Background: Human melanoma cells are able to sustain low pH conditions characterizing many solid tumors.
Results: Acidic stress in melanoma cells induces reduced nutrients uptake, inhibition of mammalian target of rapamycin, and activation of autophagy.
Conclusion: Melanoma cells activate autophagy as a protective and adaptive response to acidic stress.
Significance: Adaptation to acidosis via autophagy confirms the feasibility of anticancer therapy targeting autophagy.
Keywords: Acidosis, Autophagy, Cancer Biology, Melanoma, mTOR, AMPK, ATG5, Intracellular pH
Abstract
Cyclic hypoxia and alterations in oncogenic signaling contribute to switch cancer cell metabolism from oxidative phosphorylation to aerobic glycolysis. A major consequence of up-regulated glycolysis is the increased production of metabolic acids responsible for the presence of acidic areas within solid tumors. Tumor acidosis is an important determinant of tumor progression and tumor pH regulation is being investigated as a therapeutic target. Autophagy is a cellular catabolic pathway leading to lysosomal degradation and recycling of proteins and organelles, currently considered an important survival mechanism in cancer cells under metabolic stress or subjected to chemotherapy. We investigated the response of human melanoma cells cultured in acidic conditions in terms of survival and autophagy regulation. Melanoma cells exposed to acidic culture conditions (7.0 < pH < 6.2) promptly accumulated LC3+ autophagic vesicles. Immunoblot analysis showed a consistent increase of LC3-II in acidic culture conditions as compared with cells at normal pH. Inhibition of lysosomal acidification by bafilomycin A1 further increased LC3-II accumulation, suggesting an active autophagic flux in cells under acidic stress. Acute exposure to acidic stress induced rapid inhibition of the mammalian target of rapamycin signaling pathway detected by decreased phosphorylation of p70S6K and increased phosphorylation of AMP-activated protein kinase, associated with decreased ATP content and reduced glucose and leucine uptake. Inhibition of autophagy by knockdown of the autophagic gene ATG5 consistently reduced melanoma cell survival in low pH conditions. These observations indicate that induction of autophagy may represent an adaptation mechanism for cancer cells exposed to an acidic environment. Our data strengthen the validity of therapeutic strategies targeting tumor pH regulation and autophagy in progressive malignancies.
Introduction
The tumor microenvironment represents a complex physical and biochemical environment characterizing solid cancers. It has received renewed attention after the established association between signaling through oncogenic mutations and their role as drivers of metabolic transformation (1). Highly proliferating cancer cells create a tumor mass poor in nutrients and oxygen because of the growing distance from blood vessels. As a consequence of low oxygen tension, the stabilization of the transcription factor hypoxia inducible factor 1α represent an important process regulating the shift to glycolytic metabolism (2). In cancer cells, glucose is metabolized to pyruvate and lactate rather than entering the oxidative phosphorylation pathway, even in normal oxygen pressure, giving rise to the Warburg effect (3).
An inevitable result of up-regulated glucose metabolism is the high production rate of metabolic acids (lactate and protons) that cells need to extrude to avoid intracellular acidification. Normally, to maintain pH homeostasis cancer cells increase the activity and/or expression of several pH regulators, resulting in alkalinization of intracellular pH (pHi)2 and acidification of extracellular pH (pHe) (4). A low tumor pHe has been demonstrated in several preclinical models of human cancers, with a pHe ranging from 5.9 to 7.2 depending on the tumor type. Additionally, tumors have localized areas characterized by spatial variations in pHe (5). The acidic pH of the tumor microenvironment is an important component driving tumor invasive capacity, neoangiogenesis, anchorage-independent growth, and genetic instability, which in combination contribute to malignant progression (5–8). Moreover, low tumor pHe is a known factor responsible for the intrinsic resistance of malignant cells to chemotherapeutics (9, 10) and negatively affects immune functions (11). Recently, it was reported that the long-term culture of glioma cells in medium at pH 6.5 induces a stem cell phenotype (12). Therefore, anticancer therapies based on targeting tumor pH regulation and inducing alkalinization of tumor pHe may inhibit several tumor biological functions and are currently under investigation both in preclinical and clinical settings (13–17).
Macroautophagy (hereafter referred to as autophagy) is a catabolic process used by eukaryotic cells to recycle long lived proteins, lipids, and eliminate protein aggregates and organelles (18, 19). The process starts by the formation of a double membrane-bound autophagosome dependent on the sequential intervention of ATG (AuTophaGy-related) proteins (20, 21). In most of the cases the final destination of the autophagosome is fusion with the lysosomal compartment where sequestered cargo is degraded. Recent studies have revealed the importance of autophagy in development (20, 22), aging (23, 24), immune response (25, 26), and pathophysiology such as neurodegenerative disease, obesity, infectious disease, and cancer (20, 26–29). Autophagy has a complex role in tumor development and progression. Autophagy is a tumor suppressor pathway at the initiation stage of tumors by limiting the production of reactive oxygen species and limiting DNA damage (29–31). However, autophagy is a survival pathway for tumor cells to overcome metabolic stress and to resist cell death in response to radiation and chemotherapeutic agents (32–35). Autophagy plays a major role in metabolic adaptation in RAS-transformed cells (36, 37). Thus, autophagy has a fundamental and driving role in developing tumors and their malignant progression. Notably, recent studies performed on tissue sections of human tumors suggest the association of up-regulated autophagy with tumor hypoxia and acidity (38–40).
In the present study we evaluated the effects of acidic pH on the autophagic activity in human malignant melanoma cells. We demonstrate that exposure to acidic pH inhibits the mammalian target of rapamycin (mTOR) pathway and reduced glucose and leucine uptake. Melanoma cells under acidic stress up-regulate the autophagic flux and gene knockdown experiments showed that functional autophagy, intended as an active autophagic flux is instrumental for melanoma cells to survive acidic stress. We propose that the ability to maintain functional autophagy may characterize melanoma cells exposed to an acidic microenvironment, conferring malignant cells the capacity to overcome acidic stress-induced cell death.
EXPERIMENTAL PROCEDURES
Reagents
Bafilomycin A1 (Baf) and E64d were from Sigma. Antibiotics, phosphate-buffered saline (PBS), trypsin/EDTA, and fetal bovine serum (FBS) were from Lonza (Milano, Italy). Pepstatin A and FuGENE 6 Transfection Reagent were from Roche Applied Science. HiPerFect Transfection Reagent was from Qiagen. ON-TARGETplus SMARTpool for human ATG5 was from Dharmacon (Thermo Scientific). Leucine, l-[3,4,5-3H], was from PerkinElmer Life Sciences. 2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose (2-NBDG), 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein (BCECF-AM), and RPMI 1640 were from Invitrogen. The following antibodies were used: p70S6K, phospho-p70S6K, 4EBP1, phospho-4EBP1, mTOR, phospho-mTOR, AKT, phospho-AKT, AMP-activated protein kinase (AMPK), phospho-AMPK, ERK1/2, phospho-ERK1/2, LC3, and ATG5 (Cell Signaling Technology); p62 (BD Transduction laboratories), β-actin (Sigma), and HRP-conjugated anti-rabbit and anti-mouse antibodies (Bio-Rad). The GFP-LC3 and GFP-RFP-LC3 plasmids were a kind gift of Dr. Tamotsu Yoshimori (National Institute of Genetics, Mishima, Japan).
Cell Culture
Cell lines Me30966, Mel501, and WM793 were described before (41). The cell lines A375 and SK-Mel-28 were a kind gift from Johan Hansson (Karolinska Institute, Sweden). All cell lines were maintained in RPMI medium in the presence of 10% fetal calf serum (FCS) and antibiotics at 37 °C, 5% CO2. The different pH of RPMI media were obtained by adding different concentrations of NaHCO3 and letting the medium equilibrate in the cell culture incubator overnight.
Viability Assay
Growth curves of cell lines at different pH conditions were performed using the acid phosphatase assay. Briefly, cells were plated in standard RPMI medium in 96-well plates the day after medium was replaced with media at the indicated pH. Starting from time 0 the plates were collected every 24 h and the acid phosphatase assay was run as previously described (42).
Glucose Uptake
Melanoma cells cultured at different pH conditions were incubated with 100 μm 2-NBDG, a fluorescent derivative of 2-deoxy-d-glucose, for 45 min. Cells were collected and washed with PBS and then analyzed by flow cytometry to collect green fluorescence.
Measurements of Intracellular pH
Cytosolic pH was evaluated by fluorescence spectroscopy using the pH-sensitive fluorescent probe BCECF-AM as previously described (43). Briefly, cells were plated in black 96-well plates (Costar) at 50% confluence. The next day, cells were incubated for 30 min with 10 μm BCECF-AM in isotonic saline solution (ISS: 150 mm NaCl, 1 mm Na2HPO4, 1 mm CaCl2, 10 mm Hepes, pH 7.4), washed once in ISS, and incubated for an additional 20 min. Immediately after exposing cells to ISS at different pH values, BCECF fluorescence was recorded at 530 nm after excitation at 445 and 495 nm, in a Tecan Infinite M1000 (Tecan, Männedorf, Switzerland) fluorescence microplate reader. After background subtraction, the fluorescence ratio of BCECF-AM (495/445) was converted to pHi values by using a calibration curve with different potassium phosphate buffers in a pH range of 6.0 to 8.0 in the presence of 10 μm nigericin.
