Background: hnRNP K acts as a p53 cofactor upon DNA damage.
Results: DNA damage stimulates hnRNP K sumoylation, and this modification is required for p53 target gene expression.
Conclusion: hnRNP K sumoylation links DNA damage-induced signaling to p53 transcriptional activation.
Significance: The discovery of how different players within the p53 pathway are regulated will provide important insights into the study of chemotherapeutic drugs.
Keywords: DNA Damage, p53, Sumoylation, Transcription Regulation, Ubiquitin, Pc2/CBX4, hnRNP K
Abstract
Heterogeneous nuclear ribonucleoprotein (hnRNP) K is a nucleocytoplasmic shuttling protein that is a key player in the p53-triggered DNA damage response, acting as a cofactor for p53 in response to DNA damage. hnRNP K is a substrate of the ubiquitin E3 ligase MDM2 and, upon DNA damage, is de-ubiquitylated. In sharp contrast with the role and consequences of the other post-translational modifications, nothing is known about the role of SUMO conjugation to hnRNP K in p53 transcriptional co-activation. In the present work, we show that hnRNP K is modified by SUMO in lysine 422 within its KH3 domain, and sumoylation is regulated by the E3 ligase Pc2/CBX4. Most interestingly, DNA damage stimulates hnRNP K sumoylation through Pc2 E3 activity, and this modification is required for p53 transcriptional activation. Abrogation of hnRNP K sumoylation leads to an aberrant regulation of the p53 target gene p21. Our findings link the DNA damage-induced Pc2 activation to the p53 transcriptional co-activation through hnRNP K sumoylation.
Introduction
SUMO (small ubiquitin-related modifier)5 conjugation is a reversible, ATP-dependent process that involves an activating enzyme (E1), a conjugating enzyme (E2), and different ligases (E3) (1, 2). In the ubiquitin pathway, substrate specificity is usually provided by E3 ligases, which typically contain substrate-binding sites (3, 4). In the SUMO pathway, the sole E2 enzyme (Ubc9 in mammals) usually binds the substrate directly, but the SUMO E3 ligases seem to contribute to substrate specificity. The best-characterized SUMO E3s are the protein inhibitor of activated STAT1, PIAS1 (5), Topors (6), and the polycomb protein Pc2, also known as CBX4 (7). SUMO E3 ligases vary in their mechanism of action. While catalytic activity (SUMO transfer in sub-stoichiometric amounts) has been proven for RanBP2 (8, 9), PIAS (10), and Pc2 (7, 11), it is evident that proteins do not behave the same way in tubes as they do in living cells. Recently, elegant work from the Melchior laboratory uncovered the RanBP2 mechanism of action (12). Interestingly, the fragment known to exert the most potent E3 ligase activity is precluded to exert any activity because of the nature of its interaction with sumoylated RanGAP-Ubc9 (12). While multi-subunit E3 ligases have long been known in the ubiquitin field, work dealing with RanBP2/sumoylated RanGAP/Ubc9 as a multi-subunit SUMO E3 shows that all in vitro data should, in principle, be accompanied by a careful determination of the in vivo effect of an E3. In this line, Pc2 is known to have little effect on protein sumoylation in vitro, while its effect on substrates like CtBP1 are very robust in living cells (7, 13). Whereas the functional consequences of sumoylation vary, generally resulting from altered protein-protein interactions, sumoylated proteins are involved in a wide range of processes, including DNA repair, chromosome segregation, and gene expression (1, 14). Interestingly, one of the largest groups of SUMO-modified proteins is that of transcription factors, whose sumoylation is usually associated with transcriptional repression (15–17). This repressive role has been attributed to the SUMO-mediated recruitment of HDAC proteins (18–20). However, in a few cases, sumoylation of gene-specific transcription factors is associated with transcriptional activation (21–23). Thus, the transcriptional outcome of sumoylation of a certain protein is not predictable.
Heterogeneous nuclear ribonucleoprotein K (hnRNP K) is an evolutionarily conserved factor found in the nucleus and cytoplasm that was initially discovered as a component of hnRNP complexes (24). It is a multifaceted protein involved in different steps along mRNA metabolism (25). Interestingly, hnRNP K cooperates with p53 in transcriptional activation of cell-cycle arrest genes such as 14-3-3σ, GADD45, and p21, in response to DNA damage (26). The ATM/ATR-Chk1/Chk2-p53 axis represents the most studied DNA damage response signaling pathway, whereby p53 acts as a gatekeeper of tumor suppression (27). Several stimuli known to affect DNA integrity, and thus threatening cell survival, trigger the activation of this cascade leading to the regulation of a plethora of p53 target genes (28, 29). The outcome of p53 activation in cells ranges from a reversible cell-cycle arrest and induction of DNA repair to more drastic responses such as cell death by apoptosis or senescence (29). Importantly, hnRNP K is crucial for ionizing irradiation-induced cell cycle arrest and serves as a transcriptional cofactor for p53 (26). Recently, it has been shown that hnRNP K interacts with the large intergenic noncoding RNA (lincRNA)-p21, and this interaction is required for transcriptional regulation of p53 target genes (30). hnRNP K ubiquitylation and its role in the DNA damage response have been established (26), and hnRNP K was also reported to be a SUMO target (31). Thus, the involvement of hnRNP K in the DNA damage response is puzzling, and the mechanistic aspects that regulate its role are still poorly understood. Given current knowledge on the role of SUMO conjugation in the DNA damage response, we decided to study SUMO conjugation to hnRNP K (32).
In the present report, we confirm hnRNP K as a sumoylation substrate and map its target lysine, 422, located within the C-terminal KH3 domain. Furthermore, we characterize the polycomb protein Pc2 as the E3 ligase for hnRNP K and present evidence that SUMO and ubiquitin conjugation do not affect each other. Importantly, DNA damage, which is known to activate Pc2, stimulates hnRNP K sumoylation. Most interestingly, while DNA damage-induced hnRNP K sumoylation is enhanced by Pc2 expression, it is inhibited by the expression of the ΔSIM mutant, which has no SUMO E3 activity. To gain insight into the consequences of hnRNP K sumoylation, we analyzed p53 target gene expression as hnRNP K has been reported to be a p53 cofactor. SUMO conjugation to hnRNP K plays a key role in its cofactor activity. Intriguingly, hnRNP K-mediated transcriptional activation depends on p53. Overall, we have started to uncover a new mechanism by which DNA damage activates the p53 pathway, adding sumoylated hnRNP K downstream of Pc2 in a new DNA damage-induced, sumoylation-dependent signaling pathway.
EXPERIMENTAL PROCEDURES
Plasmids, siRNAs, and Transfection
Plasmid DNA and siRNAS were transfected in HEK 293T and HCT-116 cells with Lipofectamine 2000 as per the manufacturer's instructions (Invitrogen). The sequence of the Stealth siRNA (Invitrogen), which targeted the human hnRNP K 3′-UTR, was the following: 5′-UGUGAAGCAGUAUUCUGGAAAGUUU-3′. The Stealth RNAiTM siRNA Negative Control Low GC (Invitrogen) was used as a control.
