Abstract
Breast tumors are the second major cause of cancer-related death in women worldwide. These tumors are aggressive, leading to metastatic cancers that are heterogeneous in nature, with numerous subtypes. The basal-like tumor subtype invariably shows unfavorable prognosis and is often characterized by the lack of estrogen, progesterone and HER2 receptors. These cancer types do not respond to the current targeted therapies. Therefore, the need for the discovery of novel diagnostic markers/therapeutic targets is of paramount importance. Immortalization of breast tumor cells leading to advanced stage cancer is one of the pivotal steps in breast cancer and telomeres/telomerase play a critical role in this process. Using single telomere length analysis, cell lines with a basal-like phenotype encompassing immortalized/non-tumorigenic MCF10A and invasive/metastatic MCF10CA1 along with the MCF-7 cell line were examined for the presence of a unique class of telomere t-stumps. Telomerase activity, protein levels of telomerase and bulk telomere lengths were assessed in the above-mentioned cell lines. This is the first study describing the existence of a distinct class of extremely short telomeres termed ‘t-stumps’ in breast cancer cell lines. The cell lines MCF10A and MCF10CA1 showed distinct telomeric bands in the molecular size range of 100–1,000 bp, whereas the MCF-7 cell line showed very low levels of t-stumps. Of note is that only the highly invasive/metastatic cancer cell line MCF10CA1 exhibited an abundance of a cluster of t-stumps with a size distribution range of 500–700 bp. These unique t-stumps observed in the advanced breast cancer cell line may serve as a novel diagnostic marker and also form a key molecular target for novel anticancer therapy.
Keywords: telomere t-stumps, isogenic breast cancer cell lines, immortalized cell line, invasive/metastatic cell line, telomerase reverse transcriptase, cluster of t-stumps
Introduction
Unlimited proliferation potential is one of the hallmarks of cancer cells. Breast cancer is one of the major causes of morbidity and mortality in women worldwide, with millions diagnosed annually. Although great strides have been made in understanding and treating breast cancer, there exists a need for novel target identification and new therapeutic strategies to combat the disease (1). Breast cancer cells express a spectrum of molecular motifs encompassing various subtypes. Significant advancement has occurred in breast cancer treatment due to the introduction of hormonal therapy as well as HER2-directed immunotherapy (1). However, microarray analysis has led to the discovery of breast cancer types, including ER− (estrogen receptor-negative), PR− (progesterone receptor-negative) and HER2− (human epidermal growth factor receptor-negative) that resemble the basal-like subtype. Due to the refractory nature of this subtype of breast cancer to the presently available therapies, efforts have been intensified in the determination of novel diagnostic markers/therapeutic targets.
The transformation of a normal epithelial cell to a cancer cell is a multi-step process (2), involving pathways that harbor mutations in p53, the retinoblastoma gene pRb, BRCA1 (3,4) and changes in the Ras signaling pathway. Apart from oncogene activation and alteration in the tumor suppressor genes, the activation of a unique ribonucleoprotein termed telomerase (5), plays a pivotal role in the process of immortalization and is a key regulator of cancer cell survival. Its function is to restore telomeric sequences which are unique tandem repeats capping chromosomal termini (6). The in vitro replicative lifespan of normal cells is limited to a definite number of cell divisions due to the end replication problem, resulting in the loss of telomeric sequences (7). However, the activation of telomerase to restore the telomeric sequences has been reported in a wide array of tumors and over 80% of tumors possess telomerase activity (8). Apart from its well-known function of extending telomeres, telomerase is also reported to have a role in protecting them (9). Functionally, different forms of telomerase with processive property, leading to long telomeric sequences, and non-processive telomerase, generating short sequences, have been reported in acute myelogenous leukemia (AML) patients. Moreover, mutations in telomerase reverse transcriptase (hTERT) leading to reduced enzyme activity are reported to predispose to AML (10,11). Xu and Blackburn (12) discovered a distinct class of extremely short telomeres termed t-stumps. T-stumps are known to accumulate in telomerase-containing cells that lack checkpoint pathways involving p53 and/or pRb. Therefore, telomerase may have another role in protecting t-stumps and preventing their loss. Notably, the loss of a single telomere is known to lead to the activation of DNA damage response pathways (13), causing cell senescence/cell death. This suggests that a potent inhibitor of telomerase/telomere maintenance machinery could potentially cause loss of protection of t-stumps, resulting in cell death.
