Background: Precise chemical structure and biosynthetic pathway of the acyl chain of mycobacterial glycopeptidolipids (GPLs) is unknown.
Results: Polyketide synthases dictate biosynthesis and determine hydroxylation at C-5 position of the GPL acyl chain.
Conclusions: Retrobiosynthetic studies establish the role of bimodular polyketide synthase and fatty acyl-AMP-ligase in GPL biosynthesis.
Significance: The long-standing ambiguity on the accurate structure and the biosynthetic mechanism of GPLs was resolved.
Keywords: Acyl Carrier Protein, Fatty Acid, Lipids, Mycobacteria, Polyketides, Fatty Acyl AMP Ligase, Hydroxylation, Polyketide Synthase
Abstract
Glycopeptidolipids (GPLs) are dominant cell surface molecules present in several non-tuberculous and opportunistic mycobacterial species. GPLs from Mycobacterium smegmatis are composed of a lipopeptide core unit consisting of a modified C26-C34 fatty acyl chain that is linked to a tetrapeptide (Phe-Thr-Ala-alaninol). The hydroxyl groups of threonine and terminal alaninol are further modified by glycosylations. Although chemical structures have been reported for 16 GPLs from diverse mycobacteria, there is still ambiguity in identifying the exact position of the hydroxyl group on the fatty acyl chain. Moreover, the enzymes involved in the biosynthesis of the fatty acyl component are unknown. In this study we show that a bimodular polyketide synthase in conjunction with a fatty acyl-AMP ligase dictates the synthesis of fatty acyl chain of GPL. Based on genetic, biochemical, and structural investigations, we determine that the hydroxyl group is present at the C-5 position of the fatty acyl component. Our retrobiosynthetic approach has provided a means to understand the biosynthesis of GPLs and also resolve the long-standing debate on the accurate structure of mycobacterial GPLs.
Introduction
Mycobacteria produce diverse lipid metabolites, many of which are unique to this species (1, 2). Glycopeptidolipids (GPLs)5 are one of the most abundant cell-surface glycolipids synthesized by several non-tuberculous opportunistic mycobacteria. GPL molecules provide distinct surface properties to these organisms, like sliding motility and biofilm development (3, 4). GPLs are also known to play a significant role in pathogenesis by activating the host immune response (5). Subtle structural variability of these molecules generates phenotypic heterogeneity between the different mycobacterial species. Despite the characterization of first GPL structure (6) and the genetic cluster (7), precise chemistry as well as mechanism involved in synthesizing the acyl chain component of GPL is not clear.
GPLs are reported to have a common fatty acyl tetrapeptide core consisting of tetrapeptide amino alcohol (d-Phe-d-allo-Thr-d-Ala-l-alaninol) and amide-linked long-chain fatty acid (C26–34) (8). The fatty acyl-tetrapeptide core is glycosylated with 6-deoxy-talose and variable O-methyl-rhamnose residues (9, 10). The GPLs from Mycobacterium avium are further modified such that an additional Rha residue is added to 6-deoxy-talose. Presently 31 distinct serotype-specific GPLs have been identified, of which chemical structures for 16 have been reported (11).
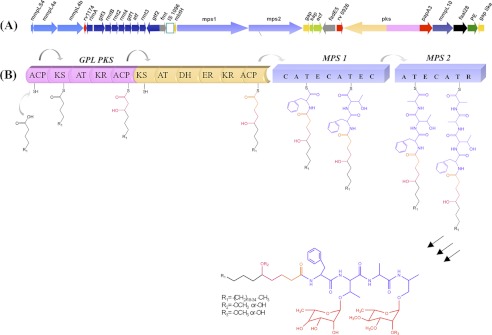
The GPL biosynthetic gene cluster has now been identified in several mycobacterial species. In Mycobacterium smegmatis, the cluster maps to a single locus of ∼65 kb in the genome, containing ∼30 open reading frames (ORF) (Fig. 1) (12). Genetic knock-out and complementation studies have established an understanding of many of the biosynthetic enzymes involved in modification of sugar moieties such as O-methyltransferase, acetyltransferase (13–15), and also of those involved in the transport of these metabolites to the cell surface (16). The disruption of genes involved in lipopeptidic core unit led to the loss of metabolites. These include three large multifunctional proteins whose biochemical function is not known. Two of these ORFs correspond to non-ribosomal peptide synthetases, designated as mps1 and mps2. Each of these proteins possesses two sets of modules that can be postulated to be responsible for synthesizing the tetrapeptide backbone, and the novel C-terminal reductase domain present on non-ribosomal peptide synthetase has been recently shown to reductively release the chain as acyl-peptidyl-alaninol (17). However, the functional relevance of the polyketide synthase (PKS) protein cannot be predicted.
FIGURE 1.
Biosynthetic pathway of GPL. A, shown is organization of GPL gene cluster in M. smegmatis. B, shown is a schematic representation of the modular organization of PKS and non-ribosomal peptide synthetase (NRPS) of GPL cluster of M. smegmatis. Bimodular GPL-PKS contains different functional domains: ACP, KS, AT, ketoreductase (KR) in module I along with dehydratase (DH) and enoyl reductase (ER) in module II. The PKS product from module II is then transferred sequentially onto the non-ribosomal peptide synthetase modules (MPS1 and MPS2) of the GPL cluster, leading to the formation of peptidolipid backbone that undergoes further modifications to form GPLs.
In recent years the involvement of PKS proteins in synthesizing unusual acyl chains of complex lipids in Mycobacterium tuberculosis has been established (18, 19). Many PKSs function along with fatty acyl-AMP ligases (FAALs), which have been demonstrated to provide starter fatty acid precursors (20–22). During polyketide biosynthesis, the final chemistry of the polyketide is dictated by the domain organization of the participating PKS and the substrate specificity of the corresponding acyltransferase (AT) domain. PKSs can catalyze chain extension by either following an iterative or a modular mechanism of ketide (-CH2CO-) unit condensation. Although the iterative enzymes catalyze successive condensations through repetitive utilization of the same set of active sites, modular proteins work in an assembly line wherein each set of active sites involved in condensation and associated reduction steps is used just once during biosynthesis. Recently, another hybrid “modularly iterative” mechanism of biosynthesis was described for the bimodular PKS12 protein in M. tuberculosis (23). Despite all these years and several structural reports, the position of hydroxyl group on the acyl chain is contentious, and at least two distinct positions of hydroxylation (β and δ) have been reported (7, 24–27).
In this study we demonstrate that the PKS protein from the GPL cluster (designated GPL-PKS) is involved in biosynthesis of the acyl component of GPLs. Based on genetic, biochemical, cell-free reconstitution, and structural analyses, we show that the PKS enzyme dictates the position of the hydroxyl group on the acyl chain. Our retrobiosynthetic studies provide an interesting approach to establish chemical structures of complex metabolites.
