Background: Maf1 is a global repressor of RNA polymerase (pol) III transcription whose function is phospho-regulated by nutrient and stress signaling pathways.
Results: We tested the hypothesis that CK2 phosphorylation of Maf1 is required for derepression of pol III transcription.
Conclusion: The hypothesis is not supported.
Significance: CK2 regulation of pol III transcription is likely to involve targets other than Maf1.
Keywords: Repressor Protein, RNA Polymerase III, Signal Transduction, Stress Response, TOR, Yeast Transcription, Maf1, Protein Kinase CK2
Abstract
Maf1 is a conserved regulator of RNA polymerase (pol) III transcription and is required for transcriptional repression under diverse stress conditions. In yeast, Maf1 function is negatively regulated at seven phosphosites by the overlapping action of protein kinase A (PKA) and the TORC1-regulated kinase Sch9. Under stress conditions, Maf1 is dephosphorylated at these sites leading to its nuclear accumulation, increased association with pol III genes and direct physical interactions with the polymerase which ultimately inhibit transcription. These changes are reversed upon return to optimal growth conditions. Transcription in this system is also regulated by protein kinase CK2. CK2 stimulates pol III transcription in yeast and human cells via phosphorylation of the initiation factor TFIIIB. Recently it was proposed that CK2 phosphorylation of Maf1 is required for reactivation of pol III transcription following growth on glycerol. We have examined this hypothesis using two Maf1 mutants (Maf1-id S388A and Maf1-ck20) which lack all of the CK2 phosphosites implicated in the response. Both mutant proteins are phosphoregulated, function normally during repression and transcription is fully restored to the wild-type level upon transfer from glycerol to glucose. Additionally, phos-tag gel analysis of Maf1 7SA, a functional mutant that cannot be phosphorylated by PKA/Sch9, did not reveal any evidence for differential phosphorylation of Maf1 during carbon source switching. Together, these data do not support the proposed requirement for CK2 phosphorylation of Maf1 during derepression of pol III transcription.
Introduction
Maf1 is a global repressor of RNA polymerase (pol)3 III transcription that is conserved in eukaryotes and functions to mediate transcriptional repression under diverse nutritional, environmental and cellular stress conditions (1–4). These conditions determine the activity of different signaling pathways (e.g. Ras/PKA, TOR, cell integrity, and DNA damage response pathways) which regulate the function of Maf1 (2, 4, 5). Studies in yeast indicate that Maf1 is essential but not sufficient for repressing pol III transcription (6). This conclusion is based on the finding that a nuclear localized form of Maf1 (a 6SA or 7SA mutant) that is able to interact with and thereby inhibit the pol III machinery is not constitutively active in yeast cells under optimal growth conditions but represses transcription normally under stress conditions (6, 7). This is explained, at least in part, by the fact that the signaling pathways regulating pol III transcription involve posttranslational control of two branches; one branch targeting Maf1 and another targeting the polymerase (8). Consistent with the fact that Maf1 binding to pol III inhibits transcription in vitro (9–12), it appears that both branches of the signaling pathway need to operate together in vivo to affect repression (8). Maf1 is negatively regulated by phosphorylation at seven sites through the overlapping action of PKA and the TORC1-regulated kinase Sch9 (6, 7, 13, 14). This mechanism of negative control is conserved in higher eukaryotes where analogous sites in human Maf1 are directly phosphorylated by mTORC1 (15–17).
Protein kinase CK2 is an important regulator of a multitude of cellular processes and plays a vital role in cell differentiation, proliferation and survival (18). In the context of this study, the important functions of CK2 concern its regulation of gene transcription, specifically by RNA pol III and its role in the cellular response to genotoxic stress (19–23). CK2 is a tetrameric enzyme comprising two catalytic α subunits and two regulatory beta subunits. It is considered to be a constitutively active enzyme since it has no known physiological regulators (18). Instead, phospho-regulation of many CK2 substrates involves a localization-based mechanism where signaling specificity is achieved by the dynamic association of CK2 with different macromolecular complexes (23). Indeed, this mechanism applies to the stimulatory function of CK2 in pol III transcription. CK2 activity is required for high levels of pol III gene transcription in both yeast and human systems (19, 20, 21). Although the details of its involvement in this process are not fully understood, CK2 interacts with and phosphorylates subunits of the initiation factor TFIIIB (TBP in yeast and Brf1 in humans) and stimulates the assembly of TFIIIB on pol III gene promoters (19, 20). CK2 is also associated with pol III genes in ChIP experiments (21, 24, 25), and therefore has the potential to regulate ongoing transcription from complexes assembled on the DNA. Previous work in human cells indicates that this is important during mitotic repression of pol III transcription where CK2 phosphorylation of the Bdp1 subunit of TFIIIB leads to its dissociation from chromatin (24). In addition, studies in yeast on the response to genotoxic stress have shown that pol III transcription and TBP-associated CK2 activity decrease in cells treated with MMS. These changes correlate with the dissociation of CK2 catalytic subunits from the regulatory subunits which remain associated with TBP (19). This dynamic interaction of CK2 with TFIIIB is unlikely to be specific to conditions of DNA damage and may occur under other circumstances where high levels of pol III transcription are subject to repression.
Maf1 was recently reported to be phosphorylated by CK2 in vitro and on CK2 sites in vivo (25). Given the known phospho-regulation of Maf1, it was hypothesized that CK2 might be the nuclear kinase responsible for dissociating Maf1-pol III interactions, directing the export of Maf1 from the nucleus and restoring high levels of pol III transcription. To test this hypothesis, yeast cells were grown in glycerol to repress transcription and then returned to glucose with or without conditional or pharmacological inhibition of CK2 activity. A consistent pattern of results were obtained for Maf1 phosphorylation, Maf1 association with tRNA genes, Maf1 interaction with Rpc160 and pol III transcription. Thus, it was concluded that CK2 phosphorylation of Maf1 triggers RNA pol III activation (25). However, this interpretation of the data has been questioned (26). Changes in Maf1 phosphorylation under the carbon source switching protocol may have occurred at PKA/Sch9 phosphosites rather than at the proposed CK2 sites. There was also a possibility that the failure to relieve Maf1-dependent repression in glucose medium containing the CK2 inhibitor tetrabromobenzotriazol (TBBt) could have resulted from a general deficiency of CK2 rather than its direct effect on Maf1.
To further explore the proposed requirement for CK2 phosphorylation of Maf1 in RNA polymerase III reactivation, we examined the recovery from glycerol-mediated repression using three Maf1 alleles: Maf1-id S388A is a functional internal deletion mutant of Maf1 that lacks all of the CK2 phosphosites implicated by Graczyk et al. (25); Maf1-ck20 is quintuple serine to alanine mutant of the same CK2 sites and Maf1 7SA is a mutant that cannot be phosphorylated at key regulatory PKA/Sch9 sites in vivo (7, 13) and thus provides a sensitive assay for detecting phosphorylation at other sites. Our findings using these mutants do not support the proposed requirement for CK2 phosphorylation of Maf1 during the recovery of transcription.
