Background: Proteasome inhibitors have been used in cancer therapy, and the search for new compounds with higher efficacy and specificity for tumor cells is imperative.
Results: Fenbendazole shows a strong antiproliferative property preferentially against cancer cells.
Conclusion: The growth-inhibitory activity of FZ can be partly attributed to interference in the proteasomal function.
Significance: This new proteasome-targeting compound may be useful in cancer therapy.
Keywords: Anticancer Drug, Apoptosis, Cell Growth, ER Stress, p53, Proteasome, Reactive Oxygen Species (ROS)
Abstract
In recent years, there has been a great deal of interest in proteasome inhibitors as a novel class of anticancer drugs. We report that fenbendazole (FZ) (methyl N-(6-phenylsulfanyl-1H-benzimidazol-2-yl)carbamate) exhibits a potent growth-inhibitory activity against cancer cell lines but not normal cells. We show here, using fluorogenic substrates, that FZ treatment leads to the inhibition of proteasomal activity in the cells. Succinyl-Leu-Leu-Val-Tyr-methylcoumarinamide (MCA), benzyloxycarbonyl-Leu-Leu-Glu-7-amido-4-MCA, and t-butoxycarbonyl-Gln-Ala-Arg-7-amido-4-MCA fluorescent derivatives were used to assess chymotrypsin-like, post-glutamyl peptidyl-hydrolyzing, and trypsin-like protease activities, respectively. Non-small cell lung cancer cells transiently transfected with an expression plasmid encoding pd1EGFP and treated with FZ showed an accumulation of the green fluorescent protein in the cells due to an increase in its half-life. A number of apoptosis regulatory proteins that are normally degraded by the ubiquitin-proteasome pathway like cyclins, p53, and IκBα were found to be accumulated in FZ-treated cells. In addition, FZ induced distinct ER stress-associated genes like GRP78, GADD153, ATF3, IRE1α, and NOXA in these cells. Thus, treatment of human NSCLC cells with fenbendazole induced endoplasmic reticulum stress, reactive oxygen species production, decreased mitochondrial membrane potential, and cytochrome c release that eventually led to cancer cell death. This is the first report to demonstrate the inhibition of proteasome function and induction of endoplasmic reticulum stress/reactive oxygen species-dependent apoptosis in human lung cancer cell lines by fenbendazole, which may represent a new class of anticancer agents showing selective toxicity against cancer cells.
Introduction
The proteasome is the major proteolytic complex, responsible for the degradation of a number of cellular proteins in eukaryotes. Proteasomes are abundant in both cytoplasm and nucleus, where they catalyze the ATP-dependent proteolysis of short-lived regulatory proteins as well as misfolded, damaged, and abnormal proteins. Cancer cells accumulate more oxidized or mutated/damaged proteins that are effectively removed by the proteasomes. Consequently, they are likely to be more dependent on proteasomal activity as compared with normal cells (1, 2). With that rationale, proteasome has been targeted for the treatment of human cancer. Bortezomib is the first proteasome inhibitor approved by the Food and Drug Administration to treat patients with advanced hematologic malignancies. Unfortunately, the efficacy of the treatment has been restricted to a narrow therapeutic window. Bortezomib as a single agent has limited activity in non-small cell lung cancer, and it has been reported earlier to have no anti-tumor activity in H460 xenografts (3, 4). Therefore, the search for new proteasomal inhibitors that can preferentially eradicate cancer cells is of high priority for effective treatment of the disease.
Fenbendazole (methyl N-(6-phenylsulfanyl-1H-benzimidazol-2-yl) carbamate) is an anthelmintic drug belonging to the benzimidazole group mainly used for the treatment of pinworm in animals (Fig. 1A). Clinically, two benzimidazole derivatives, mebendazole (methyl 5-benzoyl-2-benzimidazole-carbamate) and albendazole (methyl (5- propylthio)-1H-benzimidazol-2-yl carbamate methylester) have been used to treat human alveolar echinococcosis, a lethal pulmonary helminthic infection (5). Although echinococcosis requires long-term high dose treatment with mebendazole, the incidence of severe side effects to the host is considerably low (6, 7). Thus, benzimidazoles have a good track record as anthelmintic agents and have a wide margin of safety in humans. The selectivity of these drugs for the parasite is explained on the basis of irreversible blockade in uptake of glucose in parasite, which leads to depletion of glycogen storage and degeneration of endoplasmic reticulum in the germinal layers, resulting in cell death (8). In addition, a preferential affinity of these drugs for β-tubulin of the parasite as compared with that of the host has also been reported (9). Although FZ is known to inhibit polymerization and alter microtubule function in parasitic cells, its effect on mammalian tubulin has not been looked into (10–12). Another compound of this class, mebendazole, has been earlier reported to have a mild tubulin depolymerizing activity in human cancer cells. The structural explanation for this preferential binding for parasite tubulin has been provided on the basis of difference in certain key residues between mammals and parasites leading to “closing off” of the hydrophobic pocket into which the benzimidazoles bind, thus making it inaccessible (13). In the past, there have been a few reports regarding the use of anthelmintic benzimidazoles in human cancer (14–18), but the molecular mechanism of their inhibitory action remains elusive.
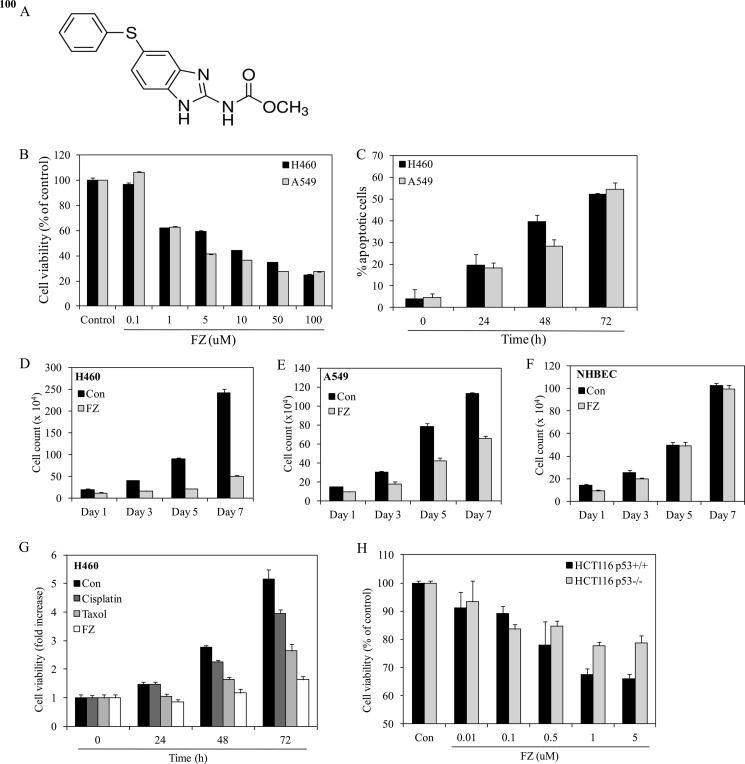
FIGURE 1.
FZ inhibits growth and induces apoptosis in human non small cell lung cancer cells. A, chemical structure of fenbendazole. B, human H460 or A549 cells were plated onto 96-well tissue culture plates. Cells were left untreated or treated with different doses of FZ for 48 h or with 1 μm FZ for different time durations (C). Cell viability was measured by MTT assay. D–F, cells were seeded at a density of 1 × 104 cells/well in 24-well culture plates and then treated with FZ. Cells were then trypsinized, and viable cells were counted by the trypan blue exclusion method every second day for 7 days. All experiments were done in triplicate. G, human H460 cells were seeded onto 96-well tissue culture plates, and the following day, cells were left untreated or treated with IC50 values of different chemotherapeutic drugs, as indicated. Cell viability was measured by an MTT assay at 0, 24, 48, or 72 h. H, human HCT116 (p53+/+) and HCT116 (p53−/−) isogenic colon cancer cell lines were treated with different doses of FZ for 48 h. Cell viability was measured by an MTT assay. Error bars, S.D.
In the present study, we report that treatment of human lung cancer cell lines with fenbendazole (FZ)3 induces apoptotic cell death, whereas primary normal cells in culture remain widely unaffected. FZ exerts its cytotoxicity possibly through interference with proteasome function and induction of unfolded protein response, ultimately resulting in cancer cell death via the intrinsic pathway of apoptosis.
EXPERIMENTAL PROCEDURES
Cell Culture and Chemicals
All cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1× penicillin/streptomycin antibiotics (100 units/ml penicillin and 100 μg/ml streptomycin). Cells were treated with fenbendazole, cycloheximide, MG132, NAC, TNFα, Z-VAD-FMK, Tiron, or colchicine as required in various experiments.
FZ; 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT); colchicine; MG132; cycloheximide; JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethyl-benzimidazoleocarbocyanine iodide); Hoechst 33342; propidium iodide; N-acetylcysteine; TNFα; Z-VAD-FMK; proteasomal and caspase substrates; Tiron; anti-β-actin; anti-ubiquitin; anti-GFP; anti-p27Kip1; anti-Bcl2; anti-mouse IgG-fluorescein isothiocyanate (FITC); horseradish peroxidase (HRP)-conjugated anti-mouse, anti-rabbit, and anti-goat IgGs; and all of the cell culture reagents were purchased from Sigma. IKK inhibitor wedelolactone was obtained from Calbiochem. Anti-p53 (Bp53–12 and DO1), anti-p21, anti MDM-2, anti-cyclin B1, anti-Bax, anti-cytochrome c, anti-IκBα, and anti-phospho-JNK were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
Cell Viability Assays
Cells (5 × 103 cells/well) were seeded into 96-well plates, and 24 h after seeding, cells were treated with different doses of fenbendazole. Cell viability was measured by an MTT assay. For the cell count assay, cells were seeded at a density of 1 × 104 cells/well in 24-well culture plates and then treated with FZ. Cells were harvested at the indicated time points, and viable cells were counted by the trypan blue exclusion method. All experiments were done in triplicate.
Hoechst/Propidium Iodide (PI) Staining
Following FZ treatment, Hoechst 33342 and PI were added into the medium at 100 ng/ml concentration for 15 min, and cells were then washed with prewarmed PBS and examined under a fluorescence microscope for detection of apoptotic cells.
FACS Analysis
After treatment with FZ for the indicated time intervals, cells were harvested, washed with PBS, and fixed in 70% ethanol overnight at 4 °C. The next day, cells were centrifuged at 1000 rpm for 5 min, the supernatant was carefully aspirated, and the pellet was resuspended in PBS containing 40 μg/ml PI and 100 μg/ml RNase A (19). FACS analysis was done on a BD FACSArray system, and the data were analyzed using FlowJo software.
TdT Assay
H460 cells were plated onto coverslips in 35-mm tissue culture plates at subconfluent density. 24 h after seeding, cells were either left untreated or exposed to 1 μm FZ or 130 nmol of colchicine for 24 h. An in situ tailing reaction was performed as described by Gold et al. (20). Briefly, following treatment, cells were washed with PBS and fixed in 1:3 acetomethanol. After fixation, the cells were rinsed with PBS and incubated with 0.25 units/ml λ exonuclease enzyme in a moist chamber at 37 °C for 10 min. Cells were again washed with PBS and incubated with 20 units/ml TdT enzyme in the presence of 5 μl of 50 nmol biotin-dUTP in a humid chamber for 1 h at 37 °C. Endogenous peroxidase activity was then blocked by incubation in 3% hydrogen peroxide in methanol for 15 min at room temperature. Detection was finally done using avidin-biotin complex from an ABC kit (Vector) and 3,3′-diaminobenzidine tetrahydrochloride substrate (Sigma).
