Background: Engineered meganucleases are one of most promising biological reagents for gene modification therapy.
Results: CpG methylation affects engineered meganuclease activity and DNA binding affinity in a position-dependent manner.
Conclusion: The activity and sequence specificity of engineered meganucleases are not the only parameters to consider for successful gene edition.
Significance: Considering epigenetic factors is crucial for designing highly active engineered meganucleases for gene editing purposes.
Keywords: Chromatin Modification, DNA-binding Protein, DNA Methylation, DNA Recombination, DNA Repair, Gene Therapy, CpG Methylation, Genome Engineering, Epigenetic, Meganuclease
Abstract
In this study, we asked whether CpG methylation could influence the DNA binding affinity and activity of meganucleases used for genome engineering applications. A combination of biochemical and structural approaches enabled us to demonstrate that CpG methylation decreases I-CreI DNA binding affinity and inhibits its endonuclease activity in vitro. This inhibition depends on the position of the methylated cytosine within the DNA target and was almost total when it is located inside the central tetrabase. Crystal structures of I-CreI bound to methylated cognate target DNA suggested a molecular basis for such inhibition, although the precise mechanism still has to be specified. Finally, we demonstrated that the efficacy of engineered meganucleases can be diminished by CpG methylation of the targeted endogenous site, and we proposed a rational design of the meganuclease DNA binding domain to alleviate such an effect. We conclude that although activity and sequence specificity of engineered meganucleases are crucial parameters, target DNA epigenetic modifications need to be considered for successful gene editions.
Introduction
Over the last decade, the field of genome engineering has been revolutionized, in part due to the discovery and optimization of biological reagents allowing precise and efficient gene editing at specific genomic loci (1). Notable among these reagents are zinc finger nucleases (2), the recently described TAL effector nucleases (3–5), and meganucleases (6, 7). Meganucleases are rare-cutting endonucleases known to induce homologous recombination and nonhomologous end-joining events by catalyzing sequence-specific double strand breaks of DNA (8). Extensive engineering of the DNA binding interface of the I-CreI meganuclease from Chlamydomonas reinhardtii led to the creation of highly active variants with modified DNA specificity (9–12). These meganucleases were successfully used for targeted insertion of exogenous DNA sequences in many human genes (13–15), for restoring the open reading frame of the dmd gene involved in the Duchenne myopathy in human cells (16), and for editing the LG1 promoter in plant cells (17).
Their high specificity, along with their significant in vivo activity, set meganucleases apart as one of the most promising reagents for genome engineering applications (7). Nevertheless, increasing the efficacy of engineered nucleases in a physiological context is still a matter of intense investigation as the factors affecting this parameter are still largely unknown. Critical to the efficiency of natural and engineered nucleases is their ability to access and cleave their endogenous target sites (15). Numerous factors may hinder cleavage, including DNA packaging into chromatin, the position of nucleosomal proteins with respect to the target site, and chemical DNA modifications such as methylation. In higher eukaryotes, DNA methylation is involved in the regulation of gene expression and predominantly occurs at the C5 position of cytosine found in the dinucleotide sequence CpG6 (18). CpG methylation may represent an important epigenetic drawback for genome engineering applications and thus understanding its influence on natural and engineered nucleases activity is of major importance.
There is a paucity of work regarding natural or engineered meganuclease activity and CpG methylation. Previous studies exploring meganuclease-DNA interactions reported only the influence of chemically induced guanine and adenine modifications (19–23). Although useful in probing studies, such work did not address the specific influence of relevant in vivo methylation in mammalian systems. Thus, to date, the effect of CpG methylation on meganuclease activity is still unclear.
In this work, we studied the effect of CpG methylation on the activity of meganucleases using I-CreI from C. reinhardtii. A combination of biochemical and structural approaches enabled us to demonstrate that CpG methylation does impair I-CreI DNA cleavage activity as well as decrease its DNA binding affinity in vitro. This inhibition depends on the position of the methylated cytosine within the DNA target and is almost total when it is located inside the central tetrabase. Crystal structures of I-CreI bound to methylated cognate target DNA suggest a molecular basis for such inhibition, although the precise mechanism still has to be specified. Finally, we demonstrate that the efficacy of engineered meganucleases can be diminished by CpG methylation of the targeted endogenous site, and we propose a rational design of the meganuclease DNA binding domain to alleviate such an effect.
MATERIALS AND METHODS
Cloning, Overexpression, and Purification of Recombinant Meganucleases
The coding sequence for I-CreI wild type and ADCY9 recombinant meganucleases was subcloned into the kanamycin-resistant pET-24 vector MCS located upstream from a His6 tag coding sequence. Recombinant proteins were overexpressed and purified according to the protocol described in Ref. 13. See the supplemental material for additional information.