EXPERIMENTAL PROCEDURES
Yeast Strains and Molecular Biology
All strains were derived from W303α (MATα leu2–3,112 trp1–1 ura3–1 can1–100 ade2–1 his3–11,15 ssd1–1, maf1Δ::KanMX4) and contained a plasmid borne Maf1 gene regulated by its cognate promoter with a carboxyl-terminal Myc epitope (6). The Maf1 7SA allele has alanine substitutions at serine positions 90, 101, 177, 178, 179, 209, and 210 (7, 13). Maf1-ck20 has alanine substitutions at serine positions 159–162 and 388. The internal deletion of Maf1 (Maf1-id) and the different point mutant alleles were generated by site-directed mutagenesis (QuikChange II; Stratagene, La Jolla, CA). The Maf1-id D248/D250A (Maf1-id DD) and Maf1-id S209/S210A alleles retain the numbering system of the full-length Maf1 protein. Strains were grown in YPD or synthetic complete media as appropriate, and where indicated, treated with rapamycin (0.2 μg/ml) or C3–1′-naphthyl-methyl PP1 (1NM-PP1, 0.1 μm, Calbiochem) for 1 h. Procedures for hot phenol RNA extraction, Northern analysis and quantitation were as reported (2). Whole cell extracts from cells harvested at room temperature by low speed centrifugation were prepared by TCA precipitation and denaturing lysis (6, 7) and resolved by high resolution SDS-PAGE (6) and Phos-tag SDS-PAGE (8). Purification of GST-kinases from yeast and their use in vitro kinase assays was as published (6, 13). Expression and native purification of full-length Maf1and Maf1-id was as described (27). The Maf1-id protein yield at 20 mg/liter was 4-fold higher than that obtained for the full-length protein. Methods for fixation of log phase cells and immunofluorescence microscopy were described previously (6, 8).
RESULTS
Construction of a Functional Saccharomyces cerevisiae Maf1 Derivative Lacking the Non-conserved Internal Domain
S. cerevisiae Maf1 (ScMaf1) has a molecular mass of 44.7 kDa and like numerous other fungal Maf1 proteins is considerably larger than its orthologs in higher eukaryotes (e.g. HsMaf1 which is 28 kDa). Biochemical studies of recombinant ScMaf1 have found that it has low solubility and is prone to aggregation (9). In contrast, the lower molecular mass Maf1 proteins from Schizosaccharomyces pombe and humans are well expressed in bacteria and are highly soluble (>20 mg/ml) (9, 11, 28). The most obvious difference between ScMaf1, SpMaf1, and HsMaf1 lies in the region between the phylogenetically conserved A and B domains: ScMaf1 contains an insertion of ∼170 amino acids between these domains (1). The sequence of ScMaf1 in this region is not well conserved among other fungal Maf1 proteins that contain insertions between domains A and B. Therefore, we explored the possibility that this internal non-conserved region of ScMaf1 is dispensible for its function in vivo.
HsMaf1 has a globular structure with the conserved A, B and C domains forming a central β sheet surrounded by α helices (28). However, because of structural flexibility/disorder in the native protein, the crystal structure that was solved was of a molecule truncated internally and at the C terminus (28). Hence, there is no information as to the organization of the C-terminal end of domain A or the adjoining region between domains A and B. Mutagenesis studies indicate that specific amino acids within the missing portion of domain A are essential for Maf1 function in vivo (6), including the highly conserved residues at positions 40–42 which are predicted to be part of an α helix (data not shown). The region between domains A and B also contains a nuclear localization motif (NtNLS), and phosphorylation sites shown to be important for the regulation of ScMaf1 function in vivo (6, 7, 13, 14). Maf1 phosphorylation in this region occurs primarily at consensus PKA sites with residues 209 and 210 accounting for ∼80% of phospho-PKA signal (13). Thus, the bulk of the phospho-regulation of ScMaf1 is likely achieved at these sites adjacent to the NtNLS. Phosphoproteomic data for ScMaf1 and HsMaf1 indicate that despite the lack of sequence conservation in the region between domains A and B, both proteins are phosphorylated at serine residues (that are positioned comparably to ScMaf1 residues 209/210) clustered upstream of conserved domain B (15–17, 29, 30). Based on these observations, an internal deletion of MAF1 (hereafter called Maf1-id, Fig. 1A) was generated that removes amino acids 64 to 194 of the non-conserved region but retains the serine cluster at positions 209/210 and the NtNLS. Maf1-id, driven from its endogenous promoter and containing a C-terminal Myc epitope was introduced into a MAF1 deletion strain and is functional in vivo as described below.
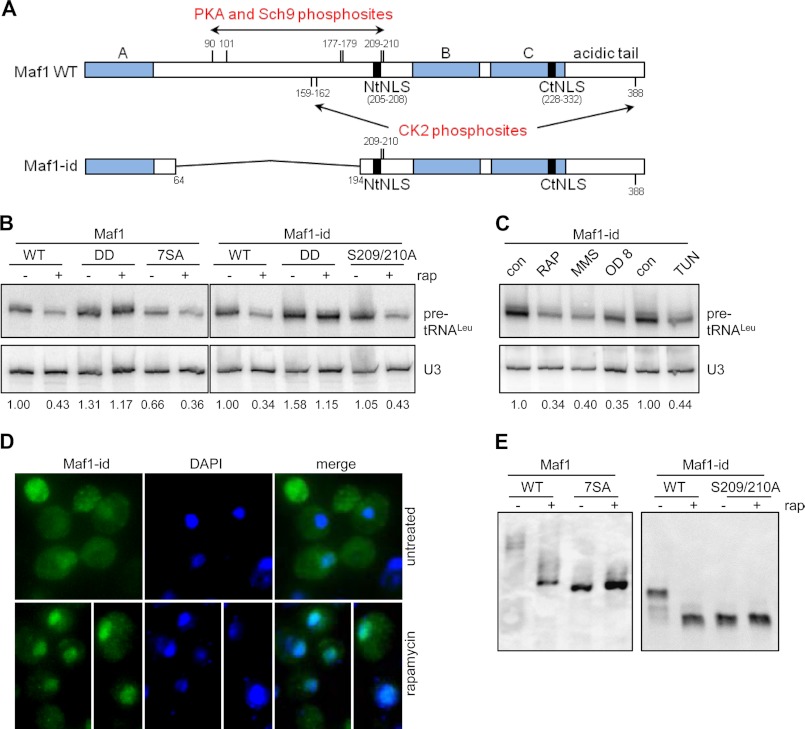
FIGURE 1.
An internal truncation of Maf1 generates a functional protein in vivo. A, schematic representation of full-length Maf1 and Maf1-id proteins drawn to scale. The conserved A, B, and C regions of Maf1 (filled blocks) and adjoining regions (open blocks) and the deleted region of Maf1-id (thin line), from amino acids 64 to 194 are annotated along with the positions of PKA, Sch9 and CK2 phosphosites ((6, 7, 13, 14, 25)). The positions of the two nuclear localization sequences in the amino (NtNLS) and carboxyl region (CtNLS) are also identified. B, Maf1-id functions in repressing pol III transcription. Northern analysis of a short-lived tRNALeu precursor and the stable U3 snRNA was performed with RNA from early log phase cultures of the indicated strains before and after treatment with rapamycin. Normalized pre-tRNALeu band intensities were used to determine the level of transcriptional repression relative to the untreated wild-type strain (full-length Maf1 or Maf1-id) and are indicated under the gel. Wild-type and mutant forms of full-length Maf1 or Maf1-id are compared including the inactive D248/D250A (DD) mutant and the PKA/Sch9 phosphosite mutants (7SA and S209/210A). Images are from non-contiguous lanes of the same gel/hybridization and are presented at the same exposure and contrast. Analysis of multiple independent RNA preparations (n = 4) showed that the untreated Maf1-id strain generated transcript levels at 1.18 ± 0.20 times the level of full-length Maf1. C, repression of pol III transcription in Maf1-id strains under different conditions: Rapamycin (200 ng/ml) and MMS (0.08%) treatment for 60 min, growth to late log phase (>OD 6) and tunicamycin (2.5 μg/ml) treatment for 3 h. The corresponding drug vehicle control (con) lanes are indicated (left, DMSO 1 h; center, MEOH 3 h). D, immunolocalization of Maf1-id was determined by indirect immunofluoresence detection of its C-terminal Myc epitope (Alexa 488, left panel) as expressed from the Maf1 promoter. Maf1-id nuclear localization is confirmed by the overlap with DAPI nuclear staining (merge). Cells were mock-treated (upper panels) or rapamycin-treated (lower panels) for 60 min. Images are representative of multiple independent experiments. E, analysis of the phosphorylation pattern of Maf1 and Maf1-id strains. Cell extracts of strains were prepared from mock-treated or rapamycin-treated strains, separated by SDS-PAGE in the presence of Phos-tag-acrylamide and immunoblotted to detect Maf1 (α-Myc). Full-length Maf1 7SA and Maf1-id 2SA extracts are included as internal references for the relative mobility of the unphosphorylated Maf1 proteins.