EXPERIMENTAL PROCEDURES
General Materials and Methods
M. smegmatis mc2155 (28) and related mutants were grown in Middlebrook 7H9 medium (Difco) supplemented with 2% (w/v) glucose and 0.05% (v/v) Tween-80 at 37 °C. Kanamycin and hygromycin were added to a final concentration of 50 and 100 μg/ml, respectively, required for mutants and overexpression constructs. The growth profile of liquid cultures was obtained by measuring the A600 at regular time intervals. The sequence analysis of mc2155 genomic DNA was done at TIGR. The restriction and modification enzymes were purchased from New England Biolabs. The Ampli taq gold enzyme was obtained from Applied Biosystems. Synthetic oligonucleotides were obtained from Microsynth. Radioactive malonyl-CoA was purchased from PerkinElmer life Sciences (1.9166 GBq/mmol), whereas radioactive methylmalonyl-CoA was purchased from American Radiolabeled Chemicals (55 mCi/mmol). XLI-blue (Stratagene) and BAP1 (29) strains of Escherichia coli were used for cloning and expression, respectively. DNA purification kits and nickel-nitrilotriacetic acid-agarose resin were procured from Qiagen. All other chemicals were of analytical grade from Sigma.
Cloning, Expression, and Purification of GPL Protein
The 10,959-bp pks gene was cloned in parts, which were assembled together by utilizing the restriction site engineered during the PCR (supplemental Fig. S1). PKS protein was expressed in BAP1 strain of E. coli. The cells were grown at 30 °C in LB medium with 50 μg/ml antibiotic (carbenicillin) to an absorbance of ∼0.6 at 600 nm. The cells were incubated at room temperature for 10 min and induced with 0.5 mm isopropyl thio-β-d-galactoside for 12–18 h at 22 °C. The cells were harvested by centrifugation (3500 × g for 15 min), resuspended in buffer A (50 mm Tris chloride (pH 8.0) containing 10% glycerol) containing 150 mm NaCl, and lysed using a French press, and cellular debris was removed by centrifugation (22,500 rpm, 40 min; Sorvall evolution RC, rotor: SA300). Polyethyleneimine was added to a final percentage of 0.1%, and the DNA pellet was removed by centrifugation (20,000 rpm, 30 min; Sorvall evolution RC, rotor: SA300). Nickel nitrilotriacetic-agarose resin was added to the supernatant (0.75 per ml of culture), and the protein was purified using affinity chromatography with an increasing concentration of imidazole in buffer A. Anion exchange chromatography was performed by using a 6-ml Resource Q column and AKTA chromatography system (GE Healthcare). The protein was purified in buffer B (100 mm phosphate buffer (pH 7.2) containing 10% glycerol) and eluted with an increasing gradient of 1 m NaCl in buffer B with a flow rate of 2 ml/min (supplemental Fig. S2). Protein concentration was determined by using BCA protein estimation kit (Pierce).
Labeling of Enzyme by [14C]Methylmalonyl- and [14C]Malonyl-CoA
Enzyme (1–5 μm) was incubated with [14C]malonyl-CoA (MCoA) and [14C]methyl malonyl-CoA (36 μm) at 4 °C for 2 min. The reaction was quenched by the addition of SDS-PAGE loading dye lacking any reducing agents like dithiothreitol or β-mercaptoethanol. Samples were directly loaded on a 6% SDS-PAGE gel, and electrophoresis was performed at 25 mA until the dye front ran out. The gel was dried and analyzed using phosphorimaging (Fuji BAS5001).
Enzymatic Assay and Product Characterization
PKS enzymatic assays induced 100 mm phosphate buffer (pH 7.2), 150 μm acyl-N-acetylcysteamine (NAC), 75 μm MCoA, 7.2 μm [14C]MCoA, 4 mm NADPH, 10% glycerol, 2 mm Tris(2-carboxyethyl)phosphine hydrochloride, and 2.5–4.5 μm protein in a 100-μl reaction volume. The reaction mixture was incubated at 30 °C for 6–12 h. The products were released with 45% KOH followed by acidification and extraction in ethyl acetate. The extract was spotted on a TLC plate that was developed using ethyl acetate:hexane:acetic acid (45:90:3.75). The radio TLC plates were analyzed by using phosphorimaging. For radio-HPLC assays, the concentration of [14C]MCoA was increased to 25 μm while keeping the total MCoA concentration as 100 μm. The band of interest was scraped from TLC, extracted in ethyl acetate, concentrated, and injected on C18 reverse-phase column (gradient: 35% B in 5 min, 100% B in 10 min, 100% B in 25 min, 20% B in 40 min, 20% B in 50 min; A: H2O, B: 5% methanol in ACN with 0.1% formic acid) on a HPLC (Shimadzu). The radioactivity was monitored online by a radioactive detector (IN/US system β-RAM Model 3) and also by using a Photodiode array detector.
Synthesis of Alkyl NAC Thioesters
Various alkyl NAC thioesters were synthesized according to a modified protocol reported earlier (20, 40).
Synthesis of 5-Hydroxydecanoic Acid
5-Hydroxydecanoic acid was synthesized by refluxing the δ-decanolactone in 50% KOH solution for 8–9 h at 80 °C. The reaction mixture was extracted with chloroform and washed with water. The organic layer was dried with sodium sulfate (anhydrous) and concentrated to dryness on rotavapor (Laborota 4001; Heidolph 2). 5-Hydroxydecanoic acid was purified on preparative thin layer chromatography and characterized by using electrospray ionization mass spectrometry (API Q- STAR pulser I, Applied Biosystems).
Synthesis of Hydroxyl Alkyl Phenylalanine
Various acyl-peptide substrates were synthesized by standard solid phase peptide synthesis using 9-fluromethoxy carbonyl (Fmoc) chemistry. Wang resin was preloaded with Fmoc-l-phenylalanine by using 1,3-diisoprylcarbodimide in the presence of 4-(dimethylamino) pyridine in dimethylformamide. The Fmoc group was deprotected by shaking with 20% piperidine in dimethylformamide (v/v) at room temperature for 30 min and then washed with dichloromethane followed by 3 washes of dimethylformamide. The hydroxy acyl acids were coupled with deprotected Fmoc-l-phenyl alanine resin using 3 eq of O-(benzotriazil-1-yl)-N-N-N-N-triamethyluranium-hexafluorophosphateand 3 eq of N-N-diisopropylethylamine with shaking at room temperature for 8 h. The resin beads were washed with dichloromethane three times followed by dimethylformamide. The alkyl peptide was cleaved from the resin by stirring with mixture of TFA and water (95:5) over a period of 3 h. The cleaved alkyl peptide was collected in chilled diethyl ether and concentrated on rotavapor. The hydroxylated alkyl phenylalanine was purified by reverse phase HPLC, column C5. The chemical identity of hydroxylated alkyl phenylalanine was confirmed by using electrospray ionization mass spectrometry.
Extraction and Purification of GPLs
The M. smegmatis mc2155 strain was grown in Middlebrook 7H9 medium till late stationary phase, and cells were harvested (5000 rpm for 20 min). Lipids were extracted with CHCl3/CH3OH (2:1, v/v) at room temperature for 24 h. The organic supernatant was dried and dissolved in CHCl3/CH3OH (2:1, v/v). The crude lipids were deacetylated by treating with an equal volume of 0.2 m NaOH in methanol at 37 °C for 1 h and neutralized with a few drops of glacial acetic acid. After drying, lipids were dissolved in CHCl3/CH3OH/H2O (4:2:1) and centrifuged. The aqueous layer was discarded, and the organic layer-containing lipids was washed with supersaturated brine and concentrated further. The deacylated lipids were spotted onto silica-coated TLC plates and developed in a CHCl3:CH3OH:H2O (90:10:1) solvent system further visualized with 10% sulfuric acid in ethanol and 5% α-naphthol/sulfuric acid in ethanol followed by charring at 120 °C for 10 min.
