Background: Matrix attachment region-binding proteins regulate gene expression through anchoring chromatin loops to nuclear scaffold.
Results: Matrix attachment region-binding protein SATB2 promotes G/Aγ-globin genes transcription, facilitates histone acetylase recruitment, and mediates physical association of G/Aγ-globin promoters.
Conclusion: SATB2 activates γ-globin gene expression as an active chromatin organizer.
Significance: The role of SATB2 as a novel γ-globin gene regulator is explored.
Keywords: Chromatin Regulation, DNA-Protein Interaction, Gene Regulation, Hemoglobin, Transcription Coactivators, γ-Globin Gene, Higher-order Chromatin Structure, MAR-binding Protein, PCAF, SATB2
Abstract
Matrix attachment region (MAR)-binding protein (MARBP) has profound influence on gene transcriptional control by tethering genes to the nuclear scaffold. MARBP SATB2 is recently known as a versatile regulator functioning in the differentiation of multiple cell types including embryonic stem cells, osteoblasts and immunocytes. Roles of SATB2 in erythroid cells and its working mechanism in orchestrating target gene expression are largely unexplored. We show here that SATB2 is expressed in erythroid cells and activates γ-globin genes by binding to MARs in their promoters and recruiting histone acetylase PCAF. Further analysis in higher-order chromatin structure shows that SATB2 affects physical proximity of human Gγ- and Aγ-globin promoters via self-association. We also found that SATB2 interacts with SATB1, which specifically activates ϵ-globin gene expression. Our results establish SATB2 as a novel γ-globin gene regulator and provide a glimpse of the differential and cooperative roles of SATB family proteins in modulating clustered genes transcription and mediating higher-order chromatin structures.
Introduction
The AT-rich matrix attachment region (MAR)4 elements that mediate the attachment of DNA to the nuclear matrix have been found to participate in the regulation of chromatin structure and gene transcription in combination with a spectrum of MAR-binding proteins, including Cux/CDP (1), SAF-A (2), Bright (3), SATB1 (4), and SATB2 (5). The pre-B cell-enriched SATB2 binds to MARs of the immunoglobulin μ locus and increases transcription of the gene (5). A study using knock-out mice showed that SATB2 regulates osteoblast function by modulating osteoblast-specific Runx2, ATF4 and several Hox genes (6). Although many genes that are targeted by SATB2 have been identified (5–7), the working mechanism of SATB2 in regulating target genes transcription has been largely unexplored.
The β-globin gene locus provides an excellent model for the study of gene regulation and chromatin structures. The human β-globin gene locus contains five functional globin genes ordered by their developmental expressions: ϵ → Gγ → Aγ → δ → β. The activation of β-like globin genes is accompanied by physical associations between the active promoters and hypersensitive sites of the locus control region (LCR) that constitute the active chromatin hub (ACH) (8, 9). Several well-characterized transcriptional factors, such as EKLF (10), GATA-1 (11), and CTCF (12), contribute to formation of this ACH. Recently, SATB1, homologue of SATB2, was found to transcriptionally regulate the ϵ-globin gene (13). Our previous study showed that SATB1 tethers ϵ-globin gene to the nuclear matrix and identified SATB1-mediated inter-MAR association in the β-globin gene locus accompanying the expression of ϵ-globin gene (14, 15), modulation of SATB1 acetylation by SIRT1 facilitates MARHS2-MARϵ association and promotes ϵ-globin expression (16). These studies on SATB1 suggest a potential role of MAR-based higher-order chromatin structures fundamental to the organization of ACH.
MAR elements have been identified in the G/Aγ-globin promoters (MAR-G/Aγ-pro) and in the intergenic region between γ- and δ-globins (MAR-IR-1, -2, -3). SATB1 was found to bind these MARs in vitro but not in vivo (13, 17–20). Here, we demonstrate that SATB2 is expressed in erythroid cells and binds to MAR-G/Aγ-pro in vivo. SATB2 activates γ-globin gene transcription through recruiting co-activator PCAF and mediating the physical proximity of G/Aγ-globin promoters. The present work, together with our previously identified SATB1-centered inter-MAR association, hints potential differentiation and cooperation of SATB family proteins in regulating the expression and higher-order chromatin structure organization of a gene cluster.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
K562 cells were maintained in RPMI 1640 medium, and 50 μm hemin (Sigma) was added to induce erythroid differentiation. 293T cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen). All media were supplemented with glutamine, penicillin/streptomycin and 10% fetal bovine serum. Primary human CD34+ cells were obtained from magnetically sorted mononuclear samples of umbilical cord blood from donors and were expanded and induced for erythroid differentiation as previously described, with some modifications (21). Cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Mice and Tissue Preparation
Specific pathogen-free C57BL/6 mice were obtained from the Laboratory Animal Center of the Chinese Academy of Military Medical Sciences. All animal experiments were performed in accordance with institutional guidelines. Yolk sac and fetal liver were collected, washed with ice-cold phosphate-buffered saline and frozen in liquid nitrogen before use. TER119-positive erythroid cells were isolated from the mouse tissues using an anti-TER119 antibody coupled to magnetic beads (Miltenyi Biotec) following the manufacturer's instruction.
Plasmids, Virus Production, and Antibodies
Full-length and serial deletion mutants of SATB2 and SATB1 were constructed in pCMV-Tag2B (Stratagene) and pcDNA3.1-myc-his plasmids (Invitrogen), respectively. Flag-PCAF was constructed in pCMV-Tag2B. The retrovirus-mediated overexpression of SATB2 was performed using the pMSCVneo system (Clontech) according to the manufacturer's instructions. The shRNA targeting SATB2 or green fluorescence protein (GFP) was inserted into pSIREN-retroQ (Clontech) with the following sequences: shSATB2, 5′-CCA GAG CAC ATT AGC CAA A-3′, and shGFP, 5′-GCA AGC TGA CCC TGA AGT T-3′. The plasmids were transfected into 293T cells along with pMD and pVSV-G, and viral supernatant was collected to infect target cells as described in the supplier's protocol.
Reporter constructs were generated by PCR amplification of G/Aγ-globin promoters (Gγ: −268 bp to +27 bp, Aγ: −264 bp to +27 bp) and cloning into the pGL3-Basic vector (Promega). The wild-type G/Aγ-globin promoter plasmids were modified using a site-directed mutagenesis kit (Promega) to generate the G/Aγ-globin promoter-ΔMAR plasmid (G/Aγ: −215 bp to +27 bp) and G/Aγ-globin promoters-mutMAR, in which the AT-rich DNA elements (Gγ: 5′-AATTAA-3′ from −230 bp to -225 bp, Aγ: 5′-AATTA-3′ from −226 bp to −222 bp) were replaced with 5′-CCG GCC-3′ and 5′-CCG GC-3′, respectively. Mutations were confirmed by sequencing. All of the indicated sites are relative to their respective transcriptional start site.
The anti-SATB2 antibody was purchased from Abcam. The anti-SATB1 antibody was purchased from BD Biosciences. Anti-Flag and anti-actin monoclonal antibodies were purchased from Sigma-Aldrich. Antibodies against GATA-1, p45 NF-E2, EKLF, CBP, p300, PCAF, and c-Myc were from Santa Cruz Biotechnology. Antibodies against AcH3, AcH4, and dimeH3K4 were from Upstate Biotechnology.