Amino Acid Uptake
Cells were incubated in RPMI buffered at different pH values (7.4 or 6.5). After 2 h [3H]leucine was added (1 μCi/ml) and incubated for 2 h. As positive control, we used C2-ceramide (C2-Cer, 50 μm), which is known to inhibit amino acids uptake (44). Cells were washed twice with ice-cold RPMI, lysed in RIPA buffer, and tritium levels were determined by scintillation counting.
ATP Determination
Quantification of cellular ATP was performed on cells cultured in 96-well plates using the ATP assay kit (Promega, Madison, WI). Cells were plated at 5000 cells/well and the ATP concentration was normalized to the cell number.
Western Blot Analysis
Cells were collected and treated with RIPA buffer (150 mm NaCl, 50 mm Tris, pH 7.4, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate) supplemented with protease and phosphatase inhibitors and protein concentration of cellular extracts was determined by Bio-Rad Protein Assay (Bio-Rad). SDS-PAGE was performed on pre-casted acrylamide gels (12–15%) using the NuPage system and loading 30 μg of proteins per lane. After electrophoresis, proteins were transferred to PVDF membranes and incubated for 1 h at room temperature in TBS-Tween with 5% nonfat dry milk. Protein loading was assessed by red Ponceau staining of the membranes. Membranes were incubated with primary antibodies diluted in TBS-Tween overnight at 4 °C, followed by appropriate HRP-conjugated secondary antibodies. Antibody staining was visualized by the ECL system (Pierce).
Evaluation of Autophagy
Me30966 cells were used to monitor GFP-LC3 fluorescence under different pH culture conditions. Twenty-four hours after plating the cells were transfected with the peGFP-LC3 plasmid by using Lipofectamine 2000 (Invitrogen). The next day, the medium was replaced with freshly prepared medium at a different pH. After an 8-h incubation with different pH media, the cells were fixed using 2% paraformaldehyde and autophagy was determined by microscopy fluorescence and quantification of the number of cells containing GFP-LC3-positive vesicles.
RNA Interference Experiments
Small interfering RNAs (siRNA) for ATG5 were synthesized by Dharmacon RNA Technologies (siGENOME SMART pool). For siRNA transfection, Me30966 and WM793 cells were seeded in standard RPMI medium at 30% confluence in 12-well tissue culture plates. The next day cells were transfected with 20 nm siRNA specific for ATG5 by using Hi-Perfect transfection reagent (Qiagen) following the manufacturer's instructions. The gene-silencing efficacy was assessed by Western blot analysis of ATG5 protein expression using polyclonal antibodies. Forty-eight hours after transfection, the medium was replaced with medium at different pH values (7.4, 6.5, and 6.2) or with EBSS. Twenty-four hours after being exposed to the new media cells were collected and evaluated for cell death using Annexin-V-FITC and propidium iodide staining (BD Pharmingen).
RESULTS
Human Melanoma Cells Survive and Proliferate Also at Very Acidic pH
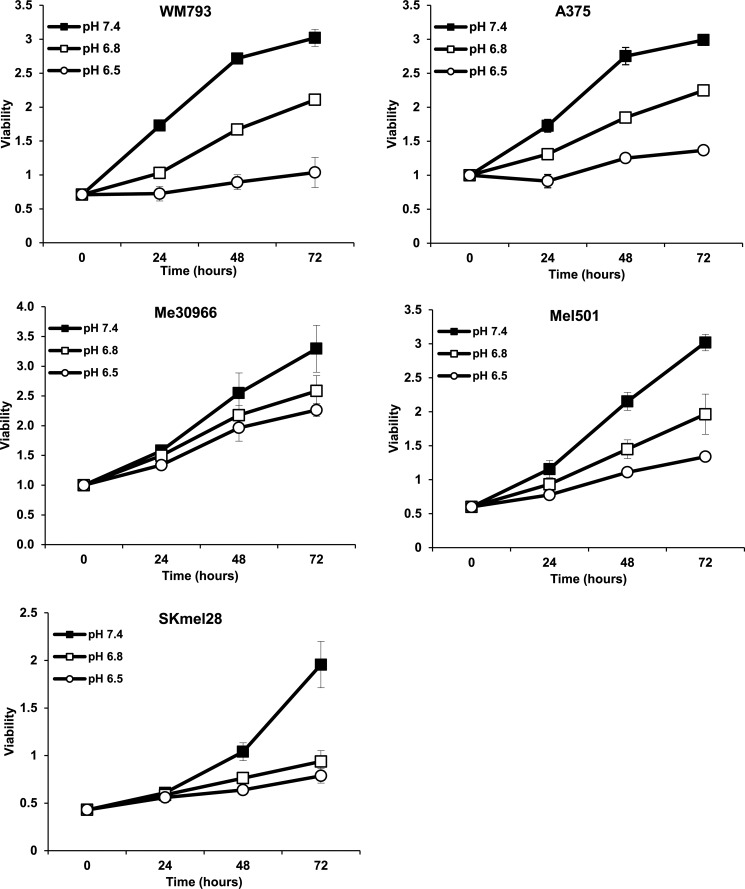
An acidic pH environment is known to delay cell growth and increase spontaneous apoptosis in cell lines derived from colon cancer and malignant glioma (45, 46). However, human melanoma cells are known to survive during acidic stress (41, 47). We cultured Me30966, WM793, A375, SK-Mel-28, and Mel501 cells at different pH conditions and evaluated cell proliferation over 72 h. As indicated in Fig. 1, melanoma cells cultured at pH 6.8 showed a reduction of 20–50% in proliferation as compared with cells cultured at pH 7.4, depending on the cell line. Interestingly, all cell lines cultured at pH 6.5 continue to slowly proliferate. Acute exposure to the pH 6.5 medium seemed to provoke a low degree of cell death (supplemental Fig. S1A) followed by recovery of proliferation of the surviving cells (Fig. 1A and supplemental Fig. S1B). Among the cell lines analyzed, Me30966 showed the highest ability to grow even at pH 6.5 as compared with other cell lines (Fig. 1, supplemental Fig. S1C), thus representing an appropriate model to study how melanoma cells adapt to survive and proliferate in very acidic conditions.
FIGURE 1.
Human melanoma cells slowly proliferate under acidic stress. The human melanoma cell lines WM793, A375, Me30966, Mel501, and SK-Mel-28 were cultured over 3 days at different pHe conditions. Cell viability was measured every day with the acid phosphatase assay. Data from two independent experiments run in triplicate wells are shown. Data are expressed as mean ± S.D.
Acidic Stress Up-regulates Autophagy in Human Melanoma
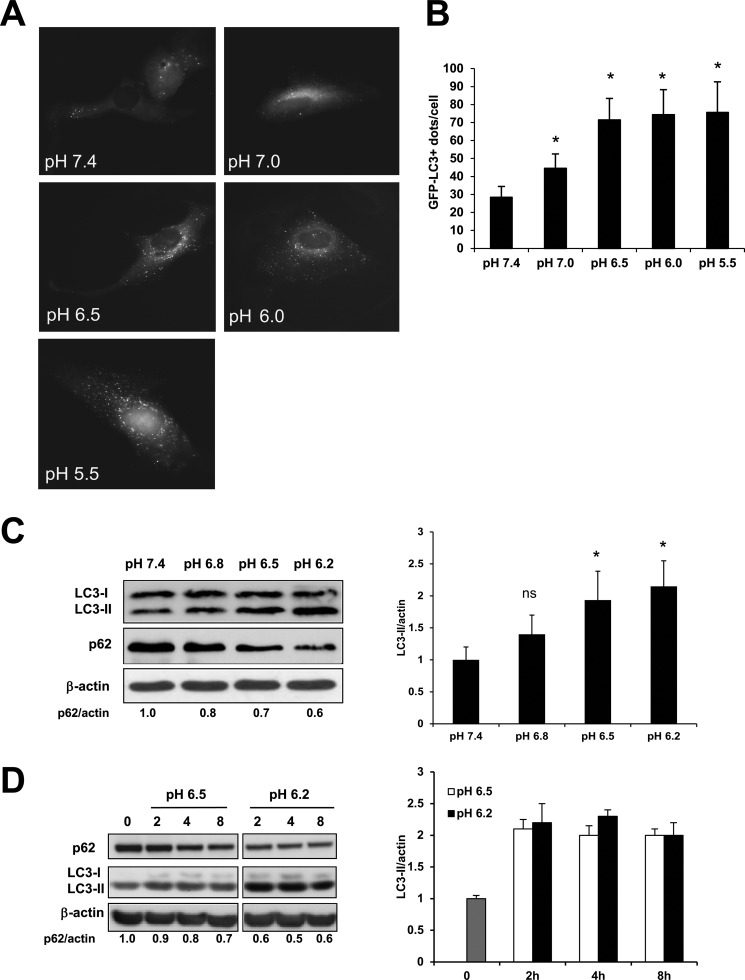
Increased autophagy linked to acidity has recently been reported as a marker of poor prognosis in melanoma and colorectal cancer (38–40). Moreover, increased autophagy was shown to represent a marker for aggressive and metastatic melanoma (48). Because autophagy is activated in cells under metabolic stress, we hypothesized that autophagy might be a protective mechanism for cancer cells acutely exposed to acidic stress. To test this hypothesis, we evaluated the effects of short-term exposure to acidic pH on basal autophagy in human melanoma cells. We used the human melanoma cell line Me30966 transiently transfected with a GFP-LC3 expressing plasmid. The day after transfection the cell culture medium was replaced with medium at different pH values (7.4 to 5.5) and GFP-LC3 fluorescence was monitored 8 h later. As depicted in Fig. 2A, cells cultured at neutral pH showed a mostly cytosolic distribution of GFP-LC3 fluorescence and around 30 LC3-positive dots/cell. With decreasing pHe, the number of LC3-positive dots per cells significantly increased 2-fold from about 30 LC3+ dots/cell at pH 7.4 to over 70 LC3+ dots/cell at low pH (p < 0.05, Fig. 2, A and B), suggesting that acidic culture conditions increase the number of autophagic vacuoles. The number of LC3+ autophagic vesicles per cell was significantly higher at pH 6.5 with respect to pH 7.0 (p < 0.05) and reached a plateau at pH 6.5 (Fig. 2B).