Site-directed Mutagenesis
Site-directed mutagenesis was performed by the DpnI method, based on Stratagene's QuickChange specifications. The primers used to mutate Lys-422 to Arg were as follows (Arg-coding triplet underlined): 5′-GTCGGGAGCTTCGATCAGAATTGATGAGCCTTTAG-3′ and 5′-CTAAAGGCTCATCAATTCTGATCGAAGCTCCCGAC-3′. The primers used to mutate Asp-424 to Ala were as follows (Ala-coding triplet underlined): 5′-CTTCGATCAAAATTGCTGAGCCTTTAGAAGGATC-3′ and 5′-GATCCTTCTAAAGGCTCAGCAATTTTGATCGAAG-3′. Bold letters represent the mutated nucleotide. Mutations were verified by sequencing the whole hnRNP K cDNA for each mutation.
Recombinant Proteins
GST, GST-hnRNP K, and GST-MDM2 were expressed in Escherichia coli in E. coli BL21(DE3) Rosetta, and GST-Ubc9 in M15(pREP4) cells by induction with 0.5 mm IPTG and purified using glutathione-Sepharose beads (GE Healthcare). T7-hnRNP K and T7-hnRNP K K422R were purified from transfected HEK 293T lysates exactly as described (33). Proteins were analyzed by SDS-PAGE and Coomassie staining for quantification and purity.
GST Pull-down Assays
Pull-down assays were performed as described (34). HEK 293T cells were lysed in 1 ml of lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 1 mm β-glycerophosphate, 1 mm KF, 5% glycerol, and 1× Complete Protease Inhibitor (Roche)) and incubated for 30 min at 4 °C. After centrifugation for 20 min at 4 °C, supernatants were used immediately for pull-down assays or kept at −80 °C. Lysates were pre-cleared with 2 μg of GST coupled to glutathione-Sepharose beads (GE Healthcare), and cleared lysates were incubated with the indicated GST-tagged recombinant protein coupled to glutathione-Sepharose beads for 1 h at room temperature. After three washes in lysis buffer and one in PBS, proteins were eluted by adding 2× Laemmli sample buffer and analyzed by Western blot.
Purification of His6-SUMO- or Ubiquitin-conjugated Proteins
HEK 293T cells were transfected in 35-mm culture wells with the indicated plasmids. After 48 h, His6-SUMO conjugates were purified under denaturing conditions using Ni-NTA-agarose beads according to the manufacturer's instructions (Qiagen). In the case of HCT-116 cells, they were transfected in 6-cm culture wells and 48 h later treated with doxorubicin (1 μm) for the indicated time points. Transfected cells were harvested in ice-cold PBS plus 100 mm iodoacetamide. An aliquot was taken as input and the remaining cells were lysed in 6 m guanidinium-HCl containing 100 mm Na2HPO4/NaH2PO4, 10 mm Tris-HCl pH 8.0, 5 mm imidazole, and 10 mm iodoacetamide. Samples were sonicated to reduce the viscosity and after centrifugation for 20 min at 12,000 × g, proteins in the supernatants were purified using Ni-NTA beads (Qiagen) according to (35). Samples were subsequently washed with wash buffer I (8 m urea, 10 mm Tris-HCl, 100 mm Na2HPO4/NaH2PO4, 5 mm imidazole, 10 mm iodoacetamide, pH 8), wash buffer II (8 m urea, 10 mm Tris-HCl, 100 mm Na2HPO4/NaH2PO4, 0.2% Triton X-100, 5 mm imidazole, 10 mm iodoacetamide, pH 6.3), and wash buffer III (8 m urea, 10 mm Tris-HCl, 100 mm Na2HPO4/NaH2PO4, 0.1% Triton X-100, 5 mm imidazole, 10 mm iodoacetamide, pH 6.3). Samples were eluted in 2× Laemmli sample buffer containing 300 mm imidazole for 5 min at 95 °C.
In Vitro Sumoylation Reactions
In vitro sumoylation reactions were performed as described (34), using T7-hnRNP K as a substrate. The sumoylation buffer was 20 mm HEPES-KOH pH 7.3, 100 mm KAcO, 0.5 mm EGTA, 1 mm DTT, 0.05% Tween-20, 0.2 mg/ml ovalbumin, and 1× Complete protease inhibitor (Roche). hnRNP K (250 ng) was incubated with 150 ng E1, 100, 250, or 500 ng of Ubc9, and 1 μg of SUMO. Reactions were stopped by addition of 2× Laemmli sample buffer. One-fourth of the reaction was run in SDS-PAGE and analyzed by Western blot. Recombinant SUMO-1, SUMO-2, SUMO-3, Ubc9 (E2 enzyme), and Aos1-Uba2 (E1 heterodimer) were purchased from Enzo Life Sciences.
In Vitro Ubiquitylation Reactions
In vitro ubiquitylation reactions were preformed in 20 μl of a buffer containing 50 mm Tris-HCl (pH 7.4), 2 mm ATP, 5 mm MgCl2, 2 mm DTT as described (26). Purified recombinant hnRNP K (500 ng) was incubated with 200 ng of Uba1 (E1), 200 ng of UbcH5b (E2), 500 ng of MDM2, and 10 μg of ubiquitin for 2 h at 30 °C and further analyzed by SDS-PAGE and Western blot. Uba1 (E1), UbcH5b (E2), MDM2, and ubiquitin were from Boston Biochem.
Luciferase Assays
Forty-eight hours after transfection, HCT-116 cells were washed with PBS and harvested in Reporter lysis buffer (Promega). Luciferase activity in cell lysates was measured as per the manufacturer's instructions.
Analysis of p21 and 14-3-3σ Expression
HCT-116 cells were plated in 6-well plates and transfected as described above with the plasmids and/or siRNA specified in each figure and treated with 1 μm doxorubicin for 24 h where indicated. RNA was extracted with TRI Reagent (Molecular Research Center) and 1 μg was reverse-transcribed using MMLV-RT (Invitrogen). cDNAs were amplified using TaqDNA polymerase (Invitrogen) with SYBR green using an Eppendorf Mastercycler and analyzed with Realplex software. Primer sequences used to amplify p21 were 5′-CTGGAGACTCTCAGGGTCGAAA-3′ (forward) and 5′-GATTAGGGCTTCCTCTTGGAGAA-3′ (reverse); and for 14-3-3σ, 5′-GCCGAACGCTATGAGGACAT-3′ (forward) and 5′-CTTCTCCACGGCGCCTT-3′ (reverse), as described (36). HSPCB was used as housekeeping with the following primers, 5′-CCAAAAAGCACCTGGAGATCA-3′ (forward) and 5′-TGTCGGCCTCAGCCTTCT-3′ (reverse).