RNA Isolation and Real-time RT-PCR
Total RNA was extracted using an RNA extraction kit (Invitrogen). Two micrograms of total RNA was reverse transcribed by extension of oligo(dT) primers (TaKaRa) using M-MLV reverse transcriptase (New England Biolabs) according to the manufacturer's protocol. Real-time PCR was performed on a Bio-Rad IQ5 cycler using a SYBR Green reaction mix (Takara). Relative expression level is calculated using the housekeeping gene GAPDH as internal control for all real-time PCR assays. Values are the means and standard deviations of the results from at least three independent experiments. All primer sets are available upon request.
Immunoprecipitation and Tandem Affinity Purification (TAP)
For immunoprecipitation, cells were collected and solubilized in IP buffer (1% Nonidet P-40, 150 mm NaCl, 50 mm Tris-HCl (pH 7.4), 0.25% sodium deoxycholate, 1 mm EDTA, and protease inhibitors) at 4 °C. Lysed protein mixtures were incubated with specific antibodies and precipitated with protein A or G-agarose (Upstate Biotechnology) at 4 °C overnight. The immunocomplex was collected, washed three to five times and probed with various antibodies. For the TAP assay, human SATB2 protein was fused to two tandem-affinity tags (SBP and CBP), and SATB2-containing complexes were isolated using the InterPlay Mammalian TAP System following the manufacturer's instructions (Stratagene). The purified complex was separated on a Bis-Tris gradient gel, visualized using silver staining and further characterized by mass spectrometry.
Biotinylated DNA Pull-down Assay and Electrophoretic Mobility Shift Assay (EMSA)
A biotinylated DNA pull-down assay was performed essentially as previously described (22), with a few modifications. Briefly, a biotinylated double-stranded oligonucleotide was incubated with nuclear proteins in binding buffer (10% glycerol, 10 mm HEPES (pH 8.0), 2.5 mm MgCl2, 40 mm KCl, 1 mm dithiothreitol, 50 μg/ml BSA, and 10 μg/ml poly(dI-dC)) at 4 °C for 4 h. Then, streptavidin-agarose beads (Merck) were added to the protein mixture and incubated at 4 °C for 2 h. Prior to this step, streptavidin-agarose beads were blocked with BSA and poly(dI-dC) to minimize nonspecific binding. The pellet was washed five times with binding buffer. Recovered proteins were separated using SDS-PAGE, cut from gel, and digested in gel for mass spectrometry.
The EMSA assay was performed using the LightShift Chemiluminescent EMSA Kit (Pierce) according to the manufacturer's instructions. Recombinant GST-SATB2 and control GST were expressed in Escherichia coli strain BL-21 using the pET42a vector system and purified on glutathione-Sepharose (GE Healthcare). Probes used in the EMSA included the following: wtMAR-Gγ-pro 5′-GAA TCG GAA CAA GGC AAA GGC TAT AAA AAA AAT TAA GCA GCA GTA TCC TCT TGG-3′ and wtMAR-Aγ-pro 5′-GAA TCG GAA CAA GGC AAA GGC TAT AAA AAA AAT TAG CAG TAT CCT CTT GG-3′.Competitor probes included the following: wtMAR (MAR from mouse IgH enhancer)/mutMAR 5′-TCT TTA ATT TCT AAT ATA TTT AGA ATT C-3′/5′- TCT TTA ATT TCT ACT GCT TTA GAA TTC-3′; wtMAR-G/Aγ-pro; serially mutated MAR-Gγ-pro (e.g. mutMAR-Gγ-pro-6) 5′-GAA TCG GAA CAA GGC AAA GGC TAT AAA AAA CCG GCC GCA GCA GTA TCC TCT TGG-3′; and mutMAR-Aγ-pro (corresponding to mutMAR-Gγ-pro-6) 5′-GAA TCG GAA CAA GGC AAA GGC TAT AAA AAA CCG GCG CAG TAT CCT CTT GG-3′.
Luciferase Reporter Assay
Luciferase assays were performed using a Dual Luciferase Reporter Assay System (Promega). Luciferase activity was normalized against that of TK-Renilla reporter (Promega) used as an internal control (30 ng/well). Results were expressed as the ratio of firefly to Renilla luciferase activity.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed essentially as previously described (23) with semi-quantitative PCR or quantitative PCR. The cross-linked chromatin DNA was sheared to an average size of 500 bp before precipitation with respective antibody. For quantification of the ChIP assay, real-time PCR was performed with SYBR Green dye on an iCycler iQ (Bio-Rad) system. All qPCR signals from IP samples were normalized to that of respective input samples. Values shown are the means and standard deviations of the results from at least three independent experiments. All primer sets are available upon request.
Chromosome Conformation Capture (3C) Assay and ChIP-3C Assay
The 3C assay was performed essentially as previously described with some modifications (23, 24). To calculate the relative crosslinking frequency between two given HindIII fragments, semi-quantitative PCR reactions were performed. A β-globin BAC clone (BAC 186D7) was digested, re-ligated and used as a control for primer efficiency. To correct for the amount of template in different samples, Ercc3–1 and Ercc3–2, primers from two HindIII restriction fragments of the housekeeping Ercc3 gene locus, were used for internal control amplification. The correction equation used was as previously described (8). The results are shown as percentages of relative proximity of the control group, which was set as 1. The ChIP-3C assay was performed essentially as previously described (14). Briefly, cross-linked chromatin was sonicated and digested using HindIII before being mixed with anti-SATB2 antibodies and precipitated using protein G-agarose beads. Subsequently, the beads were resuspended in ligation buffer for overnight ligation. After the bead washing, ligated chromatin fragments were eluted, and crosslinking was reversed for DNA purification and PCR amplification. The specificity of primer pairs was verified by sequencing their PCR products amplified from the control BAC library. All of the PCR reactions were performed in triplicate and averaged. Primer sequences and PCR conditions are available upon request.