FIGURE 2.
Autophagy response in melanoma cells upon acidic stress. A and B, Me30966 cells transiently transfected with GFP-LC3 plasmid were exposed for 8 h to media at different pH values. The presence of GFP-LC3+ dots was observed by fluorescence microscopy and quantified. C, Me30966 cells were exposed to different acidic media for 8 h and LC3 and p62 expression was evaluated by Western blot. The expression of LC3 was measured by gel densitometry using Adobe Photoshop Elements 9 and normalized to the pH 7.4 condition. D, Me30966 cells were plated overnight and culture medium was changed to pH 6.5 and 6.2. Cells were collected at time 0 (gray bar) and 2, 4, and 8 h after medium change. Expression of p62 and LC3 was analyzed by Western blot and normalized to the values at time 0. All blots shown are representative of at least two independent experiments. Where shown, * indicates p < 0.05.
To gain insight into the effects of acidic stress on basal autophagy we first cultured Me30966 cells in media at different pH values and analyzed the conversion of LC3-I into LC3-II after 8 h. Interestingly, the LC3-II form was significantly increased as the medium pH decreased (Fig. 2C). p62 is a cytoplasmic protein that shuttles ubiquitinated proteins to autophagosomes. Expression of p62 increases when autophagy is inhibited, thus the levels of p62 may reflect the autophagic flux. Analysis of p62 in the same samples indicated that p62 expression was decreased in a pH-dependent manner after short-term acidic stress (Fig. 2C). Quantification of protein expression showed that cells cultured at acidic pH have significantly higher LC3-II levels and lower p62 levels, suggesting a functional autophagy in these conditions (Fig. 2C). The accumulation of LC3-II and degradation of p62 observed in cells cultured at acidic pH was time dependent and started as early as 2 h after exposure to acidic medium (Fig. 2D). In line with data from the LC3 fluorescence analysis (Fig. 2, A and B), the increased LC3-II at pH 6.5 and 6.2 was similar.
Melanoma Cells Maintain a Functional Autophagic Flux under Acidic Stress
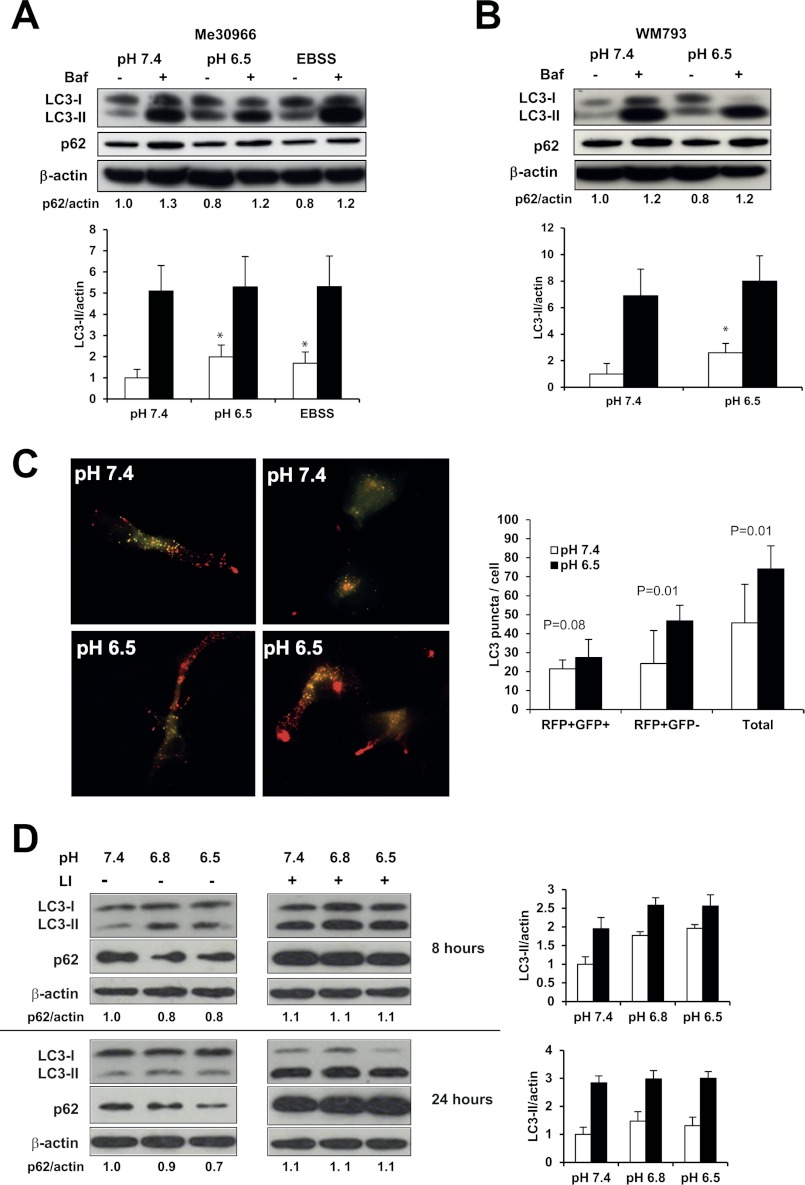
Because the accumulation of GFP-LC3+ vesicles was already detected at pH 6.5 and did not significantly change in lower pH culture conditions (Fig. 2B) we restricted further analysis to a pH range of 7.4–6.5. Moreover, in vivo MRS data indicate that pH 6.5 represents the average tumor pH (7, 12, 41). To analyze the status of the autophagic flux we performed experiments culturing Me30966 cells at pH 7.4 and 6.5 in the presence or absence of the lysosomal inhibitor bafilomycin A1, using EBSS-treated cells as positive control. In these experiments, low pH and starvation induced a significant increase in LC3-II protein (p < 0.05, Fig. 3A). Me30966 cells cultured at pH 7.4 or 6.5 were equally sensitive to the effects of bafilomycin A1 (Fig. 3A). In fact, expression of LC3-II was increased in the presence of bafilomycin A1 in both pH conditions. Similarly, accumulation of p62 in the presence of bafilomycin A1 was also increased in cells under acidic stress (pH 6.5). The same results were observed when different lysosomal inhibitors were used on Me30966 cells (supplemental Fig. S2A). Analysis of autophagic flux on WM793 (Fig. 3B), A375 (supplemental Fig. S2B), and SK-Mel-28 cells (supplemental Fig. S2C) confirmed that under acidic stress conditions melanoma cells maintain a fully functional autophagic flux. Another way to analyze the autophagic flux is to use the tandem probe RFP-GFP-LC3 (49). This probe differentiates autophagosomes (RFP+GFP+, yellow puncta) from autolysosomes (RFP+GFP−, red puncta) as the GFP fluorescence is quenched in acidic compartment. The number of autophagosomes (RFP+GFP+ puncta) tended to be slightly increased in cells under low pH conditions, whereas the number of autolysosomes (RFP+GFP-puncta) was significantly increased in Me30966 cultured in acidic pH (Fig. 3C), again indicating that the autophagic flux is maintained at acidic pH.
FIGURE 3.