Purification and Analysis of GPLs
The crude lipids were separated on a Florisil column (60–100 mesh) with increasing concentration of methanol in chloroform. Fractions were monitored by thin layer chromatography on silica-coated plates. Fractions of GPLs mixture were loaded on preparative Silica plates (20 × 20 cm) F254 and developed by using 10:90:1 (CH3OH/CHCl3/H2O). Each GPL species was eluted by scraping the bands on preparative TLC and extracted in CHCl3/CH3OH (2:1), and the purity of each GPL was monitored on TLC by spraying plates with 5% α-naphthol/sulfuric acid in ethanol followed by charring at 120 °C for 10 min (supplemental Fig. S3).
To conduct structural analysis, the four tentatively assigned de-O-acylated GPLs (dGPLs) were named dGPLI, -II, -III, and -IV, starting from the solvent front. Analysis of the dGPLs by MALDI-TOF mass spectrometry gave a signal corresponding to [M+Na]+ molecular ion. The molecular ions of dGPLI, -II, -III, and -IV were observed at m/z 1187, 1173, 1173, and 1159, respectively, and consistent with reported GPLs from Mycobacterium butyricum (30).
Acid Hydrolysis of the Glycopeptidolipids
Purified glycopeptidolipids (dGPLI, dGPLII, dGPLIII, and dGPLIV) were resuspended in a minimum amount of CHCl3/CH3OH (2:1) and refluxed at 110 °C for 24 h in a sealed tube containing 6 m HCl solution (31). The hydrolysates thus generated were cooled and extracted with chloroform, and the organic layer was washed twice with water. The organic phase was concentrated, and the hydrolyzed products were checked on a Q-TOF/MS (API Q-STAR pulser I, Applied Biosystems).
Purification of the Fatty Acyl Acids
Hydrolyzed product, having fatty acids, was purified by adsorption chromatography. Concentrated mixtures of fatty acids were loaded on an open column (silica mesh size 100–200) and washed extensively with 2% ethyl acetate in hexane and then eluted with an increment of 5–50% ethyl acetate in hexane. Fractions containing each fatty acid were concentrated and checked by TLC and mass spectrometric analysis. These partially pure fractions were injected into a preparative HPLC silica-column (Phenomenex) and purified further. An isocratic solvent system comprising 25% ethyl acetate in hexane at a flow of 0.2 ml/min was used, and elution was monitored with a refractive index detector coupled with the HPLC pumps (Waters). The acyl chain with phenylalanine group were also isolated by partial hydrolysis of GPLs and purified by column chromatography eluted in 10% chloroform in hexane. The alkyl-Phe-Ala moieties were also characterized by mass spectrometry (Q-TOF/MS (API Q-STAR pulser I, Applied Biosystems).
Disruption of M. smegmatis pks and Faal28 Genes
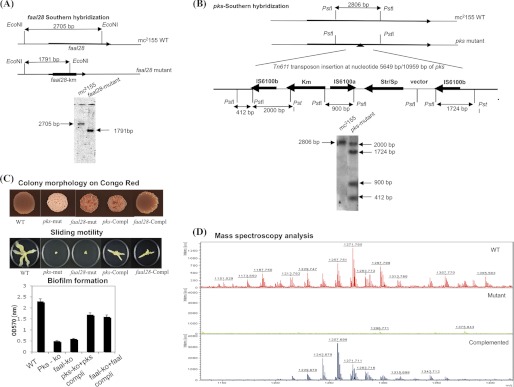
The M. smegmatis pks mutant (MYC55) was isolated as a rough mutant from a library of more than 19,000 transposition mutants (16). The site of insertion of Tn611 (32) was mapped using ligation-mediated PCR (33). Sequencing of the PCR product has shown that Tn611 has inserted in orientation 3′-tnpA-tnpR-5′ at position 5649/10,956 of the pks gene. The GPLs extracted from pks mutant was examined by both TLC and MALDI-TOF, showing no indication of GPLs as compared with wild type mc2155. The essential requirement of pks in GPL biosynthesis was confirmed by complementing the pks knock-out with a wild type M. smegmatis pks on a mycobacterial shuttle vector. The complementary strain regained its smooth phenotype and produced GPL. faal28 gene was disrupted using the homologous recombination method. Both pks and faal28 knock-out strains were confirmed by southern hybridizations.
RESULTS
Computational Analysis of the GPL-PKS
The domain organization of the PKS protein was investigated by using the non-ribosomal peptide synthetase-PKS database (34, 35), which allows easy identification of domains present in the PKS enzymes. The program predicted the GPL-PKS to encode a bimodular protein that contains two sets of domains that could bring about the extension of the starter chain by two ketide units (Fig. 1). Both the modules contain the ketosynthase (KS), AT, and acyl carrier protein (ACP) domains, which are essential for the activity of a PKS module. Among the β-carbonyl modifying domains, the first module contains only the ketoreductase domain, whereas all the three auxiliary domains (dehydratase, enoyl reductase, and ketoreductase) are present in the second module. Thus chain extension by the first module should result in the addition of a hydroxylated ketide group followed by incorporation of completely reduced 2-carbon unit by the second module. Because certain PKS modules are known to contain cryptic domains (36, 37), our prediction requires biochemical validation. The analysis of the substrate specificity of AT domain shows the presence of a crucial Phe residue in the active site pocket that is known to dictate the specificity of AT domains to utilize malonyl-CoA as the extender unit (34) (Fig. 2A). Comparative analysis of the AT domain across genomes of several mycobacterial species of GPL-PKS showed an interesting trend in their sequence conservation. The AT domains from the two modules (AT1 and AT2) cluster independently in different clades of the dendrogram, which indicates horizontal transfer of this pks gene across various species (Fig. 2A).
FIGURE 2.
Multiple sequence alignment of the PKS-AT domain and FAAL/fatty acyl-AMP (FACL) proteins. A, shown is multiple sequence alignment and dendrogram of PKS AT domain from module 1 and 2 for different mycobacterial species: M. avium paratuberculosis K10 (MAP), M. avium 104 (MAV), and Mycobacterium smegmatis (MSG). B, shown is multiple sequence alignment of M. smegmatis FAAL28 with other mycobacterial FAALs and fatty acyl-AMPs, indicating FAAL-specific insertion present in the M. smegmatis protein.
GPL-PKS possesses another ACP domain at the N terminus of the first module. Such a domain in mycobacterial PKS proteins has been demonstrated to facilitate loading of the starter unit from FAAL enzymes (22). Interestingly, an acyl activating enzyme homologue from the fadD family of enzymes could be identified in the M. smegmatis GPL cluster. Sequence analysis indicates the presence of the FAAL specific insertion sequence in this protein (Fig. 2B), which has been recently demonstrated to dictate the catalytic function of this family of proteins (39). FAAL28 is thus expected to activate starter fatty acid substrates as acyl-adenylates, which are transferred to the module 1 of GPL-PKS. As discussed above, the two rounds of condensation by the PKS enzyme would result in a hydroxyl group at the δ-position in the acyl chain of GPLs.