The activity of Maf1-id was assessed using the most stringent and sensitive assay for Maf1 function, namely its ability to repress pol III transcription in vivo (6). Specifically, we measured the level of short-lived leucine tRNA precursors in control cells and in cells treated with the TORC1 inhibitor rapamycin (Fig. 1B). Repression of pol III transcription by rapamycin is well documented in wild-type strains and is absolutely dependent on Maf1 (2). Maf1-id functioned in a quantitatively similar manner to the full-length protein: Transcription in untreated cells was comparable and rapamycin treatment caused a significant decrease in transcription (Fig. 1B). An analysis of multiple independent RNA preparations revealed transcription levels for untreated Maf1-id of 103% ± 26% (n = 6) and residual transcription after rapamycin treatment of 37 ± 1% and 40 ± 4% (n = 4) for full-length Maf1 and Maf1-id, respectively, compared with the corresponding untreated wild-type control. In addition, the Maf1-id protein was similarly sensitive to a double point mutation of the invariant acid residues (D248/D250)(Fig. 1B), which fully inactivate the protein and increase transcription in both untreated and treated cells (6). An alanine substitution mutant, Maf1-id 2SA which is non-phosphorylatable at the 209/210 PKA/Sch9 sites, exhibited normal levels of pol III transcription and the same extent of repression as full-length Maf1 (105 and 43%, respectively) (Fig. 1B). The Maf1-id protein was also capable of responding to repression signaling evoked by other stresses that repress pol III transcription (2) such as DNA damage (MMS treatment), glucose and nutrient depletion caused by growth to high cell density (A600 = 8.0) and a block to secretory pathway function (tunicamycin) (Fig. 1C).
Maf1-id retains the two NLS-like motifs (the strong phosphoregulated NtNLS and the weaker C-terminal NLS, CtNLS) that were previously shown to affect the localization of the full length protein under normal and repressing conditions (Fig. 1A), (6). However, as noted above, Maf1-id is missing five of the seven phosphosites that regulate its localization and interactions with pol III (6, 7, 13). It was unclear therefore whether Maf1-id would recapitulate the regulated localization of the full-length protein. Immunolocalization of the Maf1-id protein shows that it is distributed throughout the nucleus and the cytoplasm in untreated cells and that it is concentrated in the nucleus after rapamycin treatment (Fig. 1D). Thus, the localization of Maf1-id changes in response to rapamycin in the same way as the full-length protein (data not shown, 6, 31, 32). This implies that the regulatory phosphosites, S209/S210, which are retained in Maf1-id (Fig. 1A) are sufficient to control its localization in response to signaling via TORC1.
To examine the phosphorylation of the Maf1-id protein directly, we prepared yeast cell extracts from strains expressing Maf1-id or the double mutant S209/S210A, before and after rapamycin treatment, and analyzed the Maf1 banding pattern on denaturing phos-tag acrylamide gels (Fig. 1E). Extracts from strains expressing full-length Maf1 or its 7SA mutant (which contains alanine substitutions at all seven of the PKA/Sch9 phosphosites) were also examined since these proteins had not previously been analyzed in the phos-tag gel system. The data show that both Maf1and Maf1-id are hyperphosphorylated under normal growth conditions and become dephosphorylated under repressing conditions (Fig. 1E). Notably, the 7SA mutation reduced the complex banding pattern of the full-length protein to a single prominent band which had a similar mobility to the wild-type protein after rapamycin treatment. The phospho-pattern of Maf1-id is less complex than that of the full-length protein consistent with the internal deletion removing five of the seven PKA/Sch9 phosphosites. Untreated extracts of Maf1-id show three bands that collapse to one predominant band after rapamycin treatment. Alanine substitutions at the two remaining PKA/Sch9 phosphosites in Maf1-id (S209/S210, Maf1-id 2SA) cause the protein to migrate as a single major band with the same mobility as the rapamycin treated Maf1-id protein (Fig. 1E). Thus, S209/S210 represent the major phosphoregulated sites in Maf1-id. Quantitation of full-length and Maf1-id proteins separated by phos-tag or standard SDS-PAGE indicated that the internal deletion did not affect protein abundance in yeast under normal or repressed conditions (data not shown).
Collectively these data indicate that the Maf1-id protein is fully functional in repressing pol III gene transcription; it responds to stress signaling under multiple conditions and its accumulation in the nucleus correlates with dephosphorylation of the two remaining PKA/Sch9 sites consistent with the behavior of the full length protein.
Maf1-id Is Phosphorylated by PKA and Sch9
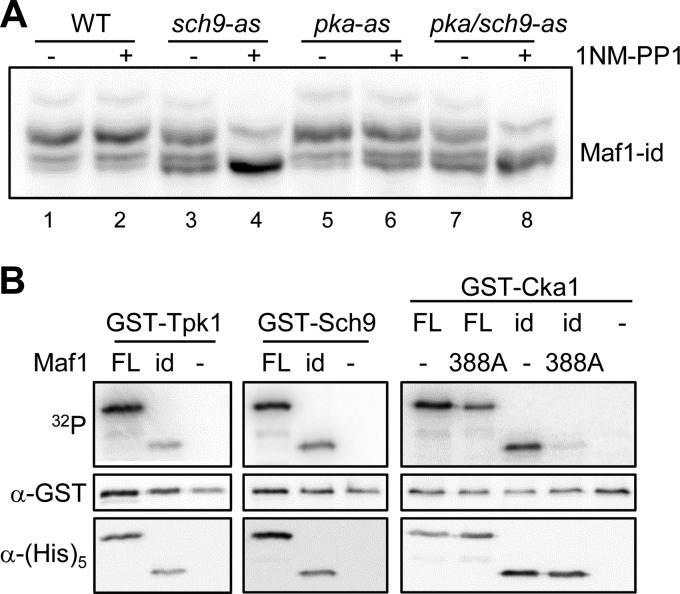
Previous studies have shown that PKA and the TORC1-regulated kinase Sch9 directly phosphorylate an overlapping set of sites in ScMaf1 (6, 7, 13, 14). In addition, cross-talk between these pathways occurs such that TORC1 activity can regulate PKA phosphorylation of ScMaf1. This cross-talk involves Sch9-dependent activation of the MAP kinase Slt2 which phosphorylates and inactivates Bcy1, the regulatory subunit of PKA (33). To investigate the role of PKA and Sch9 in Maf1-id phosphorylation, we transformed the Maf1-id construct into strains containing ATP analog-sensitive alleles of Sch9 (sch9-as), all three PKA alleles (Tpk1, -2, and -3, pka-as) or both Sch9 and PKA kinases (sch9/pka-as) (34) and analyzed the effect of kinase inhibition using phos-tag acrylamide gels. Treatment of the analog-sensitive pka-as strain with 1NM-PP1 for 1 h had only a modest effect on the phosphorylation of Maf1-id, increasing the relative amount of the fastest migrating (unphosphorylated) band compared with either the untreated pka-as strain or the analog-insensitive wild type strain (Fig. 2A, compare lane 6 to lanes 1, 2, and 5). In contrast, inhibitor treatment of the sch9-as and sch9/pka-as strains significantly diminished Maf1-id phosphorylation (Fig. 2A). Together with the data in Fig. 1E, the results suggest that residues Ser-209/S210 in Maf1-id are primarily phosphorylated by Sch9 when cells are grown in synthetic complete medium with glucose as the carbon source.