DNA Fragmentation Assay
The DNA fragmentation assay was performed as described previously (17). Control and FZ-treated cells were washed with cold PBS. The cell pellets were lysed in lysis buffer (10 mm Tris (pH 7.4), 10 mm EDTA (pH 8.0), and 0.5% Triton X-100) and incubated for 10 min at 4 °C and then incubated with 200 μg/ml RNase A for 1 h at 37 °C. After centrifugation, the supernatants were incubated with 200 μg/ml proteinase K for 30 min at 50 °C. Next, DNA fragments were precipitated with 0.5 m NaCl and 50% isopropyl alcohol, and the samples were loaded in 2% agarose TBE gel and stained with ethidium bromide.
Immunofluorescence
Cells were grown on coverslips and treated with 1 μm FZ or 130 nmol of colchicine for 24 h. Following treatment, cells were washed with PBS and fixed using 4% paraformaldehyde fixative. Cells were again washed with PBS and incubated with p53 antibody overnight at 4 °C. The following day, after several washes with PBS, the cells were incubated in FITC-conjugated secondary antibody for 1 h at 37 °C. Stained cells were visualized under a fluorescence microscope.
Measurement of Mitochondrial Membrane Potential and Cytochrome c Release
A549 cells were plated onto coverslips in 35-mm tissue culture plates at subconfluent density. 24 h later, cells were exposed to the specified doses of FZ for 24 h or 10 μm MG132 for 12 h, following which they were incubated with 5 μm JC-1 dye for 30 min in the CO2 incubator. After several washes with prewarmed PBS, mitochondrial membrane potential was evaluated qualitatively under a fluorescence microscope using a 568-nm filter.
The localization of cytochrome c was examined using immunofluorescent staining. Cells grown on coverslips were treated with FZ or MG132 as specified earlier. After treatment, cells were washed twice with PBS, fixed with 4% paraformaldehyde in PBS for 20 min, permeabilized with 0.05% Triton X-100 in PBS for 5 min, washed extensively, and then blocked with goat serum in PBS for 30 min. Primary antibody (anti-cytochrome c) incubation was carried out overnight at 4 °C. After several washings with PBS, cells were incubated with FITC-conjugated secondary antibody for 2 h at 37 °C, washed several times, and visualized under a fluorescence microscope.
Assay of Proteasome and Caspase-3-like Protease Activity
After overnight culture, cells were treated with varying doses of FZ or 10 μm MG132 for specified time periods. Cells were then isolated and processed for proteasome activity and caspase-3-like protease activity assays (21). The fluorogenic substrates succinyl-Leu-Leu-Val-Tyr-MCA, Z-Leu-Leu-Glu-7-amido-4-MCA, Boc-Gln-Ala-Arg-7-amido-4-MCA, and Ac-Asp-Glu-Val-Asp-MCA were used to determine chymotrypsin activity, peptidylglutamyl-peptide hydrolyzing activity, trypsin-like activity, and caspase-3-like protease activity, respectively. For the in vitro assay, control cell extracts were incubated with various doses of FZ for 4 h or with 5 μm FZ for different times, as indicated, and assayed for protease activity. To evaluate the direct effect of FZ on the protease activity of proteasome, pure 20S proteasome (100 ng/reaction) was used instead of cell supernatant in the protease activity assay buffer. Protease activities at a particular time point (30 min) within the linear range were used to calculate the data. The fluorescence intensity was measured at 380-nm excitation and 460-nm emission using a PerkinElmer Life Sciences Victor X3 fluorescence plate reader.
RT-PCR and Quantitative PCR
Cells were treated with FZ, and total RNA was isolated using the guanidium thiocyanate method (22). Briefly, following treatment, cells were washed with PBS and lysed and collected in 400 μl of solution D (4 m guanidium thiocyanate, 0.75 m sodium citrate, 10% N-laurylsarcosine, and 0.1 m β-mercaptoethanol). This was followed by the addition of 40 μl of 2 m sodium acetate (pH 4.0), 400 μl of water-saturated phenol, and 40 μl of 49:1 chloroform/isoamylalcohol. It was then mixed and incubated for 15 min on ice and centrifuged at 9000 rpm for 10 min at 4 °C. RNA was precipitated with an equal volume of isopropyl alcohol, resuspended in diethylpyrocarbonate-treated water, and quantified using a spectrophotometer. Finally, RNA was reverse transcribed using oligo(dT)18 primers, and RT-PCR analysis was performed. For quantitative PCR, the RealMasterMix SYBR ROX kit from Eppendorf was used. The reactions were set up according to the manufacturer's instructions in an Eppendorf Mastercycler Realplex real-time PCR machine.
Co-immunoprecipitation and Immunoblotting Experiments
24 h after transfection with pd1EGFP plasmid, cells were treated with different doses of FZ or 10 μm MG132 for 8 h. Cells were then washed with cold PBS and lysed on ice for 30 min with Nonidet P-40 lysis buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, complete protease inhibitor mixture). Cell lysates were briefly vortexed and centrifuged for 10 min at 15,000 × g at 4 °C, and the supernatants (total soluble extract) were used for immunoprecipitation. Protein concentration was measured according to the method of Bradford using bovine serum albumin as a standard (23). For each immunoprecipitation experiment, 200 μg of protein in 0.2 ml of Nonidet P-40 lysis buffer was incubated with 5 μl (2.5 μg) of GFP antibody. After overnight incubation at 4 °C with rotation, 20 μl of protein A/G-agarose beads were added, and incubation was continued at 4 °C for 5 h. The beads were washed six times with Nonidet P-40 lysis buffer. Bound proteins were eluted from the beads with SDS (1×) sample buffer, vortexed, boiled for 5 min, and analyzed by immunoblotting. The total cell lysates or the immunoprecipitated proteins were separated through 10% SDS-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride (PVDF) membranes (24). The membranes were successively incubated in blocking buffer (5% skim milk in TBST (50 mm Tris, pH 7.5, 0.15 m NaCl, 0.05% Tween-20)) with primary antibody and then with secondary antibody conjugated with horseradish peroxidase. Detection was carried out with enhanced chemiluminescence reagent from Millipore. All primary antibodies were used in 1:4000 dilutions for immunoblotting.
Histone H1 Kinase Assay
Commercially purified histone H1 (Sigma) was incubated with CDK2 immunoprecipitated from H460 whole-cell extracts equivalent to 100 μg of total protein in 50 μl of kinase reaction buffer (50 mm Tris, pH 7.2, 10 mm MgCl2, 1 mm DTT, 5 μCi of [γ-32P]ATP) for 10 min at 25 °C. Samples were analyzed by SDS-PAGE followed by autoradiography.
Pulse-Chase Experiment for the Analysis of Protein Stability
Cells were plated onto 35-mm tissue culture plates, and on the following day, they were left untreated or treated with FZ and chased in the presence of cycloheximide for the indicated times. Cells collected at each time point were then processed for immunoblotting using antibodies against p53, cyclin B1, IκBα, or Bax. For studying GFP stabilization, H1299 cells were transfected with pd1EGFP plasmid using Lipofectamine. On the following day, the cells were either left untreated or treated with 5 μm FZ and chased in the presence of cycloheximide for the indicated time durations. They were observed under a fluorescence microscope after the indicated time points or processed for immunoblotting using anti-GFP antibody as described above.
Electrophoretic Mobility Shift Assay (EMSA)
H460 cells were treated with different doses of FZ for 4 h, and nuclear extracts were then prepared according to Schreiber et al. (25). Briefly, 2 × 106 cells were washed with cold PBS and suspended in 0.4 ml of lysis buffer (10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm DTT, 0.5 mm phenylmethylsulfonyl fluoride, 2.0 μg/ml leupeptin, 2.0 μg/ml aprotinin, and 0.5 mg/ml benzamidine). The cells were allowed to swell on ice for 15 min, after which 12.5 μl of 10% Nonidet P-40 was added. The tube was then vigorously mixed on a vortex machine for 10 s, and the homogenate was centrifuged for 30 s. The nuclear pellet was resuspended in 25 μl of ice-cold nuclear extraction buffer (20 mm HEPES, pH 7.9, 0.4 m NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride, 2.0 μg/ml leupeptin, 2.0 μg/ml aprotinin, and 0.5 mg/ml benzamidine), and the tube was incubated on ice for 30 min with intermittent mixing. It was then centrifuged for 5 min at 4 °C, and the supernatant (nuclear extract) was either used immediately or stored at −70 °C for later use. The protein content was measured by the method of Bradford (23).
EMSAs were performed by incubating 6 μg of nuclear extract, with 16 fmol of 32P-end-labeled 45-mer double-stranded NFκB oligonucleotide, 5′-TTGTTACAAGGGACTTTCCGCTGGGGACTTTCCAGGGAGGCGTGG-3′ (26), for 30 min at 37 °C. The incubation mixture consisted of 2–3 μg of poly(dI-dC) in binding buffer (25 mm HEPES, pH 7.9, 0.5 mm EDTA, 0.5 mm DTT, 1% Nonidet P-40, 5% glycerol, and 50 mm NaCl). The DNA-protein complex formed was separated from free oligonucleotide on 5% native polyacrylamide gel, and then the gel was then dried. The specificity of binding was determined by using an excess of unlabeled oligonucleotide for competition.
Secreted Alkaline Phosphatase (SEAP) Assay for ER Stress
293T cells were seeded in a 96-well plate and were transfected the next day with pSEAP-con plasmid (Clontech), which constitutively expresses SEAP. 24 h after transfection, the cells were either left unexposed or exposed to different concentrations of the indicated drugs in fresh media containing 1% FBS. 24 h after incubation, 10 μl of conditioned culture media was removed and assessed for SEAP activity using 4-MUP substrate according to the protocol described by the manufacturer (Clontech, Palo Alto, CA). In brief, 10 μl of medium was mixed with 15 μl of 5× SEAP assay buffer (0.5 m Tris, pH 9.0, and 0.5% BSA) in a total volume of 50 μl in a 96-well plate and incubated at 65 °C for 30 min to inactivate endogenous SEAP activity. The plate was chilled on ice for 2 min. Then 25 μl of 1 mm 4-methylumbelliferyl phosphate substrate was added to each well and incubated at 37 °C for 2 h. The activity of SEAP was assayed using a 96-well fluorescence plate reader (PerkinElmer Multilabel reader Victor X3) with excitation set at 355 nm and emission at 460 nm. Three independent experiments were performed in triplicates.
Reactive Oxygen Species (ROS) Activity
A549 cells were grown on coverslips in 35-mm culture dishes and treated with 5 μm FZ or 10 μm MG132 for 4 h. NAC (10 mm) was added 2 h before the drug. To measure the production of reactive oxygen species, after treatment, cells were washed and incubated at 37 °C with 1 μm DCF-DA for 1 h in serum-free medium without phenol red. After washing twice, samples were illuminated by a 100-watt mercury lamp and viewed with an FITC filter on a Nikon fluorescent microscope to view DCFDA fluorescence. For quantitative analysis, fluorescence intensity was measured at 530 nm after excitation at 485 nm in a Victor 3X fluorometer (PerkinElmer Life Sciences).