Autophagic flux in melanoma cells under acidic stress. A, analysis of autophagic flux in Me30966 (A) and WM793 (B) cells was performed by evaluating the expression levels of LC3 and p62 in cells cultured in the specified pH conditions in the presence or absence of Bafilomycin A1 (30 nm) for 8 h. C, the presence of autophagosomes and autolysosomes was analyzed in Me30966 cells transfected with GFP-RFP-LC3 plasmid and observed after 8 h incubation with medium at pH 7.4 or 6.5. D, the autophagic flux was analyzed in Me30966 cells after short-term (8 h) and long-term (24 h) exposure to acidic stress in presence or absence of lysosomal inhibitors. Similar results were observed in 2 to 4 independent experiments. Where shown, * indicates p < 0.05.
Cancer cells may be exposed to sudden and dynamic changes in oxygen pressure and pH and we investigated how autophagy changed during longer exposure to low pH. To this end we cultured Me30966 cells at pH 7.4, 6.8, and 6.5 and evaluated the autophagic flux after short-time (8 h) and long-time (24 h) exposure, in the presence or absence of lysosomal inhibitors. As described above, after 8 h at different acidic media we observed LC3-II induction and decreased p62 expression, which further accumulated in the presence of lysosomal inhibitors, confirming that Me30966 cells in acidic pH maintain an active autophagic flux (Fig. 3D). Interestingly, after long-time (24 h) exposure to acidic pH cells still showed an active autophagic flux as shown by the continuous degradation of p62, which was greatly accumulated in the presence of lysosomal inhibitors (Fig. 3D). Analysis of cells kept in acidic culture conditions up to 72 h showed a similar capacity to keep an active autophagic flux (supplemental Fig. S3A). This observation was confirmed on Me30966 adapted to pH 6.5 for 2 months and showing an increased autophagic flux (supplemental Fig. S3B). Altogether, these findings suggest that melanoma cells are able to increase autophagy in response to acidic pH and maintain an active autophagic flux also during long-term exposure to low pH.
Our data show that acidic stress up-regulates autophagy in all melanoma cell lines investigated, as also recently reported in a model of metastatic breast carcinoma (50). It has also been reported that acidic stress may inhibit autophagy in mammary carcinoma cells (MCF-7) (51), as also corroborated in our experimental conditions. In fact in MCF-7 we noticed a reduction in the synthesis of LC3-II (as determined in the presence of bafilomycin A1) in acidic conditions (supplemental Fig. S4A). As the synthesis of LC3-II is correlated to the number of autophagosomes (52), this suggests that autophagy is reduced in MCF-7 maintained at acidic pH. In addition we also observed a reduction in autophagic flux in cells maintained at acidic pH as measured by the turnover of LC3-II (as determined in presence and absence of bafilomycin A1). Interestingly, whereas melanoma cells are able to slowly proliferate even at pH 6.5 (Fig. 1), viability of MCF-7 cells was dramatically decreased at acidic pH (supplemental Fig. S4B), which might be dependent on their decreased autophagic flux during acidic stress.
Acidic Stress Decreases ATP Content and Nutrients Uptake
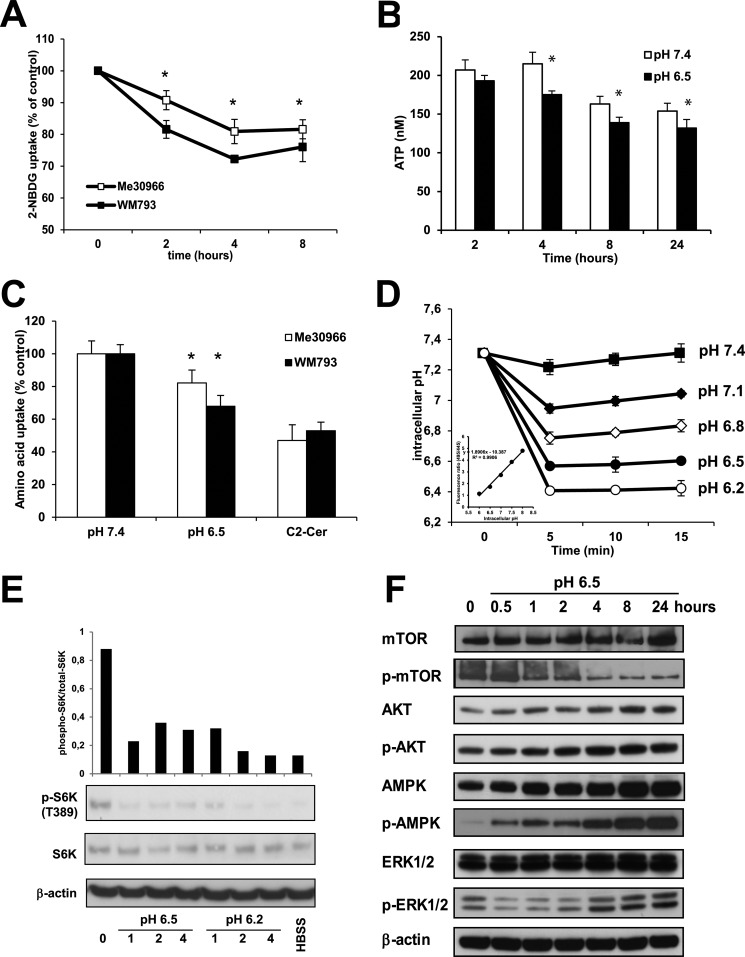
An important role of autophagy is to sustain cell survival in conditions of metabolic stress and limited nutrient availability. Therefore, we measured glucose and amino acid uptake in human melanoma cells after exposure to acidic medium. To measure glucose uptake we used 2-NBDG, a fluorescent non-metabolizable glucose analog. Me30966 and WM793 cells were exposed to medium at pH 7.4 and 6.5 and the uptake of 2-NBDG was measured at different time points. Melanoma cells cultured at acidic pH (pH 6.5) showed a time-dependent reduction in the uptake of glucose compared with cells cultured at neutral pH (Fig. 4A). The effects on glucose uptake were also pH dependent as shown for Me30966 cells (supplemental Fig. S5A). In line with this observation, both ATP (Fig. 4B) and extracellular lactate (supplemental Fig. S5B) concentrations were significantly reduced in cells cultured at acidic pH, likely as a result of decreased glucose consumption and inhibition of glycolysis.
FIGURE 4.
Effects of acidic stress on nutrients uptake and mTOR signaling. A, WM793 and Me30966 cells were exposed to pH 7.4 or 6.5 and the uptake of 2-NBDG was analyzed over time. The * shows a p < 0.05 for differences between cells cultured in acidic conditions and cells cultured at pH 7.4. B, Me30966 cells were exposed to pH 7.4 and 6.5 and cellular ATP concentration was determined over time and normalized to 100 cells. The experiments were run three times in six replicates, where * shows a p < 0.05. C, Me30966 and WM793 cells were exposed to medium at different pH values (7.4 and 6.5) or to C2-Cer (50 μm) for 4 h and the uptake of [3H]leucine was measured. The bars represent the mean ± S.D. of two experiments run in triplicate, where * shows a p < 0.05. D, the effect of medium at different pHe on intracellular pH was evaluated in A375 cells. The inset shows a representative standard curve by a ratiometric fluorescence method using BCECF. Representative data of two independent experiments run in triplicate wells are shown. E, the expression of total and phosphorylated p70S6K in Me30966 cells exposed to pH 6.5 and 6.2 was evaluated over time (hours) by Western blot and measured by densitometry using Adobe Photoshop Elements 9. F, the status of mTOR upstream signaling was analyzed in Me30966 cells exposed to pH 6.5 media at different time points (up to 24 h) by analyzing the phosphorylation of ERK, AKT, AMPK, and mTOR. The blots shown are representative of three different experiments with similar results.
Autophagy may be triggered by reduced amino acid uptake (53) and we used [3H]leucine to investigate the effects of low pH medium on amino acids uptake in Me30966 and WM793 cells. As positive control, we used C2-ceramide (C2-Cer), which is known to inhibit amino acid uptake (44). Indeed, treatment with C2-Cer significantly reduced the uptake of leucine in both cell lines (Fig. 4B). Similarly to the results from glucose uptake, short-term exposure to acidic pH induced a reduced uptake of the amino acid leucine in both Me30966 and WM793 cells (Fig. 4C). Taken together, these findings show that acidic pHe reduces nutrient uptake and causes a reduction in ATP levels.
Acidic pH Induces Cytosolic Acidification and Inhibition of mTOR Signaling
Exposure to acidic pHe was previously shown to induce acidification of pHi and inhibit glycolysis in human cancer cells (54, 55). Therefore, we first sought to investigate the effects of acidic stress on pHi in melanoma cells. The pHi was measured in A375 cells by using a ratiometric fluorescence method after incubation of cells with the pH-sensitive probe BCECF and generation of a standard curve (Fig. 4C). Although the pHi of cells exposed to pH 7.4 did not significantly change during the observation time, the pHi of cells exposed to different acidic media dropped to acidic values within minutes (Fig. 4D).