FIGURE 2.
Phosphorylation of ScMaf1, Maf1-id, and Maf1-id S388A. A, phosphorylation of Maf1-id in PKA and SCH9 ATP analog sensitive (−as) strains. Wild type and −as strains were grown to OD ∼0.5 and treated with the ATP analog 1NM-PP1 before extract preparation and separation by phos-tag SDS-PAGE as in Fig. 1E. Note the slowest migrating band in all lanes results from phosphorylation at an unknown site that is insensitive to inhibition of Sch9 and PKA. B, phosphorylation of WT Maf1 FL (full-length), Maf1-id, and S388A mutant proteins in vitro. Expression of GST-Tpk1, GST-Sch9, and GST-Cka1 kinases was induced by addition of 4% galactose to log phase yeast cells grown in YP raffinose medium. Each kinase was purified on a glutathione-Sepharose column. Recombinant WT Maf1FL, Maf1-id, and S388A mutant proteins (500 ng) were incubated with the yeast-purified kinase and [γ-32P]ATP for 1 h before separation by standard SDS-PAGE. An autoradiogram (upper panel), Western detection of each kinase using anti-GST antibody (middle panel) and Western detection of each recombinant Maf1 protein using anti-HIS antibody (lower panel) are shown for each kinase reaction.
We next tested the phosphorylation of Maf1-id by PKA (Tpk1) and Sch9 in vitro. Both kinases were purified from yeast as GST fusion proteins and showed similar activity against recombinant full-length Maf1 and Maf1-id proteins (Fig. 2B). Consistent with the removal of five consensus PKA sites in Maf1-id and the overlapping specificity of Sch9 (35), PKA and Sch9 showed comparable but lower levels of phosphorylation of Maf1-id relative to the full-length protein. These in vitro experiments demonstrate the potential for functional redundancy between PKA and Sch9 toward Maf1 in vivo.
Recent work has found that ScMaf1 can be phosphorylated in vitro by CK2 on at least one site in a serine tract from Ser-159–162 and on Ser-388 in the C terminus of the protein (25). As Maf1-id does not contain the serine tract, we generated an S388A mutation in the context of full-length Maf1 and Maf1-id to determine how efficiently CK2 can phosphorylate these proteins. Maf1 S388A which contains the serine tract but not the C-terminal site and Maf1-id which has the reciprocal combination of CK2 sites showed similar levels of phosphorylation by GST-Cka1. This phosphorylation was reduced significantly (∼50%) compared with the full length wild-type protein (Fig. 2B) confirming that both sites/regions are targeted by CK2 in vitro (25). In contrast, the phosphorylation of Maf1-id S388A was barely detectable (<5% of ScMaf1). Thus, the serine tract and S388 on Maf1 are the main sites phosphorylated by CK2 in vitro. This result suggested that the Maf1-id S388A mutant could be used to test the proposed requirement for CK2 phosphorylation of Maf1 in the reactivation of pol III transcription from the glycerol-repressed state (25).
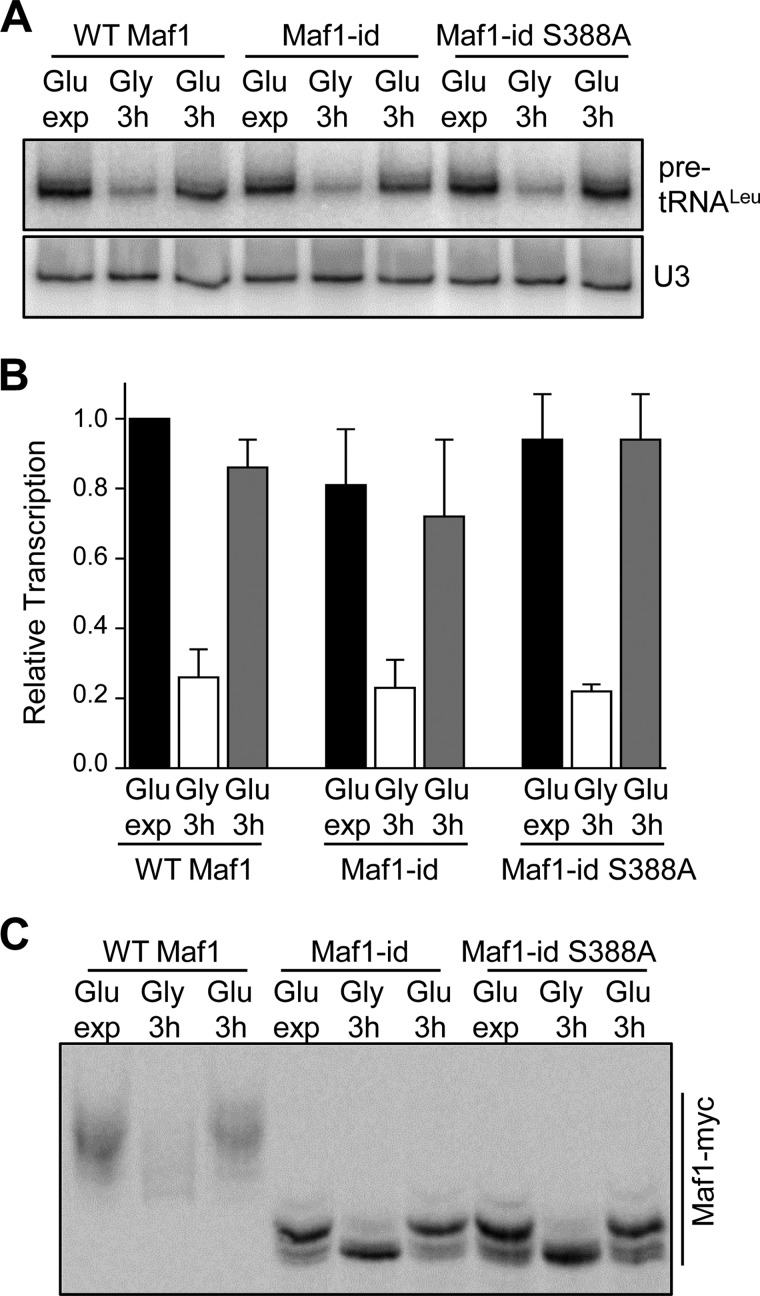
Reactivation of Pol III Transcription from the Glycerol-repressed State Occurs Normally in the Maf1-id S388A Mutant
Defects in Maf1 phosphorylation, Maf1 interactions with pol III and tRNA genes and a failure to derepress transcription are observed when yeast cells are transferred from glycerol medium to glucose medium containing tetrabromobenzotriazol (TBBt), a selective inhibitor of CK2 (25). A similar defect in Maf1 phosphorylation is also seen in a temperature-sensitive CK2 strain when the glycerol to glucose switch is performed simultaneously with a temperature shift to inactivate CK2. Thus, it was proposed that Maf1 phosphorylation by CK2 triggers activation of pol III transcription when cells growing in glycerol as a carbon source are transferred into medium containing glucose (25). In light of alternative explanations for the data (26), we decided to directly test the hypothesis using strains containing Maf1-id and Maf1-id S388A which are missing some or all of the sites implicated in the regulation by CK2 (25). These strains along with a wild-type control were grown to early log phase in glucose medium, transferred to glycerol medium for 3 h and then transferred back to glucose medium as reported by Graczyk et al. (25) with samples taken at each stage to measure pol III transcription by Northern analysis. In glycerol medium, pol III