Reporter Assays for p53 and NFκB Gene Transcription
The effect of FZ on p53- and NFκB-dependent reporter gene transcription was measured by a luciferase assay using pWWP-Luc (27) or NFκB-Luc construct, respectively. For p53 transcriptional activity, H1299 cells were co-transfected with pWWP-luc and either pCMV vector or pCMV-p53 (WT) in a 96-well plate. Transfections were done using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. 24 h post-transfection, cells were left untreated or treated with 1 μm FZ for 24 h. Similarly, for NFκB reporter assay, A549 cells were co-transfected with NFκB-Luc vector carrying NFκB binding sites and pRL-SV40. The following day, cells were treated with 5 μm FZ for 4 h, followed by 30 ng/ml TNFα for 1 h, where indicated. In both cases, the cell extracts were then prepared, and luciferase activity was monitored using a Dual Luciferase assay kit from Promega as per the manufacturer's instructions. The luciferase activity was assayed using a 96-well fluorescence plate reader (PerkinElmer Life Sciences Multilabel reader Victor X3). pRL-SV40 Renilla vector was used for co-transfection in all cases to normalize the data, and it was transfected at a lower concentration (5-fold lower than the reporter luciferase plasmid). The data were represented as relative luciferase activity (the ratio of firefly to Renilla values). The experiments were performed in triplicates.
Statistical Analysis
All results are expressed as means ± S.D. unless otherwise mentioned. Student's t test was used to calculate the significance, accepting p < 0.05 as a level of significance.
RESULTS
FZ Inhibits Growth and Induces Apoptosis in Human Non-small Cell Lung Cancer Cells
To check the effect of FZ on cell growth, NSCLC cell lines (H460 and A549) were plated onto 96-well tissue culture plates, and on the following day, the cells were exposed to different doses of FZ for 48 h. The cell viability was determined by an MTT assay. Exposure to drug decreased the viability of cells in a dose-dependent manner (Fig. 1B). The IC50 value was calculated as ∼1.2 and 1.6 μm for H460 and A549 cell lines, respectively. As shown in Fig. 1C, FZ reduced the viability of these cells in a time-dependent manner. To determine the cell viability for a longer duration, cell count assays were performed on alternate days for 7 days after 1 μm FZ treatment in H460, A549, and normal human bronchial epithelial cells (Fig. 1, D–E). Whereas H460 and A549 cells showed reduced viability when exposed to FZ, the compound showed little effect on normal human bronchial epithelial cells. We then compared the antiproliferative ability of FZ with other established anticancer drugs. The results of a time-dependent growth assay clearly indicated FZ to be a more effective cytotoxic agent as compared with cisplatin and taxol (Fig. 1F). The growth-inhibitory action of FZ in H460 and A549 cells was also compared with the Food and Drug Administration-approved proteasomal inhibitor bortezomib, and the results showed that the activities of both of the compounds were comparable (supplemental Fig. 1). Interestingly, when we treated HCT116 p53+/+ and p53−/− cells with FZ in a dose-dependent manner, p53+/+ cells showed greater sensitivity to FZ, as revealed by a relatively higher percentage of cell death (Fig. 1G). We also assessed the anti-proliferative effect of FZ on 25 cell lines derived from different tumor types, such as lung, prostate, breast, cervix, and colon, which are listed in the NCI-60 panel designed for drug screening. Most of these cell lines had IC50 values greater than 1 μm. However, FZ induced ∼50% growth inhibition at 1 μm concentration in at least seven cell lines tested (supplemental Table 1). Although FZ caused growth inhibition and apoptosis in NSCLC cells, normal short-term primary cultures or immortalized normal cell lines were relatively unaffected by its action (supplemental Fig. 2). The IC50 values for FZ were found to be severalfold higher in these normal cells than in cancer cell lines. Together, these results show that FZ acts as an inhibitor of cell proliferation in the case of cancer cells but is relatively non-toxic to normal cells.
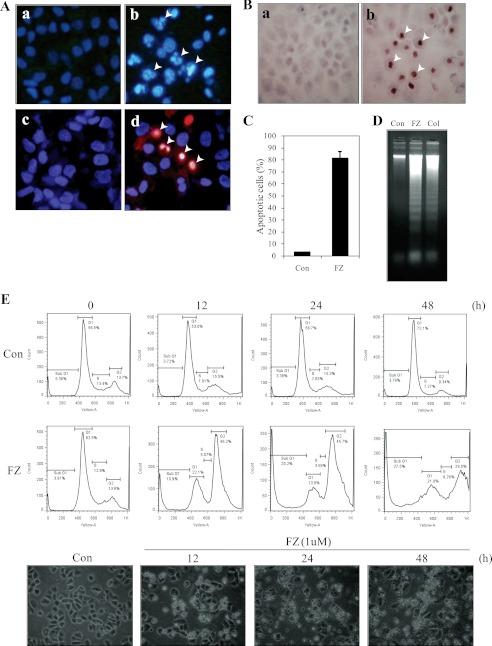
Incubation of NSCLC cells with FZ resulted in nuclear condensation and fragmentation characteristic of apoptotic nuclear morphology (Fig. 2A, a and b). Cells were double-stained with Hoechst 33342 and PI to check their nuclear morphology and plasma membrane integrity. 48-h FZ-treated cells had a significant number of apoptotic nuclei as compared with that of control untreated cells. Many of them appeared orange-red in color, characteristic of late apoptosis (Fig. 2A, c and d). The nuclei were further labeled with a TdT-mediated dUTP nick end labeling system to verify the apoptotic nature of cell death. A significant number of TUNEL-positive cells were observed 48 h post-FZ treatment (Fig. 2B), which were then quantified (Fig. 2C). DNA extracted from treated cells exhibited a distinct laddering pattern, which is a characteristic of apoptotic cell death (Fig. 2D).
FIGURE 2.
FZ results in apoptotic cell death. Cells were seeded on coverslips and then exposed to 1 μm FZ for 48 h. A, Hoechst 33342- and propidium iodide-stained images of H460 cells showing cell death following 48 h of FZ treatment. a and c, controls; b and d, FZ-treated. Top panels, stained with Hoechst 33342 only; bottom panels, stained with Hoechst 33342 and PI together. Arrowheads, apoptotic nuclei. B, the cells were processed for TdT staining and observed under a microscope. FZ-treated cells showed intense nuclear staining (b) as compared with untreated control cells (a). C, the percentage of apoptotic cells was determined by counting the number of apoptotic nucleic in control and treated cells under a microscope. D, H460 cells were left untreated or treated with 1 μm FZ or 130 nmol of colchicine for 48 h, following which they were processed for DNA fragmentation analysis, as described under “Experimental Procedures.” E, H460 cells were treated with 1 μm FZ for different time durations and processed for cell cycle analysis by FACS. Histograms depict the distribution of cells in sub-G1, G1, S, and G2 phases of the cell cycle. The bottom panel shows the corresponding images of cells under a phase-contrast microscope. Error bars, S.D.
When we performed flow cytometric analysis, results showed that FZ treatment caused an accumulation of cells in G2/M phase of cell cycle followed by a significant increase in the percentage of sub-G1 apoptotic population, indicative of DNA degradation (Fig. 2E). This could be due to FZ-induced stabilization of G2/M checkpoint regulatory molecules that contribute to the enhancement of cytotoxicity. Cell cycle delay at G2/M could be due to accelerated and prolonged CDK2 phosphorylation and stabilization of cyclin B1, p53, and p21, which have been implicated in the control of the G2/M checkpoint (28). Phosphorylation status of CDK2 kinase governs the activity of the Cdc2-cyclin B1 complex, which is responsible for the onset of mitosis. Therefore, levels of proteins involved in the G2/M transition were further examined by Western blot analysis.
FZ Treatment Results in Stability of p53 Protein
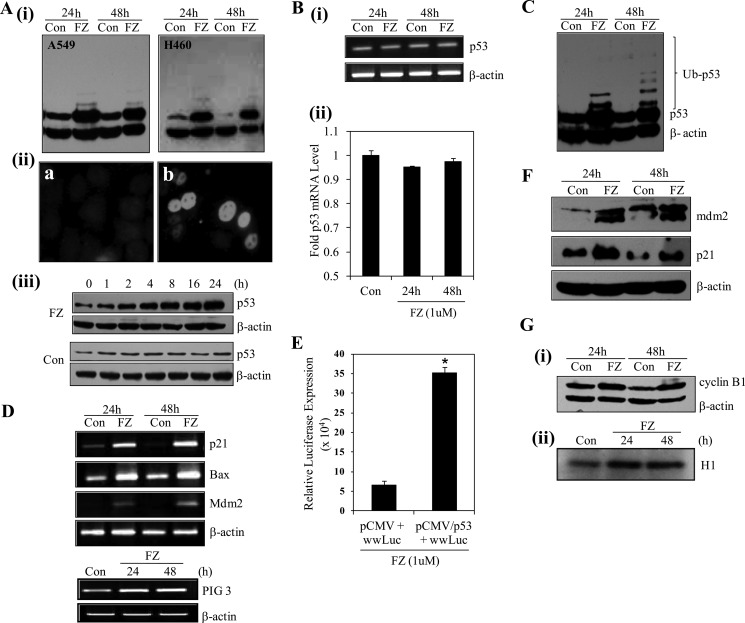
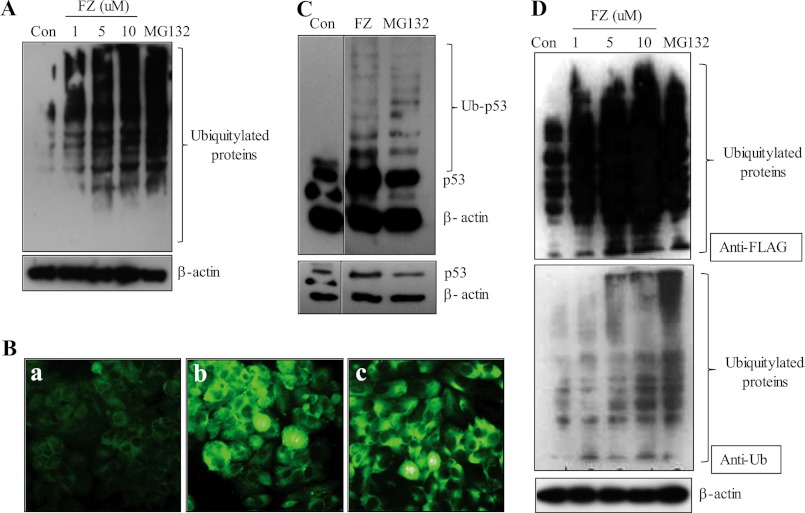
Because our earlier results indicated a possible role of p53 in sensitizing the cells toward FZ induced apoptosis, we performed Western blotting and immunofluorescence experiments for p53 following FZ treatment. Fig. 3A (i) shows a marked increase in p53 level in both H460 and A549 cells after FZ treatment for 24 and 48 h as compared with control untreated cells. Immunofluorescence for p53 in H460 cells clearly showed an increased accumulation of the protein in nuclei of treated cells (Fig. 3A (ii)). To determine the kinetics of FZ-induced p53 activation, we examined the time-dependent response after treatment with 1 μm FZ, as shown in Fig. 3A (iii). The p53 induction was observed 2 h post-treatment and reached a plateau at 24 h. We next proceeded to check if this increased level of the protein was a result of transcriptional up-regulation. For this, total RNA was isolated from control and treated cells after 24 or 48 h of FZ treatment. We used reverse transcriptase PCR to analyze the p53 gene expression. The results indicated that the accumulation of p53 protein in FZ-treated cells was not accompanied by a corresponding increase in the level of its mRNA (Fig. 3B (i)), which was confirmed by real-time PCR (Fig. 3B (ii)). Because p53 was not found to be up-regulated by FZ at the transcriptional level, these results prompted us to investigate further the events leading to its accumulation in the cell. We speculated that this could be due to inhibition of the degradation pathway, and blockage of the proteasome-mediated proteolysis step could be the cause of this stability. With this rationale, the above experiment was repeated, and interestingly, upon longer exposure of a Western blot done using p53 antibody, we could clearly detect a considerable increase in the ubiquitylated form of wild type (WT) p53 after FZ treatment, as revealed by higher molecular weight bands corresponding to the ubiquitylated form of the protein (Fig. 3C).
FIGURE 3.