RESULTS
hnRNP K Is sumoylated in Vitro and in Cells
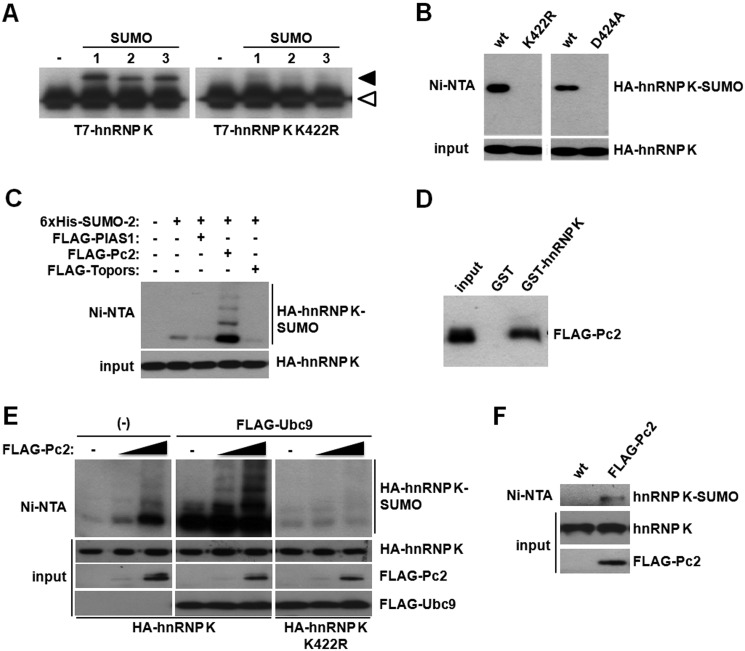
Given that hnRNP K has been reported to be a SUMO target (31), we performed a first search for SUMO consensus sites within hnRNP K using available software (SUMOsp 2.0 (37) and SUMOplot], and lysine 422, within the C-terminal KH3 domain was the highest probability site (Fig. 1A). We then wanted to corroborate this finding using HEK 293T cells expressing His6-SUMO-2 and purifying SUMO conjugates by denaturing N2+ affinity chromatography. We readily detected HA-tagged (Fig. 1B) as well as endogenous (Fig. 1C) hnRNP K sumoylation. Consistently, hnRNP K interacts with the SUMO E2 enzyme Ubc9, as demonstrated by GST pull-down assays (Fig. 1D). To further confirm this interaction functionally, hnRNP K sumoylation was evaluated by an in vitro sumoylation assay with recombinant E1, E2, SUMO-2, and hnRNP K. As shown in Fig. 1E, recombinant hnRNP K is sumoylated in vitro and increasing amounts of Ubc9 lead to increased sumoylation, roughly reaching 40% of modified hnRNP K (Fig. 1F).
FIGURE 1.
HnRNP K is a SUMO substrate in cells and in vitro. A, summary of the highest probability site according to both SUMOplot and SUMOsp, lysine 422 (top), and the schematic of hnRNP K highlighting its lysine 422 residing within the KH3 domain (bottom). B, HEK293T cells were transfected with an HA-tagged hnRNP K expression vector either with or without SUMO-2. An aliquot of cells was taken as input, and the remainder was subject to denaturing Ni2+ affinity chromatography. Both fractions were analyzed by Western blot with an anti-HA antibody. C, for the analysis of endogenous hnRNP K sumoylation, cell lysates were analyzed as in B, although an anti-hnRNP K antibody was used for the Western blot. D, pull-down assay was performed using HEK293T lysates expressing HA-hnRNP K. Samples were analyzed by Western blot with an anti-HA antibody. E, purified recombinant T7-hnRNP K was incubated with Aos1-Uba2, increasing amounts of Ubc9, and SUMO-2. Reactions were stopped by addition of 2× Laemmli sample buffer and analyzed by Western blot with an anti-T7 antibody. F, in vitro reactions as in E were performed in triplicate, and the signal was quantified with ImageJ after corroboration that all the bands in the developed films were in a linear range. Data are presented as mean ± S.E.
hnRNP K Is Sumoylated at Lysine 422 and Pc2 Functions as Its E3 Ligase
In an attempt to map the target lysine(s) for SUMO modification, we mutated the lysine within the consensus, IKID. Mutation of this lysine to arginine (K422R mutant) completely abolished SUMO conjugation to hnRNP K both in vitro (Fig. 2A) and in living cells (Fig. 2B, left panels). To rule out other putative post-translational modifications to lysine 422, we decided to mutate the aspartic acid residue 424 to alanine (D424A), altering the SUMOylation consensus while leaving the target lysine intact. The D424A hnRNP K mutant is unable to be conjugated by SUMO (Fig. 2B, right panels). SUMO conjugation does not alter hnRNP K stability as the sumoylation-deficient mutant is expressed to the same level as the wild type protein (Fig. 2B, see “input” panels). We then decided to test whether hnRNP K sumoylation could be regulated by any of the known SUMO E3 ligases. We expressed FLAG-tagged Pc2, Topors, and PIAS1 and performed chromatography purification of His6-SUMO-2 conjugates. The polycomb protein Pc2 greatly stimulated hnRNP K sumoylation, while neither Topors nor PIAS1 could augment hnRNP K sumoylation (Fig. 2C). Consistently, hnRNP K interacts with Pc2 in GST pull-down assays (Fig. 2D). Interestingly, increasing amounts of Pc2 stimulate SUMO conjugation (Fig. 2E, left panel). Moreover, in the presence of overexpressed Ubc9, a ladder of sumoylated hnRNP K isoforms becomes apparent (Fig. 2E, middle panel). The ladder represents SUMO chains attached to lysine 422 as all the bands are lost in the K422R mutant (Fig. 2E, right panel). These results show that the polycomb protein Pc2 regulates hnRNP K sumoylation and that forcing hnRNP K sumoylation to very high levels (overexpressing Ubc9 and Pc2 together) exclusively leads to the conjugation of SUMO to Lys-422. Importantly, Pc2 also enhances sumoylation of endogenous hnRNP K, as shown in Fig. 2F.
FIGURE 2.