transcription in all three strains was reduced to ∼25% of the level determined for the wild-type strain in glucose as reported previously (Fig. 3A), (25). Importantly, upon transfer of the strains into glucose-containing medium, pol III transcription was fully restored to the level observed before glycerol-mediated repression (Fig. 3A). The same carbon source switching protocol was also used to examine the phospho-pattern of Maf1-id and Maf1-id S388A in phos-tag acrylamide gels. In glycerol medium, Maf1, Maf1-id, and Maf1-id S388A were substantially dephosphorylated as expected, consistent with the repression of pol III transcription by growth in glycerol (Fig. 3B). Additionally, after transferring the cells into glucose medium, the phospho-pattern of all three proteins was restored to the pattern typical of cells in exponential phase in this medium (Fig. 3C). These results indicate that phosphorylation of the serine stretch S159–162 and S388 of Maf1 by CK2 (or any other kinase) is not necessary in order for pol III transcription to recover from glycerol-mediated repression. In addition, the data suggest that the rephosphorylation of Maf1-id that occurs as cells transition from repressive to favorable growth conditions is due to the activity of Sch9 and/or PKA at positions Ser-209/210.
FIGURE 3.
Maf1-id S388A regulates pol III transcription normally during carbon source switching. A, recovery of Pol III transcription after growth in glycerol medium in the Maf1-id S388A strain. Yeast cultures were grown to early log phase (OD ∼0.5) in glucose-containing minimal media before carbon source switching and RNA extraction. Cells were harvested after exponential growth in glucose media (Glu exp) and after a 3 h switch into glycerol-containing media (Gly 3h). An additional aliquot of glycerol-switched cells was refed glucose-containing media and grown for a further 3 h (Glu 3h). For carbon source switching cells were harvested by centrifugation and washed once in warm media containing the new carbon source before further growth. RNA extraction and Northern analysis was done as in Fig. 1B. B, quantitation of Northern data from multiple independent experiments as described in Fig. 3A. Relative transcription (normalized pre-tRNALeu band intensities) was referenced to values determined for WT Maf1 grown in glucose-containing medium. C, phosphorylation of Maf1-id S388A after recovery from growth in glycerol. Strains were grown in glucose media, switched into glycerol media for 3 h and then switched back into glucose media as in Fig. 3A. Maf1 phosphorylated forms were resolved by Phos-tag SDS-PAGE as described in Fig. 1E.
Reactivation of Transcription from the Glycerol-repressed State in the Maf1 7SA Mutant
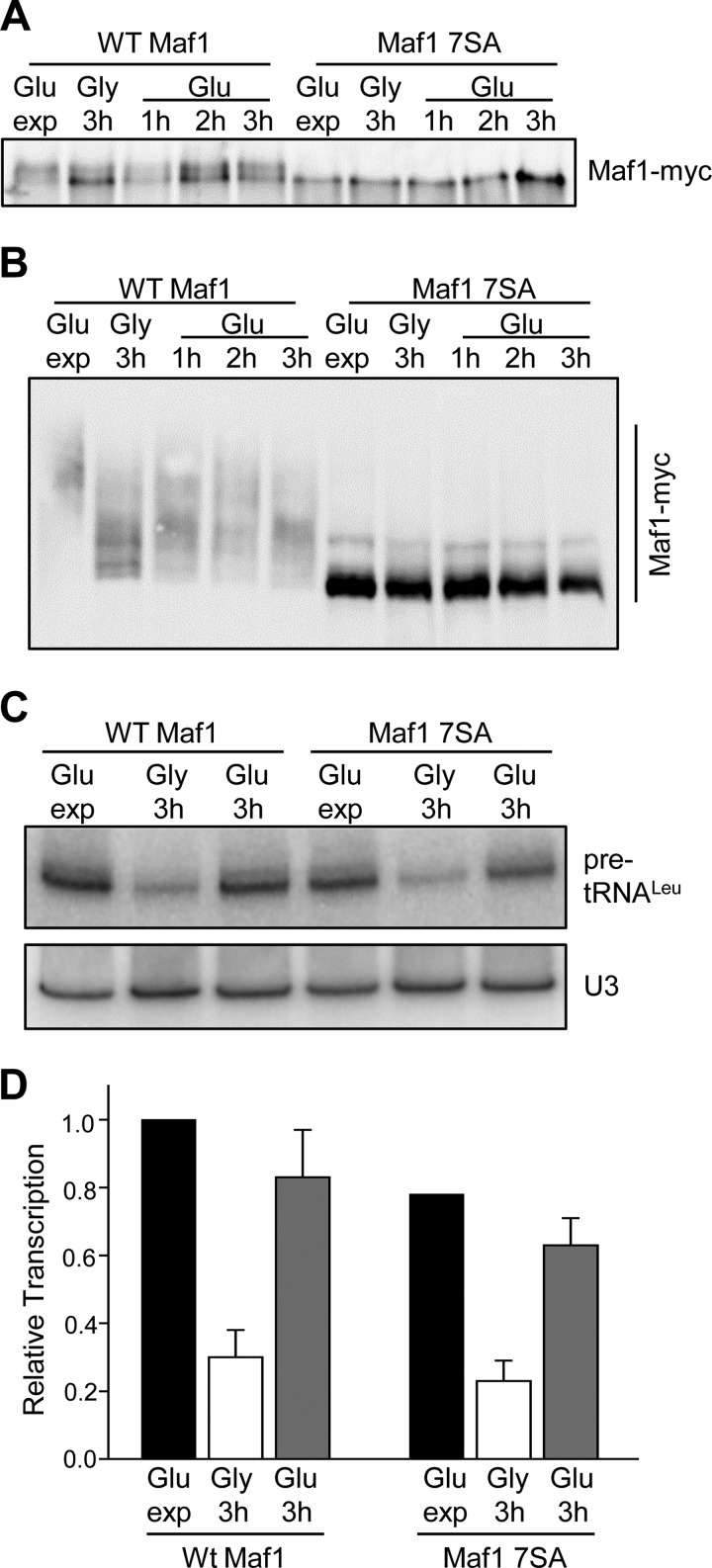
To independently assess the possibility that CK2 phosphorylates Maf1 during the recovery of transcription from the glycerol-repressed state, we employed a Maf1 7SA mutant in which all of the major sites for PKA/Sch9 phosphorylation are mutated to alanine (6, 7, 13). Importantly, these sites at serines 90, 101, 177, 178, 179, 209, and 210 do not overlap with the putative CK2 regulatory sites at Ser-159–162 and Ser-388. We prepared cell extracts from ScMaf1 and ScMaf1 7SA strains following the carbon source switching protocol, taking samples at one hour intervals after returning the strains to medium containing glucose. The extracts were analyzed initially by one-dimensional SDS-PAGE with a reduced cross-linking ratio to resolve phospho-Maf1 forms. As reported previously using this gel system (e.g. in Refs. 6, 13, 25, 31, 32), ScMaf1 runs predominantly as a slow migrating (hyperphosphorylated) form in rich medium and shifts to a fast migrating (hypophosphorylated) form under repressing conditions such as growth in glycerol (Fig. 4A). Subsequently, the relative amount of the fast migrating form decreases upon return to media containing glucose. In contrast, the ScMaf1 7SA mutant migrated exclusively as a fast migrating form; no change in mobility was observed as a result of carbon source switching (Fig. 4A). Next, we loaded the same extracts on a phos-tag acrylamide gel to provide increased resolving power for the phosphorylated Maf1 species. As before, ScMaf1 shows a change in phospho-state