RESULTS
Pull-down of SATB2 by MAR Element within Gγ-Globin Promoter from Erythroid Cells
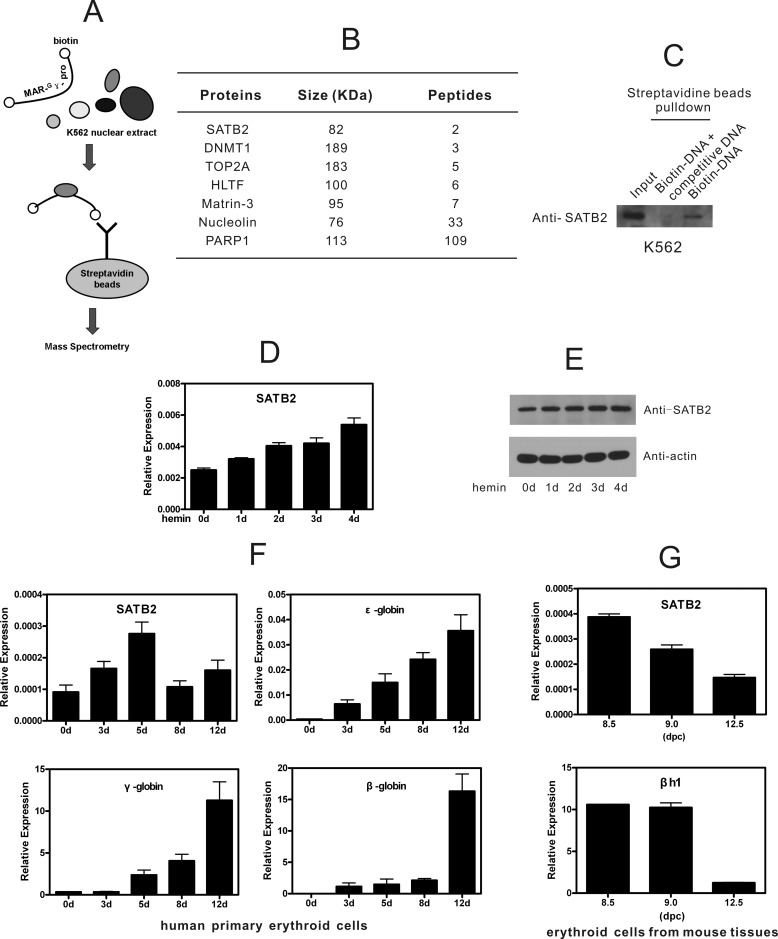
To identify MAR-binding proteins responsible for human γ-globin gene expression, we first performed biotinylated DNA pull-down screening with MAR-Gγ-pro (MAR element within the Gγ-globin promoter) using a double-stranded oligonucleotide as the probe (Fig. 1A). The associated proteins that precipitated from nuclear extracts of K562 cells were resolved using SDS-PAGE and subjected to mass spectrometry analysis after silver staining. As expected, the analysis revealed a variety of ubiquitously expressed nuclear matrix-binding proteins, including poly (ADP-ribose) polymerase I, HMG proteins, and nucleolin (Fig. 1B). Notably, the MAR-binding protein SATB2, a homolog of SATB1, was also found to bind MAR-Gγ-pro. SATB2 has been reported to bind AT-rich regulatory elements for cell type-specific transcriptional regulation in pre-B cells, osteoblasts, brain, and embryonic stem (ES) cells (5–7, 25). However, the regulatory roles of SATB2 in erythroid cells have not been investigated. Therefore, we verified the result with Western blotting using anti-SATB2 antibody following MAR-Gγ-pro pull-down in K562 cells and confirmed the binding of endogenous SATB2 to MAR-Gγ-pro (Fig. 1C). An excess of unlabeled MAR-Gγ-pro was included in the pull-down assay to confirm the binding specificity.
FIGURE 1.
MAR element in Gγ-globin promoter pulls down SATB2, which is expressed in erythroid cells. A, schematic representation of biotinylated DNA pull-down assay for MAR-Gγ-pro-associating proteins in K562 cells. The biotinylated MAR-Gγ-pro probe (−260 bp to −207 bp relative to TSS) was incubated with K562 nuclear extracts and conjugated to streptavidin-agarose beads. After bead separation and washing, bound proteins were recovered and resolved using SDS-PAGE followed by mass spectrometry identification. B, proteins identified in A are shown with sizes and the number of peptides. C, Western blot assay following MAR-Gγ-pro pull-down in K562 cells confirmed binding of endogenous SATB2 to MAR-Gγ-pro. Binding specificity was shown by adding unlabeled MAR-Gγ-pro as competitive DNA. D and E, real-time RT-PCR (D) and Western blot (E) analyses of SATB2 in hemin-treated K562 cells. Each error bar represents a standard deviation calculated from experiments performed in triplicate. F, real-time RT-PCR analysis of β-like globin genes and SATB2 during erythroid differentiation of human umbilical cord CD34+ cells. Each error bar represents a standard deviation calculated from experiments performed in triplicate. G, real-time RT-PCR analysis of SATB2 and βh1 expression in TER119+ erythroid cells of 8.5, 9.0 dpc murine yolk sacs, and 12.5 dpc murine fetal liver. Each error bar represents a standard deviation calculated from experiments performed in triplicate.
Analysis in erythroid cell lines showed that SATB2 was consistently expressed during hemin-induced differentiation of human K562 cells where fetal γ-globin was activated (Fig. 1, D and E), whereas undetectable in adult-stage murine MEL cells and G1E-ER4 cells upon DMSO- or β-estradiol-induced differentiation, where βh1, mouse counterpart of γ-globin gene, was inactive (supplemental Fig. S1A). SATB2 expression was also detected in erythropoietin (Epo) induced human umbilical CD34+ cells, which actively express γ-globin gene (Fig. 1F), and in TER 119+ cells from 8.5, 9.0 dpc murine yolk sacs expressing abundant βh1 (Fig. 1G). Both SATB2 and βh1 expression drop significantly in TER 119+ cells of 12.5 dpc murine fetal liver (Fig. 1G). The SATB2 levels are comparable to that of SATB1 in K562 cells (Fig. 1, F and G, and supplemental Fig. S1B), consistent to the magnitude of transcriptional factors. Collectively, concomitant expression of SATB2 and γ-globin genes were detected in erythroid cell lines and primary erythroid cells/tissues, suggesting potential regulatory connection between the two genes.
SATB2 Activates Human γ-Globin Gene Expression in K562 Cells and Human Primary Erythroid Cells
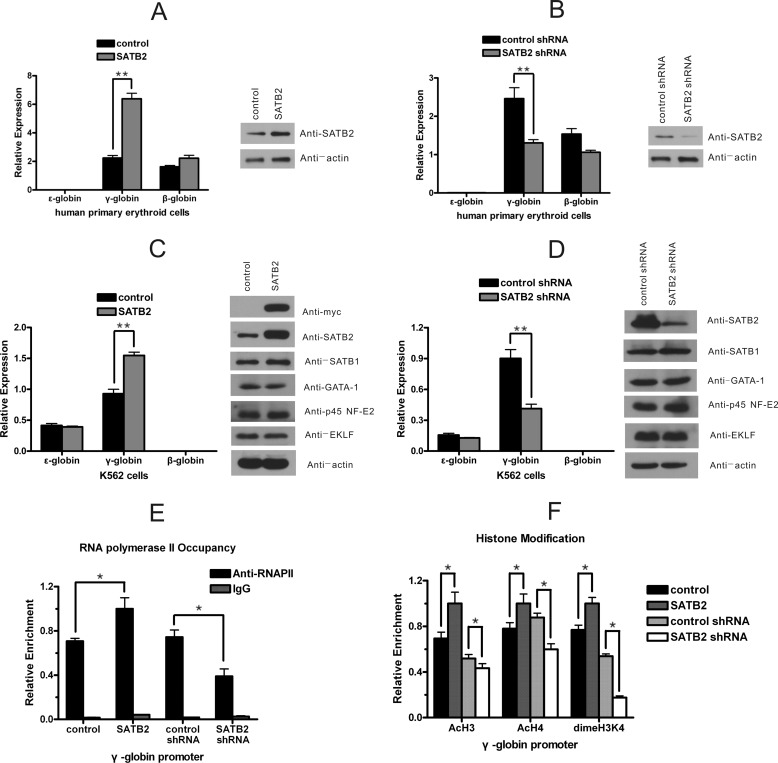
To examine the potential roles of SATB2 in the regulation of β-like globin genes, we overexpressed or knocked-down SATB2 in human erythroid cells. We first characterized the influence of SATB2 on the expression of β-like globin genes in human primary erythroid cells derived from cord blood CD34+ progenitors. Three days after infecting the cells with retroviruses to overexpress or knockdown SATB2, mRNA was collected for subsequent quantitative RT-PCR analysis. SATB2 was shown efficiently overexpressed or knocked down, γ-globin gene expression was shown positively correlated with SATB2 levels in primary erythroid cells (Fig. 2, A and B). Expression of ϵ-globin gene in the primary erythroid cells was much lower than that of γ- and β-globin genes, and was therefore invisible in the figure. Next, we infected K562 cells with retroviruses harboring Myc-tagged human SATB2 cDNA and examined the perturbations to the expression of β-like globin genes in these cells against cells infected with control retroviruses (Fig. 2C). Benzidine staining showed that hemoglobin-positive K562 cells increased from 2 to 12% in response to SATB2 overexpression (supplemental Fig. S2). Quantitative RT-PCR results further revealed that expression of γ-globin gene, but not that of ϵ-globin gene, was up-regulated in SATB2-overexpressed K562 cells, expression of β-globin gene was much lower than that of γ- and ϵ-globin genes in these cells. In addition, the expression of detected globin regulators remained largely undisturbed, as shown using Western blot analysis (Fig. 2C). This indicates that SATB2 activates γ-globin gene without activation of erythroid differentiation. We further infected K562 cells with retroviruses encoding short hairpin RNAs targeting either SATB2 or GFP. Specific down-regulation of γ-globin gene expression was observed after SATB2 knockdown. No obvious expression changes were observed for detected globin regulators (Fig. 2D). Concomitant alterations of H3 and H4 acetylation, H3K4 dimethylation and polymerase II recruitment at γ-globin gene promoters were also detected in response to the genetic modification of SATB2 expression in K562 cells (Fig. 2, E and F). Taken together, these results strongly suggest that human SATB2 serves as a transcriptional activator for the human γ-globin gene.