This study therefore aimed to identify the presence of unique t-stumps in a panel of breast cancer cell lines. Isogenic breast cancer cell lines with the immortalized MCF10A and the highly advanced invasive/metastatic cell line MCF10CA1 were selected. Furthermore, recent studies showed that the MCF10A-derived series of cell lines have the ability to acquire a basal-like phenotype (14). Utilizing the advantages offered by single telomere length analysis (STELA) in identifying very short telomeres as described by Xu and Blackburn (12), the study focused on the presence of distinct t-stumps in these basal-like breast cancer cell lines. Moreover, the highly advanced, invasive/metastatic breast cancer cell line, MCF10CA1, exhibited a characteristic abundant cluster of telomere t-stumps. These clusters were not observed in the immortalized MCF10A cell line. The t-stumps therefore form novel diagnostic markers as well as key therapeutic targets for chemotherapeutically recalcitrant breast cancer types.
Materials and methods
Cell lines
MCF10A and MCF10CA1 cell lines were obtained from Dr Fred R. Miller of the Barbara Ann Karmanos Cancer Institute, Detroit, MI, USA. The cell lines were grown in Dulbecco’s modified Eagle’s medium/F12 (1:1) with 10% fetal bovine serum, epidermal growth factor (2.5 ng/ml), insulin (10 ng/ml), cholera toxin (100 ng/ml) and hydrocortisone (0.5 μg/ml). The MCF-7 cell line was grown in Dulbecco’s modified Eagle’s medium alone containing 10% fetal bovine serum and 1% penicillin/streptomycin.
Determination of telomerase activity
Amounts of cellular proteins extracted from cells were estimated by Quick Start Bradford dye reagent protocol (Bio-Rad). Telomerase activity was monitored according to the TRAPeze telomerase detection protocol (Roche).
Extraction of genomic DNA and terminal restriction fragment (TRF) analysis
Total genomic DNA was extracted using the REDExtract-N-Amp Tissue PCR kit (Sigma). DNA amounts were determined using Smartspec (Bio-Rad). Equal amounts of DNA (20 μg) were then digested with restriction enzymes Rsa1 and Hinf1, according to the manufacturer's protocol (Roche). The digested DNA was separated on a 0.8% agarose gel, blotted onto nylon membrane and hybridized to telomeric sequences. The probe was generated by random labeling in the presence of α-P32 dCTP. The hybridized sequences were visualized by phosphorimaging (Molecular Dynamics).
Western blotting
Cells were harvested and washed with 1X PBS and suspended in RIPA lysis buffer containing protease inhibitors. The cells were then incubated at 4°C for 30 min and then sonicated prior to centrifugation at 13,500 rpm for 30 min. The samples were subjected to electrophoresis using a 10% SDS-PAGE and transferred to a PVDF membrane. The membrane was probed with a polyclonal telomerase protein (hTERT) antibody (Abcam) and then to an anti-β-actin polyclonal antibody to normalize for the levels of protein loading. The binding of the proteins to the specific antibodies was detected using a chemiluminescence protocol (Pierce) and bands were visualized by exposure to X-ray film.
Identification of t-stumps and primers
STELA was performed essentially as described by Xu and Blackburn (12). Genomic DNA was digested with EcoR1. Twenty ng of this digested DNA was incubated in a 25-μl ligation reaction along with the telorette3 linker and 1–2 units of T4 DNA ligase (Promega), and then incubated at 35°C overnight. The ligated DNA was diluted to 2 ng/μl for further PCR. Each PCR was performed using Master Mix (Qiagen) and Teltail, with XpYpE2 as primers. The conditions used for PCR and further visualization of the bands were as described by Xu and Blackburn (12). Primers used in STELA were: Telorette3: 5′-TGCTCCGTGCATCTGGCATCCCTAACC-3′; Teltail: 5′-TGCTCCGTGCATCTGGCATC-3′; XpYpE2: 5′-GGTTGT CTCAGGGTCCTAGTG-3′ and XpYpB2: 5′-TCTGAAAGT GGACCAATCAG-3′.