FZ results in p53 induction and up-regulation of p53 target genes. A, i, H460 and A549 cells were treated with FZ for 24 or 48 h, and cell lysates were made and processed for immunoblotting using p53 antibody. ii, A549 cells were either left untreated or treated with 1 μm FZ for 24 h and then subjected to immunofluorescence using p53 antibody and FITC-conjugated secondary antibody. a, control; b, FZ. iii, H460 cells were left untreated or treated with 1 μm FZ for the indicated time periods, and Western blot analysis was done using antibodies against p53 (Bp53-12) and β-actin. B, following treatment with FZ for 24 or 48 h, total RNA was isolated, and RT-PCR (i) or real-time PCR (ii) was done using p53-specific primers. C, a p53 Western blot was overexposed for the detection of high molecular weight bands representing ubiquitylated p53. D, cDNA synthesized from total RNA was used for RT-PCR using primers specific for p53 target genes. E, H1299 cells were transiently transfected with pWWP-luc and either pCMV vector or pCMV-p53 (WT) construct along with pRL-SV40 plasmids in a 96-well plate. 24 h after transfection, cells were treated with 1 μm FZ for 24 h. Cells were then processed for a dual luciferase reporter gene assay. *, p < 0.01 as compared with control vector-transfected cells. F, H460 cells were treated with FZ for 24 or 48 h, and the cell lysates were prepared and processed for immunoblotting using p21 or Mdm2 antibodies. G, i, Western blot for cyclin B1; ii, result of in vitro kinase assay for CDK2 in H460 control and treated cells. Error bars, S.D.
FZ-induced p53 Is Transcriptionally Active and Results in the Up-regulation of Its Target Genes
p53 target gene expressions were further analyzed by RT-PCR after FZ treatment. Fig. 3D shows an induction in mRNA levels of common p53 target genes like p21, BAX, MDM2, and PIG3. To ascertain the change in transcriptional activity of p53, luciferase assay was performed after co-transfecting p53 null human lung cancer H1299 cells with pWWP-Luc (containing a 2.4-kb genomic fragment from the human p21 promoter driving expression of luciferase) and pCMV-wild type p53 construct or empty vector. After 24 h of treatment with 1 μm FZ, an ∼7-fold increase in luciferase activity was observed, confirming an increase in p53 transcriptional activity (Fig. 3E). Western blot analysis of p53-regulated genes also showed an increase in the protein levels (Fig. 3F). In addition, when Western blot analysis for cyclin B1, a gene associated with the G2/M phase of cell cycle, was performed, a clear induction was observed (Fig. 3G (i)). Also, an in vitro kinase assay for CDK2 revealed an increase in its activity 24 and 48 h after treatment (Fig. 3G (ii)). Therefore, the results indicate that cell cycle regulators and pro-apoptotic genes, including cyclin B1, p53, and p21, were induced prior to cells undergoing apoptosis following FZ treatment.
FZ Inhibits the Proteasome Function
The majority of proteins in the cell are degraded by ubiquitin-proteasome pathway, and inhibition of proteasomal function can markedly increase the stability of proteins in the cell. Also, proteasomal function is known to be inhibited in response to stress inside the cells. Therefore, we sought to explore if FZ exerts its effect through inhibition of proteasome function, which could lead to a myriad of downstream consequences, many of which have been implicated in mediating cancer cell death.
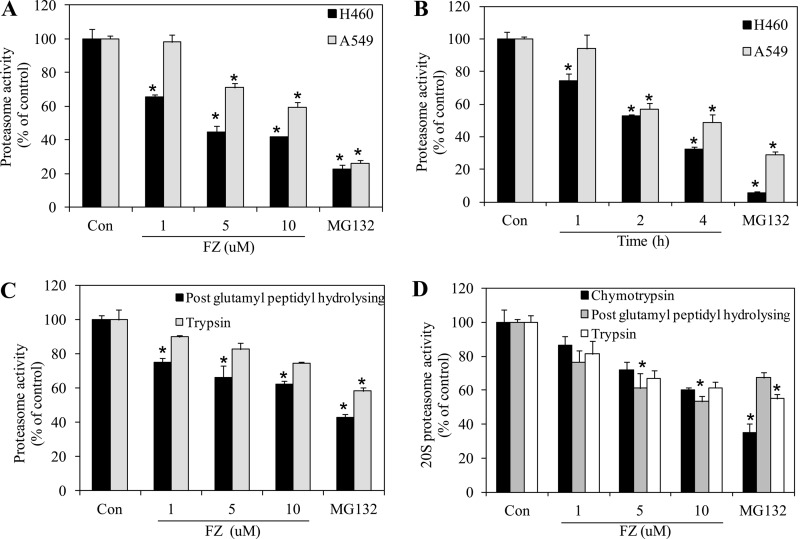
Human lung cancer cell lines H460 and A549 were exposed to FZ, and chymotrypsin-like proteasome activity was measured using a model fluorogenic substrate, succinyl-Leu-Leu-Val-Tyr-MCA, as described under “Experimental Procedures.” As shown in Fig. 4, A and B, FZ inhibited the proteasomal activity in a dose- and time-dependent manner in both of the cell lines. FZ exposure at doses of 1, 5, and 10 μm for 8 h caused 40–60% inhibition of proteasomal activity in H460 cells. Proteasomal inhibition was also noticed when cells were treated with FZ in a time-dependent manner. The direct inhibitory effect of FZ on the proteasome was further analyzed by adding it to untreated cell lysates prior to the proteasome assay (supplemental Fig. 3). The compound's biochemical effect on proteasome was further elucidated as shown in Fig. 4C. Proteasomal assays for post-glutamyl peptidyl-hydrolyzing and trypsin-like protease activities showed that FZ could effectively inhibit the protease function in a dose-dependent manner. Because FZ inhibited the proteasomal activity in cell culture, we tried to explore the possible mechanism of proteasome inhibition. We examined the direct effect of FZ on 20S proteasome activity. The purified 20S proteasome was incubated with varying doses of FZ in the presence of chymotrypsin-like, post-glutamyl peptidyl-hydrolyzing, and trypsin-like protease substrates, and protease activities were monitored. Fig. 4D shows that FZ significantly reduced the 20S proteasomal activities comparable with MG132, which was used as a positive control. Therefore, it appeared that FZ induces apoptosis possibly via inhibition of proteasomal function of the cancer cells.
FIGURE 4.
FZ inhibits proteasome activity. A, H460 and A549 cells were exposed to different doses of FZ for 8 h. Cells were then collected and processed for the proteasome activity assay (chymotrypsin like protease activity) using fluorogenic substrate Z-Leu-Leu-Glu-7-amido-4-MCA as described under “Experimental Procedures.” B, H460 and A549 cells were treated with 5 μm FZ for different time periods as indicated and processed for the proteasome assay. In A and B, the treatment groups show significant inhibition of proteasome activity as compared with control. C, H460 cells were exposed to different doses of FZ for 4 h. Cells were then collected, and cell extracts were processed for proteasome activity assays for post-glutamyl peptidyl hydrolyzing and trypsin-like protease activities using the fluorogenic substrates Z-Leu-Leu-Glu-7-amido-4-MCA and Boc-Gln-Ala-Arg-7-amido-4-MCA, respectively. D, purified 20S proteasome was incubated with different doses of FZ in the presence of chymotrypsin-like, post-glutamyl peptidyl-hydrolyzing, and trypsin-like fluorogenic substrates of proteasome, and the activities were assayed as described. MG132 was used as a positive control. *, p < 0.01 as compared with control untreated cells. Error bars, S.D.
To confirm the results further, we looked for the global ubiquitylation status of FZ-treated cells as compared with untreated control cells. Because proteasome inhibition would block the degradation of several ubiquitylated proteins, it should lead to the increased accumulation of ubiquitylated forms of numerous proteins. Fig. 5A shows the increase in the accumulation of ubiquitylated derivatives of various cellular proteins following exposure to FZ in a dose-dependent manner. This result was confirmed by immunofluorescence using ubiquitin antibody (Fig. 5B). Again, an overexposed p53 blot showed multiple higher molecular weight bands corresponding to ubiquitin-conjugated protein comparable with those seen in cells treated with known proteasomal inhibitor MG132 (Fig. 5C). To substantiate our results, we transiently transfected H460 cells with FLAG-ubiquitin plasmid and then treated them with different doses of FZ. Cell extracts were subjected to Western blot analysis using either anti-FLAG or anti-ubiquitin antibodies (Fig. 5D). Results showed an increase in the levels of ubiquitylated proteins dose-dependently.
FIGURE 5.
FZ treatment leads to an increased accumulation of ubiquitylated proteins. H460 cells were exposed to increasing concentrations of FZ for 24 h, and cell lysates were made and processed for immunoblotting (A) or the cells were processed for immunofluorescence (B) using anti-ubiquitin antibody (a, control; b, FZ; c, MG132). C, H460 cells were exposed to 1 μm FZ for 48 h or 15 μm MG132 for 12 h, and cell lysates were then prepared and processed for Western blotting using antibody against p53 (DO-1). The blot was overexposed for detection of ubiquitylated p53 bands (top, overexposure; bottom, normal exposure). D, H460 cells were transiently transfected with FLAG-ubiquitin expression plasmid. 24 h later, cells were treated with different doses of FZ. The cell lysates were then processed for immunoblotting using anti-FLAG or anti-ubiquitin antibodies. β-Actin served as an internal control.
Hence, we inferred that the substantial induction of p53 observed earlier might also be attributed to its accumulation because of the proteasomal dysfunction. Altogether, these results indicate a proteasomal inhibitory activity of FZ.
FZ Treatment Causes a Decrease in the Turnover of a Model Substrate of Proteasome
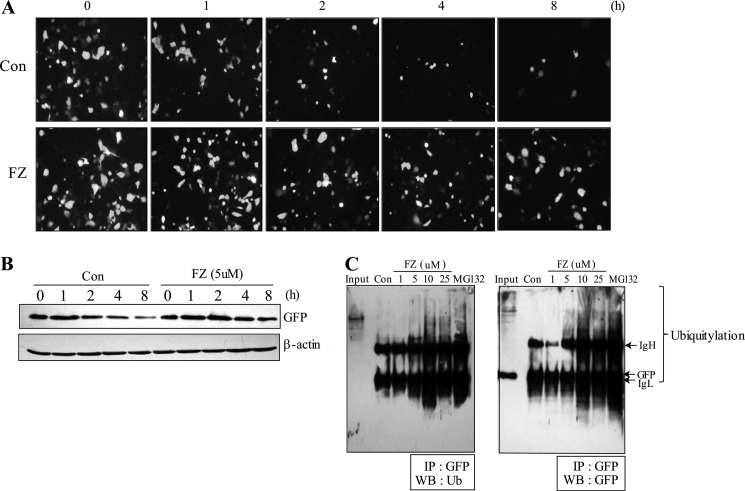
To further ascertain the activity of FZ as an inhibitor of proteasomal function, we used destabilized enhanced green fluorescent protein (d1EGFP), which is a known model substrate for proteasome with a half-life of 1 h. The pd1EGFP vector contains multiple proline, glutamic acid, serine, and threonine signal sequence in its C terminus, which identifies it as a substrate for proteasome-mediated degradation. Inhibition of the proteasome would result in an increased half-life of d1EGFP. H1299 cells were transiently transfected with the pd1EGFP expression plasmid, following which they were chased with cycloheximide in the presence or absence of 5 μm FZ. Fluorescence microscopic observation showed that treatment with FZ significantly increased the half-life of d1EGFP protein (Fig. 6A), which was confirmed by subjecting the cellular extracts to Western blotting using GFP and β-actin antibodies (Fig. 6B). This was reaffirmed by treating the cells with different doses of FZ following transfection with d1EGFP expression constructs and subjecting cell lysates to co-immunoprecipitation experiments using GFP antibody followed by Western blotting using ubiquitin or GFP antibodies. Expectedly, FZ treatment resulted in an increased accumulation of ubiquitylated d1EGFP proteins (Fig. 6C). MG132 was used as a positive control.