In Vitro Binding Assay
6-Carboxyfluorescein (FAM)-labeled DNA single strand oligonucleotides corresponding to I-CreI and ADCY9m cognate targets (C1234 and ADCY9t respectively) were synthesized and HPLC-purified by Eurogentec (supplemental Table 1). To prepare C1234 duplex corresponding to the I-CreI double strand DNA wild type target, C1234 forward (“a” strand) labeled with FAM on its 5′ end was mixed with 1 eq of C1234_reverse (“b” strand) in 100 mm Tris-HCl, 50 mm EDTA, 150 mm NaCl, pH 8. The mixture was heated to 95 °C for 2 min and then cooled down to 25 °C over 1 h. C1234 duplex was eventually purified by anion exchange chromatography using a miniQ PE column (GE healthcare) pre-equilibrated with buffer A (20 mm Tris-HCl, pH 7.4). Single-stranded oligonucleotides and other contaminants were first discarded using a 0–360 mm NaCl step gradient, and elution of pure C1234 duplex was performed with a 360–1000 mm NaCl linear gradient (5 column volumes). C1234 duplex and FAM final concentrations were assessed by spectrophotometry using their respective extinction coefficients ϵ260 nm = 62,900 m−1 cm−1 and ϵ495 nm = 83,000 m−1 cm−1. As expected, a ratio of [C1234 duplex]/[FAM] ∼1 was obtained. This procedure was used to prepare all other DNA targets used in our experiments. Oligonucleotide sequences are documented supplemental Table 1. To investigate the binding of I-CreI to the C1234 duplex, 25 nm of C1234 duplex was incubated with increasing concentrations of I-CreI (from 0 to 400 nm) in binding buffer (10 mm Tris-HCl, 400 mm NaCl, 10 mm CaCl2, 10 mm DTT, pH 8) at 25 °C. After 30 min of incubation, the fluorescence anisotropy (FA) of the mixture was recorded with a Pherastar Plus (BMG Labtech) operating in fluorescence polarization end point mode with excitation and emission wavelengths set to 495 and 520 nm, respectively. Apparent dissociation constants were determined by fitting raw data by Equation 1.
 |
where FAx is the FA value obtained at a given meganuclease concentration; FA0 and FA∞, the FA values obtained in the absence of meganuclease or in the presence of saturating concentrations of meganuclease, respectively, and Kd, the dissociation constant of the equilibrium studied. The fraction of active recombinant protein present in our preparation was determined to be 18.5% according to the protocol described by Arosio et al. (24). Variations of FA were represented as a function of active I-CreI concentration.
In Vitro Cleavage Assay
To investigate the influence of C1234 methylation on the nuclease activity of I-CreI, in vitro single turnover cleavage assays were performed with either unmethylated or methylated C1234 duplexes (unmethylated C1234, C1234_Me full, C1234_Me −6a/−5b, C1234_Me −3a/−2b, C1234_Me −3a, and C1234_Me −2b). A constant amount of C1234 duplex (50 nm) was incubated with an excess of I-CreI (1.5 μm, final concentration) in the reaction buffer (10 mm Tris-HCl, 150 mm NaCl, pH 8) at 37 °C. Cleavage reaction was triggered by the addition of MgCl2 and then quenched after different time lengths by the addition of the stop buffer (4.5% glycerol, 95 mm EDTA, 1.5% (w/v) SDS, 1.5 mg/ml proteinase K, and 0.048% (w/v) bromphenol blue, final concentrations). This was followed by a 1-h incubation at 37 °C to digest I-CreI and release free DNA molecules. Cleaved and uncleaved DNA products were separated by PAGE using a TGX Any kDa precast gel (Bio-Rad), stained with SYBR Green and then quantified using Quantity One software (Bio-Rad). The same procedure was used to investigate the influence of DNA methylation on the nuclease activity of ADCY9m.
Crystallization
The 24-bp-long target methylated DNA that almost inhibited the I-CreI catalytic activity was purchased from Proligo and consisted of two strands containing the following sequences: 5′-TCAAAACGTCGTGAGACAGTTTGG-3′ (C1234_forward) and 5′-CCAAACTGTCTCAC5mGACGTTTTGA-3′ (C1234_Me+2b_reverse), forming after incubation a 24-bp blunt-end duplex. The protein-DNA complex was formed in the presence of 2 mm noncatalytic Ca2+ or 2 mm catalytic MgCl2, by pre-warming the meganuclease and the oligonucleotide sample at 37 °C and mixing them in a 0.75:1 molar ratio (DNA/protein). The mixture was incubated for 50 min at this temperature and then spun down for 5 min to remove insoluble material. The final concentration of protein in the DNA-protein complex solution was 4 mg/ml. I-CreI:C1234_Me+2b-Ca2+ crystals were grown using the hanging-drop method at 290 K, in 2-μl droplets formed by 1 μl of the DNA-protein complex (containing 2 mm Ca2+) and 1 μl of precipitant solution consisting of 20% (v/v) PEG300, 0.2 m ammonium sulfate, 10% (v/v) glycerol in 0.1 m phosphate-citrate, pH 4.2. I-CreI:C1234_Me+2b-Mg2+ crystals were also grown from 2-μl droplets formed by 1 μl of the DNA-protein complex (containing 2 mm Mg2+) and 1 μl of precipitant solution consisting of 15% (w/v) PEG4000, 0.14 m sodium acetate, 20% (v/v) ethylene glycol, 0.35% (w/v) methanol in 0.1 m sodium citrate, pH 5.5. Crystals were directly flash-frozen in liquid nitrogen.