The mTOR has a crucial role in the regulation of metabolism and cell growth and acts as sensor for the availability of nutrients like amino acids and glucose. mTOR is present in 2 different complexes, mTORC1, which is sensitive to rapamycin and activates p70S6K, and mTORC2, which phosphorylates AKT at Ser-473 and is less sensitive to rapamycin. Inhibition of mTOR by rapamycin or other signaling pathways is known to stimulate autophagy (53, 56) and it has been recently reported that mTOR may be regulated by pH (57). While finalizing this study, an interesting study reported that niclosamide, an autophagy inducing drug causes cytosolic acidification and mTOR inhibition (58).
We analyzed the phosphorylation status of the mTOR downstream effector p70S6K following exposure of human melanoma cells to pH 6.5 and 6.2 culture conditions. As shown in Fig. 4E, the levels of phospho-p70S6K decreased significantly during acute exposure to acidic medium. Inhibition of mTOR activity was detected as early as 1 h after exposure to acidic pH. Long-term exposure of melanoma cells to acidic stress showed that the inhibition of p70S6K was stable over time and detected at least over a 16-h incubation (supplemental Fig. S5C). Moreover, analysis of another mTOR downstream effector, 4EBP1, also indicated a significantly reduced phosphorylation under acidic stress (supplemental Fig. S5D).
To evaluate the signaling molecules involved in the mTOR pathway we analyzed upstream events that might regulate the mTOR pathway in melanoma cells exposed to acidic stress (pH 6.5) for up to 24 h (Fig. 4F). In line with decreased glucose uptake and ATP content, we observed that exposure to acidic pH rapidly caused increased phosphorylation of AMPK (Fig. 4F). Moreover, the mTOR phosphorylation site Ser-2448 was clearly inhibited in cells exposed to pH 6.5 (Fig. 4F).
Analysis of phosphorylated AKT at position Ser-473 showed no effects of acidic stress, excluding a role for mTORC2. However, phosphorylation of ERK1/2 was transiently inhibited by exposure to acidic stress but promptly normalized after a 4-h incubation, suggesting that the ERK pathway is not involved in the regulation of mTOR signaling in these cells (Fig. 4F). These data suggest that inhibition of the mTOR pathway occurs very early and may drive autophagy in melanoma cells cultured at acidic pH.
Autophagy Is Crucial for Survival of Melanoma Cells under Acidic Stress
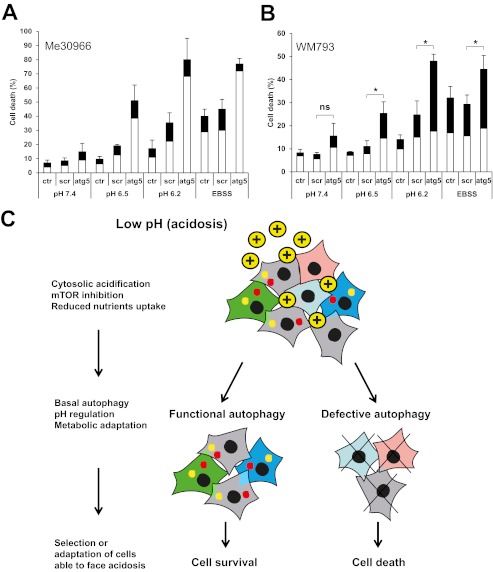
Functional autophagy represents a survival strategy for cells under metabolic stress (32–34). To determine the role of autophagy in acidic stress-induced cell death in human melanoma we used siRNA to knockdown the expression of ATG5, a crucial protein involved in the build-up of autophagosomes. Interestingly, the expression of ATG5 was increased in Me30966 and WM793 cells after an 8-h exposure to pH 6.5 (supplemental Fig. S6A). Me30966 and WM793 cells were transfected with ATG5 siRNA and after 48 h the medium was replaced with EBSS, pH 7.4, 6.5, and 6.2 media, respectively. After exposure for 24 h to these conditions cell death was evaluated and expressed as the percentage of apoptotic and necrotic cells. Knockdown of ATG5 reduced the expression of ATG5 protein and LC3-II in both cell lines (shown in supplemental Fig. S6B for WM793 cells), indicating functional inhibition of autophagy. Knockdown of ATG5 did not significantly affect survival of Me30966 cells at neutral pH, whereas it significantly increased the sensitivity to cell death induced by acidic medium in a pH-dependent manner (Fig. 5A, supplemental Fig. S6C). Cell death at pH 7.4 was similar in cells transfected with scrambled or ATG5-specific siRNA (p = 0.17). On the other hand, under acidic conditions at pH 6.5 cell death was 19 ± 3% in cells treated with scrambled siRNA and 51 ± 11 in cells treated with ATG5 siRNA (p < 0.05). Such differences were much more remarkable at pH 6.2 where cell death was 35 ± 7% in control cells and 80 ± 15% in autophagy-deficient cells. Similar results were observed on WM793 cells (Fig. 5B, supplemental Fig. S6D). In fact, at pH 7.4 cell death was 8 ± 1% in control cells, whereas it was 15 ± 5% in ATG5-KD cells (p = 0.13). On the other hand, at pH 6.5 and 6.2 cell death was 11 ± 2 and 24 ± 6% in control cells and increased to 25 ± 5 and 48 ± 3% in ATG5-KD cells (p < 0.05), respectively. As expected, expression of ATG5 and induction of autophagy was instrumental for cell survival in conditions of nutrient starvation in both cell lines.
FIGURE 5.
Autophagy protects melanoma cells following acidic stress. Me30966 cells (A) and WM793 cells (B) were transfected with siRNA for human ATG5 gene and cell death was measured 24 h after exposing the cells to different pH conditions (pH 7.4, pH 6.5, and pH 6.2). Cells exposed to nutrient starvation (EBSS) were used as positive control. Cell death is expressed as the mean percentage of apoptotic (white bars) and necrotic (black bars). Mean ± S.D. of three experiments run in duplicate wells are shown, where * shows a p < 0.05. The p value was calculated for differences between the cells treated with scrambled and ATG5-specific siRNA (paired t test). C, schematic representation of a possible model linking autophagy and acidic pH in melanoma.
Interestingly, the cell death modes induced by acidic stress in these two cell lines seemed to be different: in WM793 cells necrosis was the main cell death mode following autophagy inhibition, apoptosis was prevalent in Me30966 cells. These findings indicate that functional autophagy serves as a protective mechanism adopted by melanoma cells following acute exposure to acidic stress (Fig. 5B).
DISCUSSION
We reported here that human melanoma cells exposed to acute acidic stress rely on functional autophagy to adapt and proliferate following exposure to acidic stress. We provide evidence that acidic stress induces acidification of pHi and inhibition of mTOR, resulting in up-regulated autophagic flux. Inhibition of autophagy by knockdown expression of ATG5 dramatically sensitized melanoma cells to acidic stress-induced cell death.
Autophagy is the most important mechanism for degradation and recycling of long-lived proteins, protein aggregates, organelles, and intracellular pathogens (59). In normal physiologic conditions autophagy helps in keeping cellular homeostasis by “clearing” dispensable/damaged intracellular structures, providing a crucial tool for protection against accumulation of toxic cellular components. Thus, autophagy serves as a protective response to various stress stimuli, as suggested by the association between defective autophagy and several human pathologies, including cancer (56).
Because the identification of metabolic transformation as an important determinant of cancer progression, the role of autophagy in cancer development and progression is being debated (32). A tumor suppressive role for autophagy was first suggested by the fact that mice with allelic loss of BECLIN-1 are more prone to develop spontaneous cancers (60). Functional autophagy also contributes to reduce tissue damage, local inflammation, and DNA damage, thus suppressing tumorigenesis and tumor progression (61, 62). Moreover, constitutive activation of mTOR by oncogenic signaling via PI3K and AKT in cancer cells also supports the tumor suppressive function of autophagy (1, 19).
On the other hand, autophagy sustains tumor growth and progression in aggressive tumors. In fact, cancer cells subjected to metabolic stress and chemotherapy up-regulate the autophagic machinery to promote cell survival and/or avoid apoptosis (33, 63). Moreover, high basal autophagy is required for growth of pancreatic cancer and human cancers carrying activated K-RAS (36, 64). Intriguingly, up-regulated autophagy linked to hypoxia and acidity has been reported as a marker of poor prognosis in melanoma and colorectal cancer (39, 40). In line with this, autophagy was shown to be mostly localized in hypoxic nonvascularized tumor regions (63), suggesting that autophagy may be important to sustain tumor growth and survival in the developing, scarcely perfused tumors. Elegant experiments performed in melanoma spheroids showed that inhibition of autophagy results in cell death mostly localized in the spheroids core (48), suggesting that increased autophagy may predict invasiveness and drug resistance in malignant melanoma (48). In line with this observation, we found that multicellular spheroids from colon carcinoma cells (HCT116) displayed a strong expression of LC3 in the spheroid core (data not shown).