under repressing conditions but interestingly, the protein is not fully dephosphorylated in glycerol (Fig. 4B). This incomplete reduction in the phospho-state of Maf1 is consistent with the continued growth of the cells at a slower rate in this medium and thus a requirement for ongoing pol III transcription. As expected, increased Maf1 phosphorylation is observed upon return of the cells to glucose-containing medium (Fig. 4B). In contrast, the Maf1 7SA mutant showed a single major band consistent with the dephosphorylated protein and a minor species representing ∼6.7 ± 0.5% of the total Maf1 signal. No other bands were observed and neither the major nor the minor band changed their relative abundance during carbon source switching (Fig. 4B). These data suggest that previously reported changes in Maf1 phosphorylation during carbon source switching (25) involve the major sites phosphorylated by PKA/Sch9 rather than sites phosphorylated by CK2. To confirm that pol III transcription was responding normally in this experiment, we also analyzed precursor tRNA levels by Northern analysis (Fig. 4C). The wild-type strain showed a normal level of repression and recovery. Interestingly, the Maf1 7SA mutant showed a statistically significant reduction in transcription in untreated log phase cells. This effect was not observed in previous studies with a 6SA mutant of ScMaf1 (6) but is similar to the reported behavior of a multiply mutated hyperactive form of HsMaf1 (15). Transcriptional repression in the Maf1 7SA mutant was similar to wild-type and transcription recovered essentially to the starting level upon return to glucose (Fig. 4C). Thus, under the conditions of this experiment, pol III transcription recovers from the glycerol-repressed state in the Maf1 7SA strain without apparent phosphorylation of the protein.
FIGURE 4.
Recovery of transcription from the glycerol-repressed state in wild-type and MAF1 7SA mutant strains. A, phosphorylation of WT Maf1 and Maf1 7SA proteins as resolved by one-dimensional SDS-PAGE. Cells were grown as in Fig. 3A and cell extracts prepared at 1 h intervals after switching from glycerol into glucose medium. Maf1 proteins were separated by SDS-PAGE using a modified acrylamide:bis-acrylamide ratio of 33.5:0.3 before Western detection. B, phosphorylation of WT Maf1 and Maf1 7SA proteins resolved by Phos-tag SDS-PAGE. Extracts in Fig. 4A were separated and detected as described in Fig. 1E. C, pol III transcription in WT Maf1 and Maf1 7SA strains after carbon source switching. Cells were grown as Fig. 4A and RNA analyzed as described in Fig. 1B. D, quantitation of Northern data from multiple independent experiments as described in Fig. 4C. Relative transcription (normalized pre-tRNALeu band intensities) was referenced to values determined for untreated WT Maf1. The standard error for replicate untreated Maf17SA samples (0.78 ± 0.01) are not visible at the scale of the figure.
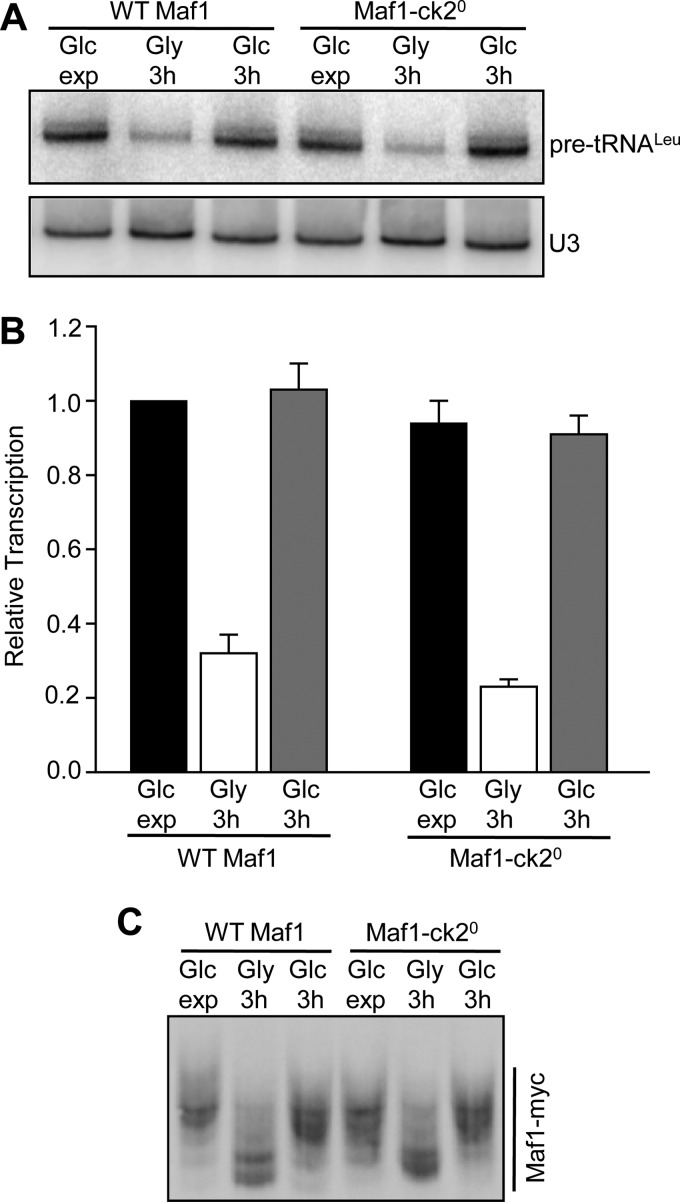
Derepression of Pol III Transcription Occurs in a Quintuple CK2 Site Mutant of Maf1
The ability of Maf1-id S388A to respond normally to derepressing conditions demonstrated that CK2 was not required for Maf1 phosphorylation is this context. However, it remained formally possible that the sequences deleted in this construct were responsible for the CK2 requirement in the first place. To examine this possibility we generated a quintuple serine to alanine mutant (Maf1-ck20) at the relevant CK2 sites (aa159–162 and 388) and tested its function under the carbon source switching protocol. As seen for Maf1-id S388A, the Maf1-ck20 mutant functioned normally to repress transcription in media containing glycerol and transcription was fully restored upon return of the cells to media containing glucose (Fig. 5, A and B). In addition, using phos-tag acrylamide, we found that the absence of the proposed regulatory CK2 sites in the Maf1-ck20 mutant did not affect the ability of the protein to be dephosphorylated or rephosphorylated during carbon switching (Fig. 5C). Considering that no change in phosphorylation was seen in the Maf1 7SA mutant under the same conditions (Fig. 4B), it appears that the sites normally phosphorylated during recovery from growth in glycerol are those targeted by PKA/Sch9.