The polycomb protein Pc2 exerts E3 ligase activity toward hnRNP K. A, in vitro sumoylation reactions were performed using SUMO-1, -2, or -3 together with wild type or the K422R mutant T7-tagged hnRNP K. The white arrowhead denotes the migration of non-sumoylated T7-hnRNP K, while the black arrowhead indicates the migration of sumoylated T7-hnRNP K. B, HEK293T cells were transfected with wild type, K422R, or D424A mutant HA-tagged hnRNP K together with His6-tagged SUMO-2. An aliquot of cells was taken as input, and the remainder was subject to denaturing Ni2+ affinity chromatography. Both fractions were analyzed by Western blot with an anti-HA antibody. C, HEK293T cells were transfected with the indicated expression vectors and analyzed as above. D, pull-down assay was performed using HEK293T lysates expressing FLAG-Pc2. Samples were analyzed by Western blot with an anti-FLAG antibody. E, HEK293T cells were transfected with the indicated expression vectors and analyzed as above. F, HEK293T cells were transfected with His6-tagged SUMO-2 either with or without Pc2 and endogenous hnRNP K sumoylation was analyzed.
hnRNP K Sumoylation and Ubiquitylation Are Independently Regulated
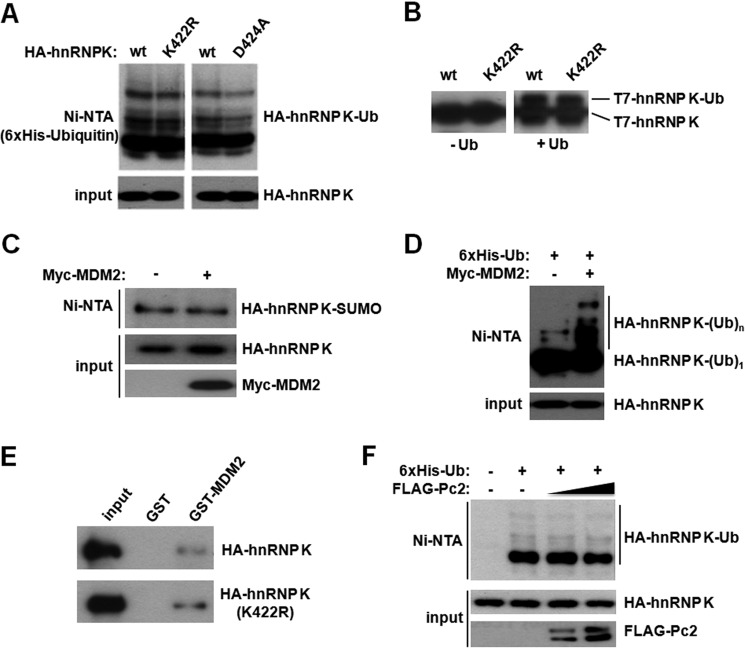
Because SUMO and ubiquitin are both conjugated to the ϵ-NH2 group of lysine side chains, we tested a putative cross talk between these two post-translational modifications. As shown above, while Lys-422 is the major (and presumably unique) sumoylation site within hnRNP K, ubiquitylation remains unaffected in both the K422R and D424A mutants (Fig. 3A). The same result was observed by performing in vitro ubiquitylation assays (Fig. 3B). To further analyze a possible cross-talk between SUMO and ubiquitin conjugation, we tested the effect of overexpressing ubiquitin E3 ligase MDM2 or the SUMO E3 ligase Pc2. MDM2 does not affect hnRNP K sumoylation (Fig. 3C), whereas poly-ubiquitylation was augmented in accordance with previous reports (Fig. 3D). Of note, mono-ubiquitylated HA-hnRNP K is not affected by MDM2 overexpression (Fig. 3D). Additionally, MDM2 interacts with both wild type and K422R hnRNP K (Fig. 3E). Conversely, while Pc2 stimulates hnRNP K sumoylation as shown above, it does not alter hnRNP K ubiquitylation (Fig. 3F). Thus, ubiquitin and SUMO conjugation occur at distinct lysine residues and are independently regulated.
FIGURE 3.
Lysine 422 is conjugated by SUMO and not ubiquitin. A, HEK293T cells were transfected either with wild type, K422R, or D424A HA-tagged hnRNP K expression vectors, together with His6-tagged ubiquitin, as indicated. Cells were subject to denaturing nickel affinity chromatography as above and analyzed by Western blot with an anti-HA antibody. B, recombinant T7-hnRNP K and T7-hnRNP K K422R were ubiquitylated in vitro in the presence of Uba1, UbcH5b, and MDM2. The left panel corresponds to reactions lacking ubiquitin. C, cells were transfected with the indicated plasmids and analyzed as in A. D, cells were transfected with HA-tagged hnRNP K together with His6-tagged ubiquitin, and 48 h later they were treated with 10 μm MG132 for 5 h before lysis and Ni2+ affinity chromatography as above. E, lysates expressing HA-hnRNP K or the K422R mutant were incubated either with GST or GST-MDM2, and the pull-down assay was performed as described under “Experimental Procedures.” Samples were analyzed by Western blot with an anti-HA antibody. F, cells were transfected with the indicated plasmids and analyzed by Ni2+ affinity chromatography as in D.
hnRNP K Sumoylation Is Increased upon DNA Damage
Given the established role for hnRNP K in the DNA damage response, we tested the hypothesis that DNA-damaging agents leading to the activation of the p53 response could regulate hnRNP K sumoylation. The p53 pathway is activated by a plethora of stimuli including numerous DNA-damaging agents. This process has been well characterized in the human colon carcinoma cell line, HCT-116, after exposure to the chemotherapeutic agent doxorubicin, which leads to DNA double-strand breaks. hnRNP K sumoylation is stimulated by doxorubicin, etoposide, UV light, and camptothecin (Fig. 4A and supplemental Fig. S1). Importantly, the effect is dependent on Lys-422 (Fig. 4B) and Asp-424 (supplemental Fig. S2) and is also observed with endogenous hnRNP K (Fig. 4C). Then, we transfected cells with His6-hnRNP K, treated them with doxorubicin, and performed a nickel pull-down followed by anti-SUMO2/3 Western blot to detect endogenous SUMO conjugated to His6-hnRNP K. Fig. 4D shows that DNA damage increases endogenous SUMO conjugation to hnRNP K. Although doxorubicin treatment does not affect hnRNP K mono-ubiquitylation at the time points tested, there is a slight decrease in poly-ubiquitylation (Fig. 4E), in agreement with published data for other DNA-damaging agents (26).