Results
Determination of telomerase activity in MCF10A, MCF10CA1 and MCF-7 cell lines
We first monitored the levels of telomerase activity in the cell-free extracts of the breast cancer cell lines. Telomerase activity was determined using the TRAPeze protocol, and the activity is represented as relative telomerase units (RTU, Fig. 1A). In this determination, the highest activity observed in the immortalized cell line, MCF10A, was considered as 100%. Activity in the other lines is presented as a percentage relative to that of MCF10A (the values were obtained from three independent experiments carried out in triplicate). Robust telomerase activity was observed in the breast cancer cell lines studied. As expected, due to the requirement of RNA for the telomerase activity, the activity was abolished by pretreatment of the extract with RNase A (Fig. 1B).
Figure 1.
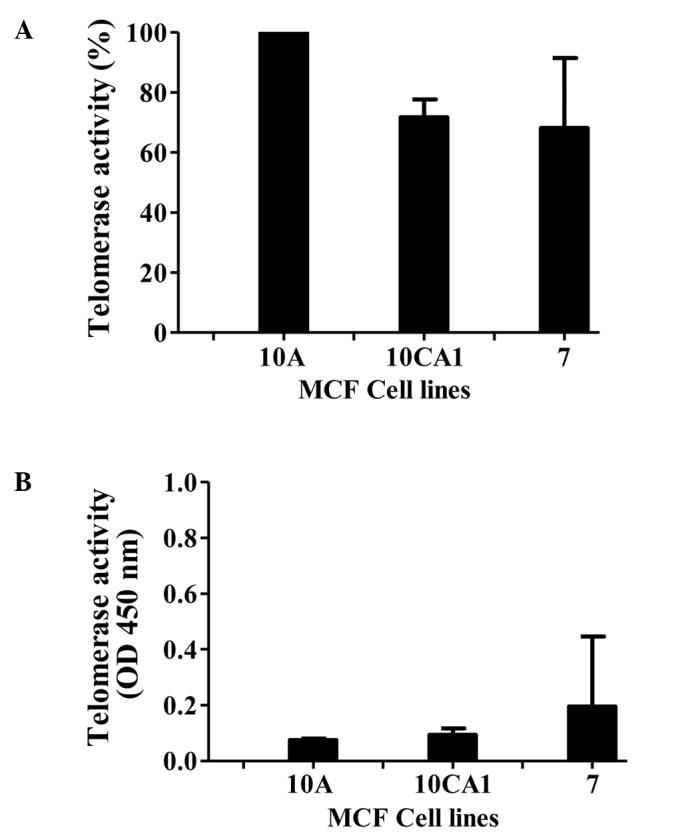
(A) Determination of telomerase activity in breast cancer cell lines MCF10A, MCF10CA1 and MCF-7. The values represented are triplicates of three independent experiments (± SD) considering MCF10A telomerase activity as 100%. (B) Telomerase activity in the presence of RNase A (values are from three independent experiments in triplicates ± SD).
Assessment of the telomerase protein and terminal telomere lengths
The amount of hTERT in the cell lines was visualized by semi-quantitative Western blotting using antibodies against hTERT. Telomerase migrated as an ~127 kDa band. When normalized to β-actin (~42 kDa band), the MCF-7 cell line exhibited low levels of hTERT when compared to hTERT levels in the advanced (invasive/metastatic) breast cancer cell line, MCF10CA1 (Fig. 2A). This assessment was made based on data from three independent experiments.
Figure 2.