FIGURE 2.
SATB2 activates human γ-globin gene transcription in erythroid cells. A and B, retroviral vectors for (A) SATB2 overexpression or (B) SATB2 knockdown were transfected into human primary erythroid cells at day 1 of differentiation. The expression of SATB2 and β-like globin genes was determined on day 4 using Western blot and real-time RT-PCR. Each error bar represents a standard deviation calculated from experiments performed in triplicate. Student's t test was used for statistical analysis and p values are indicated (**, p < 0.01). C and D, K562 cells were infected with a retrovirus harboring (C) Myc-tagged SATB2 cDNA or (D) short hairpin RNA to knockdown SATB2. Western blot analysis of SATB2, SATB1 and important erythroid transcription factors and real-time RT-PCR analysis of β-like globin genes were performed. Each error bar represents a standard deviation calculated from experiments performed in triplicate. Student's t test was used for statistical analysis, and p values are indicated (**, p < 0.01). E and F, quantitative ChIP analysis of (E) histone modifications and (F) RNA polymerase II presence at the γ-globin promoter in K562 cells after SATB2 overexpression or knockdown. Each error bar represents a standard deviation calculated from experiments performed in triplicate. Student's t test was used for statistical analysis, and p values are indicated (*, p < 0.05).
SATB2 Binds to Human γ-Globin Gene Promoters in K562 Cells
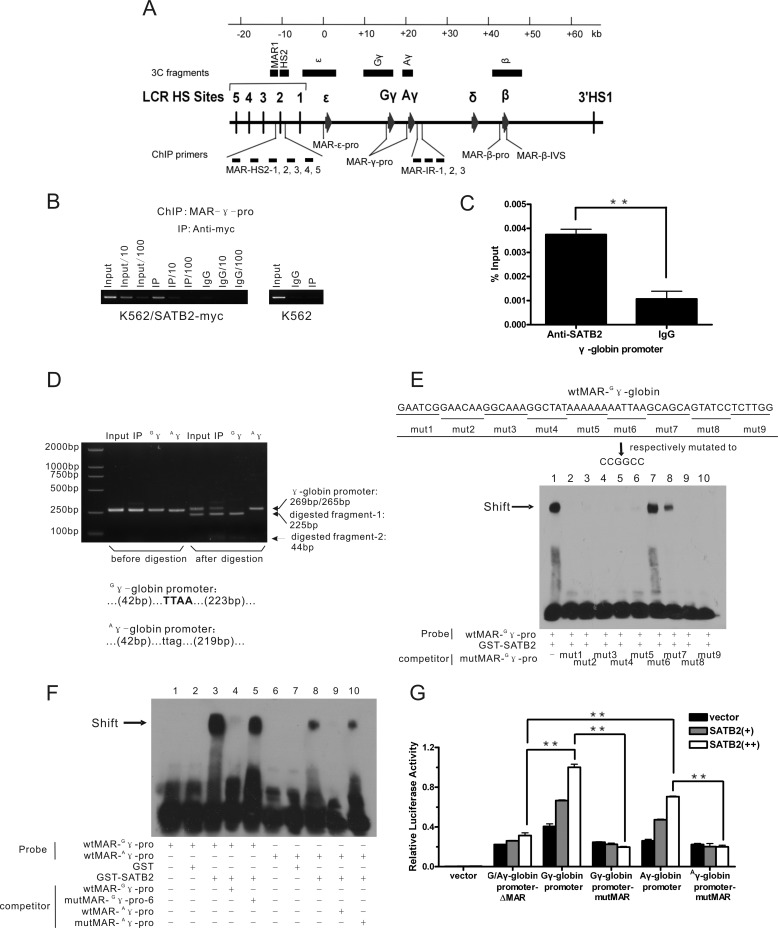
To further investigate the possible mechanism of SATB2, we measured the occupancy by human SATB2 across the β-globin gene cluster in K562 cells (the β-globin gene cluster and the primer pairs for the ChIP assay are shown in Fig. 3A). We first used K562 cells infected with a retrovirus harboring Myc-tagged human SATB2 cDNA (Fig. 2C) and measured the occupancy of myc-SATB2 using a ChIP-grade anti-Myc antibody. Multiple previously reported MAR elements within the β-globin gene cluster were investigated, and the Necdin promoter was used as a negative control. Among all the detected regions, significant enrichment of SATB2 binding was detected only at MAR-γ-pro (MAR elements of the γ-globin gene promoters) (supplemental Fig. S3, A and B). We further determined the intensity of the signal using serial dilution of the ChIP templates and verified the signal specificity by repeating the ChIP assay with extracts of control K562 cells using the anti-Myc antibody (Fig. 3B). ChIP analysis of K562 cells mildly express Flag-tagged SATB2 also showed SATB2 binding at MAR-γ-pro (supplemental Fig. S3C). Finally, binding of native SATB2 to MAR-γ-pro in K562 cells was confirmed using anti-SATB2 antibody (Ab51502) (Fig. 3C). The MAR-γ-pro region detected using the ChIP assay is composed of two homologous MAR elements located respectively at the promoters of Gγ and Aγ, the two γ-globin gene homologs. We therefore examined the binding preference of human SATB2 to the two MAR elements. To discriminate between the two MARs in the ChIP assay, PCR products amplified using MAR-γ-pro ChIP primers were subjected to direct sequencing (supplemental Fig. S3, D and E) or MseI digestion (Fig. 3D, supplemental Fig. S3E), which specifically recognizes the TTAA sequence in the Gγ-globin promoter but not the corresponding CTAA sequence in the Aγ-globin promoter. The sequencing result showed that both MAR-Gγ-pro and MAR-Aγ-pro were immunoprecipitated using the anti-Myc antibody, indicating that the Myc-tagged SATB2 binds to both MAR-γ-pro elements. MseI digestion analysis further showed that although equal amounts of MAR-Gγ-pro and MAR-Aγ-pro were found in the input, MAR-Gγ-pro was more enriched in the precipitated DNA, indicating the preferential binding of SATB2 to MAR-Gγ-pro.
FIGURE 3.