Genetic Studies with pks and faal Genes
Computational analysis suggests the possible involvement of GPL-pks and faal28 genes in the synthesis of the lipopeptide core of GPLs. We, therefore, predicted that knock-out of these genes would not be able to synthesize GPLs. faal28 gene was disrupted using a homologous recombination method (40, 41), the positive clones were selected on kanamycin plates, and resistant colonies were then confirmed by southern hybridization (Fig. 3A). The pks mutant was isolated as a rough phenotype from a library of more than 19,000 transposition mutants. The site of mutation was first mapped by using ligation-mediated PCR and then confirmed by performing a southern hybridization (Fig. 3B). Both faal28 and pks mutant strains showed altered phenotypic morphology, as demonstrated by Congo Red staining, sliding motility, and biofilm formation experiments (Fig. 3C). Mass spectrometric analysis of the total lipid extract of these two strains also showed the absence of GPL (Fig. 3D). The essential requirement of these two genes was proved unambiguously by complementing the mutant strains with cognate wild type genes cloned in a mycobacterial shuttle vector. The mass spectrometric analysis of these complemented strains indeed confirmed the presence of GPLs (Fig. 3D). These complemented strains also substantially regained their phenotypic GPL(+) characteristics.
FIGURE 3.
Construction and characterization of the M. smegmatis pks and faal28 mutant and complemented strains. A, shown is a schematic representation of the wild type and mutant faal28 gene and the strategy for confirming the mutants by southern hybridization. For Southern blotting, DNA was isolated and digested with EcoNI restriction enzyme. Fragments were separated on 0.8% agarose gel, transferred, and hybridized with 32P-radiolabeled probe covering 584 bp upstream of the fadD28 gene. Restriction sites are indicated as a solid line. B, shown is a schematic representation of the wild type and mutant pks gene. The position where Tn611 inserted in the pks gene is indicated with a black triangle (at nucleotide 5649/10,959). Transposition of Tn611 results in the insertion of two copies of IS6100. For Southern blotting, DNA was isolated and digested with PstI restriction enzyme. Fragments were separated on 0.8% agarose gel, transferred, and hybridized with 32P-radiolabeled probe covering the 450-bp internal fragment of IS6100 plus 409 bp corresponding to the coding sequence of pks gene. Restriction endonuclease sites are shown as a solid line. C, shown is phenotypic analysis of M. smegmatis mc2155 (wt), pks-mutant, faal28 mutant, and complemented strains. Phenotype on Congo Red plates, sliding motility, and biofilm formation of wild type, mutant, and complemented strains is compared. D, shown is MALDI-TOF analysis of crude lipid fractions of the M. smegmatis mc2155 (wt), pks-mutant, and the pks complemented strains.
Biochemical Characterization of the GPL-PKS
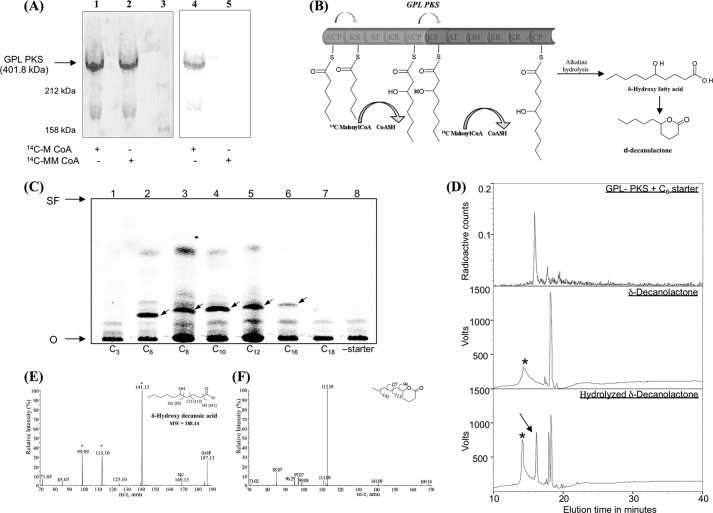
The 10,959-bp pks gene was cloned in a T7 expression vector such that the protein expressed with a C-terminal His6 tag. The protein was expressed in the holo-form by phosphopantetheinylation of the ACP domains by the surfactin phosphopantetheinyl transferase (sfp) from Bacillus subtilis. This was achieved by expressing the protein in the BAP1 strain of E. coli, which has a single copy of the sfp gene integrated into the genome (29). Purification by nickel-nitrilotriacetic acid chromatography followed by anion exchange chromatography showed a protein band migrating above the 212-kDa marker (Fig. 4A, lane 2). The size of the protein band of ∼400 kDa was confirmed by simultaneously loading PKS12 protein from M. tuberculosis on the same gel, which had been confirmed earlier to be of ∼456 kDa (supplemental Fig. S2) (23). The protein could also be detected by an anti-histidine antibody, specific to the His6 tag. Moreover, mass spectrometry sequencing of the protein showed peptides covering the entire protein sequence of GPL-PKS. The specificity of the AT domains was probed by performing SDS-PAGE gel binding assays using MCoA and [14C]methylmalonyl-CoA, two common extenders used by majority of the PKS enzymes. As can be seen in Fig. 4A, lane 3 and 4, the PKS enzyme showed specificity only for the malonate extender, and no labeling could be detected with the methyl malonate extender unit. The catalytic activity of the protein was further investigated by incubating various starters (C3 to C18) acyl-NACs with radiolabeled MCoA and NADPH. Because the protein does not contain a chain-releasing domain, the enzyme-bound product was released by alkali hydrolysis (Fig. 4B) and was detected on TLC by autoradiography. Autoradiography analysis showed a product band running at an Rf of ∼0.2 for the reaction primed with hexanoyl-NAC as starter (Fig. 4C, lane 2). The Rf of this band progressively increased as the starter chain length increased from C6 to C16 (Fig. 4C, lanes 2–6). The other bands observed in the autoradiogram varied from assay to assay and could be a result of degradation of acyl-CoAs used in the reaction. According to PKS enzymology, the bimodular PKS protein would be expected to add four carbons to the starter chain, leading to the formation of a δ-hydroxy fatty acid that can also spontaneously cyclize to form δ-decanolactone (Fig. 4B).
FIGURE 4.
