 | 0.16 | 0.05 | 3.08 |
| Y338F | 0.08 | 0.06 | 0.15 | 0.04 | 0.71 | 0.15 | 1.85 | 8.83 |
| F103L | 0.61 | 0.12 | 0.28 | 0.01 | 1.22 | 0.11 | 0.46 | 2.00 |
| F103W | 0.57 | 0.11 | 0.24 | 0.01 | 0.61 | 0.04 | 0.42 | 1.07 |
| F103Y | 0.33 | 0.07 | 0.21 | 0.03 | 0.36 | 0.06 | 0.64 | 1.09 |
| F103I | 0.27 | 0.06 | 0.03 | 0.01 | 0.44 | 0.04 | 0.11 | 1.63 |
a Data are given as -fold change from wild-type expression ± S.D.; n ≥ 3.
b Data are given as -fold wild-type RPE65 isomerase activity ± S.D.; n ≥ 3.
FIGURE 4.
Mutations of Phe-526 and Tyr-338 modulate product isomer specificity of RPE65 isomerase activity. Substitution of Phe-526 and Tyr-338 with alanine, leucine, or aromatic residues modulates RPE65 isomerase product specificity by raising 13-cis relative to 11-cis production. A and B, RPE65 isomerase activity in Phe-526 (A) and Tyr-338 (B) mutants. 11-cis- and 13-cis-ROL production in HEK293-F cells transfected with constructs expressing wild type and mutants of dog RPE65 Phe-526 or Tyr-338 was determined. Mutant activities are expressed as the percentage of wild-type RPE65 activity ± S.D. (n ≥ 4).
Modeling of Substrate and Intermediate Docking in Wild-type RPE65
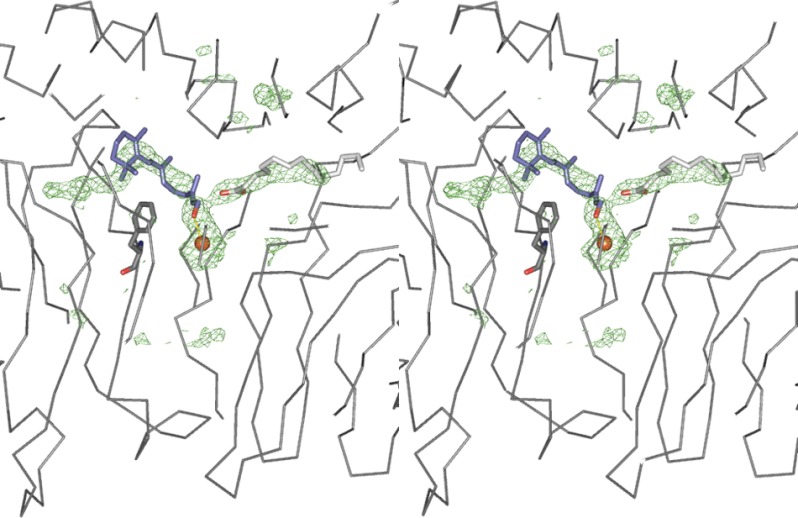
The bovine RPE65 crystal structure, derived from native material, contains residual densities in cleft pockets. When we docked the products, 11-cis-ROL and palmitate, they traced the density areas (Fig. 5), suggesting that these residual densities may represent the presence of the products in the crystal. There is a bend in the residual electron density going around Phe-418 (highlighted in Fig. 5), indicating that this residue helps shape the cavity. To evaluate the docking simulations, two assumptions were made: first, that the retinyl ester had to be positioned in the phenylalanine arch prior to cleavage, and second, that the retinyl ester O-alkyl bond should be positioned over the iron catalytic center in a specific orientation. Docking of all-trans-retinyl palmitate, however, was unable to meet the basic criteria as it was never docked appropriately over the iron center in any simulation. However, when all-trans-retinyl palmitate was docked on the mouse RPE65 model based on the open VP14 crystal, the highest affinity docking placed it in a cleft between the two loops carrying Phe-103 and Tyr-338 (Fig. 6A). Use of the mouse model was required to model the loop (carrying Phe-103), which was unresolved in the bovine RPE65 crystal structure, as indicated previously (15). (This loop is almost perfectly conserved between bovine and mouse RPE65s with only one conservative substitution at amino acid 124 (Arg in bovine replaced by Lys in mouse)). In contrast, both 11-cis-retinyl palmitate and 13-cis-retinyl palmitate docked in the bovine RPE65 crystal structure in the presence of water modeled into coordination position 5 of the iron (Fig. 6, B and C). However, if dioxygen was modeled into position 6 (as in the VP14 crystal), no retinyl palmitate docking within the phenylalanine arch was obtained (not shown). These results suggest that the early steps of isomerization of retinyl esters occur in the capture and binding of the substrate, thus allowing it to fit appropriately over the iron center for cleavage.