SATB2 occupies G/Aγ-globin promoters in K562 cells. A, schematic representation of the human β-globin locus indicating the locations of ChIP primers (see also supplemental Fig. S3) and 3C fragments (HindIII fragments) used in this study (see Fig. 5, E and F). B, ChIP analysis of myc-SATB2 binding status at the γ-globin gene promoter in K562 cells ectopically expressing Myc-tagged SATB2 using an anti-Myc antibody. Serial dilution was employed for semi-quantitative assay. C, ChIP analysis using anti-SATB2 antibody (Ab51502) shows native SATB2 binding at the γ-globin gene promoter in K562 cells. (**, p < 0.01). D, restriction enzyme assay to show proportion of G/Aγ-globin promoters in the SATB2 ChIP samples before and after immunoprecipitation. The result indicates relative enrichment of the Gγ promoter after immunoprecipitation. The PCR product amplified from the Gγ promoter was digested by MseI into 225-bp and 44-bp bands, whereas that of the Aγ promoter formed an uncut 265-bp band in the assay. Bottom: MAR- G/Aγ-pro sequence across the MseI site. E and F, EMSA analysis of in vitro SATB2 binding at MAR-G/Aγ-pro. The biotinylated DNA probes derived from MAR-G/Aγ-pro were incubated with purified GST-SATB2 in the absence or presence of unlabeled competitors, with GST as a negative control. E, competitors were serially mutated MAR-Gγ-pro with underlined bases changed to CCGGCC or (F) wild-type and mutated MAR-A/Gγ-pro corresponding to mut6 of MAR-Gγ-pro. The arrows indicate the band shift position of the SATB2-MAR complex. G, luciferase reporter analysis of the SATB2 regulatory effect on transcriptional activities of wild-type and mutated G/Aγ- promoters in 293T cells. Each error bar represents a standard deviation calculated from experiments performed in triplicate. Student's t test was used for statistical analysis, and p values are indicated (**, p < 0.01).
An EMSA assay was performed to analyze in vitro binding abilities and the exact binding site of human SATB2 at MAR elements in the Gγ-globin and Aγ-globin promoters. We incubated recombinant human SATB2 with biotinylated oligonucleotides containing either MAR-Gγ-pro or MAR-Aγ-pro and resolved the complex on native polyacrylamide gels. Both probes from the shifted bands were completely competed by the cold probe derived from the MAR element of the mouse IgH gene enhancer but not by the mutated one. Moreover, in accordance with the ChIP results, we found that the same amount of SATB2 shifted more MAR-Gγ-pro probes than the MAR-Aγ-pro probes using the EMSA assay (supplemental Fig. S3F). To further determine the specific regions of the two MAR-γ-pro elements that interacted with SATB2, we generated differentially mutated MAR-Gγ-pro probes by sequentially substituting the indicated six bases with the CCG GCC sequence and used them as cold probes to compete with the wild-type MAR-Gγ-pro probe in an EMSA assay (Fig. 3E). The results demonstrated that the mutation of the AAT TAA sequence abolished the ability of the cold probe to compete with the labeled wild-type probe for SATB2 binding, indicating that the AATTAA sequence is critical for SATB2 association with MAR-Gγ-pro. In addition, the GCA GCA sequence next to the AAT TAA sequence at the 3′-end of MAR-Gγ-pro is important for SATB2 binding because its mutation inhibited the cold probe in the competition test. MAR-Aγ-pro also lost affinity for SATB2 when the homologous motif was mutated (AAT TA to CCG GC) (Fig. 3F). Therefore, we conclude that human SATB2 makes direct contact with both MAR-Gγ-pro and MAR-Aγ-pro, and that MAR-Gγ-pro binds human SATB2 with higher avidity than MAR-Aγ-pro.
Reporter gene assays in 293T cells showed that SATB2 activated both Gγ-globin and Aγ-globin promoters, whereas the regulatory effect was abolished with either deletion or the AAT TAA/AAT TA→CCG GCC/CCG GC mutation of the MAR-γ-pro element in the corresponding γ-globin promoter (Fig. 3G). Taken together, our results demonstrate that SATB2 regulates γ-globin gene expression through direct binding to the MAR-γ-pro elements of human G/Aγ-globin promoters.
PCAF Cooperates with SATB2 in Human γ-Globin Gene Activation
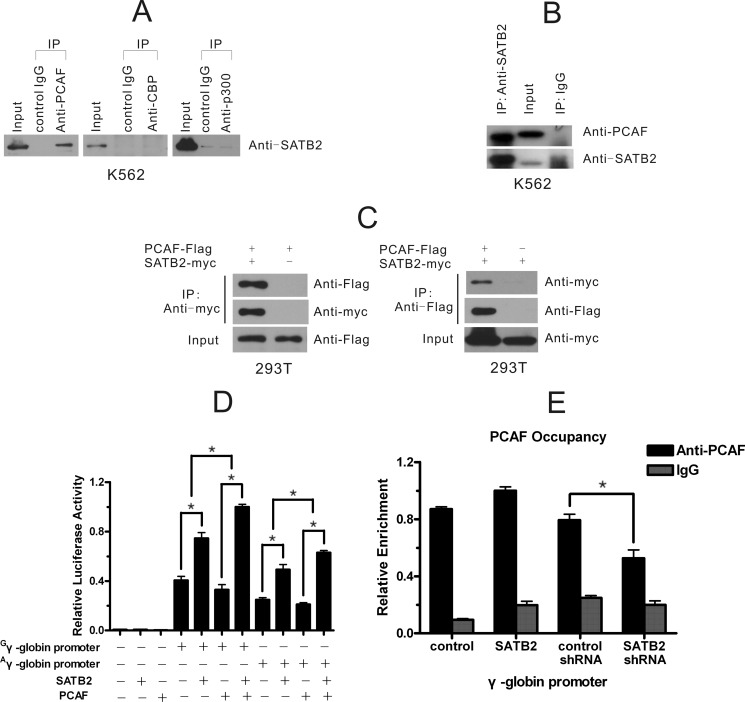
To gain further insight into the molecular basis for the activation of the γ-globin gene by SATB2, we investigated whether histone acetylases participated in the regulation. We immunoprecipitated protein complexes from K562 cell extracts using antibodies against human PCAF, CBP, and p300 with normal rabbit IgG as the negative control and detected SATB2 protein in the precipitates (Fig. 4A). SATB2 co-precipitated with PCAF but not with CBP and p300. The SATB2-PCAF interaction was further confirmed by detecting of PCAF in the immunoprecipitate of SATB2 in K562 cells (Fig. 4B). Mutual co-immunoprecipitation of exogenously expressed SATB2-myc and PCAF-Flag were also detected in 293T cells (Fig. 4C). We next determined the functional relevance of the SATB2-PCAF interaction. Luciferase reporter assay was performed in 293T cells where endogenous SATB2 is low. We showed that PCAF enhanced the activities of G/Aγ-globin promoters only when co-transfected with exogenous SATB2, suggesting the involvement of PCAF in SATB2 activation of the γ-globin promoter (Fig. 4D). Next, we assessed the recruitment of PCAF at the γ-globin promoter in K562 cells and its change after SATB2 overexpression or knockdown (Fig. 4E). ChIP results showed a strong enrichment of PCAF at the γ-globin promoter in K562 cells and that the recruitment was positively correlated with the cellular SATB2 level. What's more, PCAF knockdown in K562 cells leads to significant decrease of H4 acetylation at γ-globin promoter (supplemental Fig. S4A) and treatment of K562 cells with HDAC inhibitor sodium butyrate (NaB) overcomes the inhibitory effect to γ-globin transcription observed for SATB2 knockdown (supplemental Fig. S4B). Together, we conclude that PCAF participates and facilitates SATB2-mediated γ-globin gene activation.
FIGURE 4.