Biochemical characterization of M. smegmatis GPL-PKS. A, gel binding assays using purified PKS protein were performed with radiolabeled MCoA and [14C]methyl malonyl-CoA (MMCoA). Coomassie-stained protein bands in lanes 1 and 2 correspond to the purified GPL-PKS that migrates above the 212-kDa marker protein (lane 3). The PKS protein was radiolabeled in the presence of [14C]MCoA (lane 4) and not in presence of 14C- methyl malonyl-CoA (lane 5). B, a schematic representation of the extension of hexanoyl diketide from PKS protein is shown. The substrate is transferred from KS domain of module 1 to module 2. Furthermore, two-carbon chain extension and chemical hydrolysis leads to cyclization to form δ-decanolactone as the final product. C, an PKS enzymatic assay with [14C]malonyl-CoA as the extender unit and different starter units as their NAC thioesters (mimics of fatty acyl-CoA; hexanoyl-SNAC (lane 2), octanoyl-SNAC (lane 3), decanoyl-SNAC (lane 4), dodecanoyl-SNAC (lane 5), and palmitoyl-SNAC (lane 6)) indicates an increase in the Rf of product formed from C6 (0.23), C8 (0.26), C10 (0.27), and C12 (0.28) to C16 (0.29) with increasing chain lengths. PKS protein did not show any activity with propinoyl-SNAC (lane 1) and stearoyl-SNAC (lane 7) as starters. Lane 8 indicates negative control where no starter unit was added. SF, solvent front. D, the radioactive product synthesized by PKS protein was analyzed on reverse phase HPLC, and chromatograms were recorded at 220 nm by simultaneously using photomultiplier and radioactive detector. To verify the PKS product formed, chemical standards were also injected. The 18.5-min peak corresponds to standard δ-decanolactone (Sigma) (middle panel). The 15.7-min HPLC peak corresponded to the retention time of hydrolyzed δ-decanolactone or 5-hydroxydecanoic acid (arrow, lower panel). *, corresponds to contamination. E, shown is electrospray ionization-MS/MS characterization of a HPLC peak with a retention time of 15.7 min. A molecular ion peak at m/z 187.13 indicates the eluate to be 5-hydroxydecanoic acid. The fragmentation pattern obtained by tandem MS of [M-H]−1 187.13 peak shows fragments at m/z 169 corresponding to the loss of water molecule and at m/z 125 corresponding to the loss of water and CO2 molecules from the hydrolyzed fatty acid. Further putative cleavage fragments are indicated for three peaks with a mass shift of 2 Da (*). The possible explanation for this is described under “Results.” F, shown is electrospray ionization-MS/MS characterization of the HPLC peak with a retention time of 18.5 min. A molecular ion peak at m/z 169 indicates the eluate to be δ-decanolactone. Tandem MS of [M-H]−1 169 produced peaks at m/z 99 (-C5H8O2) and 113 (-C3H5O2), in concordance with the fragmentation of hydrolyzed δ-decanolactone.
To unambiguously establish the identity of the products, HPLC-based assays were performed with appropriate standards. Enzyme assays included purified PKS protein and a radiolabeled extender unit, and hexanoyl-NAC was used as a starter unit. The assay yielded one major peak at retention time of 15.7 min (Fig. 4D, upper panel). This peak could either correspond to 5-hydroxydecanoic acid or its corresponding δ-decanolactone. The standard δ-decanolactone showed a retention time of 18.5 min under identical conditions at 220 nm (Fig. 4D, middle panel). To obtain 5-hydroxydecanoic acid, we performed alkali hydrolysis of the standard (±)δ-decanolactone and subjected it to HPLC analysis. Surprisingly, we observed four peaks in chromatogram (Fig. 4D, bottom panel). One of the peaks eluted with a retention time identical to δ-decanolactone, and another showed identical retention time to the GPL-PKS product. To ascertain the chemical nature of this peak, we resorted to MS/MS analysis using electrospray tandem mass spectrometry. MS/MS fragmentation revealed a dehydration peak at 169 atomic mass unit and a peak at 125 atomic mass unit, which is characteristic of loss of water and a CO2 molecule from hydroxyl fatty acids (Fig. 4E). Some other fragments observed in this spectrum could not be explained based on predicted fragmentation of 5-hydroxydecanoic acid and were shifted by 2 mass units. Interestingly, many of these peaks could be observed in the MS/MS fragmentation of the δ-decanolactone (Fig. 4f). It is possible that the hydroxyl fatty acid partially lactonizes in the gas phase during mass spectrometric analysis, and thus we observe this pattern. Such a fragmentation pattern could also be explained through mechanisms involving charge remote fragmentation, which results in the formation of terminally unsaturated product ions. The charge remote fragmentation mechanism has been demonstrated earlier for collision-induced ionization of fatty acids in the negative ion fragmentation to produce terminally unsaturated anions that show a mass difference of 2 Da (42, 43). Our studies thus demonstrate that the major product observed in the GPL-PKS enzymatic assay corresponds to 5-hydroxydecanoic acid.
Structural Elucidation of Acyl Chain of GPL
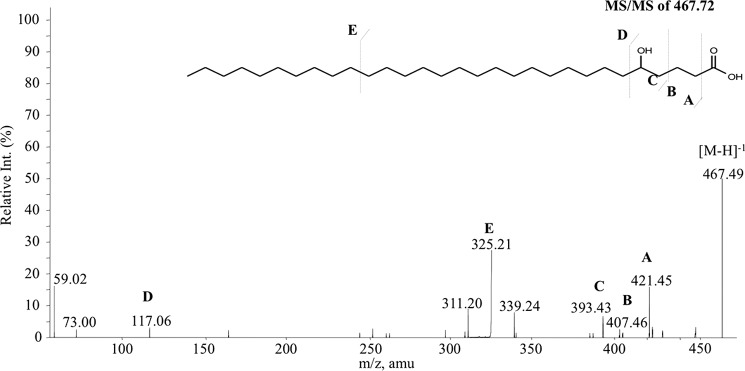
Because in vitro analysis clearly demonstrated that the GPL-PKS would result in the generation of the hydroxyl group at δ-position in GPLs, we decided to confirm this in the GPL extracted from M. smegmatis cultures. Mass spectrometric analysis of intact GPLs did not reveal any information about the nature of the acyl subsistent as the majority of the peaks in the MS/MS spectra corresponded to fragments obtained by cleavage across the more labile sugar and peptide moieties on GPLs. To perform a structural analysis of the GPL acyl chain, four main GPL variants produced by M. smegmatis were purified using combination of silica chromatography and preparative TLC (supplemental Fig. S3). These compounds were confirmed by using MALDI-TOF/MS analysis. To perform structural analysis of the acyl chain, the purified GPLs were subjected to extensive hydrolysis with 6 m HCl at 110 °C. Purified C30 fatty acyl chain on mass spectrometry analysis showed the expected [M-H]−1 at m/z 467 Da. MS/MS analysis of fatty acid first showed a loss of the [M-46]− peak. As discussed in the earlier section, this fragment could be ascribed to the loss of carboxylate anion and the formation of terminal unsaturation through charge remote fragmentation mechanism or through the fragmentation of the lactone product. Further fragmentation gives 407.46 and 393.43 ions that correspond to subsequent loss of one and two methylene group from 421.45 fragments (Fig. 5). These fragmentations clearly suggest that the position of the hydroxyl group has to be after C-β carbon atom. Another fragment at 117.06 could also be observed in the spectrum, and this could correspond to the C-terminal fragment resulting from the cleavage between the C5 and C6. This would indicate that the hydroxyl group could be either at C4 or C5.
FIGURE 5.
ESI-MS/MS analysis of the fatty acyl chain of purified GPL. Tandem MS/MS spectra of saturated hydroxy fatty acid of mass 468 Da together with the observed cleavage pattern is indicated.