FIGURE 5.
Product docking in RPE65. Docking of 11-cis-ROL (purple) and palmitate (light gray) in the cleft of the bovine RPE65 crystal structure is shown. The darker green mesh represents σ-A-weighted difference density (contoured at 3σ) as observed in the 3KVC crystal structure. Phe-418 (dark gray stick), 11-cis-retinol (blue), palmitate (light gray), and iron (orange) are depicted.
FIGURE 6.
Substrate and intermediate docking in RPE65. A, docking of all-trans-retinyl palmitate in the mouse RPE65 model based on the VP14 open structure. The dioxygen and water present in the VP14 structure are shown (red spheres). The membrane is shown as blue dotted plane. B, docking of 11-cis- and 13-cis-retinyl RE in the presumed substrate cleft of the mouse RPE65 model with water in the fifth position. C, enlarged view of B, showing overlay of 11-cis- and 13-cis-retinyl palmitate and residues mutated in this study. 11-cis-Retinyl palmitate (cyan), 13-cis-retinyl palmitate (yellow), iron (orange), water (red), histidines His-180, His-241, His-313, and His-527 (green carbons and blue nitrogens), phenylalanines Phe-312, Phe-526, and Phe-61 (sticks in magenta), and tyrosine Tyr-338 (green carbons and red oxygen).
Modeling of Substrate and Intermediate Docking in Mutant RPE65
Docking simulations were also carried out on mutants of the various residues studied. Both F61L and F312L showed docked substrates with the ester bond significantly off position when compared with wild type (Fig. 7, A and C). Leucines occur naturally in paralogous positions to Phe-61 and Phe-312 in some carotenoid oxygenases of plants and in the carotenoid isomero-oxygenases of insects (25), respectively. However, in RPE65, mutations to these residues render RPE65 inactive, which suggests that the role of these residues is to orient the substrate in such a way that the ester cleavage activity of the enzyme can occur. F418L showed docked substrate with distortions around the β-ionone to 9-carbon position stretch of the retinyl moiety, thus indirectly affecting the positioning of the ester bond (Fig. 7, A and C). In the case of F526A, which allowed 13-cis-ROL production whereas inhibiting 11-cis-ROL production, our docking results suggest that the consequence of increased space in the mutant probably allows for the 13-cis-RE to be positioned in the effective orientation, thus allowing 13-cis product formation (Fig. 7, B and D). The reduction of 11-cis isomerization activity in this mutant could reflect the requirement of an aromatic residue to stabilize the 11-cis isomer as the docking simulation appears to be feasible. Docking of both 11-cis-retinyl palmitate and 13-cis-retinyl palmitate in F526L is also essentially the same as in the wild type. However, given the complete loss of activity with respect to 11-cis-ROL production, it is probable that formation and/or stabilization of the 11-cis isomer is compromised by a leucine mutation in this member of the phenylalanine arch. Likewise, the F103L mutation and the Y338A mutation showed apparently normal docking for both isomers (Fig. 7, B and D), suggesting a failure of isomerization to 11-cis-ROL in these mutants. Overall, the dockings show a clustering of the substrate conformations in inactivating mutants and modulating mutants into two respective groups.