Data Collection, Structure Solution, Model Building, and Refinement
All data were collected at 100 K, using synchrotron radiation at the PX beam line (SLS, Villigen, Switzerland). The diffraction pattern was recorded on 6 m Pilatus detector. Data processing and scaling were accomplished with XDS (25) or MOSFLM (26) (see Table 2). The structures were solved by molecular replacement as implemented in the programs MOLREP (27) or PHASER (28). The search models was based on a polyalanine backbone derived from the PDB codes 1G9Y (I-CreI-DNA-Ca2+) and 1G9Z (I-CreI-DNA-Mg2+), respectively. The structures were then subjected to iterative cycles of model building and refinement with O (29), Coot (30), and PHENIX (31). The identification and analysis of the protein-DNA hydrogen bonds and van der Waals contacts was done with the Protein Interfaces, Surfaces, and Assemblies service PISA at the European Bioinformatics Institute.
TABLE 2.
Crystallographic statistics and refinement
| I-CreI:C1234_Me+2b-Ca2+(PDB code 4AQU) | I-CreI:C1234_Me+2b-Mg2+(PDB code 4AQX) | |
|---|---|---|
| Data collection | ||
| Space group | P21 21 2 | P21 21 2 |
| Cell dimensions | ||
| a, b, c | 45.35; 71.58; 172.33 Å | 45.84; 71.40; 179.05 Å |
| α, β, γ | 90, 90, 90° | 90, 90, 90° |
| Resolution | 45.35 to 2.30 Å (2.42 to 2.30 Å) | 45.84 to 2.20 Å (2.32 to 2.20 Å) |
| Rsym or Rmerge | 0.12 (0.37) | 0.12 (0.38) |
| I/σI | 4.0 (1.9) | 2.6 (1.9) |
| Completeness | 99.9% (99.8%) | 99.9% (99.9%) |
| Redundancy | 5.4 (5.1) | 4.4 (4.4) |
| Refinement | ||
| Resolution | 44.81 to 2.30 Å | 45.79 to 2.20 Å |
| No. of reflections | 25,711 | 30,701 |
| Rwork/Rfree | 0.20/0.25 | 0.20/0.24 |
| No. of atoms | ||
| Protein | 2471 | 2471 |
| Ions | 2 | 3 |
| Nucleic acids | 974 | 980 |
| Water | 102 | 135 |
| Root mean square deviations | ||
| Bond lengths | 0.010 Å | 0.010 Å |
| Bond angles | 1.227° | 1.279° |
Cells Culture and Transfections
Human 293H cells (Invitrogen) and hamster CHO-KI cells (ATCC) were cultured according to the protocol described in Refs. 13, 15.
5-Aza-2-deoxycytidine Treatment, Bisulfite Treatment, and DNA Sequencing
To investigate the influence of 5-aza-2-deoxycytidine (5-aza-dC) on the methylation status of genomic DNA and on meganuclease activity, 293H cells were pretreated every day and for 2 days with 0.2 or 1 μm 5-aza-dC before transfection, and this treatment was maintained up to 48 h post-transfection. To determine the level of DNA methylation, genomic DNA was extracted, treated by bisulfite according to the manufacturer's protocol (EZ DNA methylation-Gold Kit, Zymo Research), and then amplified by PCR using primers that were specific for the flanking regions of the bisulfite-treated meganuclease locus (supplemental Table 3). PCR amplicons were then analyzed by deep sequencing.
Monitoring Meganuclease Extrachromosomal Activity and Meganuclease-induced TM and HGT
Intrinsic meganuclease activity was measured in yeast and mammalian CHO-KI cells using an extrachromosomal target as reported previously (13, 15). Meganuclease-induced TM and HGT were monitored in 293H cells as described previously (13, 15). See the supplemental material for additional information.
RESULTS
Influence of CpG Methylation on the DNA Binding Affinity of I-CreI
To investigate the influence of DNA methylation on the binding affinity of I-CreI, dissociation constants (Kd) with unmethylated and methylated target DNA (C1234, Fig. 1) were determined in vitro. To prevent DNA cleavage, catalytic Mg2+ was substituted by the noncatalytic metal ion Ca2+ (8). Quantitative binding of I-CreI to C1234 was assayed using fluorescence anisotropy (FA) of FAM- labeled DNA. According to the Perrin equation, FA correlates to the volume and rotational time of the labeled DNA (32). Thus, upon binding to I-CreI, we expected a variation of FAM-labeled DNA FA that can be exploited to determine dissociation constants.
FIGURE 1.
Nucleotide sequence of C1234, the cognate target of I-CreI. The central tetrabase is displayed in gray; overhangs induced by I-CreI endonuclease activity are indicated by dashed lines, and base numbering and positions of methylated cytosines are indicated at the top and bottom of strands a and b, respectively.
For unmethylated C1234, we observed an increase in FA that leveled off at saturating concentration of I-CreI (Fig. 2A, open circles). This pattern was consistent with a tight binding equilibrium between I-CreI and C1234 with dissociation constant estimated to ≤2.5 nm on the basis of Equation 1 (see under “Material and Methods”). For fully methylated C1234, FA remained almost constant in the presence of up to 15-fold excess I-CreI with respect to DNA (Fig. 2A, closed squares, data shown up to 80 nm), with a binding isotherm similar to that obtained with a randomized 24-bp DNA duplex used as a negative control (Fig. 2A, closed triangles). As complex formation between I-CreI and methylated C1234 could not be accomplished under the experimental conditions used, a binding constant could not be accurately determined. Nevertheless, using Equation 1 (see “Material and Methods”), a base-line estimate of Kd ≥1000 nm was applied, resulting in a >400-fold difference in binding affinity between unmethylated and fully methylated C1234 DNA. Therefore, CpG methylation of C1234 strongly affected the I-CreI DNA binding affinity.