Identification of PCAF as a SATB2 functional partner. A, co-immunoprecipitation of SATB2 with PCAF but not CBP and p300 in K562 cells. B, PCAF co-immunoprecipitates with SATB2 in K562 cells. C, mutual co-immunoprecipitation of exogenously expressed SATB2-myc and PCAF-Flag in 293T cells. D, PCAF synergistically combines with SATB2 in transactivating the G/Aγ-globin promoter in 293T cells. Each error bar represents a standard deviation calculated from experiments performed in triplicate. Student's t test was used for statistical analysis and p values are indicated (*, p < 0.05). E, quantitative ChIP assay shows alteration of PCAF occupancy at the γ-globin gene promoter in K562 cells after SATB2 overexpression or knock down. Each error bar represents a standard deviation calculated from experiments performed in triplicate. Student's t test was used for statistical analysis, and p value is indicated (*, p < 0.05).
SATB2 Regulates the Physical Association between Gγ- and Aγ-Globin Genes
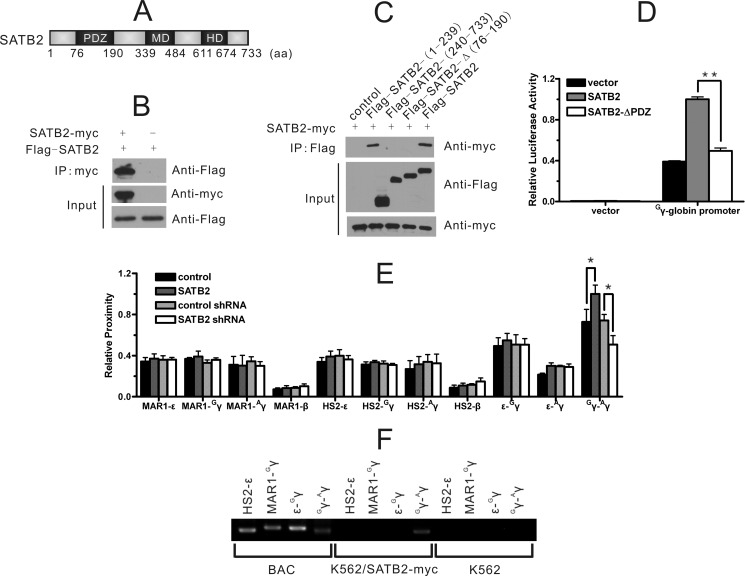
To further address the molecular basis of SATB2-mediated transcriptional activation, we employed a tandem-affinity purification (TAP) assay to obtain protein partners of SATB2. As a ubiquitously expressed protein, SATB2 may use a common set of partners and mechanisms when functioning in different types of cells. We therefore transfected the tandem-tagged SATB2 into 293T cells and investigated SATB2-binding proteins. Affinity-purified SATB2-containing protein complexes were analyzed using mass spectrometry (supplemental Fig. S5A). Notably, SATB1, the homolog of SATB2, was identified as one of the SATB2-binding proteins. The interaction was confirmed using co-immunoprecipitation (co-IP) assay for exogenous SATB1/SATB2 in non-erythroid 293T cells and the endogenous SATB1/SATB2 in erythroid K562 cells (supplemental Fig. S5B). An immunofluorescence assay also revealed partially overlapping distributions of the exogenously expressed SATB1 and SATB2 in 293T cell nuclei (supplemental Fig. S5C). Perinuclear localization of SATB2 has been shown to be led by sumoylation (5). We further showed that the SATB1-SATB2 interaction was dependent on their PDZ-domains using the co-IP assay (supplemental Fig. S5, D and E). Interestingly, SATB1 has been reported to have self-association ability through its PDZ domain (26). Consequently, we explored whether SATB2 exhibited the same property (Fig. 5A). The co-IP assay showed that SATB2-myc could precipitate Flag-SATB2 in 293T cells (Fig. 5B), and further analysis with truncated SATB2 revealed the importance of the PDZ-domain for self-association and the transcriptional activity of SATB2 (Fig. 5, C and D). The comprehensive self-association and hetero-dimerization between SATB1 and SATB2 may provide a multi-protein platform for MAR attachment and the regulation of gene expression through facilitating higher-order chromatin organization.
FIGURE 5.
SATB2 regulates the physical proximity between Gγ- and Aγ-globin genes. A, SATB2 protein is depicted with domains, including a PDZ domain, a MAR-binding domain (MD) and a homeodomain (HD). B and C, co-IP assay in 293T cells depicting (B) the interaction between differently tagged SATB2 proteins and (C) the PDZ domain dependence of the interaction. D, luciferase reporter assay in 293T cells showed that deletion of the PDZ domain of SATB2 abolished the transcriptional activity of SATB2. Each error bar represents a standard deviation calculated from experiments performed in triplicate. Student's t test was used for statistical analysis, and p value is indicated (**, p < 0.01). E, 3C assay of HindIII fragments covering the indicated regulatory elements of the β-globin locus in K562 cells. Perturbation of SATB2 expression specifically affected the physical proximity between the Gγ- and Aγ-globin promoters. Each error bar represents a standard deviation calculated from experiments performed in triplicate. Student's t test was used for statistical analysis, and p values are indicated (*, p < 0.05). F, ChIP-3C assay with anti-Myc antibody in K562/SATB2-myc cells confirmed that SATB2 mediated the physical association of the Gγ- and Aγ-globin promoters.
We therefore investigated influence of human SATB2 on the higher-order chromatin structure of the β-globin gene cluster (Fig. 5E). Primers were designed from individual HindIII fragments (HS2, MAR1, ϵ, Gγ, Aγ and β, as depicted in Fig. 3A) containing HS2, MAR-HS2–1 (SATB1-binding site) and the ϵ-globin, Gγ-globin, Aγ-globin, and β-globin promoters, respectively. All the PCR products were confirmed as expected ligation products through sequencing. Digestion efficiency of 3C templates was verified by quantitative PCR across the HindIII digestion sites (supplemental Fig. S6). Unexpectedly, neither HS2 nor the MAR-HS2–1 fragment changed in their relative proximity to any of the globin gene promoters after SATB2 was overexpressed or knocked down. The only remarkable spatial proximity change as a result of SATB2 genetic modulation was between the Gγ-globin and Aγ-globin promoter regions. Follow-up ChIP-3C analysis (fragments were the same as in the 3C assay) of ectopically expressed SATB2-myc in K562 cells further supported that SATB2 bridged the Gγ-globin and Aγ-globin promoter regions (Fig. 5F).
DISCUSSION
We show here that the MAR-binding protein SATB2, a homolog of SATB1, transcriptionally activates the Gγ and Aγ-globin genes in erythroid cells by directly binding to MAR elements at their promoters, recruiting co-activator PCAF and modulating the spatial proximity between the two γ-globin promoters. Our findings, to our knowledge, are the first to establish the MAR-binding protein SATB2 as a γ-globin gene activator and higher-order chromatin structure organizer in the β-globin gene locus, and suggest SATB2 as a potential target for β-thalassemia therapy.