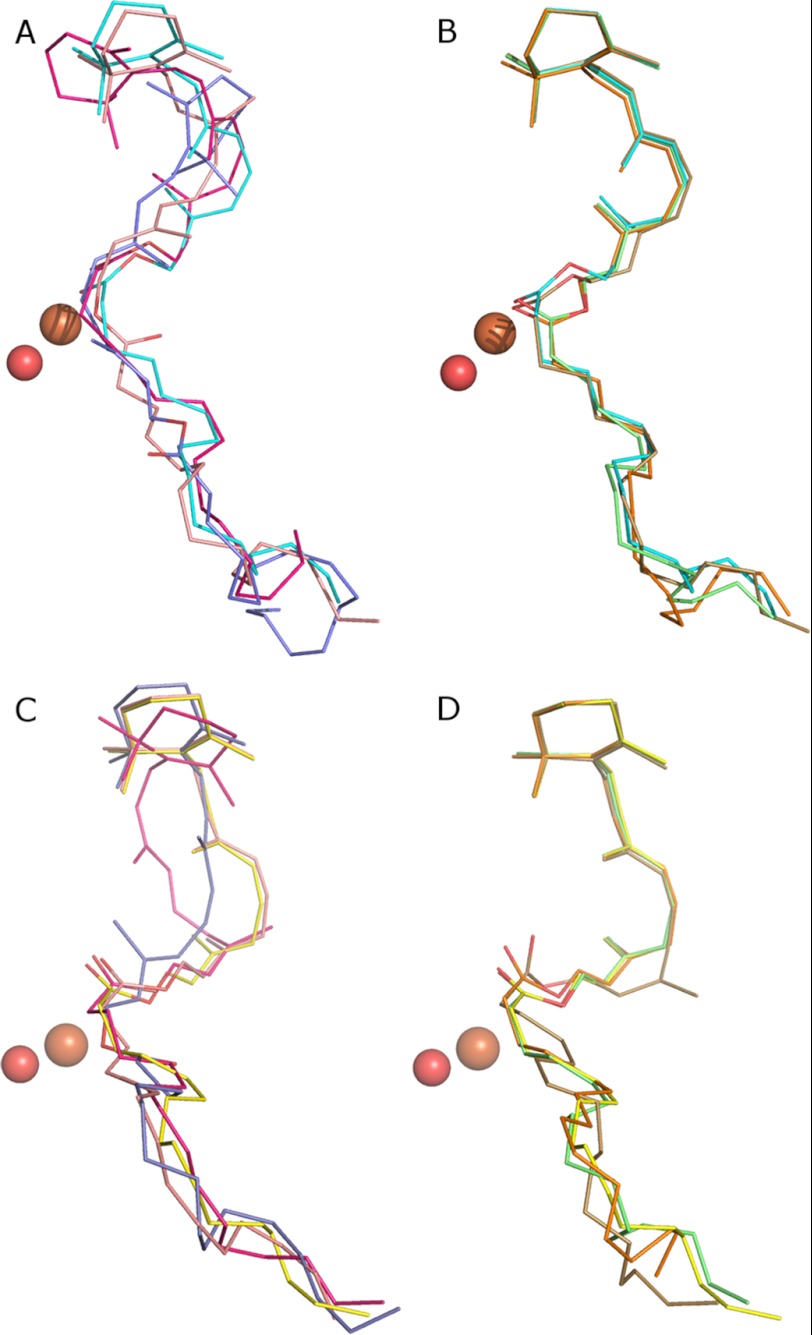
FIGURE 7.
Overlay of docking simulations on wild-type and mutant models of the bovine RPE65 crystal structure. A, 11-cis-palmitate in inactive mutants. B, 11-cis-palmitate in modulating mutants. C, 13-cis-palmitate in inactive mutants. D, 13-cis-palmitate in modulating mutants. Inactive mutants are: F61L (peach), F312L (purple), F418L (magenta); modulating mutants are: F103L (green), F526A (orange), Y338A (olive green), WT 11-cis-palmitate (cyan), and WT 13-cis-palmitate (yellow).
DISCUSSION
In this study, we demonstrate, using site-directed mutagenesis and structure-based modeling, that an assemblage of aromatic residues in the substrate-binding cleft of RPE65 governs the progression and specificity of retinoid isomerization by this enzyme. These data extend our previous conclusions (8) that RPE65 utilizes a cationic intermediate (carbocation or radical cation) in its mechanism by showing that multiple aromatic residues contribute to the retinyl moiety-binding cavity within which isomerization occurs. This environment is a result of contributions from the various aromatic side chains we have studied and appears to be crucial to isomerization. Furthermore, we interpret the residual unresolved density in the RPE65-binding cleft as being due to the products of the reaction, not only fatty acid, as indicated previously (15), but also 11-cis-retinol. Finally, based on docking of various modeled substrates, we can discern a temporal sequence for the progression of retinoid isomerization by RPE65.