FIGURE 6.
FZ exposure decreases the turnover of the destabilized EGFP, a model substrate of proteasome. A, H1299 cells were transiently transfected with pd1EGFP plasmid as described under “Experimental Procedures.” 24 h post-transfection, cells were treated with cycloheximide (50 μg/ml) and chased in the presence or absence of 5 μm FZ for different time periods. GFP fluorescence was observed under a Nikon fluorescence microscope. B, following treatment, the cells were processed for Western blotting using GFP and β-actin antibodies. C, H1299 cells were transiently transfected as described in A and treated with different doses of FZ. Cell lysates were subjected to immunoprecipitation (IP) using anti-GFP antibody followed by Western blot analysis (WB) using GFP and ubiquitin antibodies.
Exposure to FZ Increases the Half-life of Various Cellular Proteasomal Substrates
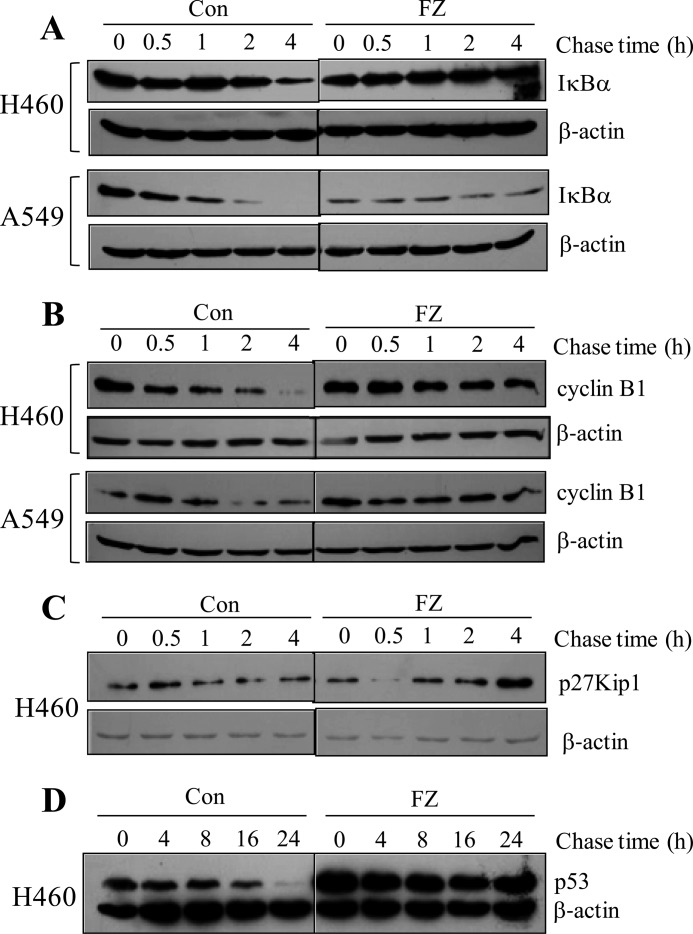
Our results indicate that FZ acts as a potent proteasome inhibitor. This observation led us to examine more closely the stability of some important target molecules involved in cell signaling, cell cycle regulation, and apoptosis. We measured the half-lives of IκBα, p27Kip1, p53, and cyclin B1.
First, we checked the effect of FZ on the accumulation and degradation of IκBα, a known target for proteasomal degradation. As shown in Fig. 7A, exposure to FZ caused an increase in the half-life of IκBα in A549 and H460 cells. A similar increase was observed in the half-lives of other proteasomal targets cyclin B1, p27Kip1, and p53 (Fig. 7, B–D). These findings were consistent with our earlier results wherein we showed stabilization of p53 with increased half-life.
FIGURE 7.
FZ treatment increases the half-life of known proteasomal substrates IκBα, cyclin B1, p27Kip1, and p53. H460 and A549 cells were treated with 50 μg/ml cycloheximide and chased in the presence and absence of 1 μm FZ for different time periods. Cell extracts were prepared, and Western blotting experiments were done using IκBα (A), cyclin B1 (B), p27Kip1 (C), p53 (D), and β-actin antibodies. In case of p53, cycloheximide was added 24 h after FZ treatment. FZ treatment significantly increased the accumulation of p53, cyclin B1, p27Kip1, and IκBα in comparison with control at all of the time points tested.
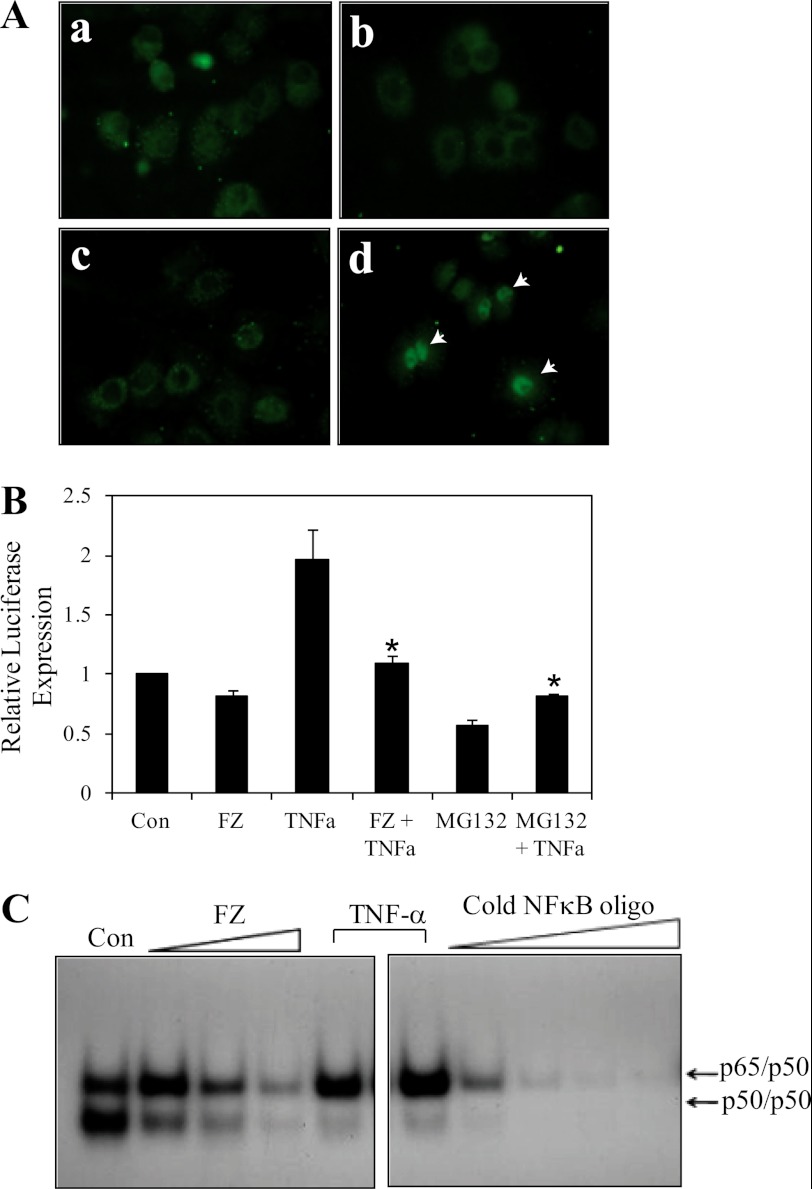
The increase in half-life of IκBα is expected to result in the down-regulation of NFκB activity. Immunofluorescence for NFκB in H460 cells pretreated with FZ showed an inhibition in the nuclear translocation of NFκB after TNFα exposure (Fig. 8A). Consequently, the transcriptional activity of NFκB was reduced upon FZ treatment, as determined by luciferase assay. Pretreatment with FZ resulted in an inhibition of TNFα-induced NFκB activity in A549 cells (Fig. 8B). Next, we performed EMSA for assessing NFκB DNA binding activity. Treatment with FZ led to a dose-dependent decrease in NFκB levels (Fig. 8C). Taken together, these results suggest that FZ exposure results in a down-regulation of NFκB activity, which, in turn, is likely to be mediated via stabilization of IκBα. However, IKK inhibitor wedelolactone, which inhibits NFκB-mediated gene transcription in cells by blocking the phosphorylation and degradation of IκBα, had no significant effect on FZ-induced apoptosis (supplemental Fig. 4). Therefore, it appeared that the hindrance of the prosurvival NFκB pathway may not be completely responsible for inhibition of cell proliferation by FZ.
FIGURE 8.
FZ blocks nuclear translocation of NFκB and results in down-regulation of its transcriptional activity. A, H460 cells were either left untreated (a), exposed to 5 μm FZ for 4 h (b), exposed to FZ followed by 30 ng/ml TNFα for 1 h (c), or exposed to TNFα only (d). Following treatment, the cells were subjected to immunofluorescence using NFκB (p65) antibody. B, A549 cells were transiently transfected with NFκB luciferase and pRL-SV40 plasmids. 24 h after transfection, cells were treated with 5 μm FZ or 10 μm MG132 for 4 h, followed by 30 ng/ml TNFα for 1 h, where indicated. Cells were then processed for dual luciferase reporter gene assay as described. *, p < 0.01 as compared with cells treated with TNFα only. C, H460 cells were treated with different doses of FZ for 4 h or TNFα for 30 min, and nuclear extracts were prepared and processed for EMSA as described under “Experimental Procedures,” using 32P-end-labeled oligonucleotide encoding an NFκB-binding sequence as probes. A competition assay was performed using a 5–100-fold excess of unlabeled competitor. Error bars, S.D.
FZ Increases ROS Generation in A549 Cells
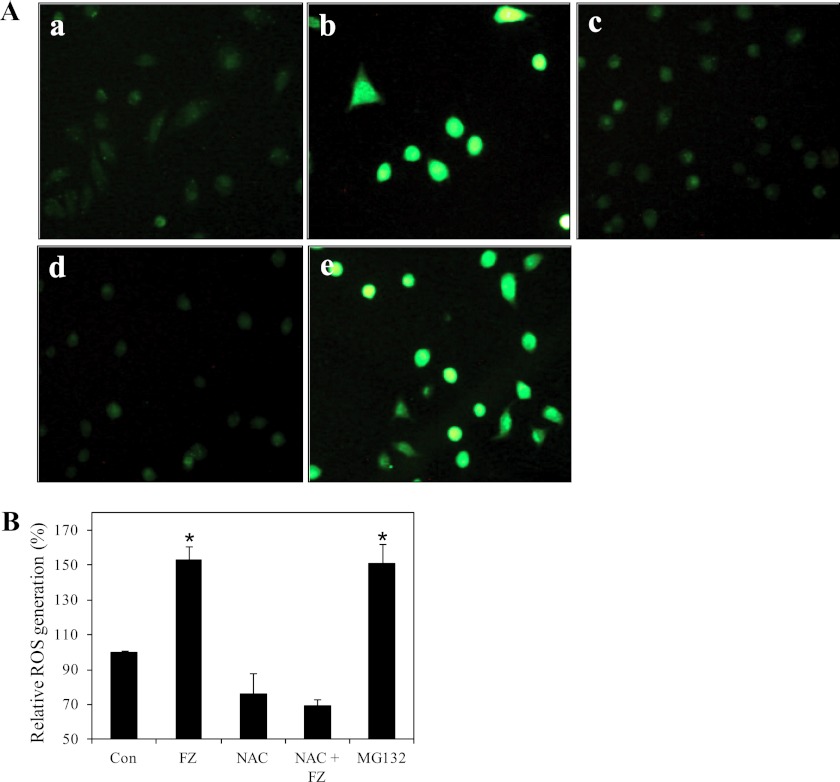
It has been reported that proteasome inhibition increases intracellular ROS, which play a key role in apoptosis in some cancer cells. Apoptotic signals may disrupt the mitochondrial electron transport chain, leading to ROS production (29). We examined ROS production in A549 cells treated with FZ by 2-dichlorodihydrofluoresein diacetate staining. As shown in Fig. 9, A and B, 6 h of FZ treatment resulted in a marked increase in ROS production as compared with control untreated cells. Significantly, a 2-h pretreatment with the antioxidant NAC was able to inhibit the ROS production by FZ, suggesting that ROS production may have some role in FZ- induced apoptosis in these cells.