SATB2 Recruits a Co-activator and Activates Human γ-Globin Gene Expression in Erythroid Cells
Multiple transcriptional factors have previously been described to specifically regulate γ-globin gene expression and contribute to the β-globin switch from the fetal to the adult stage, such as the recently identified Ikaros (27) and BCL11A factors (21). Here, we observed the specific activation of γ-globin gene expression by the MAR-binding protein SATB2. We show that SATB2 directly binds to the γ-globin gene promoter in K562 cells and preferentially associates with the Gγ-globin promoter rather than the Aγ-globin promoter. The binding preference of SATB2 is consistent with and may provide structural basis for the observation that Gγ-globin gene is more actively transcribed than the Aγ-globin gene in K562 cells. We further identified that SATB2 interacted with PCAF and promoted its recruitment to the γ-globin gene promoter, which is consistent to the concomitant changes in histone acetylation at the promoter and partially explains the mechanism of SATB2 activation of the γ-globin gene promoter. Recruitment of histone deacetylase has been shown as typical functional characteristics of MAR-binding proteins (28). Other γ-globin regulators like FKLF2 and NF-E4 have been reported to interact with and serve as substrates of PCAF (29, 30), therefore may also contribute to the strong PCAF recruitment to γ-globin promoter observed in K562 cells.
Looping Formation Mediated by SATB2
While their nature of tethering genes to the nuclear scaffold suggests role of MAR-binding protein in higher-order chromatin structure organization, experimental evidences are available only for CTCF and SATB1 (31, 32). Studies on multiple loci have shown that SATB1 anchors MAR elements and governs transcription by establishing chromatin loop architecture and recruiting cofactors to form a densely active transcriptional complex (33, 34). As the homolog of SATB1, SATB2 has been reported to regulate Immunoglobulin and Hoxa2 genes, and cooperate with SATB1 in regulating ES cell balancing and initiation of X inactivation (35), role of SATB2 in higher-order chromatin structure organization remains unexplored. Here, we show that SATB2 mediates the local association of gene promoter regions by self-association. Unlike the case of SATB1, which facilitates the proximity of LCR and ϵ-globin gene by mediating the inter-MAR association (14), SATB2 does not affect physical proximity of LCR and β-like globin genes. Thus, SATB2 seems to facilitate the formation of a sub-structure in the actively looped architecture of the β-globin gene locus. PCAF has been shown to acetylate SATB1 and regulate its DNA binding (36), whether PCAF may similarly affect SATB2 acetylation, hence regulate SATB2 transcriptional activity and the association between the two γ-globin promoters presents an interesting issue for further investigation.
Relationship between SATB2 and SATB1 in Gene Regulation and Genome Organization
The binding of SATB1 and SATB2 at ϵ-globin and γ-globin promoters, respectively, provides a basis for different expression modes of ϵ-globin and γ-globin genes. A recent study on the Nanog locus showed that both SATB2 and SATB1 regulated Nanog gene expression (7). We have previously shown that SATB1 interference in K562 cells also mildly decreases γ-globin gene expression and the association between LCR and γ-globin promoter (14), furthermore, the type III deacetylase SIRT, which deacetylates SATB1 in erythroid cells, also affects γ-globin gene expression (16), whereas ChIP and ChIP-3C assays failed to confirm close connection of SATB1 to γ-globin gene (14). Here we show that the γ-globin regulator SATB2 interacts with SATB1 in K562 cells, providing the possibility that SATB1 affects γ-globin expression through interacting with SATB2. Hence we propose a differential and cooperative relationship of SATB proteins in organizing the higher-order chromatin structure of the whole β-globin gene locus, namely, SATB1 mediates inter-MAR association across β-globin gene locus, whereas SATB2 mediates regional MAR association of Gγ-globin and Aγ-globin promoters. SATB1 has been shown globally reprograms gene expression and directly binds to SATB1-dependent genes in metastatic breast cancer cells (37). Genome-wide analyses like ChIP-seq and ChIA-PET will be required for a comprehensive understanding to the relationship between SATB1 and SATB2 in coordinating global gene co-regulation.
Active G/Aγ-MAR Association Model
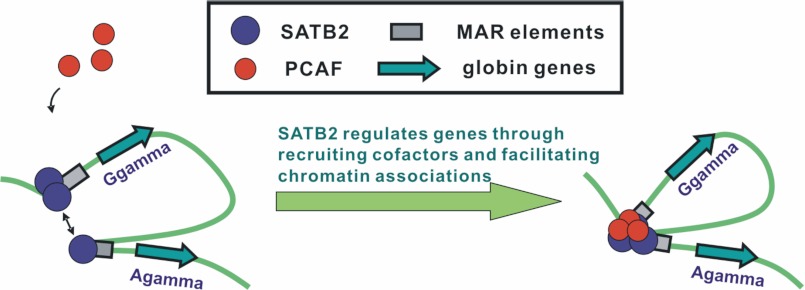
Based on our results, we present an active G/Aγ-MAR association model in which SATB2 forms an active inter-MAR bridge between Gγ-globin and Aγ-globin promoters by recruiting co-activators and facilitating associations between its binding sites (Fig. 6). Incorporating with our previous SATB1-centered inter-MAR association model (14), we illustrate the potential mechanism through which SATB proteins regulate higher-order chromatin structure and gene transcription at the β-globin gene locus and suggest that the higher-order chromatin structure in the β-globin gene locus involves the substructure of active chromatin interactions (supplemental Fig. S7).
FIGURE 6.
Active G/Aγ-MAR association model in β-globin gene locus in embryonic-fetal-stage erythroid cells. The diagram illustrates that MAR-binding protein SATB2 promotes γ-globin gene expression and active chromatin structure formation by recruiting the co-activator PCAF and bridging the physical association of the two γ-globin promoters.
Acknowledgments
We thank Prof. Mitchell J. Weiss for kindly providing the G1E-ER4 cells used in this study and Prof. Ann Dean for valuable discussion on the manuscript.
This work was supported by the National Natural Science Foundation of China (Grant 31030026), the National Basic Research Program (Grants 2011CB964803 and 2011CB965203), the National Natural Science Foundation of China (Grant 31021091), the National Laboratory of Medical Molecular Biology (Grant 2060204), and Grant YB20081002301 from the Beijing municipal government.

This article contains supplemental Figs. S1–S7.
- MAR
- matrix attachment region
- LCR
- locus control region
- ACH
- active chromatin hub
- TAP
- tandem affinity purification.