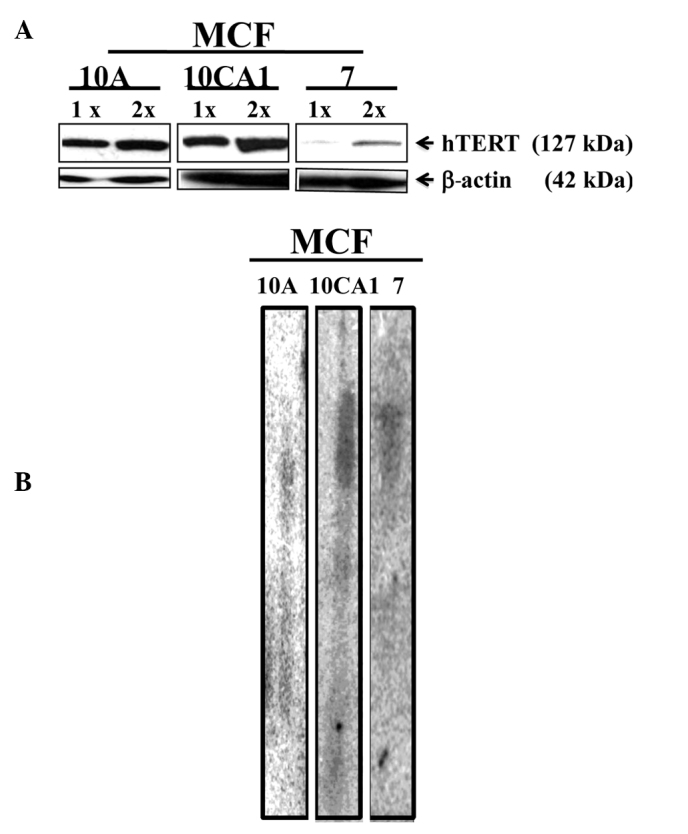
(A) Levels of telomerase reverse transcriptase (hTERT) in the breast cancer cell lines studied. Two different concentrations of protein were used in the analysis; 1x, low and 2x, high, as observed by the levels of β-actin (~42 kDa). Arrows indicate specific bands for telomerase (~127 kDa). (B) Terminal restriction fragment (TRF) analysis. Bulk telomere lengths were measured by Southern hybridization using equal amounts of genomic DNA (20 μg). The exposure of each lane varied, so as to visualize the long, bulk telomeres in each of the cell lines.
The lengths of the terminal bulk telomeres were measured by terminal restriction fragment (TRF) analysis after restriction enzyme digestion using the enzymes Rsa1 and Hinf1. When measurements were conducted using the TRF analysis, bulk telomere lengths were identified in the cell lines studied (Fig. 2B).
Single telomere length analysis
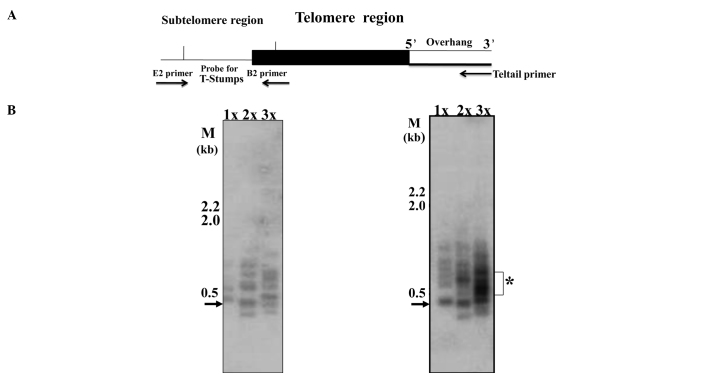
For a better resolution of telomere lengths and to determine the distribution of single extremely short telomeres, we performed STELA as previously described (12,15). This methodology involves the ligation of the telorette3 linker to the 5′ overhang which exists at the end of the ‘X’ chromosome; a schematic representation of the assay is shown in Fig. 3A. The PCR product was identified by Southern blotting and hybridizing with a 500-bp telomere-specific probe (this probe also encompasses ~400 bp of subtelomeric region). These analyses showed the existence of extremely short telomeres in the two basal-like breast carcinoma lines, MCF10A and MCF10CA1, with a molecular size range of 100–1,000 bp (Fig. 3B and C). The 500-bp (0.5 kb shown by arrow) band actually represents t-stumps only 100 bp in size due to the obligatory amplification of 400-bp subtelomeric region (Fig. 3A).
Figure 3.
Identification of extremely short telomeres, t-stumps in the panel of breast cancer cells lines. T-stumps were identified using STELA. (A) Schematic representation of the analysis (12,15). Varying concentrations of EcoR1-digested genomic DNA 1× (0.66 ng/μl), 2× (1 ng/μl), 3× (2 ng/μl) were PCR amplified, resolved on gels, blotted and hybridized as described in the Materials and methods section. (B) MCF10A; (C) MCF10CA1; *cluster of t-stumps.