FIGURE 5.
Recovery of transcription from the glycerol-repressed state in a quintuple CK2 phosphosite mutant. A, recovery of Pol III transcription after growth in glycerol medium in wild-type and Maf1-ck20 strains. Maf1-ck20 contains serine to alanine substitutions at five sites, Ser-159–162 and Ser-388. The same mutant was designated 5StA in ref (25). The experimental design and procedures were the same as in Fig. 3. The figure shows a representative Northern blot. B, quantitation of Northern data from panel A and a second independent experiment performed under the same conditions. Relative transcription (normalized pre-tRNALeu band intensities) was referenced to values determined for WT Maf1 grown in glucose-containing medium. C, phosphorylation of Maf1-ck20 after recovery from growth in glycerol. Strains were grown in glucose media, switched into glycerol media for 3 h and then switched back into glucose media as in Fig. 3. Maf1 phosphorylated forms were resolved in gels containing Phos-tag acrylamide as described in Fig. 1E.
DISCUSSION
We initiated this work with the goal of generating a smaller functional version of ScMaf1 that was internally-deleted for the non-conserved region (∼14 kDa) between domains A and B in the protein. Based on the biochemical properties of SpMaf1 and HsMaf1 which do not contain the non-conserved region, it was anticipated that a structurally similar ScMaf1 protein would be well-expressed as a native protein in bacteria and would have improved solubility. Our analysis of the internally deleted MAF1 allele, MAF1-id, showed that it is functional in repressing pol III gene transcription in yeast cells. The Maf1-id protein responds to stress signaling under multiple conditions and its accumulation in the nucleus correlates with dephosphorylation of the two (out of seven) remaining PKA/Sch9 sites (Ser-209/210), consistent with the behavior of the full-length protein. In addition, the recombinant Maf1-id protein is well expressed, highly soluble and can be phosphorylated in vitro by PKA and Sch9 kinases. Thus, Maf1-id provides a simplified and better behaved protein for biological and biochemical studies of Maf1 in budding yeast.
Our finding that Maf1-id was functional in vivo and the fact that the protein lacked the CK2 serine cluster at Ser-159–162 suggested a simple approach to test the requirement for CK2 phosphorylation of Maf1 during derepression of transcription. Only a single point mutation at Ser-388 was necessary in the context of Maf1-id to eliminate all of the sites implicated in the proposed response. We found that not only was the Maf1-id S388A mutant as effective as wild-type Maf1 in repressing transcription during growth on glycerol but transcription was derepressed to the same extent as wild-type Maf1 upon return of the cells to medium containing glucose (Fig. 3). Equivalent findings were obtained with the quintuple CK2 site mutant, Maf1-ck20 (Fig. 5). In addition, we employed the Maf1 7SA mutant (in which all of the PKA/Sch9 phosphosites known to regulate Maf1 function are changed to alanine) and phos-tag acrylamide gel electrophoresis as a sensitive assay for Maf1 phosphorylation by other kinases. Regardless of the growth conditions that were examined, ∼93% of the Maf1 had a mobility equivalent to the unphosphorylated protein with a single minor species accounting for the remainder (Fig. 4). Thus, aside from the seven known PKA/Sch9 sites there is no significant, sustained phosphorylation of Maf1 by other kinases during log phase growth in glucose or under repressing conditions in glycerol. We should emphasize that these experiments do not exclude the possibility that a short lived phosphoform of Maf1 may exist under some circumstances. As for the minor species, its nature is unknown but its abundance does not vary under the growth conditions studied here. A similar minor species was seen in phos-tag gels of Maf1-id S388A (Fig. 3B) suggesting that it does not result from CK2 phosphorylation at the sites preferred by this enzyme in vitro (Fig. 2B). Together our results do not support the proposed requirement for CK2 phosphorylation of Maf1 in pol III reactivation. Rather, it seems that the rephosphorylation of Maf1 during the glycerol to glucose carbon source switch occurs at PKA/Sch9 sites. Moreover, this phosphorylation is not required for recovery of transcription since the Maf1 7SA mutant shows a normal response. We infer therefore that during derepression of transcription, the dissociation of Maf1 from pol III may occur in response to changes in the polymerase. Part of this response could involve the dephosphorylation of Rpc53 (8).
In light of the present findings, some explanations are needed for the previously published observations (25). With respect to the changes observed in Maf1 phosphorylation when CK2 activity is inhibited (e.g. by TBBt or in a temperature-sensitive CK2 strain), as noted above, our data suggest that these changes occur at one or more PKA/Sch9 sites. Phosphorylation of these sites is known to affect the mobility of Maf1 in SDS gels in an unusual way. In rich medium, mutation of two sites, serines 177 and 178 (that together account for only ∼20% of the total phosphorylation at PKA/Sch9 sites), abolishes the slow migrating forms of Maf1 whereas mutations of two other sites, serines 209 and 210 (that account for ∼80% of the PKA/Sch9 site phosphorylation), have no effect (13). Thus, the mobility of Maf1 in this gel system is especially sensitive to subtle changes in the phosphorylation of serines 177 and 178. Perhaps this sensitivity underlies the observed effect of combining mutations at the CK2 sites (i.e. the 5StA mutant of the serine cluster, Ser-159–162, and Ser-388) (25). Besides altering Maf1 phosphorylation, inhibition of CK2 at the time of carbon source switching led to defects in other activities and functions associated with Maf1, most importantly, a failure to derepress transcription. These effects may reflect a role for CK2 in the phosphorylation of a target other than Maf1 that directly or indirectly impacts the recovery of pol III transcription. Such a role for CK2 could be the same as the already known requirement of CK2 for high levels of pol III transcription (19). Alternatively, the loss of CK2 catalytic activity, which is essential for viability, could constitute a form of cellular stress that does not permit pol III transcription to be derepressed, despite the presence of an optimal carbon source. Clearly, a more detailed understanding of the mechanisms by which CK2 function impacts pol III transcription would help to address these issues.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM085177 (to I. M. W.).
- pol
- polymerase
- CK2
- protein kinase CK2
- TF
- transcription factor
- tetrabromobenzotriazol
- TBBt
- TBP
- TATA-binding protein
- Brf1
- TFIIB-related factor
- rap
- rapamycin
- Sp
- S. pombe
- Sc
- S. cerevisiae
- r
- recombinant
- 1NM-PP1
- C3–1′-naphthyl-methyl-PP1.