Based on our previous findings that we could modulate cis isomer specificity by mutation of single residues (8), we determined that isomerization of all-trans-ROL to 11-cis-ROL occurs via a cationic intermediate with loss of polyene bond order as this alternative was the only one that could accommodate possible formation of isomers besides 11-cis. Although a carbocation intermediate was considered originally based on prior studies (9, 11, 26), we concluded that the data could just as easily support a radical cation intermediate. Support for the latter possibility was provided by our finding that aromatic lipophilic spin traps such as N-tert-butyl-α-phenylnitrone (PBN) inhibit RPE65 activity (26), and this was corroborated by others (27).
Phe-103 was one of the original residues that we showed to modulate isomer specificity (8). Although it is itself resolved, Phe-103 is located on a flexible loop that is not completely modeled in the RPE65 crystal structure (15). Paralogous to Phe-113 in ACO (12, 13), Phe-103 is one of the aromatic residues of the substrate-binding tunnel of RPE65. It is part of the conserved EFG triplet motif in metazoan carotenoid oxygenases, of which the glycine is invariantly conserved throughout the family. Herein we show that two other tunnel aromatic residues, Tyr-338 and Phe-526, are also required for 11-cis product specificity. Of these, Tyr-338 is also located on a loop, what we term the metazoan loop because it is conserved only among animal carotenoid oxygenases. This loop is observed in the RPE65 crystal structure, and the aromatic side chain of Tyr-338 inserts into the tunnel. Phe-526 is part of what we have termed the phenylalanine arch. A similar arrangement of phenylalanines also occurs in VP14, where it is proposed to be important for “caging” the substrate over the iron (14), a role we predict to be reprised in RPE65 by the paralogous residues Phe-61, Phe-312, and Phe-526. The opening up of space around residues 526 and 338, in their respective Ala mutants, probably allows for the more energetically favorable 13-cis product to be formed. This is similar to the F103L mutant, which also affects product specificity. The varying effects of the Phe arch residues, located at the core of RPE65, point to their critical role in the RPE65 retinol isomerase mechanism. Along with Phe-103, we conclude that Tyr-338 and Phe-526 also constrain cleft shape and volume to favor production of 11-cis-ROL. Such a key role for phenylalanine in binding polyene chains has previously been shown for carotenoid binding in light-harvesting complex where García-Martín et al. (28) showed that about 25% of total protein-carotenoid contacts are with phenylalanine residues. Conversely, phenylalanines, unlike tyrosines, are rarely involved in catalysis (29).
An important finding of the cavity modeling of RPE65, ACO, and VP14 is the observation of the relative constriction of the RPE65 cavity when compared with those of ACO and VP14. We propose that this may partly reflect the closed state of the RPE65 crystal (15) and the mechanistic requirements for the assured conformational molding of the delocalized retinyl moiety to the 11-cis conformation. Selective mutational alteration of the cavity, such as we have performed here with residues Tyr-338 and Phe-526, extends our findings on Phe-103 (and Thr-147 (8)). These alterations disturb the predicted molecular molding, allowing isomerization to the more thermodynamically favorable 13-cis isomer. Therefore, in all these mutants, of Phe-103, Phe-526, and Tyr-338, we conclude that the dominant effect is reduced stringency disfavoring 11-cis isomerization rather than preferential selection for 13-cis-RE binding.