FIGURE 4.
hnRNP K sumoylation is stimulated by DNA damage. A, HCT-116 cells were transfected with HA-tagged hnRNP K together with His6-tagged SUMO-2, and 48 h later they were left untreated or treated with 1 μm doxorubicin, 1 μm Etoposide, or UV light (40 J/m2) for 4 h. SUMO conjugation to hnRNP K was analyzed by Ni2+ affinity chromatography as above. B, doxorubicin-induced hnRNP K sumoylation depends on Lys-422. Cells were transfected and processed as above. C, endogenous hnRNP K sumoylation is induced by doxorubicin. D, cells were transfected with His6-tagged hnRNP K, and 48 h later they were treated with doxorubicin and hnRNP K was purified by Ni2+ affinity chromatography and endogenous SUMO2/3 conjugation was assessed by Western blot. E, cells were transfected with HA-tagged hnRNP K together with His6-tagged ubiquitin; 48 h later they were treated with 1 μm doxorubicin for 3 h in the presence of 10 μm MG132 and ubiquitin conjugation to hnRNP K was analyzed by Ni2+ affinity chromatography as above.
hnRNP K Sumoylation Is Regulated by the HIPK2-Pc2 Axis
Interestingly, while we found that Pc2 functions as an hnRNP K SUMO E3 ligase, others have shown that Pc2 activity is regulated by doxorubicin: upon treatment with the DNA-damaging stimuli, Pc2 is phosphorylated by HIPK2, in turn, stimulating its E3 ligase activity (39). Thus, we wanted to test whether HIPK2 expression regulated hnRNP K sumoylation. Fig. 5A shows that HIPK2 overexpression stimulates hnRNP K sumoylation and HIPK2 and Pc2 co-expression show a synergism in augmenting hnRNP K sumoylation. We then decided to test the effect of doxorubicin in the presence of overexpressed wild type Pc2 or the SUMO E3 activity-deficient mutant Pc2 ΔSIM (13, 40). While expression of wild type Pc2 potentiates doxorubicin-induced hnRNP K sumoylation, expression of Pc2 ΔSIM behaved like a dominant-negative, strongly inhibiting doxorubicin-triggered sumoylation (Fig. 5B). Interestingly, a similar effect was observed with HIPK2: overexpression of wild type HIPK2 enhanced hnRNP K sumoylation (in both basal and DNA damage conditions) while overexpression of the kinase inactive mutant (K221A, (39)) abrogates doxorubicin-induced sumoylation (Fig. 5C). We then decided to test whether Pc2 and HIPK2 regulate hnRNP K-mediated p53 transcriptional activation. While both Pc2 and HIPK2 enhanced hnRNP K-induced p53 transcriptional activity (Fig. 5D), expression of both Pc2 ΔSIM as well as K221A HIPK2 mutants inhibited hnRNP K-triggered p53-dependent transcription (Fig. 5E).
FIGURE 5.
hnRNP K sumoylation is required for p53 transcriptional co-activation. A, HEK 293T cells were transfected with the indicated plasmids and then analyzed by Ni2+ affinity chromatography. B and C, Pc2 and HIPK2 regulate hnRNP sumoylation. HEK 293T cells were transfected with the indicated plasmids, and 48 h later they were treated with 1 μm doxorubicin (Dox) for 4 h. Then, HA-hnRNP K sumoylation was analyzed by Ni2+ affinity chromatography. D, cells were transfected with a plasmid carrying 16 tandem p53 binding sites upstream of the firefly luciferase coding sequence, together with HA-hnRNP K, Pc2, or HIPK2 and lysed 48 h later for luciferase activity measurement. E, cells were transfected with a plasmid carrying 16 tandem p53 binding sites upstream of the firefly luciferase coding sequence, together with HA-hnRNP K, Pc2 ΔSIM, or HIPK2 K221A and analyzed as in D. Data are represented as mean ± S.E., and p values of Student's t test are shown.
Pc2-mediated hnRNP K Sumoylation Enhances p53 Transcriptional Activity
Given the involvement of hnRNP K and the Pc2 activator (HIPK2) in the p53 pathway, we then wanted to analyze the contribution of SUMO-conjugated hnRNP K in the p53 transcriptional response. As previously reported, hnRNP K leads to increased p53 transcriptional activity as measured by a transcriptional reporter (Fig. 6A). Most strikingly, mutation of Lys-422 to Arg abrogates this transcriptional stimulation (Fig. 6A, white bars). Further supporting a critical role for hnRNP K sumoylation on p53 transcriptional activity, Pc2 overexpression potentiates hnRNP K transcriptional co-activator activity, and this effect is abolished in the K422R mutant (Fig. 6A, black bars). A similar behavior was observed when co-expressing hnRNP K and HIPK2 (Fig. 6B). As reporter constructs do not necessarily reproduce the scenario of endogenous promoters, we then analyzed the role of hnRNP K sumoylation in endogenous p21 expression. Doxorubicin treatment leads to enhanced p21 expression, and knocking down hnRNP K by means of an siRNA (reducing its level to less than 40%) lead to inhibition of doxorubicin-induced p21 expression (Fig. 6C). We then performed a rescue experiment by transfecting cells with an siRNA directed against hnRNP K 3′-UTR and expressing siRNA-resistant wild type and K422R hnRNP K constructs. In hnRNP K-depleted cells, expression of wild type HA-tagged hnRNP K rescues doxorubicin-induced p21 expression, while the sumoylation-defective K422R mutant does not (Fig. 6D). Overall, the results from Fig. 6 show that sumoylated hnRNP K is a key player of the DNA damage-induced pathway that results in p21 expression.
FIGURE 6.
hnRNP K sumoylation is required for p53 transcriptional co-activation. Pc2-mediated sumoylation of hnRNP K at lysine 422 activates p53-dependent transcription. A, HCT-116 cells were transfected with a plasmid carrying 16 tandem p53 binding sites upstream of the firefly luciferase coding sequence, together with HA-hnRNP K, or HA-hnRNP K K422R and FLAG-Pc2. Data are represented as mean ± S.E. B, HCT-116 cells were transfected with a plasmid carrying 16 tandem p53 binding sites upstream of the firefly luciferase coding sequence, together with HA-hnRNP K, or HA-hnRNP K K422R and FLAG-HIPK2. C, hnRNP K is required for proper p21 expression upon DNA damage. HCT-116 cells were transfected either with a control (Ctl) or an hnRNP K siRNA and 48 hs later they were treated with 1 μm doxorubicin (Dox) for 24 h before RNA extraction and real-time PCR analysis. D, HCT-116 cells were transfected either with a control (Ctl.) or an hnRNP K siRNA, and 24 h later they were transfected with the indicated hnRNP K construct. The next day, cells were treated with 1 μm doxorubicin (Dox) for 24 h before RNA extraction and real-time PCR analysis as in C. Real-time PCR experiments were always performed in triplicate, and data are represented as mean ± S.E. In parallel, HA-hnRNP K and β-actin expression was monitored by Western blot. All HA (upper) and β-actin (lower) panels correspond to cropped images of the same blot. Luciferase reporter assays were performed in duplicate and real-time PCR experiments in triplicate. p values of the Student's t test are shown.
hnRNP K Transcriptional Activation Depends on p53
To confirm that the cofactor function of hnRNP K was sumoylation-dependent, we made use of the p53-deficient HCT-116 cells, in order to test that the sumoylated hnRNP K-induced transcriptional activity was indeed p53-dependent. We measured the well-characterized p53 target genes, p21 and 14-3-3σ expression levels in both wild type and p53-deficient cells. HA-hnRNP K expression leads to a 2-fold increase in endogenous p21 mRNA levels in wild type p53 cells, and FLAG-Pc2 expression potentiated this effect to nearly 4-fold (Fig. 7A). This effect is 100% dependent on p53, since it is completely abolished in p53-null HCT-116 cells (Fig. 7B). To further confirm this finding, we analyzed 14-3-3σ mRNA levels and found that hnRNP expression increased it by 1.5-fold by itself and nearly 3-fold when co-expressed with Pc2 (Fig. 7C). Once again, these effects are p53-dependent (Fig. 7D). These results were further confirmed with a p21 promoter reporter assay. We used HCT-116 p53-deficient cells transfected either with empty vector or a p53 encoding vector, together with wild type or sumoylation-deficient (D424A mutant) hnRNP K. Fig. 7E confirms that hnRNP sumoylation (and not other lysine post-translational modification) is required for p53-dependent transcription.