FIGURE 9.
FZ increases ROS generation. A, A549 cells were treated with 1 μm FZ or 10 μm MG132 for 4 h. NAC (10 mm) was added 2 h before drug treatment. The cells were then stained with DCFDA for detection of ROS by fluorescence microscopy. a, control; b, FZ; c, NAC; d, NAC followed by FZ; e, MG132. B, for quantitative analysis, fluorescence intensity was measured at 530 nm after excitation at 485 nm in a PerkinElmer Life Sciences Victor 3X fluorometer. *, p < 0.01 as compared with control untreated cells. Error bars, S.D.
ER Stress Response following FZ Treatment
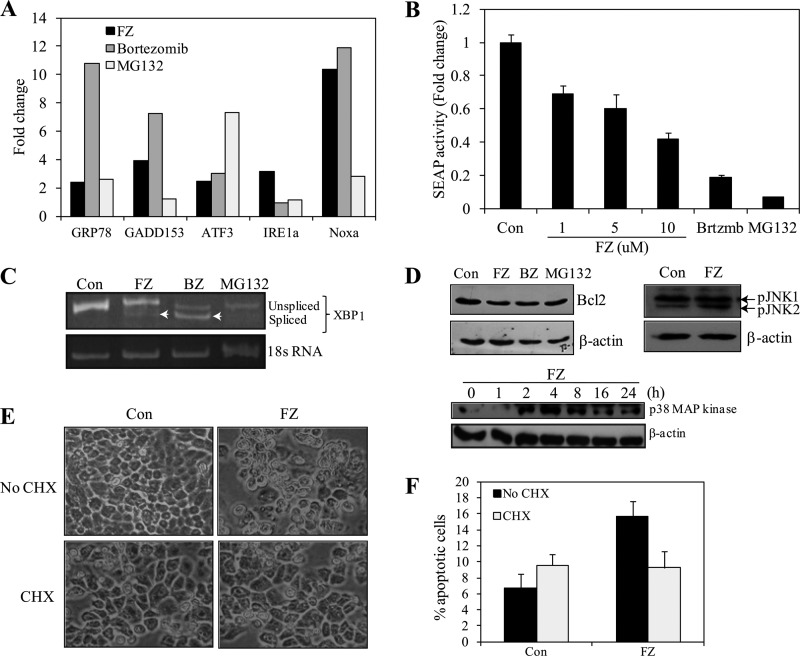
Proteasome inhibitors have been shown to cause cell death via the ER stress-ROS pathway (30). We examined the expression of a set of known unfolded protein response genes, namely BiP/GRP78, CHOP/GADD153, IRE1α, ATF3, and NOXA, by RT-PCR (data not shown) and quantitative PCR. Analysis of real-time PCR data indicated that there was an ∼2–4-fold increase in expression levels of most of these genes in response to 24-h FZ treatment, whereas NOXA showed an ∼11-fold increase (Fig. 10A). Bortezomib and MG132 were used as positive controls. Because the level of SEAP production has been shown to be inversely correlated with the induction of ER stress, the assay has been successfully used to detect and quantify ER stress in real time (31). SEAP traverses the secretory pathway, and its activity can be detected at very low levels (0.2 pg/ml). Because SEAP is a secreted protein, medium supernatant can be assayed for SEAP activity. Most importantly, transfection of cells with SEAP does not, by itself, cause ER stress. When 293T cells were transiently transfected with SEAP vector and exposed to FZ for 24 h, there was a dose-dependent decrease in the accumulation of SEAP protein in the medium (Fig. 10B).
FIGURE 10.
FZ induces ER stress response in human cancer cells. A, H460 cells were treated with FZ for 24 h, total RNA was isolated, and quantitative PCR was performed for analyzing relative gene expression for ER stress-specific genes. B, 293T cells were transfected with pSEAP-con plasmid, and on the following day fresh media containing 1% serum and specified concentrations of FZ, bortezomib, or MG132 were added. SEAP activities were determined as described under “Experimental Procedures” after incubation for 24 h. C, H460 cells were treated with 1 μm FZ, 1 μm bortezomib, or 10 μm MG132 for 24 h, following which RNA was isolated and reverse transcribed, and semi-quantitative PCR was performed using specific primers for the detection of two different splice variants of XBP1. D, Western blotting for BCL2, phospho-JNK (24 h) and p38 MAPK in H460 cells at different time points after FZ treatment. E, phase-contrast images of H460 cells after treatment with FZ alone or in the presence of cycloheximide (CHX). H460 cells were seeded at a density of 2 × 104 cells/well in 24-well culture plates and then left untreated or treated with 5 μm FZ either alone or in the presence of 10 μg/ml cycloheximide for 24 h. F, cells were then trypsinized, and viable cells were counted by the trypan blue exclusion method. All experiments were done in triplicate. Error bars, S.D.
IRE1 is a transmembrane receptor involved in the initiation phase of ER stress-induced apoptosis. Upon activation, the endonuclease activity of IRE1 removes a 26-nucleotide intron from the XBP1 mRNA, generating a frameshift splice variant (sXBP1), which encodes a stable, active transcription factor (32). We detected this splicing of XBP1 mRNA by semiquantitative RT-PCR using primers specific for XBP1 that can detect both unspliced and spliced isoforms. Our results show the two isoforms, although the spliced variant was not as marked as in the case of bortezomib, which was included as a positive control along with MG132 (Fig. 10C).
Upon ER stress, IRE1 stimulates p38, causing activation of JNK. We observed an increase in phosphorylated JNK and p38 MAPK levels and a reduction in BCL2 levels after FZ treatment (Fig. 10D). Up-regulation of CHOP/GADD153, as well as JNK activation by ER stress, is known to repress BCL2 gene expression and execute apoptosis by the activation of proapoptotic BCL2 proteins in the cell (33). Taken together, these results suggest that ER stress may be involved in FZ-mediated cell death. Interestingly, treatment of cells with translational inhibitor cycloheximide, which relieves ER load, was able to abrogate FZ-induced cytotoxicity (Fig. 10, E and F). This finding is consistent with earlier reports showing a protective effect of cycloheximide on cells under ER stress (34, 35) and further supports our notion that FZ-induced cell death is ER stress-dependent.
FZ Induces Mitochondrial Membrane Depolarization, Cytochrome c Release, and Activation of Caspases
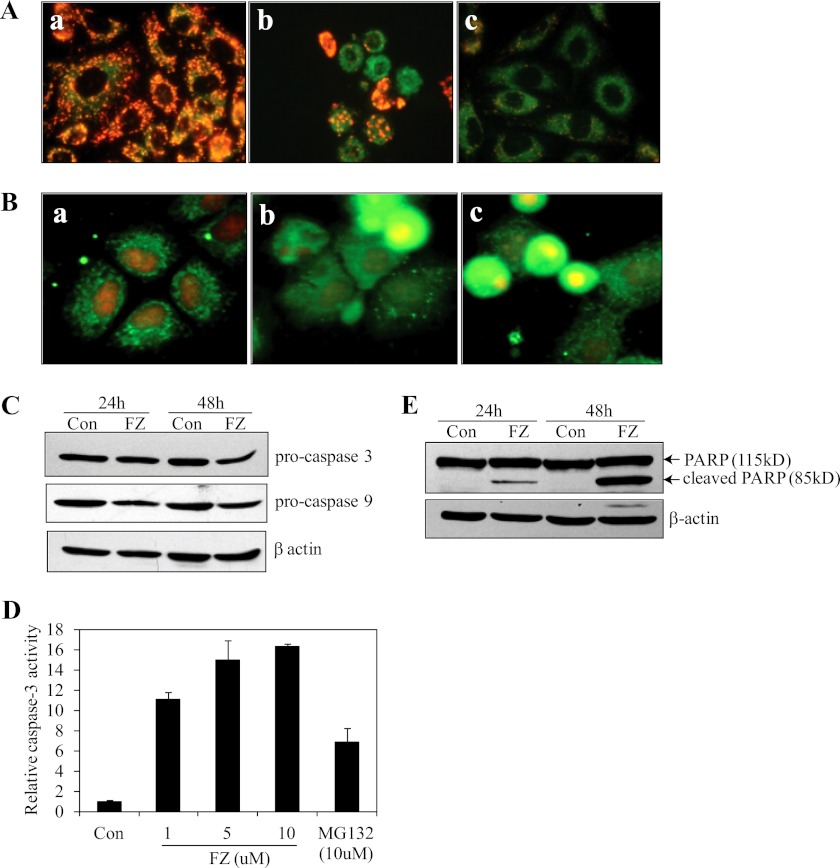
Proteasomal inhibition is known to induce the mitochondrial pathway of apoptosis, and because we also observed a marked induction of p53 by FZ, we proceeded to check whether the intrinsic pathway of apoptosis was activated in response to FZ. To monitor the effect on mitochondrial membrane potential, we used JC-1, a voltage-sensitive fluorescence dye that forms aggregates in the polarized mitochondria, which fluoresce red but become green as monomers, which diffuse out in the case of depolarized membrane. The treatment with FZ resulted in a decrease in the mitochondrial membrane potential, as evident from the loss of red-dotted mitochondrial staining (Fig. 11A). FZ also induced the cytochrome c release from the mitochondria, as detected by immunofluorescence using cytochrome c antibody (Fig. 11B).
FIGURE 11.
FZ treatment induces cytochrome c release from mitochondria, activates caspase-3, and causes PARP cleavage. A, H460 cells were treated with either 1 μm FZ for 24 h or 10 μm MG132 for 12 h and then processed for JC-1 staining. a, control; b, FZ; c, MG132. B, after treatment as described in A, the cells were subjected to immunofluorescence using cytochrome c antibody. C, H460 cells were plated onto 35-mm tissue culture plates, and on the following day, the cells were treated with different doses of FZ for 24 or 48 h. The cells were then collected and processed for immunoblotting using caspase-3 and caspase-9 antibodies. D, H460 cells were treated with different doses of FZ or 10 μm MG132 for 48 h, and cell extracts were then prepared and processed for the caspase-3 activity assay using fluorogenic substrate as described under “Experimental Procedures.” E, H460 cell extracts were prepared as described in C, and Western blotting was performed using anti-PARP antibody. Error bars, S.D.