FIGURE 2.
Influence of CpG methylation on the DNA binding affinity of I-CreI in vitro; spectrofluorometric titration of FAM-labeled C1234 by I-CreI wild type. 25 nm unmethylated or methylated C1234 duplex was incubated with increasing concentrations of I-CreI (from 0 to 400 nm) in binding buffer (10 mm Tris-HCl, 400 mm NaCl, 10 mm CaCl2, 10 mm DTT, pH 8) at 25 °C. After 30 min of incubation, the FA of the mixture was recorded with a Pherastar Plus (BMG Labtech) operating in fluorescence polarization end point mode with excitation and emission wavelengths set to 495 and 520 nm, respectively. Normalized FA is plotted as a function of active I-CreI concentration. Apparent dissociation constants were determined by fitting raw data by the quadratic Equation 1. Left panel, A–C, titration curves obtained with unmethylated and different methylated forms of C1234. Right panel, schemes representing the different forms of C1234 used for titrations. Methyl moiety positions are indicated, and the central tetrabase is shown in gray.
To decipher how CpG methylation inhibited the I-CreI-DNA interaction, we used a systematic approach. We first asked whether this inhibition was strand-dependent and second position-dependent. To investigate the strand dependence of this inhibition, we first compared the affinity of I-CreI for different hemimethylated C1234 targets. DNA targets were synthesized wherein either the a (positions −6a and −3a) or b (positions +5b and +2b respectively) strands were methylated (Fig. 2B). FA assays demonstrated that the effects of 5mC modifications were strand-dependent (Fig. 2B and Table 1, compare C1234_Me-6a/-3a to C1234_Me+5b/+2b). Indeed, whereas full methylation of the C1234 a strand was found to have no effect, full methylation of the C1234 b strand decreased the DNA binding affinity of I-CreI as observed for fully methylated C1234 and the randomized control target. An absence of quantifiable results allowed only for estimating a base-line Kd ≥1000 nm (Table 1). Second, to further determine whether one or both 5mCs were responsible for this inhibition, we compared the affinities of I-CreI for C1234 methylated at either position +5b or +2b (Fig. 2C). Results indicated that methylation of either position decreased I-CreI binding affinity by a factor ≥10 (Table 1). Taken together, in vitro binding assays demonstrated that I-CreI-DNA interactions are impaired by 5mC modification of the two CpGs located on the C1234 b strand.
TABLE 1.
Thermodynamic and kinetic parameters of I-CreI meganuclease toward unmethylated and methylated C1234 DNA targets
Top, 25 nm of different methylated forms of fluorescent C1234 DNA targets was incubated with increasing amounts of I-CreI at 25 °C. The FA of the resulting mixture was determined and plotted as a function of I-CreI total concentration. Apparent dissociation constant (Kd) was determined by fitting raw data by the quadratic Equation 1 described under “Materials and Methods.” The dissociation constants obtained from the best fits are reported. Bottom, 50 nm unmethylated or different methylated forms of C1234 DNA targets was incubated with 1.5 μm I-CreI in the reaction buffer containing 50 mm MgCl2. The reaction was carried out at 37 °C and then quenched after different lengths of time. The substrate disappearance was monitored as a function of time and was fit with a mono-exponential model. The rate constants (kcat) obtained from the best fits are reported (bottom). Results obtained with a random target are included as a negative control. ND means not done.
Influence of CpG Methylation on the Endonuclease Activity of I-CreI
To assess the influence of 5mC modification on I-CreI DNA cleavage activity, in vitro single turnover kinetic assays were performed using methylated and unmethylated C1234 targets (Fig. 3). In these assays, the C1234 target was premixed with a large excess of recombinant meganuclease prior to the addition of MgCl2 to initiate catalysis. Reactions were quenched at various time points by addition of a stop buffer, and the resulting products were separated and quantified. Under these experimental conditions, substrate disappearance is a first-order process, and its rate constant corresponds to the turnover number (kcat) of I-CreI. Turnover number measurements should not be affected by affinity differences between methylated and unmethylated DNA because of the following: (i) I-CreI is in large excess with respect to C1234 (1500 and 25 nm, respectively), and (ii) reaction buffer contains low amounts of NaCl (150 mm). Thus, in these conditions, the totality of C1234 DNA target is bound by the meganuclease at the beginning of reaction as verified by FA experiments (data not shown). In addition, these measurements are not affected by a rate-limiting step of product release because protein-DNA complexes are artificially disrupted by proteinase K and SDS present in the stop buffer (23).