FIGURE 7.
p53 is required for hnRNP K transcriptional co-activation. hnRNP K and Pc2 activate p53 target gene expression synergistically. Wild type (p53 +/+) and p53-null (p53 −/−) HCT-116 cells were transfected with the indicated plasmids, and RNA was extracted 48 h later. p21 (A and B) and 14-3-3σ (C and D) mRNA levels were quantified by real-time PCR. In all cases, HSPCB was used as a housekeeping gene. Real-time PCR experiments were performed in triplicates. E, HCT-116 p53 −/− cells were transfected with a p21 reporter plasmid together with hnRNP K or hnRNP K D424A, and a p53 expression vector. Data are represented as mean ± S.E. p values of the Student's t test are shown.
DISCUSSION
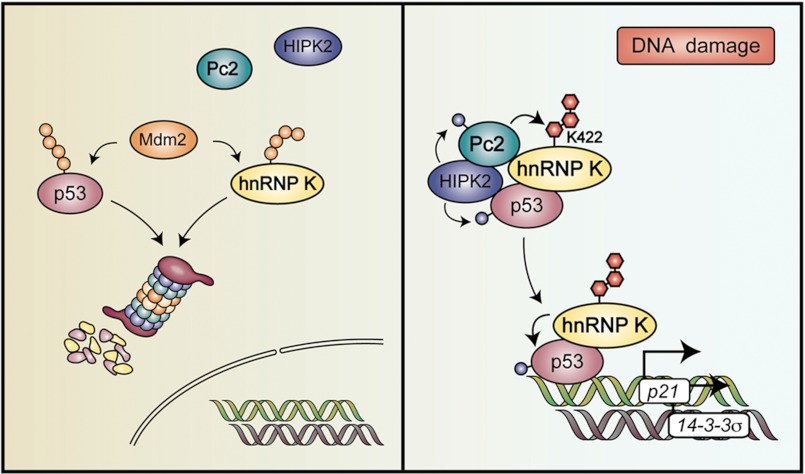
Our results are summarized in the model depicted in Fig. 8. First we confirmed hnRNP K as a sumoylation substrate and went on to map its target lysine, 422, located within the C-terminal KH3 domain. Furthermore, we characterized Pc2 as the E3 ligase for hnRNP K and present evidence that SUMO and ubiquitin conjugation do not seem to affect each other. Importantly, DNA damage, which is known to activate Pc2, stimulates hnRNP K sumoylation. Most interestingly, while DNA damage-induced hnRNP K sumoylation is enhanced by Pc2 expression, it is inhibited by the expression of the ΔSIM mutant, which has no SUMO E3 activity. To gain insight into the consequences of hnRNP K sumoylation, we analyzed p53 target gene expression as hnRNP K has been reported to be a p53 cofactor. Two lines of evidence suggest that SUMO conjugation to hnRNP K plays a key role in its cofactor activity: 1) while wild type hnRNP K activates a p53-dependent transcriptional reporter, the K422R mutant does not and, furthermore, Pc2 potentiates hnRNP K cofactor activity in a K422-dependent manner; 2) Knocking down hnRNP K inhibits doxorubicin-induced p21 expression and rescuing hnRNP K-depleted cells with siRNA-resistant wild type but not with the K422R mutant restores full DNA damage-induced p21 expression. Overall, we have started to uncover a new mechanism by which DNA damage activates the p53 pathway, adding sumoylated hnRNP K downstream of HIPK2 and Pc2 in a new DNA damage-induced, sumoylation-dependent pathway.
FIGURE 8.
Model illustrating the role of hnRNP K sumoylation upon DNA damage. Under normal conditions, p53 and hnRNP K are ubiquitylated and degraded by the proteasome. Upon treatment with DNA-damaging agents, HIPK2 phosphorylates and activates Pc2, which in turn, leads to hnRNP K sumoylation. Sumoylated hnRNP K then serves as a cofactor for p53-mediated gene expression. The model incorporates the new data obtained in the present report with data from previous studies.
While pioneer work from the Jackson laboratory started to uncover the role of hnRNP K in the p53 transcriptional program upon ionizing and UV irradiation through de-ubiquitylation and stabilization (26), we found that SUMO conjugation to hnRNP K does not seem to affect its stability. Furthermore, we found that as opposed to poly-ubiquitylation, mono-ubiquitylation is not affected neither by MDM2 nor DNA damage in the cell lines tested. Our results are consistent with a lack of cross talk between hnRNP K ubiquitylation and sumoylation, raising the possibility that these two post-translational modifications may act in combination with other post-translational modifications to decode different stimuli into different responses.
The role of SUMO modification in DNA repair has been extensively studied (32) and some examples include (i) Rad52 recombination activity is regulated by double strand break-induced sumoylation (41), (ii) the SUMO protease SENP6 has been shown to be involved in the regulation of DNA repair through homologous recombination (42), (iii) SUMO localizes to DNA damage sites (43, 44). Very recently, Pc2 has been reported to be recruited to DNA damage sites and to mediate DNA damage-induced BMI1 sumoylation and the consequent accumulation at DNA damage sites. Although our present work reveals hnRNP K as a new link between SUMO and DNA damage, the contribution of the HIPK2-Pc2-hnRNP K pathway to the DNA damage response is actually more complex, as HIPK2 also stimulates p53 transcriptional activity (45) and HIPK2 and MDM2 have been reported to regulate each other (46–48). Although future work will determine the precise mechanism by which sumoylated hnRNP K influences p53 transcriptional activity, we speculate that hnRNP K, Pc2, HIPK2, and p53 might be part of a complex formed or stabilized upon certain DNA damaging signals. Interestingly, it was reported that Axin forms a complex with HIPK2 and p53, resulting in p53 Ser-46 phosphorylation (49). Thus, sumoylated hnRNP K, like Axin, might be acting as a tumor suppressor by facilitating p53 function through integration of multiple factors.