Structural Analysis of GPL Fragment by Using NMR
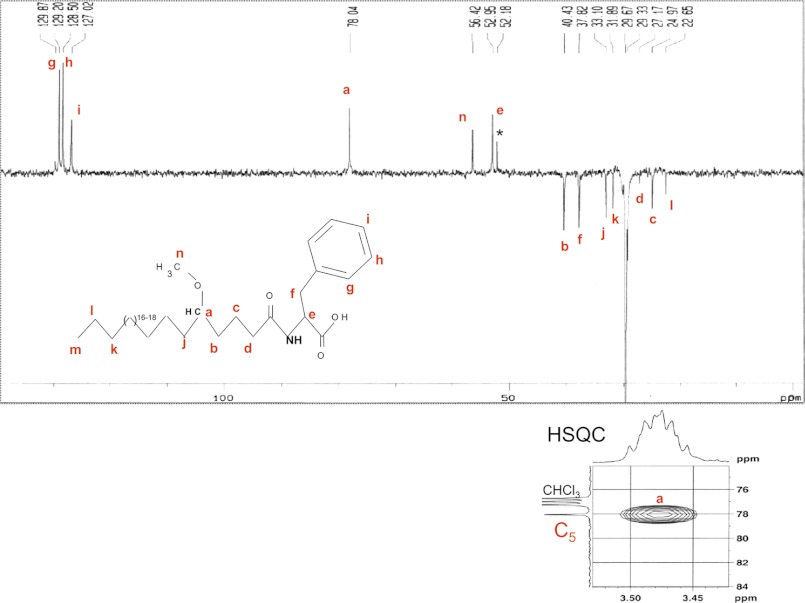
The partial acid hydrolysis of GPLs generates another fragment that could be assigned to fatty acylphenylalanine (FaPhe). In some of the GPLs, hydroxyl group is modified to methoxy. We performed NMR studies with the methoxy-form of this GPL fragment (FaPhe) that facilitated spectroscopic characterization. The chemical shift of the methine proton is deterministic of a C5 methoxylation, which however cannot be distinguished from others in a 1H spectrum. To perform unambiguous assignments, we first acquired the edited 13C subspectra to know the number of protons attached to each carbon. The DEPT-135 (distortionless nhancement by polarization transfer) edited 13C subspectra, which distinguish carbon bearing odd and even protons, identified two methine carbons at δppm 52.95 and 78.04 (labeled a and e in Fig. 6). This also identified aromatic and methoxy carbons. Furthermore, the corresponding single-bonded protons coupling to the respective carbons were determined by using a heteronuclear spin-quantum correlation experiment (supplemental Fig. S4). A heteronuclear spin-quantum correlation experiment spectrum of the methoxy peptidolipid confirmed the protons of the Phe together with that of the methoxy group (δppm 3.26/56.42 1H/13C) (Fig. 6). Moreover, it also helped to unambiguously assign the two methines of the phenylalanine Cα (δppm 4.88/52.95 1H/13C) and that on the fatty acyl chain (δppm 3.46–3.5/78.04 1H/13C). Finally, the correlations obtained from the COSY spectrum (supplemental Fig. S5) helped to build connectivity between the protons of the consecutive carbons. A cross-peak due to scalar coupling between methine protons of C5 and the diastereotopic methylene protons of C4 at δppm 3.46/2.35 was observed. The C3 protons at δppm 1.65 were further coupled to the C4 protons at δppm 2.35 and C2 protons at δppm 1.32. These NMR studies provided proof that the position of hydroxyl/methoxy group is present at the C-5 position.
FIGURE 6.
NMR spectra of methoxy GPL fragment. 13C DEPT-135 spectra of saturated methoxy phenyl fatty acyl in CDCl3 are shown. The inset shows the heteronuclear spin-quantum correlation (HSQC) experiment spectra of the methine carbon. *, contaminating peak.
Confirmation of the C-5 Position of the Hydroxyl Group on the Fatty Acyl Chain of GPL
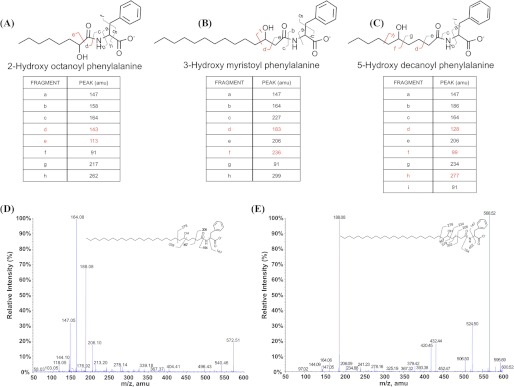
The FaPhe fragment of GPL ionizes much better than the fatty acyl fragment in the electrospray ionization. To determine the position of hydroxyl group, we synthesized several FaPhe analogs that possess hydroxylation at the 2, 3, and 5 position of the fatty acyl chain. The fragmentation patterns of these synthetic analogs were systematically investigated by using ESI-MS/MS. All these three compounds showed several common fragments as shown in Fig. 7. At the same time, few characteristic fragments were also detected during these studies, which correlate with the position of hydroxy substituent (marked in red in Fig. 7, A–C). This signature of fragmentation was then used to unambiguously determine the position of hydroxyl group in GPLs. The two GPL FaPhe fragments corresponding to m/z 572.5 (C27) and 600.5 (C29) were purified and subjected to MS/MS analysis (Fig. 7, D and E). Clearly, the mass differences observed in the two FaPhe GPL fragments in MS/MS spectra correspond to the mass differences observed for the standard 5-hydroxy-analog. Together, our studies thus establish that hydroxylation is located at the 5 position of the acyl chain.
FIGURE 7.
ESI-MS/MS analysis of chemically synthesized fatty acylphenyl analogs and fatty acyl chain of GPL molecule in negative ion mode. A, a–h indicate the fragmentation ion profile on tandem MS analysis of [M-H]−1 306 characteristic of 2-hydroxyoctanoyl phenylalanine. B, a–h indicate the fragmentation ion profile on tandem MS analysis of [M-H]−1 390 characteristic of 3-hydroxymyristoyl phenylalanine. C, a–i indicate the fragmentation ion profile on tandem MS analysis of [M-H]−1 334 characteristic of 5-hydroxy decanoyl phenylalanine. D and E, fragmentation patterns were obtained by tandem MS/MS for C27 and C29 fatty acyl chain of GPL molecules. respectively. amu, atomic mass units.
DISCUSSION
The cell envelope of mycobacteria contains several unique complex lipid metabolites. Other than defining unique morphological features, many of these molecules possess a variety of biological activities. The fatty acyl component of these lipid molecules is unusually long, and carbon chain length of C72 has been reported for mycolic acids. Some of these metabolites also contain methyl-branching that arise from the incorporation of methyl malonyl-CoA instead of commonly used malonyl-CoA (30). Although chemical structures of many of these molecules have been known for years, the molecular mechanisms underlying their biosynthesis were elucidated more recently after the availability of complete genome sequences. Studies with M. tuberculosis has led to the identification of a new paradigm, wherein the complex acyl chains of lipids are produced by the biochemical cross-talk between PKSs and FAALs. We demonstrate here that the same theme is also relevant in the biosynthesis of GPLs, an abundant glycolipid found on non-tuberculous and opportunistic mycobacterial species. Cell-free constitution studies of GPL-PKS have also led to the precise identification of the position of the hydroxylation on the acyl chain of GPL.