A major consequence of the abnormal tumor metabolism is the accumulation of metabolic acids within the tumor microenvironment, characterized by low pHe heterogeneously distributed within the tumor mass (4, 7). Exposure of cancer cells to an acidic environment plays a crucial function in promoting invasive capacity, metastasis, and resistance to apoptotic-inducing agents. Intriguingly, autophagy was shown to represent a major survival mechanism for cytogenetically abnormal neoplastic ductal carcinoma in situ cells (65). The capacity of these cells to form spheroids and generate tumors in SCID mice was abrogated upon treatment with the autophagy inhibitor chloroquine. Because the contribution of hypoxia and acidity to tumor progression from ductal carcinoma in situ to invasive carcinoma has been well established, this observation supports a direct link between tumor acidic pH and autophagy.
We hypothesized that exposure of cancer cells to an acidic environment would stimulate autophagy. We used human melanoma cell lines to investigate how exposure to acidic stress affected autophagy and study the underlying mechanisms. Human melanoma cells exhibit a peculiar capacity to easily adapt to low pH culture conditions (35, 41, 66). Moreover, autophagy was recently suggested as an important therapeutic target for treatment of human melanoma (67). The major finding of our study is that acidic stress stimulates a protective autophagic response in melanoma cells. We observed that melanoma cells up-regulated the conversion of LC3-I into LC3-II upon short-term exposure to acidic medium, with the appearance of numerous LC3-positive vesicles. Analysis of the autophagic flux indicated the presence of functional autophagy in melanoma cells exposed to acidic stress. Our data are well in agreement with a recent study performed on breast carcinoma showing that the acidic tumor microenvironment promotes autophagy and that chronic autophagy represents a survival adaptation to acidic stress (50). However, the relationship between pH and autophagy in breast cancer seems more complex because it has also been reported that acidic stress may indeed inhibit autophagy in MCF7 cells (51). To exclude differences in experimental settings we analyzed the response of MCF7 cells to acidic conditions and confirmed that synthesis of autophagosomes and the autophagic flux are reduced in MCF-7 cells under acidic stress (supplemental Fig. S3A). Interestingly, whereas all melanoma cell lines tested were able to slowly proliferate at pH 6.5, MCF-7 cells did not show this capacity. MCF-7 cells are known to have low basal autophagic activity likely because of the low expression of BECLIN-1 (68) and this might explain differences in the capacity of maintaining a functional autophagic flux under acidic stress. Interestingly, acidic stress caused inhibition of the mTOR pathway also in MCF-7 cells (data not shown), suggesting that the capacity of cancer cells to modulate autophagy following acidic stress may not be dependent only on mTOR inhibition but likely on their ability to restore their physiological pHi by extruding acids via pH-regulating systems. On the other hand, human melanoma cells rely on functional autophagy for survival and invasion (48) and increased autophagy in several human cancers, including melanoma is associated to metastasis and poor outcome (69). Interestingly, the melanoma cell line Me30966 maintained an increased autophagic flux even after a long-term (24–72 h) culture in acidic conditions. Moreover, we have developed Me30966 cells adapted for 2 months to grow at pH 6.5 and observed that these cells maintain a fully functional autophagic flux, strongly supporting a recent observation reported on breast cancer cells (50). In line with the clinical studies so far available, our data support a crucial role of autophagy in survival of melanoma cells under acidic stress. We reckon that the ability to maintain a functional autophagic flux in response to pHe changes may vary among different types of cancer, possibly depending on the mutational status affecting the metabolic requirements to sustain cell growth and survival in stress conditions. Moreover, it has also been reported that alkaline stress induces autophagy by inhibition of mTOR (70). Because exposure to alkaline medium is expected to induce partial alkalinization of pHi, this data would support a role for cytoplasmic alkalinization in induction of autophagy through mTOR inhibition (71).
However, Balgi and colleagues (57) recently demonstrated that exposure to acidic pHe causes a rapid inhibition of mTOR signaling, supporting a crucial role for pH in modulation of mTOR activity. These findings indicate that mTOR signaling may be sensitive to cytosolic pH variations. To further validate this hypothesis, we analyzed the status of the mTOR pathway in response to acute extracellular acidification in human melanoma by investigating the phosphorylation of p70S6K, a downstream effector of the mTOR signaling pathway. We consistently found that acidic stress induces very early inhibition of mTOR activity, which occurred within 1 h. Conversely to what is described in MCF7 cells (57), we found that in association with the decreased ATP levels, acidic pH induced phosphorylation of AMPK, an essential positive regulator of autophagy (56). Because levels of phospho-AMPK remained elevated, whereas both phospho-ERK1/2 and phospho-AKT were unchanged, our observation suggests that melanoma cells undergo an autophagic response and at the same time maintain the capacity to proliferate under acidic stress (Fig. 1). Acidic pHe was reported to inhibit glycolysis via decreased glucose uptake in Ehrlich ascites tumor cells (72) and our data indicate that melanoma cells exposed to acidic pH show signs of glycolysis inhibition because they reduced glucose uptake and lactate release. Similarly, glycolysis inhibition by 2-deoxy-d-glucose was reported to induce cytoprotective autophagy in endothelial and melanoma cells (73, 74). Because it is known that active mTOR stimulates glucose uptake via increased membrane expression of glucose transporters (75) it is likely that inhibition of mTOR by acidic pH may be the upstream event leading to reduced glycolysis and autophagy induction. In line with this, we reported that cytosolic acidification mediated by proton pump inhibitors and associated with mTOR inhibition induces a protective autophagy in melanoma cells (35). Moreover, we also observed that acidic stress inhibited uptake of the amino acid leucine. Intriguingly, mTOR signaling can regulate nutrients transporters, as shown in the case of leucine transporter (76). Because leucine is a potent inhibitor of autophagy (56) it is possible that acidic pH-mediated inhibition of mTOR stimulates autophagy in melanoma cells via decreased leucine uptake. With this regard, a combination of leucine deprivation and autophagy inhibition has been recently suggested as an anticancer strategy against human melanoma (67).
To get insight on the functional role of autophagy under acidic stress, we analyzed acidic stress-induced cell death in human melanoma after knockdown of ATG5, a crucial gene for the formation of autophagosomes. We observed that defective autophagy achieved by ATG5 knockdown significantly increased the sensitivity to acidic stress-induced cell death in human melanoma cells. Intriguingly, whereas in Me30966 cells apoptosis was the main cell death mechanism, WM793 cells died mostly through necrosis. Recently, the increased autophagic index has been associated to the malignant behavior of aggressive melanoma cells (48). An important observation made by Ma and colleagues (48) was that autophagy inhibition via ATG5 knockdown induced selective killing of metabolically stressed cells localized in the core of multicellular spheroids. Moreover, the authors suggest a direct link between up-regulated autophagy and invasive potential of melanoma cells. Because acidic pH stimulates melanoma cell invasion, our findings support a role for autophagy in melanoma progression. We have reported that human melanoma cells exposed to acidic medium and/or starvation face off nutrient deprivation by increasing dramatically their ability to cannibalize other cells (47). It is well established that functional autophagy is required for digestion of intracellular pathogens (59), thus an intriguing hypothesis is that a functional autophagy may participate in the cannibal activity of malignant cells exposed to acidic stress (77).
Our study provides further support to therapeutic strategies targeting autophagy to kill tumor cells, directly or in combination with chemotherapy (36, 37, 78). Recently, we showed that melanoma cells under acidic conditions up-regulate autophagy as a protective mechanism following treatment with proton pump inhibitors (35). Inhibition of autophagy in these cells increased their sensitivity to proton pump inhibitor-induced cell death. Because targeting pH regulation in tumor cells represents a novel and attractive therapeutic strategy (13, 79, 80), the finding that functional autophagy may protect cells exposed to acidic stress, thus contributing to malignant progression, further supports the validity of therapeutic targeting autophagy and pH regulation in cancer.
Supplementary Material
Acknowledgments
We thank Dr. Francesco Lozupone (Istituto Superiore di Sanità, Italy) and Dr. Emma Hernlund (Karolinska Institute, Sweden) for helpful discussion and Dr. Tamotsu Yoshimori (Graduate School of Medicine, Osaka University, Osaka, Japan) for kindly providing the LC3 plasmids.
This work was supported by Italian Association for Cancer Research Grant 5940, Association for International Cancer Research Grant 11-0522, the Alex and Eva Wallströms Foundation, the Sigurd and Elsa Goljes Foundation, the Axel and Signe Lagermans Donation Fondations, and grants from INSERM and the University Paris-Sud 11 (to P. C.).

This article contains supplemental Figs. S1–S6.
- pHi
- intracellular pH
- pHe
- extracellular pH
- 2-NBDG
- 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose
- mTOR
- mammalian target of rapamycin
- AMPK
- AMP-activated protein kinase
- BCECF-AM
- 2′,7′-bis-(2-carboxyethyl)-5-(and -6)-carboxyfluorescein
- EBSS
- Earle's balanced salt solution.