Consideration of these data allows us to put forward a temporal sequence for isomerization by RPE65. We propose that: 1) all-trans-RE enters the open ready state conformation of RPE65, perhaps via initial recognition of the β-ionone ring; 2) the bound all-trans-RE undergoes a single electron oxidation by an as yet unidentified means, resulting in the delocalization of the retinyl bond order; 3) the delocalized retinyl moiety in the substrate-binding cleft is constrained by the geometry of the closing pocket to adopt the 11-cis conformation as RPE65 transitions to the state captured by the RPE65 crystal structure (15); 4) the O-alkyl bond is positioned over and interacts with the iron, which catalyzes cleavage; 5) attack by water completes hydrolysis and restores retinyl bond order; 6) 11-cis-retinol is released; and 7) RPE65 returns to the open ready state. This chronology is different from the mechanism proposed by Kiser et al. (15) in a number of respects. These authors proposed that initially, the oxygen of the all-trans-RE acyl bond interacts with the iron, resulting in cleavage of the O-alkyl bond. They proposed that the leaving of the palmitate anion results in formation of a retinyl carbocation, which can then be constrained to the 11-cis conformation. However, our docking studies indicate that this is too late for delocalization to occur as the predicted binding energy of all-trans-RE is quite unfavorable in the closed state modeled in the RPE65 crystal structure. Instead, we postulate that the retinyl moiety must be in a delocalized state prior to approach of the O-alkyl bond to the iron center and its ultimate cleavage. We propose that single-electron oxidation generating a radical cation intermediate is the source of the delocalized state. An earlier loss of bond order allows the retinyl moiety to be constrained to the 11-cis conformation, which has a favorable binding energy in the closed state, whereas the O-alkyl bond is brought into register for cleavage. This point is supported by the inhibitory effect of aromatic lipophilic spin traps (such as PBN) on RPE65 activity, an effect that would be less likely if delocalization via radical formation occurs later as the putative PBN-retinyl ester adduct is not predicted to be able to access the binding cleft in its later closed state (26). Consequently, we propose that O-alkyl cleavage occurs later. However, we do concur with the proposal of Kiser et al. (15, 30) that the bond-cleaving activity of RPE65 has become that of O-alkyl cleavage, with retinol isomerization an acquired function. Thus, by strategic placement of aromatic residues to modulate the geometry and chemistry of its catalytic cleft, RPE65 has evolved from a carotenoid oxygenase to an isomerohydrolase role to drive the vertebrate retinal visual cycle.
Supplementary Material
Acknowledgments
We thank Dr. John Saari for the gift of mouse monoclonal antibody to CRALBP. In addition, we thank Dr. Danielle Gutierrez for critical reading of the manuscript.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program of the NEI.

This article contains supplemental Figs. S1 and S2 and Tables S1 and S2.
- RPE
- retinal pigment epithelium
- ACO
- Synechocystis apocarotenoid oxygenase
- CRALBP
- cellular retinaldehyde-binding protein
- PBN
- N-tert-butyl-α-phenylnitrone
- RDH5
- retinol dehydrogenase 5
- RE
- retinyl ester
- ROL
- retinol
- VP14
- viviparous14 (Zea mays 9-cis-epoxycarotenoid dioxygenase).
REFERENCES
- 1. Saari J. C. (2000) Biochemistry of visual pigment regeneration: the Friedenwald lecture. Invest. Ophthalmol. Vis. Sci. 41, 337–348 [PubMed] [Google Scholar]
- 2. Lamb T. D., Pugh E. N., Jr. (2004) Dark adaptation and the retinoid cycle of vision. Prog. Retin. Eye. Res. 23, 307–380 [DOI] [PubMed] [Google Scholar]
- 3. Hamel C. P., Tsilou E., Pfeffer B. A., Hooks J. J., Detrick B., Redmond T. M. (1993) Molecular cloning and expression of RPE65, a novel retinal pigment epithelium-specific microsomal protein that is post-transcriptionally regulated in vitro. J. Biol. Chem. 268, 15751–15757 [PubMed] [Google Scholar]
- 4. Redmond T. M., Yu S., Lee E., Bok D., Hamasaki D., Chen N., Goletz P., Ma J. X., Crouch R. K., Pfeifer K. (1998) Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. Nat. Genet. 20, 344–351 [DOI] [PubMed] [Google Scholar]