The catalytic and mechanistic versatility polyketide biosynthetic machinery has been utilized by variety of microorganisms to produce tremendous chemical diversity of secondary metabolites. As of now mycolactones produced by Mycobacterium ulcerans is the only example where PKSs produce macrolide-like molecules in mycobacteria (44). In this study we also show that a FAAL28 protein is essential for GPL biosynthesis. FAAL28 activates fatty acids as acyladenylates and transfers them on to the N terminus ACP domain of PKSs. The bimodular PKS extends the acyl chain by four carbons, and the position of the hydroxyl group is dictated by the absence of dehydratase and enoyl reductase domains in the module 1 of this PKS. The second module of GPL-PKS contains all reductive domains and thus results in a saturated fatty acyl chain. Previously, we showed that diols in the phthiocerol chain of phthiocerol dimycocerosate are produced by PpsA and PpsB proteins, where again the dehydratase and enoyl reductase domains are absent from these modules. Mycobacterial biosynthetic machinery has cleverly used PKSs to generate functional acyl chains that may be crucial to the functional requirement of these lipids. The hydroxyl groups in phthiocerol dimycocerosates are esterified by methyl-branched fatty acyl chain, whereas in GPL the hydroxyl group is sometimes modified to methoxy analog. Such a modification can generate chemical diversity to the cell envelope that may be crucial in generating phenotypic heterogeneity or for remodeling of mycobacterial cell wall during specific conditions. Although the spatiotemporal mechanisms involved in modifying lipids during various physiological niches are unclear, the ability of microbes to reprogram the outer coat must be an important mechanism of survival or pathogenesis. For example, Azotobacter during formation of dormant cysts has been shown to replace its outer cell envelope by resorcinolic and pyrone lipids (38).
In conclusion, our studies resolve long-standing ambiguity in terms of chemical structure of GPL and also provide understanding of the involvement of FAAL28 and GPL-PKS in biosynthesis of GPLs. By understanding exact mechanism of biosynthesis, it is now feasible to generate variants of GPLs at the cell surface by suitably engineering PKSs.
Supplementary Material
Acknowledgment
We thank Dr. S. Raghothama from the Indian Institute of Science, Bangalore, India, for useful discussions and help in NMR experiments.
This work was supported by Le Centre Indo-Français Pour la Recherche Avancée (CEFIPRA)/Indo-French Centre for the Promotion of Advanced Research (IFCPAR) Grant 3403-03 (to J.-M. R. and R. S. G.)

This article contains supplemental Figs. S1–S5.
- GPL
- glycopeptidolipid
- dGPL
- de-O-acylated GPL
- PKS
- polyketide synthase
- FAAL
- fatty acyl-AMP
- AT
- acyltransferase
- MCoA
- malonyl-CoA
- NAC
- N-acetylcysteamine
- SNAC
- thio-NAC
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- KS
- ketosynthase
- ACP
- acyl carrier protein
- FaPhe
- fatty acylphenylalanine.
REFERENCES
- 1. Neyrolles O., Guilhot C. (2011) Recent advances in deciphering the contribution of Mycobacterium tuberculosis lipids to pathogenesis. Tuberculosis 91, 187–195 [DOI] [PubMed] [Google Scholar]
- 2. Gokhale R. S., Sankaranarayanan R., Mohanty D. (2007) Versatility of polyketide synthases in generating metabolic diversity. Curr. Opin. Struct. Biol. 17, 736–743 [DOI] [PubMed] [Google Scholar]
- 3. Recht J., Martínez A., Torello S., Kolter R. (2001) Sliding motility and biofilm formation in mycobacteria. Acta Cient. Venez. 52, 45–49 [PubMed] [Google Scholar]
- 4. Deshayes C., Perrodou E., Gallien S., Euphrasie D., Schaeffer C., Van-Dorsselaer A., Poch O., Lecompte O., Reyrat J. M. (2007) Interrupted coding sequences in Mycobacterium smegmatis. Authentic mutations or sequencing errors? Genome Biol. 8, R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaufmann S. H. (2001) How can immunology contribute to the control of tuberculosis? Nat. Rev. Immunol. 1, 20–30 [DOI] [PubMed] [Google Scholar]
- 6. Smith D. W., Randall H. M., Maclennan A. P., Lederer E. (1960) Mycosides. A new class of type-specific glycolipids of mycobacteria. Nature 186, 887–888 [DOI] [PubMed] [Google Scholar]
- 7. Billman-Jacobe H., McConville M. J., Haites R. E., Kovacevic S., Coppel R. L. (1999) Identification of a peptide synthetase involved in the biosynthesis of glycopeptidolipids of Mycobacterium smegmatis. Mol. Microbiol. 33, 1244–1253 [DOI] [PubMed] [Google Scholar]
- 8. Brennan P. J., Goren M. B. (1979) Structural studies on the type-specific antigens and lipids of the Mycobacterium avium. Mycobacterium intracellulare, Mycobacterium scrofulaceum serocomplex, Mycobacterium intracellulare serotype 9. J. Biol. Chem. 254, 4205–4211 [PubMed] [Google Scholar]
- 9. Aspinall G. O., Chatterjee D., Brennan P. J. (1995) The variable surface glycolipids of mycobacteria. Structures, synthesis of epitopes, and biological properties. Adv. Carbohydr. Chem. Biochem. 51, 169–242 [DOI] [PubMed] [Google Scholar]
- 10. Belisle J. T., Brennan P. J. (1989) Chemical basis of rough and smooth variation in mycobacteria. J. Bacteriol. 171, 3465–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chatterjee D., Khoo K. H. (2001) The surface glycopeptidolipids of mycobacteria. Structures and biological properties. Cell. Mol. Life Sci. 58, 2018–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ripoll F., Deshayes C., Pasek S., Laval F., Beretti J. L., Biet F., Risler J. L., Daffé M., Etienne G., Gaillard J. L., Reyrat J. M. (2007) Genomics of glycopeptidolipid biosynthesis in Mycobacterium abscessus and M. chelonae. BMC Genomics 8, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jeevarajah D., Patterson J. H., McConville M. J., Billman-Jacobe H. (2002) Modification of glycopeptidolipids by an O-methyltransferase of Mycobacterium smegmatis. Microbiology 148, 3079–3087 [DOI] [PubMed] [Google Scholar]
- 14. Patterson J. H., McConville M. J., Haites R. E., Coppel R. L., Billman-Jacobe H. (2000) Identification of a methyltransferase from Mycobacterium smegmatis involved in glycopeptidolipid synthesis. J. Biol. Chem. 275, 24900–24906 [DOI] [PubMed] [Google Scholar]
- 15. Recht J., Kolter R. (2001) Glycopeptidolipid acetylation affects sliding motility and biofilm formation in Mycobacterium smegmatis. J. Bacteriol. 183, 5718–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sonden B., Kocincova D., Deshayes C., Euphrasie D., Rhayat L., Laval F., Frehel C., Daffe M., Etienne G., Reyrat J. M. (2005) Gap, a mycobacterial specific integral membrane protein, is required for glycolipid transport to the cell surface, Mol. Microbiol. 58, 426–440 [DOI] [PubMed] [Google Scholar]