Single telomere length analysis in MCF-7 cell line and in different passage cells of MCF10A and MCF10CA1
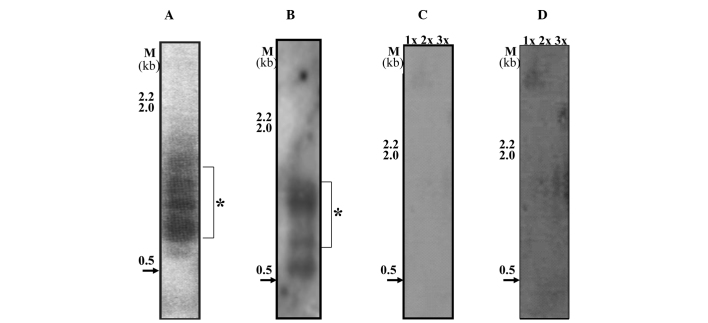
In contrast to the MCF10A and MCF10CA1 cell lines, MCF-7 showed a low abundance of t-stumps under the same exposure and conditions (Fig. 4C) with minor bands being visible after long exposure (Fig. 4D). On the other hand, the highly invasive/metastatic breast cancer cell line, MCF10CA1 (Fig. 3C), exhibited an abundance of a cluster of t-stumps with a range of 500–700 bp in three independent batches of cells. In an earlier passage of cells to the one shown in Fig. 3C, the amount of 100 bp t-stumps were minimal, but the 500–700 bp cluster was abundant (indicated by the star in Fig. 4A). However, in the batches of cells that were further subcultured, t-stumps with a size range of 100 bp were observed along with the distinct and abundant cluster of 500–700 bp (Fig. 4B, the star indicates the abundant cluster). This consistent presence of the abundant cluster of distinct t-stumps in the MCF10CA1 (invasive/metastatic) cell line, leads to the conclusion that it may form a key molecular target.
Figure 4.
(A) MCF10CA1− identification of t-stumps in early passage cells when compared to the cells analyzed in Fig 3C. (B) MCF10CA1− t-stumps identified in the later passage of cells when compared to the cells analyzed in Fig 3C. *The position of the distinct cluster. (C) MCF-7; (D) MCF-7 long exposure of phosphorimaging. HindIII-digested LDNA was used as a marker (M).
Discussion
Breast cancers are heterogeneous and harbor various subtypes, leading to poor clinical outcomes (16). Moreover, certain subtypes do not respond to currently available treatment regimens. The breast cancer cell lines (MCF10A and MCF10CA1) studied have the same genetic background and therefore are isogenic. These cell lines exhibit characteristics of basal-like breast cancer types (17). Xu and Blackburn (12), reported on the existence of t-stumps in human cancer cells. These authors also suggested that telomerase plays a role in the protection of this distinct class of extremely short telomeres.
This study, for the first time, reports on the existence of the unique t-stumps in breast cancer cell lines. These cell lines exhibited robust telomerase activity and high levels of hTERT, with the exception of the MCF-7 breast cancer cell line which had low levels of hTERT, thereby correlating with a low abundance of t-stumps. This observation is in agreement with studies carried out by Xu and Blackburn (12) who showed that the overexpression of hTERT leads to an increase in the amount of t-stumps. However, the differences we observed between the MCF-7 and the MCF10A and MCF10CA1 cell lines may also relate to cell type specificity. This was an observation made by Xu and Blackburn (12) with the HeLa cell line which exhibited very low levels of t-stumps.
Of note is that STELA reveals dynamic changes at single telomeres that occur at the chromosomal termini. The presence of a distinct cluster of telomere t-stumps with a size range of 500–700 bp, which we identified in the highly advanced cell line MCF10CA1, may be due to the elongation of the shortest telomeres by telomerase. This observation was recently made in a study using human (18) and budding yeast cells, in which early replication of short telomeres was noted (19). We cannot disregard the fact that the cluster of t-stumps observed in the MCF10CA1 line may also be due to the recombination of the shortest telomeres as reported recently by Morrish and Greider (20).
The cell lines selected for our study have the same genetic background. Therefore, it should be noted that during the progression of tumorigenesis from an immortalized state, i.e., MCF10A, to the highly advanced stage, which is the invasive/metastatic state represented by MCF10CA1, we observed dynamic changes at the single telomere level. Recently, it was reported that the elongation of short telomeres can occur in small size increments in human cells (18). The extension and accumulation of the short telomeres which occurred in small increments from 100 bp to 500–700 bp in the MCF10CA1 cell line were noted in the present study. Notably, this increment in short telomere elongation resulting in a distinct cluster was observed in the invasive/metastatic cell line and was distinct from the immortalized cell line (MCF10A) telomere pattern. Therefore, these unique clusters of t-stumps form signature markers and in particular molecular targets to treat advanced breast cancer types. It has been reported that single telomere loss results in cell senescence (13). Therefore, these very short telomeres, or t-stumps, may form key targets for chemotherapeutic intervention especially for the class of breast cancer types which do not respond to the currently available therapeutic regimens.