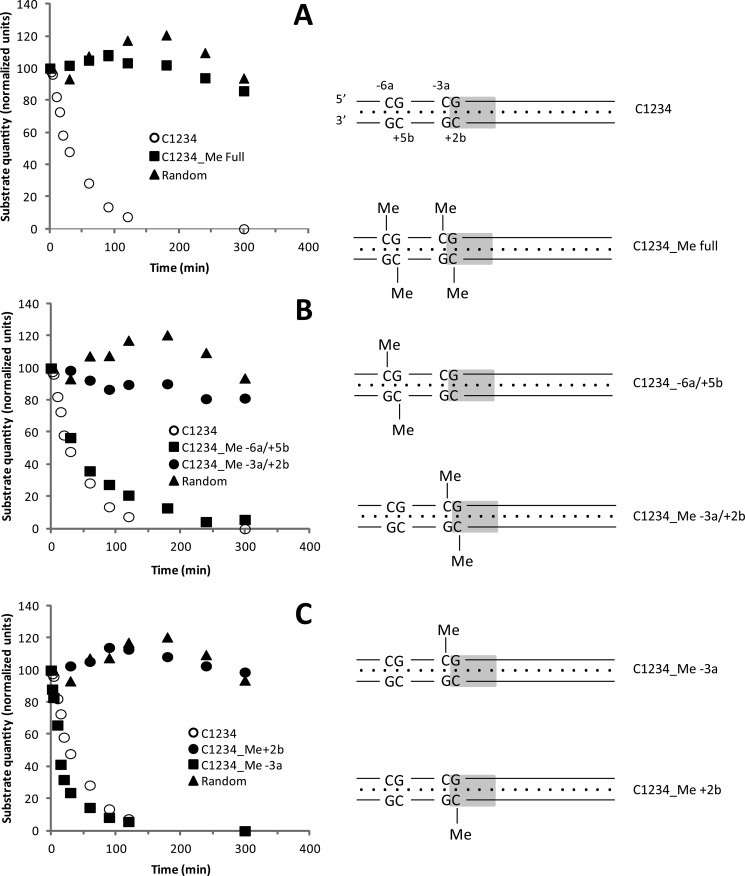
FIGURE 3.
Influence of CpG methylation on the nuclease activity of I-CreI in vitro. A constant amount of C1234 duplex (50 nm) was incubated with an excess of I-CreI (1.5 μm, final concentration) in the reaction buffer (10 mm Tris-HCl, 150 mm NaCl, pH 8) at 37 °C. Cleavage reaction was triggered by the addition of MgCl2 and then stopped after different time lengths by the addition of the stop buffer. This was followed by an hour of incubation at 37 °C to digest I-CreI and release free DNA molecules. Cleaved and uncleaved DNA products were separated by PAGE using a TGX Any kDa precast gel (Bio-Rad), stained with SYBR Green, and then quantified using Quantity One software (Bio-Rad). Disappearance of substrate (uncleaved DNA) is plotted as a function of time. Left panel, A–C, kinetic of DNA cleavage obtained with unmethylated and different methylated forms of C1234. Right panel, schemes representing the different forms of C1234 used for kinetics experiments. Methyl moiety positions are indicated, and the central tetrabase is shown in gray.
In the case of unmethylated C1234, the disappearance of C1234 substrate followed a mono-exponential trend characteristic of a first-order process (Fig. 3A). The rate constant of this process (kcat) was determined to be kcat = 0.025 min−1 (Table 1), consistent with previous reports (0.03 min−1 (33)). However, when fully methylated C1234 was assayed, a much slower process was observed with kcat estimated to be <0.0001 min−1, indicating that C1234 methylation significantly inhibited the nuclease activity of I-CreI (Fig. 3A and Table 1).
To investigate in more detail the cleavage inhibition by target methylation, we once again used a systematic approach. The effects of methylated CpGs located outside the central tetrabase (C1234 methylated in positions −6a/+5b, Fig. 1) were first compared with those located within the central tetrabase (C1234 methylated in positions −3a and +2b, Fig. 1). Results showed that methylation of CpGs located outside the central tetrabase did not affect I-CreI catalytic activity as no significant kcat difference could be detected when compared with the kcat obtained with unmethylated C1234 (Fig. 3B, filled squares and open circles, kcat = 0.02 ± 0.007 and 0.025 ± 0.002, respectively, and Table 1). Methylation of CpGs located within the central tetrabase, however, strongly affected the catalytic activity of I-CreI (Fig. 3B, filled circles, kcat = 0.0002 ± 0.0003, and Table 1). Next, the inhibitory role of each single 5mC modification within the central tetrabase (Fig. 1, position −3a or in position +2b) was investigated. Our results showed that although methylation of position −3a did not affect I-CreI catalytic activity, methylation of position +2b was almost completely inhibitory (Fig. 3C, filled squares and filled circles, kcat = 0.057 ± 0.0006 and <0.0001 respectively, and Table 1).
Crystal Structure of I-CreI:C1234_Me+2b
To gain more insight into the molecular mechanisms underlying I-CreI sensitivity to CpG methylation, we solved the structure of I-CreI bound to C1234_Me+2b DNA target in the presence of catalytic (Mg2+, PDB code 4AQX, Table 2) and noncatalytic ions (Ca2+, PDB code 4AQU, Table 2).