- 17. Chhabra A., Haque A. S., Pal R. K., Goyal A., Rai R., Joshi S., Panjikar S., Pasha S., Sankaranarayanan R., Gokhale R. S. (2012) Nonprocessive [2 + 2]e- off-loading reductase domains from mycobacterial nonribosomal peptide synthetases. Proc. Natl. Acad. Sci. U.S.A. 109, 5681–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gokhale R. S., Saxena P., Chopra T., Mohanty D. (2007) Versatile polyketide enzymatic machinery for the biosynthesis of complex mycobacterial lipids. Nat. Prod. Rep. 24, 267–277 [DOI] [PubMed] [Google Scholar]
- 19. Jackson M., Stadthagen G., Gicquel B. (2007) Long-chain multiple methyl-branched fatty acid-containing lipids of Mycobacterium tuberculosis. Biosynthesis, transport, regulation, and biological activities. Tuberculosis 87, 78–86 [DOI] [PubMed] [Google Scholar]
- 20. Portevin D., de Sousa-D'Auria C., Montrozier H., Houssin C., Stella A., Lanéelle M. A., Bardou F., Guilhot C., Daffé M. (2005) The acyl-AMP ligase FadD32 and AccD4-containing acyl-CoA carboxylase are required for the synthesis of mycolic acids and essential for mycobacterial growth. Identification of the carboxylation product and determination of the acyl-CoA carboxylase components. J. Biol. Chem. 280, 8862–8874 [DOI] [PubMed] [Google Scholar]
- 21. Trivedi O. A., Arora P., Sridharan V., Tickoo R., Mohanty D., Gokhale R. S. (2004) Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature 428, 441–445 [DOI] [PubMed] [Google Scholar]
- 22. Mohanty D., Sankaranarayanan R., Gokhale R. S. (2011) Fatty acyl-AMP ligases and polyketide synthases are unique enzymes of lipid biosynthetic machinery in Mycobacterium tuberculosis. Tuberculosis 91, 448–455 [DOI] [PubMed] [Google Scholar]
- 23. Chopra T., Banerjee S., Gupta S., Yadav G., Anand S., Surolia A., Roy R. P., Mohanty D., Gokhale R. S. (2008) Novel intermolecular iterative mechanism for biosynthesis of mycoketide catalyzed by a bimodular polyketide synthase. PLoS Biol. 6, e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ojha A. K., Varma S., Chatterji D. (2002) Synthesis of an unusual polar glycopeptidolipid in glucose-limited culture of Mycobacterium smegmatis. Microbiology 148, 3039–3048 [DOI] [PubMed] [Google Scholar]
- 25. Villeneuve C., Etienne G., Abadie V., Montrozier H., Bordier C., Laval F., Daffe M., Maridonneau-Parini I., Astarie-Dequeker C. (2003) Surface-exposed glycopeptidolipids of Mycobacterium smegmatis specifically inhibit the phagocytosis of mycobacteria by human macrophages. Identification of a novel family of glycopeptidolipids. J. Biol. Chem. 278, 51291–51300 [DOI] [PubMed] [Google Scholar]
- 26. Mukherjee R., Gomez M., Jayaraman N., Smith I., Chatterji D. (2005) Hyperglycosylation of glycopeptidolipid of Mycobacterium smegmatis under nutrient starvation: structural studies. Microbiology 151, 2385–2392 [DOI] [PubMed] [Google Scholar]
- 27. Miyamoto Y., Mukai T., Nakata N., Maeda Y., Kai M., Naka T., Yano I., Makino M. (2006) Identification and characterization of the genes involved in glycosylation pathways of mycobacterial glycopeptidolipid biosynthesis. J. Bacteriol. 188, 86–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Snapper S. B., Melton R. E., Mustafa S., Kieser T., Jacobs W. R., Jr. (1990) Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4, 1911–1919 [DOI] [PubMed] [Google Scholar]
- 29. Pfeifer B. A., Admiraal S. J., Gramajo H., Cane D. E., Khosla C. (2001) Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 291, 1790–1792 [DOI] [PubMed] [Google Scholar]
- 30. Khoo K. H., Suzuki R., Morris H. R., Dell A., Brennan P. J., Besra G. S. (1995) Structural definition of the glycopeptidolipids and the pyruvylated, glycosylated acyltrehalose from Mycobacterium butyricum. Carbohydr. Res. 276, 449–455 [DOI] [PubMed] [Google Scholar]
- 31. López Marin L. M., Lanéelle M. A., Promé D., Daffé M., Lanéelle G., Promé J. C. (1991) Glycopeptidolipids from Mycobacterium fortuitum, A variant in the structure of C-mycoside. Biochemistry 30, 10536–10542 [DOI] [PubMed] [Google Scholar]
- 32. Guilhot C., Otal I., Van Rompaey I., Martìn C., Gicquel B. (1994) Efficient transposition in mycobacteria. Construction of Mycobacterium smegmatis insertional mutant libraries. J. Bacteriol. 176, 535–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Prod'hom G., Lagier B., Pelicic V., Hance A. J., Gicquel B., Guilhot C. (1998) A reliable amplification technique for the characterization of genomic DNA sequences flanking insertion sequences. FEMS Microbiol. Lett. 158, 75–81 [DOI] [PubMed] [Google Scholar]
- 34. Yadav G., Gokhale R. S., Mohanty D. (2003) Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 328, 335–363 [DOI] [PubMed] [Google Scholar]
- 35. Ansari M. Z., Yadav G., Gokhale R. S., Mohanty D. (2004) NRPS-PKS. A knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 32, W405–W413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koglin A., Walsh C. T. (2009) Structural insights into nonribosomal peptide enzymatic assembly lines. Nat. Prod. Rep. 26, 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tang Y., Lee H. Y., Tang Y., Kim C. Y., Mathews I., Khosla C. (2006) Structural and functional studies on SCO1815. A β-ketoacyl acyl carrier protein reductase from Streptomyces coelicolor A3(2). Biochemistry 45, 14085–14093 [DOI] [PubMed] [Google Scholar]
- 38. Funa N., Ozawa H., Hirata A., Horinouchi S. (2006) Phenolic lipid synthesis by type III polyketide synthases is essential for cyst formation in Azotobacter vinelandii. Proc. Natl. Acad. Sci. U.S.A. 103, 6356–6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Arora P., Goyal A., Natarajan V. T., Rajakumara E., Verma P., Gupta R., Yousuf M., Trivedi O. A., Mohanty D., Tyagi A., Sankaranarayanan R., Gokhale R. S. (2009) Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis. Nat. Chem. Biol. 5, 166–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van Kessel J. C., Hatfull G. F. (2007) Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152 [DOI] [PubMed] [Google Scholar]
- 41. Medjahed H., Reyrat J. M. (2009) Construction of Mycobacterium abscessus defined glycopeptidolipid mutants. Comparison of genetic tools. Appl. Environ. Microbiol. 75, 1331–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Griffiths W. J., Jonsson A. P., Liu S., Rai D. K., Wang Y. (2001) Electrospray and tandem mass spectrometry in biochemistry. Biochem. J. 355, 545–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Griffiths W. J. (2003) Tandem mass spectrometry in the study of fatty acids, bile acids, and steroids. Mass Spectrom. Rev. 22, 81–152 [DOI] [PubMed] [Google Scholar]
- 44. Stinear T. P., Hong H., Frigui W., Pryor M. J., Brosch R., Garnier T., Leadlay P. F., Cole S. T. (2005) Common evolutionary origin for the unstable virulence plasmid pMUM found in geographically diverse strains of Mycobacterium ulcerans. J. Bacteriol. 187, 1668–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.