REFERENCES
- 1. Cairns R. A., Harris I. S., Mak T. W. (2011) Regulation of cancer cell metabolism. Nat. Rev. Cancer 11, 85–95 [DOI] [PubMed] [Google Scholar]
- 2. Chiche J., Brahimi-Horn M. C., Pouysségur J. (2010) Tumor hypoxia induces a metabolic shift causing acidosis. A common feature in cancer. J. Cell Mol. Med. 14, 771–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Warburg O. (1956) On the origin of cancer cells. Science 123, 309–314 [DOI] [PubMed] [Google Scholar]
- 4. Gatenby R. A., Gillies R. J. (2004) Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4, 891–899 [DOI] [PubMed] [Google Scholar]
- 5. Gillies R. J., Robey I., Gatenby R. A. (2008) Causes and consequences of increased glucose metabolism of cancers. J. Nucl. Med. 49, 24S-42S [DOI] [PubMed] [Google Scholar]
- 6. Morita T., Nagaki T., Fukuda I., Okumura K. (1992) Clastogenicity of low pH to various cultured mammalian cells. Mutat Res. 268, 297–305 [DOI] [PubMed] [Google Scholar]
- 7. Gillies R. J., Raghunand N., Karczmar G. S., Bhujwalla Z. M. (2002) MRI of the tumor microenvironment. J. Magn. Reson. Imaging 16, 430–450 [DOI] [PubMed] [Google Scholar]
- 8. Orive G., Reshkin S. J., Harguindey S., Pedraz J. L. (2003) Hydrogen ion dynamics and the Na+/H+ exchanger in cancer angiogenesis and antiangiogenesis. Br. J. Cancer 89, 1395–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. De Milito A., Fais S. (2005) Tumor acidity, chemoresistance, and proton pump inhibitors. Future Oncol. 1, 779–786 [DOI] [PubMed] [Google Scholar]
- 10. Tannock I. F., Rotin D. (1989) Acid pH in tumors and its potential for therapeutic exploitation. Cancer Res. 49, 4373–4384 [PubMed] [Google Scholar]
- 11. Fischer K., Hoffmann P., Voelkl S., Meidenbauer N., Ammer J., Edinger M., Gottfried E., Schwarz S., Rothe G., Hoves S., Renner K., Timischl B., Mackensen A., Kunz-Schughart L., Andreesen R., Krause S. W., Kreutz M. (2007) Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 109, 3812–3819 [DOI] [PubMed] [Google Scholar]
- 12. Hjelmeland A. B., Wu Q., Heddleston J. M., Choudhary G. S., MacSwords J., Lathia J. D., McLendon R., Lindner D., Sloan A., Rich J. N. (2011) Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ. 18, 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fais S. (2010) Proton pump inhibitor-induced tumor cell death by inhibition of a detoxification mechanism. J. Intern. Med. 267, 515–525 [DOI] [PubMed] [Google Scholar]
- 14. Harguindey S., Arranz J. L., Wahl M. L., Orive G., Reshkin S. J. (2009) Proton transport inhibitors as potentially selective anticancer drugs. Anticancer Res. 29, 2127–2136 [PubMed] [Google Scholar]
- 15. Huber V., De Milito A., Harguindey S., Reshkin S. J., Wahl M. L., Rauch C., Chiesi A., Pouysségur J., Gatenby R. A., Rivoltini L., Fais S. (2010) Proton dynamics in cancer. J. Transl. Med. 8, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lou Y., McDonald P. C., Oloumi A., Chia S., Ostlund C., Ahmadi A., Kyle A., Auf dem Keller U., Leung S., Huntsman D., Clarke B., Sutherland B. W., Waterhouse D., Bally M., Roskelley C., Overall C. M., Minchinton A., Pacchiano F., Carta F., Scozzafava A., Touisni N., Winum J. Y., Supuran C. T., Dedhar S. (2011) Targeting tumor hypoxia. Suppression of breast tumor growth and metastasis by novel carbonic anhydrase IX inhibitors. Cancer Res. 71, 3364–3376 [DOI] [PubMed] [Google Scholar]
- 17. Neri D., Supuran C. T. (2011) Interfering with pH regulation in tumors as a therapeutic strategy. Nat. Rev. Drug Discov. 10, 767–777 [DOI] [PubMed] [Google Scholar]
- 18. Mehrpour M., Esclatine A., Beau I., Codogno P. (2010) Overview of macroautophagy regulation in mammalian cells. Cell Res. 20, 748–762 [DOI] [PubMed] [Google Scholar]
- 19. Yang Z., Klionsky D. J. (2010) Eaten alive. A history of macroautophagy. Nat. Cell Biol. 12, 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mizushima N., Komatsu M. (2011) Autophagy. Renovation of cells and tissues. Cell 147, 728–741 [DOI] [PubMed] [Google Scholar]
- 21. Weidberg H., Shvets E., Elazar Z. (2011) Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 80, 125–156 [DOI] [PubMed] [Google Scholar]
- 22. Cecconi F., Levine B. (2008) The role of autophagy in mammalian development. Cell makeover rather than cell death. Dev. Cell 15, 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cuervo A. M. (2008) Autophagy and aging. Keeping that old broom working. Trends Genet. 24, 604–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rubinsztein D. C., Mariño G., Kroemer G. (2011) Autophagy and aging. Cell 146, 682–695 [DOI] [PubMed] [Google Scholar]
- 25. Deretic V. (2011) Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol. Rev. 240, 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Levine B., Mizushima N., Virgin H. W. (2011) Autophagy in immunity and inflammation. Nature 469, 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harris H., Rubinsztein D. C. (2012) Control of autophagy as a therapy for neurodegenerative disease. Nat. Rev. Neurol. 8, 108–117 [DOI] [PubMed] [Google Scholar]
- 28. Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. White E., DiPaola R. S. (2009) The double-edged sword of autophagy modulation in cancer. Clin. Cancer Res. 15, 5308–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janku F., McConkey D. J., Hong D. S., Kurzrock R. (2011) Autophagy as a target for anticancer therapy. Nat. Rev. Clin. Oncol. 8, 528–539 [DOI] [PubMed] [Google Scholar]
- 31. Kimmelman A. C. (2011) The dynamic nature of autophagy in cancer. Genes Dev. 25, 1999–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mathew R., White E. (2011) Autophagy in tumorigenesis and energy metabolism. Friend by day, foe by night. Curr. Opin. Genet. Dev. 21, 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rabinowitz J. D., White E. (2010) Autophagy and metabolism. Science 330, 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Song J., Guo X., Xie X., Zhao X., Li D., Deng W., Song Y., Shen F., Wu M., Wei L. (2011) Autophagy in hypoxia protects cancer cells against apoptosis induced by nutrient deprivation through a Beclin 1-dependent way in hepatocellular carcinoma. J. Cell Biochem. 112, 3406–3420 [DOI] [PubMed] [Google Scholar]
- 35. Marino M. L., Fais S., Djavaheri-Mergny M., Villa A., Meschini S., Lozupone F., Venturi G., Della Mina P., Pattingre S., Rivoltini L., Codogno P., De Milito A. (2010) Proton pump inhibition induces autophagy as a survival mechanism following oxidative stress in human melanoma cells. Cell Death Dis. 1, e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guo J. Y., Chen H. Y., Mathew R., Fan J., Strohecker A. M., Karsli-Uzunbas G., Kamphorst J. J., Chen G., Lemons J. M., Karantza V., Coller H. A., Dipaola R. S., Gelinas C., Rabinowitz J. D., White E. (2011) Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 25, 460–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang S., Wang X., Contino G., Liesa M., Sahin E., Ying H., Bause A., Li Y., Stommel J. M., Dell'antonio G., Mautner J., Tonon G., Haigis M., Shirihai O. S., Doglioni C., Bardeesy N., Kimmelman A. C. (2011) Pancreatic cancers require autophagy for tumor growth. Genes Dev. 25, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Giatromanolaki A., Koukourakis M. I., Harris A. L., Polychronidis A., Gatter K. C., Sivridis E. (2010) Prognostic relevance of light chain 3 (LC3A) autophagy patterns in colorectal adenocarcinomas. J. Clin. Pathol. 63, 867–872 [DOI] [PubMed] [Google Scholar]
- 39. Koukourakis M. I., Giatromanolaki A., Sivridis E., Pitiakoudis M., Gatter K. C., Harris A. L. (2010) Beclin 1 over- and underexpression in colorectal cancer. Distinct patterns relate to prognosis and tumor hypoxia. Br J. Cancer 103, 1209–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sivridis E., Koukourakis M. I., Mendrinos S. E., Karpouzis A., Fiska A., Kouskoukis C., Giatromanolaki A. (2011) Beclin-1 and LC3A expression in cutaneous malignant melanomas. A biphasic survival pattern for beclin-1. Melanoma Res. 21, 188–195 [DOI] [PubMed] [Google Scholar]
- 41. De Milito A., Canese R., Marino M. L., Borghi M., Iero M., Villa A., Venturi G., Lozupone F., Iessi E., Logozzi M., Della Mina P., Santinami M., Rodolfo M., Podo F., Rivoltini L., Fais S. (2010) pH-dependent antitumor activity of proton pump inhibitors against human melanoma is mediated by inhibition of tumor acidity. Int. J. Cancer 127, 207–219 [DOI] [PubMed] [Google Scholar]
- 42. Yang T. T., Sinai P., Kain S. R. (1996) An acid phosphatase assay for quantifying the growth of adherent and nonadherent cells. Anal. Biochem. 241, 103–108 [DOI] [PubMed] [Google Scholar]