Work from the Yeh laboratory has recently uncovered an interesting example of an interaction involving SUMO, Pc2 and transcriptional repression. In SENP2 null embryos, sumoylated Pc2 occupancy on the promoters of PcG target genes is markedly increased, leading to repression of Gata4 and Gata6 transcription, which are involved in cardiac development (50). In this case, sumoylated Pc2 binds K27 trimethylated histone H3 repressing transcription. Interestingly, hnRNP K has been implicated in the p53 pathway many years ago (26, 51). Most recently, a large intergenic non-coding RNA (lincRNA), lincRNA-p21 has been shown to regulate p53-dependent transcriptional responses, and the observed transcriptional effect is mediated through the physical association with hnRNP K (30). In this scenario, it will be interesting to determine whether lincRNA-p21 might play a role in sumoylated hnRNP K-mediated p53 transcriptional activation. Furthermore, recent work by Rosenfeld and co-workers shows that the ncRNA NEAT2 regulates Pc2-dependent E2F1 sumoylation, by binding to unmethylated Pc2 (52). Thus, it is tempting to speculate that a complex network of ncRNAs, Pc2, and hnRNP K could be involved in the fine-tuning of the p53-triggered DNA damage response. Moreover, as both Pc2 and hnRNP K are modified by phosphorylation, methylation, and sumoylation, these modifications provide an extra layer of regulation, which makes this hypothesis even more provocative. We hypothesize that upon certain DNA insults, hnRNP K and Pc2 form a complex with HIPK2 and with p53 that is necessary for hnRNP K sumoylation. Interestingly, it was recently shown by a SILAC/mass spectrometry approach that DNA damage stimulates hnRNP K phosphorylation in serine 420, located two amino acids N-terminal to the SUMO target lysine, by ∼2-fold (53). It will certainly be interesting to identify the kinase responsible for this phosphorylation and to analyze a possible cross-talk between these two post-translational modifications.
Interestingly, hnRNP K is not the only hnRNP involved in the DNA damage response because hnRNP H/F depletion compromises p53 pre-mRNA 3′-end processing, protein expression, and p53-mediated apoptosis (54). Knowing that hnRNP F is a SUMO substrate (31) as well as many other hnRNPs (55), it will be interesting to analyze if this family of proteins and their sumoylation might contribute to the regulation of the DNA damage response. Once again, these results strengthen the hypothesis of an intricate network of RNAs, RNA-binding proteins, and post-translational modifications, all contributing to the DNA damage response. Overall, Pc2-triggered hnRNP K sumoylation provides a new link between hnRNP K and the central players in the p53 pathway, providing another target for future studies involving the mechanistic action of chemotherapeutic drugs.
Supplementary Material
Acknowledgments
We thank Valeria Buggiano for technical help, Ignacio Schor, Jimena Druker, and Luciana Giono for critical reading of the manuscript, and Luciana Giono for the artwork. We acknowledge Ron Hay, Vanesa Gottifredi, David Wotton, Carol Prives, Ze'ev Ronai, Karol Bomsztyk, Lienhard Schmitz, Ling Qi, Stephen Jackson, Alfredo Fusco, and Moshe Oren for providing reagents.
This work was supported by EURSNET (to A. S.) and ANPCyT and University of Buenos Aires (to F. P. and A. S.).

This article contains supplemental Figs. S1 and S2.
- SUMO
- small ubiquitin-related modifier
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- lincRNA
- large intergenic noncoding RNA.
REFERENCES
- 1. Geiss-Friedlander R., Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 2. Hay R. T. (2005) SUMO: a history of modification. Mol. Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 3. Bernassola F., Karin M., Ciechanover A., Melino G. (2008) The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14, 10–21 [DOI] [PubMed] [Google Scholar]
- 4. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 5. Kahyo T., Nishida T., Yasuda H. (2001) Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8, 713–718 [DOI] [PubMed] [Google Scholar]
- 6. Weger S., Hammer E., Heilbronn R. (2005) Topors acts as a SUMO-1 E3 ligase for p53 in vitro and in vivo. FEBS Lett. 579, 5007–5012 [DOI] [PubMed] [Google Scholar]
- 7. Kagey M. H., Melhuish T. A., Wotton D. (2003) The polycomb protein Pc2 is a SUMO E3. Cell 113, 127–137 [DOI] [PubMed] [Google Scholar]
- 8. Pichler A., Knipscheer P., Saitoh H., Sixma T. K., Melchior F. (2004) The RanBP2 SUMO E3 ligase is neither HECT- nor RING-type. Nat. Struct. Mol. Biol. 11, 984–991 [DOI] [PubMed] [Google Scholar]
- 9. Pichler A., Gast A., Seeler J. S., Dejean A., Melchior F. (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108, 109–120 [DOI] [PubMed] [Google Scholar]
- 10. Yunus A. A., Lima C. D. (2009) Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA. Mol. Cell 35, 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kagey M. H., Melhuish T. A., Powers S. E., Wotton D. (2005) Multiple activities contribute to Pc2 E3 function. EMBO J. 24, 108–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Werner A., Flotho A., Melchior F. (2012) The RanBP2/RanGAP1 *SUMO1/Ubc9 Complex Is a Multisubunit SUMO E3 Ligase. Mol. Cell 46, 287–298 [DOI] [PubMed] [Google Scholar]
- 13. Merrill J. C., Melhuish T. A., Kagey M. H., Yang S. H., Sharrocks A. D., Wotton D. (2010) A role for non-covalent SUMO interaction motifs in Pc2/CBX4 E3 activity. PLoS One 5, e8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ulrich H. D. (2009) The SUMO system: an overview. Methods Mol. Biol. 497, 3–16 [DOI] [PubMed] [Google Scholar]
- 15. Garcia-Dominguez M., Reyes J. C. (2009) SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim. Biophys. Acta 1789, 451–459 [DOI] [PubMed] [Google Scholar]
- 16. Gill G. (2005) Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 15, 536–541 [DOI] [PubMed] [Google Scholar]
- 17. Zhao J. (2007) Sumoylation regulates diverse biological processes. Cell Mol. Life Sci 64, 3017–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Girdwood D., Bumpass D., Vaughan O. A., Thain A., Anderson L. A., Snowden A. W., Garcia-Wilson E., Perkins N. D., Hay R. T. (2003) P300 transcriptional repression is mediated by SUMO modification. Mol. Cell 11, 1043–1054 [DOI] [PubMed] [Google Scholar]
- 19. Shiio Y., Eisenman R. N. (2003) Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. U.S.A. 100, 13225–13230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang S. H., Sharrocks A. D. (2004) SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell 13, 611–617 [DOI] [PubMed] [Google Scholar]