REFERENCES
- 1. Scheuermann R. H., Chen U. (1989) A developmental-specific factor binds to suppressor sites flanking the immunoglobulin heavy-chain enhancer. Genes Dev. 3, 1255–1266 [DOI] [PubMed] [Google Scholar]
- 2. Romig H., Fackelmayer F. O., Renz A., Ramsperger U., Richter A. (1992) Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J. 11, 3431–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Herrscher R. F., Kaplan M. H., Lelsz D. L., Das C., Scheuermann R., Tucker P. W. (1995) The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev. 9, 3067–3082 [DOI] [PubMed] [Google Scholar]
- 4. Dickinson L. A., Joh T., Kohwi Y., Kohwi-Shigematsu T. (1992) A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell 70, 631–645 [DOI] [PubMed] [Google Scholar]
- 5. Dobreva G., Dambacher J., Grosschedl R. (2003) SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev. 17, 3048–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dobreva G., Chahrour M., Dautzenberg M., Chirivella L., Kanzler B., Fariñas I., Karsenty G., Grosschedl R. (2006) SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 125, 971–986 [DOI] [PubMed] [Google Scholar]
- 7. Savarese F., Dávila A., Nechanitzky R., De La Rosa-Velazquez I., Pereira C. F., Engelke R., Takahashi K., Jenuwein T., Kohwi-Shigematsu T., Fisher A. G., Grosschedl R. (2009) Satb1 and Satb2 regulate embryonic stem cell differentiation and Nanog expression. Genes Dev. 23, 2625–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tolhuis B., Palstra R. J., Splinter E., Grosveld F., de Laat W. (2002) Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10, 1453–1465 [DOI] [PubMed] [Google Scholar]
- 9. Palstra R. J., Tolhuis B., Splinter E., Nijmeijer R., Grosveld F., de Laat W. (2003) The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 35, 190–194 [DOI] [PubMed] [Google Scholar]
- 10. Drissen R., Palstra R. J., Gillemans N., Splinter E., Grosveld F., Philipsen S., de Laat W. (2004) The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 18, 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vakoc C. R., Letting D. L., Gheldof N., Sawado T., Bender M. A., Groudine M., Weiss M. J., Dekker J., Blobel G. A. (2005) Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell 17, 453–462 [DOI] [PubMed] [Google Scholar]
- 12. Splinter E., Heath H., Kooren J., Palstra R. J., Klous P., Grosveld F., Galjart N., de Laat W. (2006) CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 20, 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wen J., Huang S., Rogers H., Dickinson L. A., Kohwi-Shigematsu T., Noguchi C. T. (2005) SATB1 family protein expressed during early erythroid differentiation modifies globin gene expression. Blood 105, 3330–3339 [DOI] [PubMed] [Google Scholar]
- 14. Wang L., Di L. J., Lv X., Zheng W., Xue Z., Guo Z. C., Liu D. P., Liang C. C. (2009) Inter-MAR association contributes to transcriptionally active looping events in human beta-globin gene cluster. PLoS One 4, e4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gong H., Wang Z., Zhao G. W., Lv X., Wei G. H., Wang L., Liu D. P., Liang C. C. (2009) SATB1 regulates beta-like globin genes through matrix related nuclear relocation of the cluster. Biochem. Biophys. Res. Commun. 383, 11–15 [DOI] [PubMed] [Google Scholar]
- 16. Xue Z., Lv X., Song W., Wang X., Zhao G. N., Wang W. T., Xiong J., Mao B. B., Yu W., Yang B., Wu J., Zhou L. Q., Hao D. L., Dong W. J., Liu D. P., Liang C. C. (2012) SIRT1 deacetylates SATB1 to facilitate MARHS2-MAR{varepsilon} interaction and promote {varepsilon}-globin expression. Nucleic Acids Res. 40, 4804–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cunningham J. M., Purucker M. E., Jane S. M., Safer B., Vanin E. F., Ney P. A., Lowrey C. H., Nienhuis A. W. (1994) The regulatory element 3' to the A gamma-globin gene binds to the nuclear matrix and interacts with special A-T-rich binding protein 1 (SATB1), an SAR/MAR-associating region DNA-binding protein. Blood 84, 1298–1308 [PubMed] [Google Scholar]
- 18. Case S. S., Huber P., Lloyd J. A. (1999) The γPE complex contains both SATB1 and HOXB2 and has positive and negative roles in human γ-globin gene regulation. DNA Cell Biol. 18, 805–817 [DOI] [PubMed] [Google Scholar]
- 19. Jackson C. E., O'Neill D., Bank A. (1995) Nuclear factor binding sites in human beta globin IVS2. J. Biol. Chem. 270, 28448–28456 [DOI] [PubMed] [Google Scholar]
- 20. Lloyd J. A., Case S. S., Ponce E., Lingrel J. B. (1994) Positive transcriptional regulation of the human γ-globin gene. γPE is a novel nuclear factor with multiple binding sites near the gene. J. Biol. Chem. 269, 19385–19393 [PubMed] [Google Scholar]
- 21. Sankaran V. G., Menne T. F., Xu J., Akie T. E., Lettre G., Van Handel B., Mikkola H. K., Hirschhorn J. N., Cantor A. B., Orkin S. H. (2008) Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 322, 1839–1842 [DOI] [PubMed] [Google Scholar]
- 22. Ragione F. D., Cucciolla V., Criniti V., Indaco S., Borriello A., Zappia V. (2003) p21Cip1 gene expression is modulated by Egr1: a novel regulatory mechanism involved in the resveratrol antiproliferative effect. J. Biol. Chem. 278, 23360–23368 [DOI] [PubMed] [Google Scholar]
- 23. Zhou G. L., Xin L., Song W., Di L. J., Liu G., Wu X. S., Liu D. P., Liang C. C. (2006) Active chromatin hub of the mouse α-globin locus forms in a transcription factory of clustered housekeeping genes. Mol. Cell Biol. 26, 5096–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dekker J., Rippe K., Dekker M., Kleckner N. (2002) Capturing chromosome conformation. Science 295, 1306–1311 [DOI] [PubMed] [Google Scholar]
- 25. Gyorgy A. B., Szemes M., de Juan Romero C., Tarabykin V., Agoston D. V. (2008) SATB2 interacts with chromatin-remodeling molecules in differentiating cortical neurons. Eur. J. Neurosci. 27, 865–873 [DOI] [PubMed] [Google Scholar]
- 26. Galande S., Dickinson L. A., Mian I. S., Sikorska M., Kohwi-Shigematsu T. (2001) SATB1 cleavage by caspase 6 disrupts PDZ domain-mediated dimerization, causing detachment from chromatin early in T-cell apoptosis. Mol. Cell Biol. 21, 5591–5604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lopez R. A., Schoetz S., DeAngelis K., O'Neill D., Bank A. (2002) Multiple hematopoietic defects and delayed globin switching in Ikaros-null mice. Proc. Natl. Acad. Sci. U.S.A. 99, 602–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yasui D., Miyano M., Cai S., Varga-Weisz P., Kohwi-Shigematsu T. (2002) SATB1 targets chromatin remodeling to regulate genes over long distances. Nature 419, 641–645 [DOI] [PubMed] [Google Scholar]
- 29. Song C. Z., Keller K., Murata K., Asano H., Stamatoyannopoulos G. (2002) Functional interaction between coactivators CBP/p300, PCAF, and transcription factor FKLF2. J. Biol. Chem. 277, 7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao Q., Cumming H., Cerruti L., Cunningham J. M., Jane S. M. (2004) Site-specific acetylation of the fetal globin activator NF-E4 prevents its ubiquitination and regulates its interaction with the histone deacetylase, HDAC1. J. Biol. Chem. 279, 41477–41486 [DOI] [PubMed] [Google Scholar]
- 31. Phillips J. E., Corces V. G. (2009) CTCF: master weaver of the genome. Cell 137, 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Galande S., Purbey P. K., Notani D., Kumar P. P. (2007) The third dimension of gene regulation: organization of dynamic chromatin loopscape by SATB1. Curr. Opin. Genet. Dev. 17, 408–414 [DOI] [PubMed] [Google Scholar]
- 33. Cai S., Han H. J., Kohwi-Shigematsu T. (2003) Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 34, 42–51 [DOI] [PubMed] [Google Scholar]
- 34. Cai S., Lee C. C., Kohwi-Shigematsu T. (2006) SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 38, 1278–1288 [DOI] [PubMed] [Google Scholar]
- 35. Agrelo R., Souabni A., Novatchkova M., Haslinger C., Leeb M., Komnenovic V., Kishimoto H., Gresh L., Kohwi-Shigematsu T., Kenner L., Wutz A. (2009) SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev. Cell 16, 507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pavan Kumar P., Purbey P. K., Sinha C. K., Notani D., Limaye A., Jayani R. S., Galande S. (2006) Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol. Cell 22, 231–243 [DOI] [PubMed] [Google Scholar]
- 37. Han H. J., Russo J., Kohwi Y., Kohwi-Shigematsu T. (2008) SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature 452, 187–193 [DOI] [PubMed] [Google Scholar]