A comparison of the two structures with their corresponding unmethylated counterparts (1G9Z and 1G9Y respectively (34)) showed that despite its small size, the presence of the methyl group affects both the protein and DNA to alter their conformation. Fig. 4 shows the general perturbation surrounding the region of the methyl group leading to the shift of valine 73 by about 1 Å in both Mg2+- and Ca2+-containing structures. Interestingly, the methylated target was found cleaved in the presence of Mg2+ indicating that the methylation in this base does not significantly affect catalysis under our crystallization conditions.
FIGURE 4.
Comparison of I-CreI:C1234_Me+2b crystal structures obtained in the presence of Mg2+ (left panel, green, PDB code 4AQX) or Ca2+ (right panel, green, PDB code 4AQX) with their respective unmethylated structures obtained in the presence of Mg2+ (left panel, orange, PDB code 1G9Z) or Ca2+ (right panel, orange, PDB code 1G9Y).
Influence of CpG Methylation on Nuclease Activity of the Engineered Meganuclease ADCY9m in Vitro and in Vivo
In the previous sections, we showed that the nuclease activity of I-CreI meganuclease could be impaired by CpG methylation and especially when this modification occurs at the cytosine located in the central tetrabase of its cognate target (position +2b/−3a). Then we asked whether this property was also observed in vitro and in vivo for engineered meganucleases used for genome engineering applications. For that purpose, we chose an engineered meganuclease model named ADCY9m. ADCY9m, a single chained variant of I-CreI harboring different mutations in its DNA binding domain, was originally designed to cleave the 3′ end of the adenylate cyclase 9 gene (15). Its cognate target named ADCY9t is a 22-bp DNA sequence containing one single CpG dinucleotide located within the central tetrabase at positions −3a/+2b (gray box, supplemental Fig. 1). Bisulfite treatment of genomic DNA followed by specific amplification of adcy9 locus showed that more than 90% of the detected ADCY9t were methylated at these positions thus presaging a possible effect of DNA methylation on ADCY9m activity (supplemental Fig. 2).
To challenge this hypothesis, we first investigated the influence of CpG methylation on ADCY9m activity in vitro by performing single turnover experiments with unmethylated and fully methylated ADCY9t, according to the protocol described earlier for I-CreI. Our results showed that methylation of ADCY9t strongly inhibited the nuclease activity of ADCY9m as seen by the absence of cleavage of ADCY9_Me −3a/+2b target (supplemental Fig. 3, kcat < 0.0001, and supplemental Table 2).
We then evaluated ADCY9m activity in vivo and compared it with a meganuclease control named CAPNS1m (15). CAPNS1m was used as a control as it displayed similar nuclease activity to ADCY9m on an extrachromosomal unmethylated target (supplemental Fig. 4) and as its endogenous locus contains two unmethylated CpGs (data not shown). To assess the ability of ADCY9m and CAPNS1m to induce TM via nonhomologous end-joining events pathways (35), at their respective loci, 293H cells were transfected with meganuclease-expressing plasmids and cultivated for 2 days. Genomic DNA was extracted; adcy9 and capns1 loci were amplified by PCR, and amplicons were then analyzed by deep sequencing to determine the frequency of TM (consisting in 90% of deletions and 10% of insertions of multiple base pairs) induced by both meganucleases. In contrast to the similarity observed between ADCY9m and CAPNS1m activities toward their extrachromosomal unmethylated target (supplemental Fig. 4), the frequency of TM induced by ADCY9m was more than 10-fold smaller than the one induced by CAPNS1m (0.47 and 6% respectively, Fig. 5A). Thus, these results suggested that methylation of ADCY9t at positions −3a/+2b could inhibit ADCY9m activity in vivo.
FIGURE 5.
Influence of CpG methylation on the nuclease activity of ADCY9m activity in vivo. A, determination of TM frequencies induced by ADCY9m and CAPNS1m in 293H cells. 293H cells were transfected with 5 μg of meganuclease-expressing vector or empty vector. Two days post-transfection, genomic DNAs were extracted, and the loci of interest were amplified by PCR and then analyzed by deep sequencing. B, effect of 5-aza-dC treatment on the methylation pattern of ADCY9t. 293H cells were pretreated every day and for 2 days with or without 0.2 or 1 μm 5-aza-dC before transfection, and this treatment was maintained up to 48 h post-transfection. Genomic DNA was extracted, treated with bisulfite, and then amplified by PCR using primers specific for methylated and unmethylated ADCY9t (supplemental Table 3). PCR amplicons were then analyzed by deep sequencing, and the percentage of methylated ADCY9t was determined. C, effect of 5-aza-dC treatment on the frequencies of TM induced by ADCY9m and CAPNS1m. D, influence of 5-aza-dC on the ADCY9 level of expression observed by Western blot using anti-ADCY9m and anti-tubulin antibodies. Total proteins were extracted from 5-aza-dC-treated 293H cells and analyzed by Western blot (Wb) using anti I-CreI and β-tubulin antibodies. E and F, influence of ADCY9m level of expression on the frequency of TM in 293H cells. 293H cells were transfected with 0, 5, and 10 μg of plasmid encoding ADCY9m and cultured for 2 days. Cells were then recovered and split into two equivalent samples. E, first sample was used to determine the ADCY9m level of expression by Western blot analysis using anti-ADCY9m and anti-tubulin antibodies. F, second sample was used to determine the frequency of TM induced by ADCY9m according to the protocol described in B.