Acknowledgements
This study was partially supported by grants from NIH (NCI R01CA095317) and from the University Research Committee of the Emory University to H.C.J. We thank Rohith B. Pai for help during the preparation of the manuscript.
References
- 1.Chu D, Lu J. Novel therapies in breast cancer: what is new from ASCO 2008. J Hematol Oncol. 2008;1:16. doi: 10.1186/1756-8722-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pai SB, Steele VE, Nettesheim P. Neoplastic transformation of primary tracheal epithelial cell cultures. Carcinogenesis. 1983;4:369–374. doi: 10.1093/carcin/4.4.369. [DOI] [PubMed] [Google Scholar]
- 3.Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, Popescu NC, Hahn WC, Weinberg RA. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosen EM, Fan S, Isaacs C. BRCA1 in hormonal carcinogenesis: basic and clinical research. Endocr Relat Cancer. 2005;12:533–548. doi: 10.1677/erc.1.00972. [DOI] [PubMed] [Google Scholar]
- 5.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 6.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 7.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 8.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 9.Zhu J, Wang H, Bishop JM, Blackburn EH. Telomerase extends the lifespan of virus-transformed human cells without net telomere lengthening. Proc Natl Acad Sci USA. 1999;96:3723–3728. doi: 10.1073/pnas.96.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pai RB, Pai SB, Kukhanova M, Dutschman GE, Guo X, Cheng YC. Telomerase from human leukemia cells: properties and its interaction with deoxynucleoside analogues. Cancer Res. 1998;58:1909–1913. [PubMed] [Google Scholar]
- 11.Calado RT, Regal JA, Hills M, Yewdel WT, Dalmazoo LF, Zago MF, Zago MA, Lansdorp PM, Hogge D, Chanock SJ, Estey EJ, Falcao RP, Young NS. Constitutional hypomorphic telomerase mutations in patients with acute myeloid leukemia. Proc Natl Acad Sci USA. 2009;106:1187–1192. doi: 10.1073/pnas.0807057106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu L, Blackburn EH. Human cancer cells harbor T-stumps, a distinct class of extremely short telomeres. Mol Cell. 2007;28:315–327. doi: 10.1016/j.molcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 14.Sarrio D, Rodriguez-Pimilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 15.Baird DM, Rowson J, Wynford-Thomas D, Kipling D. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat Genet. 2003;33:203–207. doi: 10.1038/ng1084. [DOI] [PubMed] [Google Scholar]
- 16.Liu ZB, Liu GY, Yang WT, Di GH, Lu JS, Shen KW, Shen ZZ, Shao ZM, Wu J. Triple-negative breast cancer types exhibit a distinct poor clinical characteristic in lymph node-negative Chinese patients. Oncol Rep. 2008;20:987–994. [PubMed] [Google Scholar]
- 17.Hu M, Yao J, Carroll DK, Weremowicz S, Chen H, Carrasco D, Richardson A, Violette S, Nikolskaya T, Nikolsky Y, Bauerlein EL, Hahn WC, Gelman RS, Allred C, Bissell MJ, Schnitt S, Polyak K. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13:394–406. doi: 10.1016/j.ccr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Britt-Compton B, Capper R, Rowson J, Baird DM. Short telomeres are preferentially elongated by telomerase in human cells. FEBS Lett. 2009;583:3076–3080. doi: 10.1016/j.febslet.2009.08.029. [DOI] [PubMed] [Google Scholar]
- 19.Bianchi A, Shore D. Early replication of short telomeres in budding yeast. Cell. 2007;128:1051–1062. doi: 10.1016/j.cell.2007.01.041. [DOI] [PubMed] [Google Scholar]
- 20.Morrish TA, Greider CW. Short telomeres initiate telomere recombination in primary and tumor cells. PLoS Genet. 2009;5:e1000357. doi: 10.1371/journal.pgen.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]