- 43. Grant R. L., Acosta D. (1997) Ratiometric measurement of intracellular pH of cultured cells with BCECF in a fluorescence multiwell plate reader. In Vitro Cell Dev. Biol. Anim. 33, 256–260 [DOI] [PubMed] [Google Scholar]
- 44. Guenther G. G., Peralta E. R., Rosales K. R., Wong S. Y., Siskind L. J., Edinger A. L. (2008) Ceramide starves cells to death by down-regulating nutrient transporter proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 17402–17407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reichert M., Steinbach J. P., Supra P., Weller M. (2002) Modulation of growth and radiochemosensitivity of human malignant glioma cells by acidosis. Cancer 95, 1113–1119 [DOI] [PubMed] [Google Scholar]
- 46. Williams A. C., Collard T. J., Paraskeva C. (1999) An acidic environment leads to p53 dependent induction of apoptosis in human adenoma and carcinoma cell lines. Implications for clonal selection during colorectal carcinogenesis. Oncogene 18, 3199–3204 [DOI] [PubMed] [Google Scholar]
- 47. Lugini L., Matarrese P., Tinari A., Lozupone F., Federici C., Iessi E., Gentile M., Luciani F., Parmiani G., Rivoltini L., Malorni W., Fais S. (2006) Cannibalism of live lymphocytes by human metastatic but not primary melanoma cells. Cancer Res. 66, 3629–3638 [DOI] [PubMed] [Google Scholar]
- 48. Ma X. H., Piao S., Wang D., McAfee Q. W., Nathanson K. L., Lum J. J., Li L. Z., Amaravadi R. K. (2011) Measurements of tumor cell autophagy predict invasiveness, resistance to chemotherapy, and survival in melanoma. Clin. Cancer Res. 17, 3478–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kimura S., Noda T., Yoshimori T. (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 [DOI] [PubMed] [Google Scholar]
- 50. Wojtkowiak J., Rothberg J. M., Kumar V., Schramm K. J., Haller E., Proemsey J. B., Lloyd M. C., Sloane B. F., Gillies R. J. (2012) Chronic autophagy is a cellular adaptation to tumor acidic pH microenvironments. Cancer Res. doi: 10.1158/0008-5472.CAN-11-3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu T., Su H., Ganapathy S., Yuan Z. M. (2011) Modulation of autophagic activity by extracellular pH. Autophagy 7, 1316–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tanida I., Minematsu-Ikeguchi N., Ueno T., Kominami E. (2005) Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 1, 84–91 [DOI] [PubMed] [Google Scholar]
- 53. Meijer A. J. (2008) Amino acid regulation of autophagosome formation. Methods Mol. Biol. 445, 89–109 [DOI] [PubMed] [Google Scholar]
- 54. Newell K. J., Tannock I. F. (1989) Reduction of intracellular pH as a possible mechanism for killing cells in acidic regions of solid tumors. Effects of carbonylcyanide-3-chlorophenylhydrazone. Cancer Res. 49, 4477–4482 [PubMed] [Google Scholar]
- 55. Rotin D., Robinson B., Tannock I. F. (1986) Influence of hypoxia and an acidic environment on the metabolism and viability of cultured cells. Potential implications for cell death in tumors. Cancer Res. 46, 2821–2826 [PubMed] [Google Scholar]
- 56. Meijer A. J., Codogno P. (2009) Autophagy, regulation and role in disease. Crit. Rev. Clin. Lab. Sci. 46, 210–240 [DOI] [PubMed] [Google Scholar]
- 57. Balgi A. D., Diering G. H., Donohue E., Lam K. K., Fonseca B. D., Zimmerman C., Numata M., Roberge M. (2011) Regulation of mTORC1 signaling by pH. PLoS One 6, e21549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fonseca B. D., Diering G. H., Bidinosti M. A., Dalal K., Alain T., Balgi A. D., Forestieri R., Nodwell M., Rajadurai C. V., Gunaratnam C., Tee A. R., Duong F., Andersen R. J., Orlowski J., Numata M., Sonenberg N., Roberge M. (2012) Structure-activity analysis of niclosamide reveals potential role for cytoplasmic pH in control of mammalian target of rapamycin complex 1 (mTORC1) signaling. J. Biol. Chem. 287, 17530–17545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Orvedahl A., Levine B. (2009) Eating the enemy within. Autophagy in infectious diseases. Cell Death Differ. 16, 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E. L., Mizushima N., Ohsumi Y., Cattoretti G., Levine B. (2003) Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112, 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mathew R., Karp C. M., Beaudoin B., Vuong N., Chen G., Chen H. Y., Bray K., Reddy A., Bhanot G., Gelinas C., Dipaola R. S., Karantza-Wadsworth V., White E. (2009) Autophagy suppresses tumorigenesis through elimination of p62. Cell 137, 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mathew R., Kongara S., Beaudoin B., Karp C. M., Bray K., Degenhardt K., Chen G., Jin S., White E. (2007) Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 21, 1367–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Degenhardt K., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gélinas C., Fan Y., Nelson D. A., Jin S., White E. (2006) Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Karantza-Wadsworth V., Patel S., Kravchuk O., Chen G., Mathew R., Jin S., White E. (2007) Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 21, 1621–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Espina V., Mariani B. D., Gallagher R. I., Tran K., Banks S., Wiedemann J., Huryk H., Mueller C., Adamo L., Deng J., Petricoin E. F., Pastore L., Zaman S., Menezes G., Mize J., Johal J., Edmiston K., Liotta L. A. (2010) Malignant precursor cells pre-exist in human breast DCIS and require autophagy for survival. PLoS One 5, e10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wahl M. L., Owen J. A., Burd R., Herlands R. A., Nogami S. S., Rodeck U., Berd D., Leeper D. B., Owen C. S. (2002) Regulation of intracellular pH in human melanoma. Potential therapeutic implications. Mol. Cancer Ther. 1, 617–628 [PubMed] [Google Scholar]
- 67. Sheen J. H., Zoncu R., Kim D., Sabatini D. M. (2011) Defective regulation of autophagy upon leucine deprivation reveals a targetable liability of human melanoma cells in vitro and in vivo. Cancer Cell 19, 613–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 69. Lazova R., Camp R. L., Klump V., Siddiqui S. F., Amaravadi R. K., Pawelek J. M. (2012) Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin. Cancer Res. 18, 370–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Suk J., Kwak S. S., Lee J. H., Choi J. H., Lee S. H., Lee D. H., Byun B., Lee G. H., Joe C. O. (2011) Alkaline stress-induced autophagy is mediated by mTORC1 inactivation. J. Cell Biochem. 112, 2566–2573 [DOI] [PubMed] [Google Scholar]
- 71. Korolchuk V. I., Rubinsztein D. C. (2011) Regulation of autophagy by lysosomal positioning. Autophagy 7, 927–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kaminskas E. (1978) The pH dependence of sugar-transport and glycolysis in cultured Ehrlich ascites tumor cells. Biochem. J. 174, 453–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang Q., Liang B., Shirwany N. A., Zou M. H. (2011) 2-Deoxy-d-glucose treatment of endothelial cells induces autophagy by reactive oxygen species-mediated activation of the AMP-activated protein kinase. PLoS One 6, e17234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Giammarioli A. M., Gambardella L., Barbati C., Pietraforte D., Tinari A., Alberton M., Gnessi L., Griffin R. J., Minetti M., Malorni W. (2012) Differential effects of the glycolysis inhibitor 2-deoxy-d-glucose on the activity of pro-apoptotic agents in metastatic melanoma cells, and induction of a cytoprotective autophagic response. Int. J. Cancer 131, E337–347 [DOI] [PubMed] [Google Scholar]
- 75. Edinger A. L., Thompson C. B. (2002) Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol. Biol. Cell 13, 2276–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Roos S., Jansson N., Palmberg I., Säljö K., Powell T. L., Jansson T. (2007) Mammalian target of rapamycin in the human placenta regulates leucine transport and is down-regulated in restricted fetal growth. J. Physiol. 582, 449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fais S. (2010) Moulding the shape of a metastatic cell. Leuk. Res. 34, 843–847 [DOI] [PubMed] [Google Scholar]
- 78. Amaravadi R. K., Yu D., Lum J. J., Bui T., Christophorou M. A., Evan G. I., Thomas-Tikhonenko A., Thompson C. B. (2007) Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J. Clin. Invest. 117, 326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Robey I. F., Baggett B. K., Kirkpatrick N. D., Roe D. J., Dosescu J., Sloane B. F., Hashim A. I., Morse D. L., Raghunand N., Gatenby R. A., Gillies R. J. (2009) Bicarbonate increases tumor pH and inhibits spontaneous metastases. Cancer Res. 69, 2260–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Supuran C. T. (2008) Carbonic anhydrases. Novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 7, 168–181 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.