To confirm this hypothesis, we used 5-aza-dC as the demethylating agent and evaluated its effect on ADCY9m activity in vivo. We first verified the ability of 5-aza-dC to hypomethylate 293H cells at the adcy9 locus. 293H cells were cultivated in the presence of 0, 0.2, and 1 μm 5-aza-dC, and the methylation status of ADCY9t was evaluated by bisulfite treatment and deep sequencing. We found that demethylation of ADCY9t was promoted by 5-aza-dC in a dose-dependent manner, and we observed, at best, 40% of reduction of methylation in the presence of 1 μm 5-aza-dC (Fig. 5B). However, at this concentration, 5-aza-dC was found to be toxic for 293H cells. Thus, to avoid any experimental bias due to cell mortality, the experiments described in the following were all performed in the presence of 0.2 μm 5-aza-dC. This dose was sufficient to reduce the methylation status of ADCY9t by 20% while preserving cellular integrity. Interestingly, the presence of 0.2 μm of 5-aza-dC increased by about 3-fold the amount of targeted modification events at the adcy9 locus as shown by the difference of TM observed between untreated and treated cells (0.17 and 0.49%, respectively, Fig. 5C). This result strongly suggested that CpG methylation could inhibit ADCY9m activity in vivo. Interestingly, 5-aza-dC treatment was specific to the adcy9 locus. Indeed, this compound did not show any effect on the frequency of TM induced by the meganuclease control CAPNS1m targeting an unmethylated CpG-rich target (Fig. 5C).
DNA methyltransferase inhibitors such as 5-azacytidine and 5-aza-2-deoxycytidine have pleiotropic effects (36, 37). They are known to modify cell growth and gene expression. We thus verified that the enhancement of TM frequency described above was not due to an increase of ADCY9m expression and that it was specific to the adcy9 locus. The influence of 5-aza-dC on ADCY9m expression was verified by performing a Western blot analysis of 5-aza-dC-treated 293H cell extracts with anti I-Cre antibodies. Our results showed a slight increase in ADCY9m expression upon 5-aza-dC treatment (Fig. 5D). However, this variation could not account for the increase of TM frequency observed in Fig. 5C as a 1.5–2-fold increase of ADCY9m expression did not influence TM at the adcy9 locus in untreated cells (Fig. 5, E and F). Taken together, our results showed that 5-aza-dC increased ADCY9m activity via its capacity to reduce the methylation status of its cognate target.
To strengthen this assumption, we also evaluated the ability of 5-aza-dC to stimulate HGT induced by ADCY9m in 293H cells. Our results showed that treatment of 293H cells by 0.2 μm 5-aza-dC increased by 4-fold the frequency of HGT with respect to untreated cells (Fig. 6). In agreement with the results described above, demethylation of ADCY9t increased ADCY9m activity in vivo.
FIGURE 6.
Effect of 5-aza-dC treatment on HGT induced by ADCY9m. 293H cells were pretreated for 2 days with or without 0.2 μm 5-aza-dC and then co-transfected with or without 5 μg of meganuclease expressing vector and 2 μg of DNA matrix (see “Material and Methods”). 5-aza-dC treatment was maintained for 2 days, and the transfected cells were then plated at low density. Two weeks after plating, individual colonies were picked and analyzed by PCR screening to determine the frequencies of HGT induced by ADCY9m. Frequencies reported in this figure were corrected from variations of transfection efficiency.
Engineering of Methyl-insensitive Meganuclease Variants
Our structural and biochemical data shed light on the involvement of valine 73 in I-CreI sensitivity to methylation, and it unmasked interesting perspectives to engineer meganuclease variants that are insensitive to methylation. Indeed, we reasoned that the steric hindrance observed between the methyl moiety of cytosine +2b and the side chain of valine 73 could be alleviated by substituting valine 73 by smaller hydrophobic amino acids. Thus, an ADCY9m variant harboring an alanine in position 73 was generated (ADCY9m_V73A) and assayed in yeast and mammalian cells in the presence of unmethylated extrachromosomal ADCY9t (see under “Material and Methods”). Our results showed that the substitution of valine 73 to alanine did not change the intrinsic nuclease activity of ADCY9m (Fig. 7, A and B). In contrast, when the nuclease activity of ADCY9m_V73A was assessed at its endogenous methylated locus, the frequency of TM induced by this new protein variant was on average 10-fold higher than the one induced by ADCY9m (∼0.6 and 0.06% respectively, mean of two independent experiments, p value experiments 1 and 2 = 0.37, p value experiment 1 = 3.1e-05, and p value experiment 2 = 7.8e-4, see Fig. 7C and supplemental Table 4). This improvement was not due to expression or processing differences between the ADCY9m and ADCY9m_V73A variant as shown by their similar Western blot patterns (Fig. 7D).
FIGURE 7.
Effect of valine 73 to alanine substitution on the nuclease activity of ADCY9m toward extrachromosomal unmethylated ADCY9t in yeast (A) and in mammalian cells (B). Effect of valine 73 to alanine substitution on the frequency of TM induced by ADCY9m in 293H cells (C) and on the expression of ADCY9m in 293H cells (D).