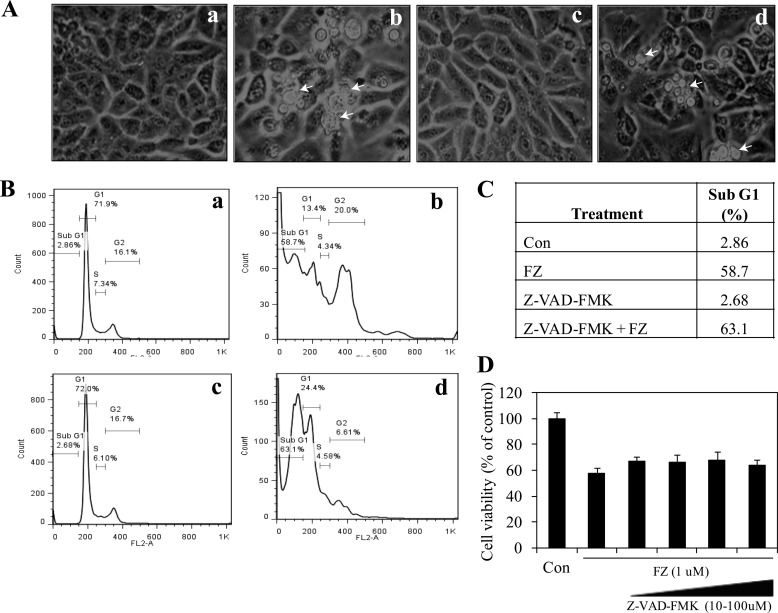
Because the release of cytochrome c from mitochondria finally results in the activation of caspase-3, we next examined its level by Western blotting. As shown in Fig. 11C, Western blot for pro-caspase-3 upon FZ exposure showed a decrease in its level that could be correlated with a corresponding increase in its activity measured using a fluorogenic substrate (Fig. 11D). Further, treatment with 1 μm FZ for 24 and 48 h resulted in cleavage of the 115-kDa PARP into an 85-kDa product (Fig. 11E). The above results lead to the conclusion that FZ activates the cytochrome c-mediated intrinsic pathway of apoptosis in these cells. However, pretreatment with broad-spectrum caspase inhibitor Z-VAD-FMK had no effect on FZ-induced cell death. Cells pretreated with Z-VAD-FMK could not be morphologically distinguished from cells exposed to FZ alone (Fig. 12, A–D). Because caspase inhibition has been shown to be ineffective in preventing ER stress-mediated cell death (36, 37), this result is in agreement with our data indicating the role of ER stress in FZ-induced apoptosis.
FIGURE 12.
Z-VAD-FMK pretreatment does not rescue the cells from FZ-induced apoptosis. A, phase-contrast images of H460 cells after 48 h of FZ treatment alone or cells pretreated with 50 μm Z-VAD-FMK for 2 h are shown (a, control; b, FZ; c, Z-VAD-FMK only; d, Z-VAD-FMK followed by FZ). Arrowheads, apoptotic nuclei. B, cell cycle analysis of H460 cells indicating the percentage of sub-G1 apoptotic cells (C) after different treatment regimes as in A. D, human H460 cells were treated with increasing concentrations of Z-VAD-FMK (10, 25, 50, and 100 μm) and FZ as indicated, and cell viability was measured after 48 h. Error bars, S.D.
DISCUSSION
FZ belongs to a class of broad-spectrum anthelmintics, the benzimidazoles, which have a high therapeutic index. The key factor to which the success of benzimidazoles as effective anthelmintic agents is attributable is their selective toxicity to helminths and high degree of safety toward normal human cells. This has been explained previously on the basis of two different phenomena: by selective targeting of parasitic tubulin by benzimidazoles due to their structural refinement or by differences in the metabolic and detoxification pathways between mammals and target parasites (10–12, 38). Earlier, a few compounds belonging to this class have been shown to act as effective antitumor agents, although the molecular mechanism remains elusive. In this study, we show that fenbendazole acts as a growth inhibitor and causes apoptotic cell death in human lung cancer cell lines.
Appropriate, targeted proteolysis is essential for cellular function and homeostasis. In eukaryotic cells, the ubiquitin proteasome pathway is the central non-lysosomal pathway for protein degradation (39). The vast majority of proteins are selectively degraded by the ubiquitin-proteasome system, which, in turn, makes it crucial to the cell's existence. The dynamic balance of substrate ubiquitylation and degradation plays an important role in cell cycle progression and cell survival (39, 40). It is, therefore, self-evident that the dysfunction of this pathway will promote cell death. Predictably, the pharmacological inhibition of proteasome function has been found to induce dual apoptotic signaling pathways, depending on the specific cellular context, and various proteasome inhibitors have been propelled toward therapeutic applications, bortezomib (Velcade) being the first clinically approved small molecule proteasome inhibitor (41). However, bortezomib has been reported to have no anti-tumor activity in a non-small cell lung cancer-derived xenograft mouse model (3). After bortezomib, several second generation proteasome-targeting agents like NPI-0052 (salinosporamide A), MLN9708, CEP18770 (phase I), argyrin A, and carfilzomib (phase III) have entered clinical trials (1, 41–45). Our results indicated that FZ acts as an inhibitor of proteasomal function and results in cell death by activating the mitochondrial pathway.
First, we have shown that the exposure to FZ inhibits the cellular proteasome function dose- and time-dependently and leads to accumulation of ubiquitylated derivatives of various cellular proteins, including p53, which, in turn, leads to apoptosis via the mitochondrial pathway. This was further confirmed by inhibition of degradation of destabilized enhanced green fluorescence proteins, a model substrate of proteasome, and also various other cellular substrates of proteasome like IκBα, p53, and cyclin B1. Finally, we have demonstrated that FZ treatment results in changes in mitochondrial membrane potential, release of cytochrome c from mitochondria into cytosol, and activation of downstream caspases followed by PARP cleavage. However, the broad-spectrum caspase inhibitor Z-VAD-FMK did not substantially rescue the cells from FZ-induced cell death, suggesting the existence of an alternative, caspase-independent mechanism that is instrumental for the cell-killing effect of FZ.
Our results reveal that in response to FZ treatment, the cells first undergo G2/M arrest followed by apoptosis. p53 activation has been previously reported to be a key event in cell death caused by proteasomal inhibitors (46). Because FZ treatment results in marked induction of p53 and corresponding up-regulation of its target genes, this could contribute to the cells undergoing cell death via the mitochondrial pathway. Therefore, FZ-induced proteasomal dysfunction could indirectly cause mitochondrial cytochrome c release. Besides p53 and its target genes, proteasome inhibition can also modulate the expression of other proteins implicated in the apoptotic pathway as well as in cell growth and survival pathways (47, 48). This alteration in the degradation of various cellular proteins would result in a change in their expression levels, which, in turn, is likely to affect cell survival and promote apoptosis. In addition, we also report inhibition of NFκB transcriptional activity after FZ treatment. This can possibly be attributed to the stabilization and increase in half-life of IκBα, which is known to inhibit the NFκB cell survival pathway. We provide evidence that FZ-induced proteasomal dysfunction prevents the degradation of IκBα and thereby blocks the nuclear translocation and transactivation of NFκB. In this context, it might be noted that the NFκB pathway, which may play a key role in protecting cells from oxidative stress is known to be disabled by proteasome inhibitors (49, 50).
FZ-induced ROS generation could be associated with the transcriptional activation of ROS-related genes. Our data show PIG3 induction upon FZ treatment. PIG3 is a p53-inducible gene homolog of oxidoreductase and is reported to be associated with p53-induced elevation of ROS production (51).
Proteasomal inhibition is likely to accumulate mitochondrial damage and generate oxidative stress inside the cell (52). Oxidative stress can, in turn, affect proteasome function either by producing excessive levels of damaged proteins, causing unfolded protein response or by reducing the levels of ATP production. Proteasome inhibitors have been previously reported to elicit ER stress response (53, 54). Certain compounds selectively kill cancer cells by induction of ER stress-ROS, because cancer cells are expected to rely strongly on the ROS stress response pathway as compared with normal cells (55–58). Altogether, our results indicate that FZ induces cytotoxicity in cancer cells via impairment of proteasomal function and induction of unfolded protein response.
The ubiquitin-proteasome system is integral to cellular protein homeostasis. The growing recognition of the fundamental importance of this pathway to normal cell function and in disease has prompted an in depth search for small-molecule inhibitors that selectively block the function of these pathways. Therefore, we propose FZ as a novel drug candidate that targets the ubiquitin-proteasome pathway and effectively kills cancer cells. The in vivo efficacy of FZ as an anti-tumor agent is being studied.
Supplementary Material
Acknowledgments
We greatly appreciate the gift of the pWWP-Luc construct and H1299 and HCT116 (p53−/− and p53+/+) cell lines from Dr. Bert Vogelstein; the FLAG-ubiquitin construct from Dr. Caixia Guo (University of Texas Southwestern Medical Center, Dallas, TX); and pNFκB-Luc kindly provided by Prof. Macus T. Kuo (University of Texas MD Anderson Cancer Center, Houston, TX).
This work was supported by grants from the University Grants Commission and Indian Council of Agricultural Research, Government of India (to T. M.).

This article contains supplemental Table 1 and Figs. 1–4.
- FZ
- fenbendazole (methyl N-(6-phenylsulfanyl-1H-benzimidazol-2-yl)carbamate)
- Z
- benzyloxycarbonyl
- MG132
- Z-Leu-Leu-Leu-al
- NAC
- N-acetylcysteine
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NSCLC
- non-small cell lung carcinoma
- JC-1
- 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolocarbocyanine iodide
- EGFP
- enhanced green fluorescence protein
- MCA
- methylcoumarinamide
- DCFDA
- 6-carboxy-2′,7′-dichlorodihydrofluorescein diacetate
- FMK
- fluoromethyl ketone
- PI
- propidium iodide
- SEAP
- secreted alkaline phosphatase
- ER
- endoplasmic reticulum
- ROS
- reactive oxygen species
- PARP
- poly(ADP-ribose) polymerase.