- 5. Jin M., Li S., Moghrabi W. N., Sun H., Travis G. H. (2005) Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell 122, 449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moiseyev G., Chen Y., Takahashi Y., Wu B. X., Ma J. X. (2005) RPE65 is the isomerohydrolase in the retinoid visual cycle. Proc. Natl. Acad. Sci. U.S.A. 102, 12413–12418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Redmond T. M., Poliakov E., Yu S., Tsai J. Y., Lu Z., Gentleman S. (2005) Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc. Natl. Acad. Sci. U.S.A. 102, 13658–13663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Redmond T. M., Poliakov E., Kuo S., Chander P., Gentleman S. (2010) RPE65, visual cycle retinol isomerase, is not inherently 11-cis-specific: support for a carbocation mechanism of retinol isomerization. J. Biol. Chem. 285, 1919–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maeda A., Maeda T., Imanishi Y., Golczak M., Moise A. R., Palczewski K. (2006) Aberrant metabolites in mouse models of congenital blinding diseases: formation and storage of retinyl esters. Biochemistry 45, 4210–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parker R. O., Crouch R. K. (2010) Retinol dehydrogenases (RDHs) in the visual cycle. Exp. Eye. Res. 91, 788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McBee J. K., Kuksa V., Alvarez R., de Lera A. R., Prezhdo O., Haeseleer F., Sokal I., Palczewski K. (2000) Isomerization of all-trans-retinol to cis-retinols in bovine retinal pigment epithelial cells: dependence on the specificity of retinoid-binding proteins. Biochemistry 39, 11370–11380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kloer D. P., Ruch S., Al-Babili S., Beyer P., Schulz G. E. (2005) The structure of a retinal-forming carotenoid oxygenase. Science 308, 267–269 [DOI] [PubMed] [Google Scholar]
- 13. Kloer D. P., Schulz G. E. (2006) Structural and biological aspects of carotenoid cleavage. Cell Mol. Life Sci. 63, 2291–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Messing S. A., Gabelli S. B., Echeverria I., Vogel J. T., Guan J. C., Tan B. C., Klee H. J., McCarty D. R., Amzel L. M. (2010) Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. Plant Cell 22, 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kiser P. D., Golczak M., Lodowski D. T., Chance M. R., Palczewski K. (2009) Crystal structure of native RPE65, the retinoid isomerase of the visual cycle. Proc. Natl. Acad. Sci. U.S.A. 106, 17325–17330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Landers G. M., Olson J. A. (1988) Rapid, simultaneous determination of isomers of retinal, retinal oxime, and retinol by high-performance liquid chromatography. J. Chromatogr 438, 383–392 [DOI] [PubMed] [Google Scholar]
- 17. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 18. Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T. (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37, D387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sanner M. F., Olson A. J., Spehner J. C. (1996) Reduced surface: an efficient way to compute molecular surfaces. Biopolymers 38, 305–320 [DOI] [PubMed] [Google Scholar]
- 20. van Aalten D. M., Bywater R., Findlay J. B., Hendlich M., Hooft R. W., Vriend G. (1996) PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J. Comput Aided Mol. Des. 10, 255–262 [DOI] [PubMed] [Google Scholar]
- 21. Schüttelkopf A. W., van Aalten D. M. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 22. Trott O., Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kleywegt G. J., Harris M. R., Zou J. Y., Taylor T. C., Wählby A., Jones T. A. (2004) The Uppsala Electron-Density Server. Acta Crystallogr. D Biol. Crystallogr. 60, 2240–2249 [DOI] [PubMed] [Google Scholar]
- 24. Fischer M., Zhang Q. C., Dey F., Chen B. Y., Honig B., Petrey D. (2011) MarkUs: a server to navigate sequence-structure-function space. Nucleic Acids Res. 39, W357–W361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oberhauser V., Voolstra O., Bangert A., von Lintig J., Vogt K. (2008) NinaB combines carotenoid oxygenase and retinoid isomerase activity in a single polypeptide. Proc. Natl. Acad. Sci. U.S.A. 105, 19000–19005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poliakov E., Parikh T., Ayele M., Kuo S., Chander P., Gentleman S., Redmond T. M. (2011) Aromatic lipophilic spin traps effectively inhibit RPE65 isomerohydrolase activity. Biochemistry 50, 6739–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mandal M. N., Moiseyev G. P., Elliott M. H., Kasus-Jacobi A., Li X., Chen H., Zheng L., Nikolaeva O., Floyd R. A., Ma J. X., Anderson R. E. (2011) α-Phenyl-N-tert-butylnitrone (PBN) prevents light-induced degeneration of the retina by inhibiting RPE65 protein isomerohydrolase activity. J. Biol. Chem. 286, 32491–32501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. García-Martín A., Pazur A., Wilhelm B., Silber M., Robert B., Braun P. (2008) The role of aromatic phenylalanine residues in binding carotenoid to light-harvesting model and wild-type complexes. J. Mol. Biol. 382, 154–166 [DOI] [PubMed] [Google Scholar]
- 29. Holliday G. L., Mitchell J. B., Thornton J. M. (2009) Understanding the functional roles of amino acid residues in enzyme catalysis. J. Mol. Biol. 390, 560–577 [DOI] [PubMed] [Google Scholar]
- 30. Kiser P. D., Palczewski K. (2010) Membrane binding and enzymatic properties of RPE65. Prog. Retin. Eye Res. 29, 428–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Golczak M., Kiser P. D., Lodowski D. T., Maeda A., Palczewski K. (2010) Importance of membrane structural integrity for RPE65 retinoid isomerization activity. J. Biol. Chem. 285, 9667–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.