- 21. Guo B., Sharrocks A. D. (2009) Extracellular signal-regulated kinase mitogen-activated protein kinase signaling initiates a dynamic interplay between sumoylation and ubiquitination to regulate the activity of the transcriptional activator PEA3. Mol. Cell Biol. 29, 3204–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lyst M. J., Stancheva I. (2007) A role for SUMO modification in transcriptional repression and activation. Biochem. Soc. Trans. 35, 1389–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rosonina E., Duncan S. M., Manley J. L. (2010) SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev. 24, 1242–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matunis M. J., Michael W. M., Dreyfuss G. (1992) Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol. Cell Biol. 12, 164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bomsztyk K., Denisenko O., Ostrowski J. (2004) hnRNP K: one protein multiple processes. Bioessays 26, 629–638 [DOI] [PubMed] [Google Scholar]
- 26. Moumen A., Masterson P., O'Connor M. J., Jackson S. P. (2005) hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell 123, 1065–1078 [DOI] [PubMed] [Google Scholar]
- 27. Vogelstein B., Lane D., Levine A. J. (2000) Surfing the p53 network. Nature 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 28. Beckerman R., Prives C. (2010) Transcriptional regulation by p53. Cold Spring Harb. Perspect. Biol. 2, a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vousden K. H., Prives C. (2009) Blinded by the Light: The Growing Complexity of p53. Cell 137, 413–431 [DOI] [PubMed] [Google Scholar]
- 30. Huarte M., Guttman M., Feldser D., Garber M., Koziol M. J., Kenzelmann-Broz D., Khalil A. M., Zuk O., Amit I., Rabani M., Attardi L. D., Regev A., Lander E. S., Jacks T., Rinn J. L. (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li T., Evdokimov E., Shen R. F., Chao C. C., Tekle E., Wang T., Stadtman E. R., Yang D. C., Chock P. B. (2004) Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc. Natl. Acad. Sci. U.S.A. 101, 8551–8556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bergink S., Jentsch S. (2009) Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458, 461–467 [DOI] [PubMed] [Google Scholar]
- 33. Cazalla D., Sanford J. R., Cáceres J. F. (2005) A rapid and efficient protocol to purify biologically active recombinant proteins from mammalian cells. Protein Expr. Purif. 42, 54–58 [DOI] [PubMed] [Google Scholar]
- 34. Pelisch F., Gerez J., Druker J., Schor I. E., Muñoz M. J., Risso G., Petrillo E., Westman B. J., Lamond A. I., Arzt E., Srebrow A. (2010) The serine/arginine-rich protein SF2/ASF regulates protein sumoylation. Proc. Natl. Acad. Sci. U.S.A. 107, 16119–16124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tatham M. H., Rodriguez M. S., Xirodimas D. P., Hay R. T. (2009) Detection of protein SUMOylation in vivo. Nat. Protoc. 4, 1363–1371 [DOI] [PubMed] [Google Scholar]
- 36. Gomes N. P., Bjerke G., Llorente B., Szostek S. A., Emerson B. M., Espinosa J. M. (2006) Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20, 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ren J., Gao X., Jin C., Zhu M., Wang X., Shaw A., Wen L., Yao X., Xue Y. (2009) Systematic study of protein sumoylation: Development of a site-specific predictor of SUMOsp 2.0. Proteomics 9, 3409–3412 [DOI] [PubMed] [Google Scholar]
- 38.Deleted in proof
- 39. Roscic A., Möller A., Calzado M. A., Renner F., Wimmer V. C., Gresko E., Lüdi K. S., Schmitz M. L. (2006) Phosphorylation-dependent control of Pc2 SUMO E3 ligase activity by its substrate protein HIPK2. Mol. Cell 24, 77–89 [DOI] [PubMed] [Google Scholar]
- 40. Yang S. H., Sharrocks A. D. (2010) The SUMO E3 ligase activity of Pc2 is coordinated through a SUMO interaction motif. Mol. Cell Biol. 30, 2193–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sacher M., Pfander B., Hoege C., Jentsch S. (2006) Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 8, 1284–1290 [DOI] [PubMed] [Google Scholar]
- 42. Dou H., Huang C., Singh M., Carpenter P. B., Yeh E. T. (2010) Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol. Cell 39, 333–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Galanty Y., Belotserkovskaya R., Coates J., Polo S., Miller K. M., Jackson S. P. (2009) Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462, 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morris J. R., Boutell C., Keppler M., Densham R., Weekes D., Alamshah A., Butler L., Galanty Y., Pangon L., Kiuchi T., Ng T., Solomon E. (2009) The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462, 886–890 [DOI] [PubMed] [Google Scholar]
- 45. Puca R., Nardinocchi L., Givol D., D'Orazi G. (2010) Regulation of p53 activity by HIPK2: molecular mechanisms and therapeutical implications in human cancer cells. Oncogene 29, 4378–4387 [DOI] [PubMed] [Google Scholar]
- 46. Rinaldo C., Prodosmo A., Siepi F., Moncada A., Sacchi A., Selivanova G., Soddu S. (2009) HIPK2 regulation by MDM2 determines tumor cell response to the p53-reactivating drugs nutlin-3 and RITA. Cancer Res. 69, 6241–6248 [DOI] [PubMed] [Google Scholar]
- 47. Rinaldo C., Prodosmo A., Mancini F., Iacovelli S., Sacchi A., Moretti F., Soddu S. (2007) MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol. Cell 25, 739–750 [DOI] [PubMed] [Google Scholar]
- 48. Di Stefano V., Mattiussi M., Sacchi A., D'Orazi G. (2005) HIPK2 inhibits both MDM2 gene and protein by, respectively, p53-dependent and independent regulations. FEBS Lett. 579, 5473–5480 [DOI] [PubMed] [Google Scholar]
- 49. Rui Y., Xu Z., Lin S., Li Q., Rui H., Luo W., Zhou H. M., Cheung P. Y., Wu Z., Ye Z., Li P., Han J., Lin S. C. (2004) Axin stimulates p53 functions by activation of HIPK2 kinase through multimeric complex formation. EMBO J. 23, 4583–4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kang X., Qi Y., Zuo Y., Wang Q., Zou Y., Schwartz R. J., Cheng J., Yeh E. T. (2010) SUMO-specific protease 2 is essential for suppression of polycomb group protein-mediated gene silencing during embryonic development. Mol. Cell 38, 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Enge M., Bao W., Hedström E., Jackson S. P., Moumen A., Selivanova G. (2009) MDM2-dependent downregulation of p21 and hnRNP K provides a switch between apoptosis and growth arrest induced by pharmacologically activated p53. Cancer Cell 15, 171–183 [DOI] [PubMed] [Google Scholar]
- 52. Yang L., Lin C., Liu W., Zhang J., Ohgi K. A., Grinstein J. D., Dorrestein P. C., Rosenfeld M. G. (2011) ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147, 773–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Beli P., Lukashchuk N., Wagner S. A., Weinert B. T., Olsen J. V., Baskcomb L., Mann M., Jackson S. P., Choudhary C. (2012) Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell 46, 212–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Decorsière A., Cayrel A., Vagner S., Millevoi S. (2011) Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3′-end processing and function during DNA damage. Genes Dev. 25, 220–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vassileva M. T., Matunis M. J. (2004) SUMO modification of heterogeneous nuclear ribonucleoproteins. Mol. Cell Biol. 24, 3623–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.