REFERENCES
- 1. Adams J. (2004) The proteasome. A suitable antineoplastic target. Nat. Rev. Cancer 4, 349–360 [DOI] [PubMed] [Google Scholar]
- 2. Hoeller D., Dikic I. (2009) Targeting the ubiquitin system in cancer therapy. Nature 458, 438–444 [DOI] [PubMed] [Google Scholar]
- 3. Williamson M. J., Silva M. D., Terkelsen J., Robertson R., Yu L., Xia C., Hatsis P., Bannerman B., Babcock T., Cao Y., Kupperman E. (2009) The relationship among tumor architecture, pharmacokinetics, pharmacodynamics, and efficacy of bortezomib in mouse xenograft models. Mol. Cancer Ther. 8, 3234–3243 [DOI] [PubMed] [Google Scholar]
- 4. Davies A. M., Lara P. N., Jr., Mack P. C., Gandara D. R. (2007) Incorporating bortezomib into the treatment of lung cancer. Clin. Cancer Res. 13, s4647–s4651 [DOI] [PubMed] [Google Scholar]
- 5. Eckert J., Conraths F. J., Tackmann K. (2000) Echinococcosis. An emerging or re-emerging zoonosis? Int. J. Parasitol. 30, 1283–1294 [DOI] [PubMed] [Google Scholar]
- 6. Ammann R. W., Ilitsch N., Marincek B., Freiburghaus A. U. (1994) Effect of chemotherapy on the larval mass and the long-term course of alveolar echinococcosis. Swiss Echinococcosis Study Group. Hepatology 19, 735–742 [DOI] [PubMed] [Google Scholar]
- 7. Müller E., Akovbiantz A., Ammann R. W., Bircher J., Eckert J., Wissler K., Witassek F., Wüthrich B. (1982) Treatment of human echinococcosis with mebendazole. Preliminary observations in 28 patients. Hepatogastroenterology 29, 236–239 [PubMed] [Google Scholar]
- 8. Marriner S. E., Morris D. L., Dickson B., Bogan J. A. (1986) Pharmacokinetics of albendazole in man. Eur. J. Clin. Pharmacol. 30, 705–708 [DOI] [PubMed] [Google Scholar]
- 9. El-On J. (2003) Benzimidazole treatment of cystic echinococcosis. Acta Trop. 85, 243–252 [DOI] [PubMed] [Google Scholar]
- 10. Lacey E., Watson T. R. (1985) Structure-activity relationships of benzimidazole carbamates as inhibitors of mammalian tubulin, in vitro. Biochem. Pharmacol. 34, 1073–1077 [DOI] [PubMed] [Google Scholar]
- 11. Laclette J. P., Guerra G., Zetina C. (1980) Inhibition of tubulin polymerization by mebendazole. Biochem. Biophys. Res. Commun. 92, 417–423 [DOI] [PubMed] [Google Scholar]
- 12. Gull K., Dawson P. J., Davis C., Byard E. H. (1987) Microtubules as target organelles for benzimidazole anthelmintic chemotherapy. Biochem. Soc. Trans. 15, 59–60 [DOI] [PubMed] [Google Scholar]
- 13. Robinson M. W., McFerran N., Trudgett A., Hoey L., Fairweather I. (2004) A possible model of benzimidazole binding to β-tubulin disclosed by invoking an interdomain movement. J. Mol. Graph. Model. 23, 275–284 [DOI] [PubMed] [Google Scholar]
- 14. Morris D. L., Jourdan J. L., Pourgholami M. H. (2001) Oncology 61, 42–46 [DOI] [PubMed] [Google Scholar]
- 15. Gao P., Dang C. V., Watson J. (2008) Unexpected antitumorigenic effect of fenbendazole when combined with supplementary vitamins. J. Am. Assoc. Lab. Anim. Sci. 47, 37–40 [PMC free article] [PubMed] [Google Scholar]
- 16. Pourgholami M. H., Cai Z. Y., Badar S., Wangoo K., Poruchynsky M. S., Morris D. L. (2010) Potent inhibition of tumoral hypoxia-inducible factor 1α by albendazole. BMC Cancer 10, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mukhopadhyay T., Sasaki J., Ramesh R., Roth J. A. (2002) Mebendazole elicits a potent antitumor effect on human cancer cell lines both in vitro and in vivo. Clin. Cancer Res. 8, 2963–2969 [PubMed] [Google Scholar]
- 18. Sasaki J., Ramesh R., Chada S., Gomyo Y., Roth J. A., Mukhopadhyay T. (2002) The anthelmintic drug mebendazole induces mitotic arrest and apoptosis by depolymerizing tubulin in non-small cell lung cancer cells. Mol. Cancer Ther. 1, 1201–1209 [PubMed] [Google Scholar]
- 19. Nakamura S., Gomyo Y., Roth J. A., Mukhopadhyay T. (2002) C terminus of p53 is required for G2 arrest. Oncogene 21, 2102–2107 [DOI] [PubMed] [Google Scholar]
- 20. Gold R., Schmied M., Giegerich G., Breitschopf H., Hartung H. P., Toyka K. V., Lassmann H. (1994) Differentiation between cellular apoptosis and necrosis by the combined use of in situ tailing and nick translation techniques. Lab. Invest. 71, 219–225 [PubMed] [Google Scholar]
- 21. Jana N. R., Zemskov E. A., Wang G., Nukina N. (2001) Altered proteasomal function due to the expression of polyglutamine-expanded truncated N-terminal huntingtin induces apoptosis by caspase activation through mitochondrial cytochrome c release. Hum. Mol. Genet. 10, 1049–1059 [DOI] [PubMed] [Google Scholar]
- 22. Chomczynski P., Sacchi N. (2006) The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Twenty-something years on. Nat. Protoc. 1, 581–585 [DOI] [PubMed] [Google Scholar]
- 23. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 24. Nakamura S., Roth J. A., Mukhopadhyay T. (2000) Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Mol. Cell. Biol. 20, 9391–9398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schreiber E., Matthias P., Müller M. M., Schaffner W. (1989) Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maxwell S. A., Mukhopadhyay T. (1995) A novel NF-κB p65 spliced transcript lacking exons 6 and 7 in a non-small cell lung carcinoma cell line. Gene 166, 339–340 [DOI] [PubMed] [Google Scholar]
- 27. el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 28. Dash B. C., El-Deiry W. S. (2005) Phosphorylation of p21 in G2/M promotes cyclin B-Cdc2 kinase activity. Mol. Cell. Biol. 25, 3364–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen Q., Chai Y. C., Mazumder S., Jiang C., Macklis R. M., Chisolm G. M., Almasan A. (2003) The late increase in intracellular free radical oxygen species during apoptosis is associated with cytochrome c release, caspase activation, and mitochondrial dysfunction. Cell Death Differ. 10, 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fribley A., Zeng Q., Wang C. Y. (2004) Proteasome inhibitor PS-341 induces apoptosis through induction of endoplasmic reticulum stress-reactive oxygen species in head and neck squamous cell carcinoma cells. Mol. Cell. Biol. 24, 9695–9704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hiramatsu N., Kasai A., Hayakawa K., Yao J., Kitamura M. (2006) Real-time detection and continuous monitoring of ER stress in vitro and in vivo by ES-TRAP. Evidence for systemic, transient ER stress during endotoxemia. Nucleic Acids Res. 34, e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 [DOI] [PubMed] [Google Scholar]
- 33. Szegezdi E., Logue S. E., Gorman A. M., Samali A. (2006) Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 7, 880–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Harding H. P., Zhang Y., Bertolotti A., Zeng H., Ron D. (2000) Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5, 897–904 [DOI] [PubMed] [Google Scholar]
- 35. Fels D. R., Ye J., Segan A. T., Kridel S. J., Spiotto M., Olson M., Koong A. C., Koumenis C. (2008) Preferential cytotoxicity of bortezomib toward hypoxic tumor cells via overactivation of endoplasmic reticulum stress pathways. Cancer Res. 68, 9323–9330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Egger L., Madden D. T., Rhême C., Rao R. V., Bredesen D. E. (2007) Endoplasmic reticulum stress-induced cell death mediated by the proteasome. Cell Death Differ 14, 1172–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Egger L., Schneider J., Rhême C., Tapernoux M., Häcki J., Borner C. (2003) Serine proteases mediate apoptosis-like cell death and phagocytosis under caspase-inhibiting conditions. Cell Death Differ. 10, 1188–1203 [DOI] [PubMed] [Google Scholar]
- 38. Lacey E., Gill J. H. (1994) Biochemistry of benzimidazole resistance. Acta Trop. 56, 245–262 [DOI] [PubMed] [Google Scholar]
- 39. Almond J. B., Cohen G. M. (2002) The proteasome. A novel target for cancer chemotherapy. Leukemia 16, 433–443 [DOI] [PubMed] [Google Scholar]
- 40. Bernassola F., Ciechanover A., Melino G. (2010) The ubiquitin proteasome system and its involvement in cell death pathways. Cell Death Differ. 17, 1–3 [DOI] [PubMed] [Google Scholar]
- 41. Bedford L., Lowe J., Dick L. R., Mayer R. J., Brownell J. E. (2011) Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat. Rev. Drug Discov. 10, 29–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Orlowski R. Z., Kuhn D. J. (2008) Proteasome inhibitors in cancer therapy. Lessons from the first decade. Clin. Cancer Res. 14, 1649–1657 [DOI] [PubMed] [Google Scholar]
- 43. Adams J., Kauffman M. (2004) Development of the proteasome inhibitor Velcade (Bortezomib). Cancer Invest. 22, 304–311 [DOI] [PubMed] [Google Scholar]
- 44. Adams J. (2004) The development of proteasome inhibitors as anticancer drugs. Cancer Cell 5, 417–421 [DOI] [PubMed] [Google Scholar]
- 45. Nickeleit I., Zender S., Sasse F., Geffers R., Brandes G., Sörensen I., Steinmetz H., Kubicka S., Carlomagno T., Menche D., Gütgemann I., Buer J., Gossler A., Manns M. P., Kalesse M., Frank R., Malek N. P. (2008) Argyrin a reveals a critical role for the tumor suppressor protein p27Kip1 in mediating antitumor activities in response to proteasome inhibition. Cancer Cell 14, 23–35 [DOI] [PubMed] [Google Scholar]
- 46. Lopes U. G., Erhardt P., Yao R., Cooper G. M. (1997) p53-dependent induction of apoptosis by proteasome inhibitors. J. Biol. Chem. 272, 12893–12896 [DOI] [PubMed] [Google Scholar]
- 47. Mitsiades N., Mitsiades C. S., Poulaki V., Chauhan D., Fanourakis G., Gu X., Bailey C., Joseph M., Libermann T. A., Treon S. P., Munshi N. C., Richardson P. G., Hideshima T., Anderson K. C. (2002) Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc. Natl. Acad. Sci. U.S.A. 99, 14374–14379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mitsiades N., Mitsiades C. S., Poulaki V., Chauhan D., Richardson P. G., Hideshima T., Munshi N., Treon S. P., Anderson K. C. (2002) Biologic sequelae of nuclear factor-κB blockade in multiple myeloma. Therapeutic applications. Blood 99, 4079–4086 [DOI] [PubMed] [Google Scholar]
- 49. Lezoualc'h F., Sagara Y., Holsboer F., Behl C. (1998) High constitutive NF-κB activity mediates resistance to oxidative stress in neuronal cells. J. Neurosci. 18, 3224–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hideshima T., Chauhan D., Richardson P., Mitsiades C., Mitsiades N., Hayashi T., Munshi N., Dang L., Castro A., Palombella V., Adams J., Anderson K. C. (2002) NF-κB as a therapeutic target in multiple myeloma. J. Biol. Chem. 277, 16639–16647 [DOI] [PubMed] [Google Scholar]
- 51. Polyak K., Xia Y., Zweier J. L., Kinzler K. W., Vogelstein B. (1997) A model for p53-induced apoptosis. Nature 389, 300–305 [DOI] [PubMed] [Google Scholar]
- 52. Lomonosova E., Ryerse J., Chinnadurai G. (2009) BAX/BAK-independent mitoptosis during cell death induced by proteasome inhibition? Mol. Cancer Res. 7, 1268–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nawrocki S. T., Carew J. S., Pino M. S., Highshaw R. A., Dunner K., Jr., Huang P., Abbruzzese J. L., McConkey D. J. (2005) Bortezomib sensitizes pancreatic cancer cells to endoplasmic reticulum stress-mediated apoptosis. Cancer Res. 65, 11658–11666 [DOI] [PubMed] [Google Scholar]
- 54. Nawrocki S. T., Carew J. S., Dunner K., Jr., Boise L. H., Chiao P. J., Huang P., Abbruzzese J. L., McConkey D. J. (2005) Bortezomib inhibits PKR-like endoplasmic reticulum (ER) kinase and induces apoptosis via ER stress in human pancreatic cancer cells. Cancer Res. 65, 11510–11519 [DOI] [PubMed] [Google Scholar]
- 55. Raj L., Ide T., Gurkar A. U., Foley M., Schenone M., Li X., Tolliday N. J., Golub T. R., Carr S. A., Shamji A. F., Stern A. M., Mandinova A., Schreiber S. L., Lee S. W. (2011) Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature 475, 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56. Trachootham D., Alexandre J., Huang P. (2009) Targeting cancer cells by ROS-mediated mechanisms. A radical therapeutic approach? Nat. Rev. Drug. Discov. 8, 579–591 [DOI] [PubMed] [Google Scholar]
- 57. Trachootham D., Zhou Y., Zhang H., Demizu Y., Chen Z., Pelicano H., Chiao P. J., Achanta G., Arlinghaus R. B., Liu J., Huang P. (2006) Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by β-phenylethyl isothiocyanate. Cancer Cell 10, 241–252 [DOI] [PubMed] [Google Scholar]
- 58. Huang P., Feng L., Oldham E. A., Keating M. J., Plunkett W. (2000) Superoxide dismutase as a target for the selective killing of cancer cells. Nature 407, 390–395 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.