REFERENCES
- 1. Pluta K., Lefebvre O., Martin N. C., Smagowicz W. J., Stanford D. R., Ellis S. R., Hopper A. K., Sentenac A., Boguta M. (2001) Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 5031–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Upadhya R., Lee J., Willis I. M. (2002) Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol. Cell 10, 1489–1494 [DOI] [PubMed] [Google Scholar]
- 3. Willis I. M., Moir R. D. (2007) Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem. Sci. 32, 51–53 [DOI] [PubMed] [Google Scholar]
- 4. Wei Y., Zheng X. S. (2010) Maf1 regulation: a model of signal transduction inside the nucleus. Nucleus 1, 162–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Willis I. M., Desai N. A., Upadhya R. (2004) Signaling repression of transcription by RNA polymerase III in yeast Prog. Nucleic Acids Res. Mol. Biol. 77, 323–353 [DOI] [PubMed] [Google Scholar]
- 6. Moir R. D., Lee J., Haeusler R. A., Desai N., Engelke D. R., Willis I. M. (2006) Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. Proc. Natl. Acad. Sci. U.S.A. 103, 15044–15049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huber A., Bodenmiller B., Uotila A., Stahl M., Wanka S., Gerrits B., Aebersold R., Loewith R. (2009) Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis Genes Dev. 23, 1929–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee J., Moir R. D., McIntosh K. B., Willis I. M. (2012) TOR signaling regulates ribosome and tRNA synthesis via LAMMER/Clk and GSK-3 family kinases. Mol. Cell 45, 836–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Desai N., Lee J., Upadhya R., Chu Y., Moir R. D., Willis I. M. (2005) Two steps in Maf1-dependent repression of transcription by RNA polymerase III. J. Biol. Chem. 280, 6455–6462 [DOI] [PubMed] [Google Scholar]
- 10. Reina J. H., Azzouz T. N., Hernandez N. (2006) Maf1, a New Player in the Regulation of Human RNA Polymerase III Transcription. PLoS. ONE. 1, e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cabart P., Lee J., Willis I. M. (2008) Facilitated recycling protects human RNA polymerase III from repression by Maf1 in vitro. J. Biol. Chem. 283, 36108–36117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goodfellow S. J., Graham E. L., Kantidakis T., Marshall L., Coppins B. A., Oficjalska-Pham D., Gérard M., Lefebvre O., White R. J. (2008) Regulation of RNA polymerase III transcription by Maf1 in mammalian cells. J. Mol. Biol. 378, 481–491 [DOI] [PubMed] [Google Scholar]
- 13. Lee J., Moir R. D., Willis I. M. (2009) Regulation of RNA polymerase III transcription involves SCH9-dependent and SCH9-independent branches of the target of rapamycin (TOR) pathway. J. Biol. Chem. 284, 12604–12608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wei Y., Zheng X. F. (2009) Sch9 partially mediates TORC1 signaling to control ribosomal RNA synthesis Cell Cycle 8, 4085–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Michels A. A., Robitaille A. M., Buczynski-Ruchonnet D., Hodroj W., Reina J. H., Hall M. N., Hernandez N. (2010) mTORC1 directly phosphorylates and regulates human MAF1. Mol. Cell. Biol. 30, 3749–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shor B., Wu J., Shakey Q., Toral-Barza L., Shi C., Follettie M., Yu K. (2010) Requirement of the mTOR kinase for the regulation of Maf1 phosphorylation and control of RNA polymerase III-dependent transcription in cancer cells. J. Biol. Chem. 285, 15380–15392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kantidakis T., Ramsbottom B. A., Birch J. L., Dowding S. N., White R. J. (2010) mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc. Natl. Acad. Sci. U.S.A. 107, 11823–11828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. St Denis N. A., Litchfield D. W. (2009) Protein kinase CK2 in health and disease: From birth to death: the role of protein kinase CK2 in the regulation of cell proliferation and survival. Cell Mol Life Sci. 66, 1817–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghavidel A., Schultz M. C. (2001) TATA binding protein-associated CK2 transduces DNA damage signals to the RNA polymerase III transcriptional machinery. Cell 106, 575–584 [DOI] [PubMed] [Google Scholar]
- 20. Johnston I. M., Allison S. J., Morton J. P., Schramm L., Scott P. H., White R. J. (2002) CK2 forms a stable complex with TFIIIB and activates RNA polymerase III transcription in human cells. Mol. Cell. Biol. 22, 3757–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu P., Wu S., Hernandez N. A. (2003) Minimal RNA polymerase III transcription system from human cells reveals positive and negative regulatory roles for CK2. Mol. Cell 12, 699–709 [DOI] [PubMed] [Google Scholar]
- 22. Clelland B. W., Schultz M. C. (2010) Genome stability control by checkpoint regulation of tRNA gene transcription. Transcription 1, 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Filhol O., Cochet C. (2009) Protein kinase CK2 in health and disease: Cellular functions of protein kinase CK2: a dynamic affair. Cell Mol. Life Sci. 66, 1830–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu P., Samudre K., Wu S., Sun Y., Hernandez N. (2004) CK2 phosphorylation of Bdp1 executes cell cycle-specific RNA polymerase III transcription repression. Mol. Cell 16, 81–92 [DOI] [PubMed] [Google Scholar]
- 25. Graczyk D., Debski J., Muszyńska G., Bretner M., Lefebvre O., Boguta M. (2011) Casein kinase II-mediated phosphorylation of general repressor Maf1 triggers RNA polymerase III activation. Proc. Natl. Acad. Sci. U.S.A. 108, 4926–4931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Willis I. M., Moir R. D., Lee J. (2011) Does casein kinase II phosphorylation of Maf1 trigger RNA polymerase III activation? Proc. Natl. Acad. Sci. U.S.A. 108, E300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nika H., Lee J., Willis I. M., Angeletti R. H., Hawke D. H. (2012) Phosphopeptide characterization by mass spectrometry using reversed-phase supports for solid-phase beta-elimination/michael addition J. Biomol. Tech. 23, 51–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vannini A., Ringel R., Kusser A. G., Berninghausen O., Kassavetis G. A., Cramer P. (2010) Molecular basis of RNA polymerase III transcription repression by Maf1. Cell 143, 59–70 [DOI] [PubMed] [Google Scholar]
- 29. Gnad F., Ren S., Cox J., Olsen J., Macek B., Oroshi M., Mann M. (2007) PHOSIDA (phosphorylation site database): management, structural, and evolutionary investigation, and prediction of phosphosites. Genome Biol. 8, R250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bodenmiller B., Campbell D., Gerrits B., Lam H., Jovanovic M., Picotti P., Schlapbach R., Aebersold R. (2008) PhosphoPep[mdash]a database of protein phosphorylation sites in model organisms. Nat. Biotech. 26, 1339–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roberts D. N., Wilson B., Huff J. T., Stewart A. J., Cairns B. R. (2006) Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol. Cell 22, 633–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oficjalska-Pham D., Harismendy O., Smagowicz W. J., Gonzalez, de Peredo A., Boguta M., Sentenac A., Lefebvre O. (2006) General repression of RNA polymerase III transcription is triggered by protein phosphatase Type 2A-mediated dephosphorylation of Maf1. Mol. Cell 22, 623–632 [DOI] [PubMed] [Google Scholar]
- 33. Soulard A., Cremonesi A., Moes S., Schütz F., Jeno P., Hall M. N. (2010) The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol. Biol. Cell 21, 3475–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yorimitsu T., Zaman S., Broach J. R., Klionsky D. J. (2007) Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 4180–4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mok J., Kim P. M., Lam H. Y. K., Piccirillo S., Zhou X., Jeschke G. R., Sheridan D. L., Parker S. A., Desai V., Jwa M., Cameroni E., Niu H., Good M., Remenyi A., Ma J. L. N., Sheu Y. J., Sassi H. E., Sopko R., Chan C. S. M., De Virgilio C., Hollingsworth N. M., Lim W. A., Stern D. F., Stillman B., Andrews B. J., Gerstein M. B., Snyder M., Turk B. E. (2010) Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 3, ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]