DISCUSSION
Our goal was to investigate the influence of CpG dinucleotide methylation on the nuclease activity of natural and engineered meganucleases used for genome engineering application. Using I-CreI and ADCY9m as meganuclease models, we found that methylation of CpG dinucleotides present in their cognate targets affect their DNA binding affinity and cleavage efficiency.
CpG methylation prevents I-CreI DNA binding especially when this modification occurs on the b strand of its cognate target. Indeed, we found that full methylation of C1234 “b” strand decreased the DNA binding affinity of I-CreI by more than 400-fold (data not shown), whereas full methylation of C1234 a strand remained effectless. The relative influence of cytosine +5b and +2b methylation on I-CreI DNA binding affinity could not be deciphered with our original experimental conditions containing low NaCl concentrations (150 mm). Indeed, at such a low NaCl concentration, the effect of single cytosine methylation on I-CreI DNA binding capacity was undetectable. To tackle this issue, we increased the sensibility of our binding experiments by raising the NaCl concentration to 400 mm and found that both methylated cytosines contributed equally well to this inhibition (Fig. 2). These findings are consistent with our structural data showing that the presence of a methyl moiety in position +2b constrained the neighboring amino acids to adopt new conformations. A similar perturbation is likely to occur between the methyl moiety of 5mC +5b and isoleucine 24. Although this assumption has to be confirmed, it could explain the negative effect of such a modification on the DNA binding affinity of I-CreI. Finally, this DNA binding affinity is unlikely to be affected by 5mC −6a and 5mC −3a as no obvious steric clash could be envisioned between them and I-CreI backbone.
CpG methylation also decreases the enzymatic activity of I-CreI especially when this modification occurs in the central tetrabase of its cognate targets. Such findings are consistent with the work of Molina et al. (38), which showed that the sequence of the central tetrabase significantly affects I-CreI meganuclease activity in vivo. Surprisingly, the methylated target was cleaved in the I-CreI:C1234_Me+2b structure obtained in the presence of Mg2+ (Fig. 4, PDB code 4AQX). The cleavage observed in the methylated target could be due to the high concentration of protein, DNA, and metal in the crystallization condition that is likely to force the formation of a catalytically proficient complex. In addition, the crystallization process (>1 day) could be slow enough to allow the reaction to proceed to completion, even though kcat was shown to be greatly reduced by methylation (supplemental Fig. 3 and supplemental Table 2). Altogether, our results showed that 5mC modifications could influence different aspects of the I-CreI catalytic mechanism, i.e. DNA binding and cleavage (compare +5b binding data with +5b cleavage results). Such differences highlighted the relevance of 5mC modifications, such as at position +2b, that are refractory to both the binding and cleavage mechanisms.
Although I-CreI meganuclease is known to be highly specific toward its cognate target, it is also known to accommodate single or multiple base pair changes without being significantly inhibited (39, 40). Our data add another layer of complexity to this fine balance by showing that an additional single methyl moiety in the central tetrabase region is sufficient to inhibit I-CreI activity. Such sensitivity to DNA methylation might play a role in the regulation of the homing process induced by I-CreI. Interestingly, the methylation pattern of C. reinhardtii nuclear and chloroplastic genome has been recently reported by Feng et al. (41). Analysis of their dataset revealed that the homing site of I-CreI was methylated on CpG dinucleotide and 5′-cytosine-phosphoadenosine-phosphoguanine trinucleotide (5′-AAAACGTCGTGAGACAGTTTGG-3′, after bisulfite treatment (41)). Such findings show that methylations of the I-CreI homing site could take place in a relevant physiological context and suggests that methylation might prevent it from being cleaved in vivo by I-CreI.
I-CreI may not be the only natural meganuclease sensitive to CpG methylation. So far, 14 different I-CreI homologues have been identified in mitochondrial and chloroplastic genomes of over 75 green algae (42). Interestingly, all those homologues were shown to target DNA sequences containing two CpG dinucleotides located at the same positions as the ones identified in I-CreI cognate target (positions −3a/−2a and −6a/−5a, Fig. 1). In addition, these homologues bear strictly conserved (valine) or similar amino acids (isoleucine and methionine) at position 73 that was identified in this work to play a key role in the sensitivity of I-CreI to CpG methylation. These 14 different I-CreI homologues could thus also be sensitive to CpG methylation, although such a feature still has to be demonstrated.
In a genome-engineering prospect, our work also provides evidence that CpG methylation does impact engineered meganuclease activity in a physiological context. Even though the engineered meganuclease ADCY9m was highly active toward its nonmethylated target (supplemental Fig. 4), it displayed low activity toward its methylated endogenous locus (0.1% TM, Fig. 5A). Such residual activity (probably due to the editing of the remaining 10% of unmethylated ADCY9t, Fig. 5B) could be enhanced by demethylation of ADCY9t using 5-aza-dC, thus showing that methylation inhibited meganuclease activity in vivo.
Engineered meganuclease sensitivity to CpG methylation represents a hurdle that needs to be minimized. Utilization of 5-aza-dC could be an option to overcome such sensitivity, although its cytotoxicity and potential pleiotropic effects are redhibitory for precise therapeutic genome engineering applications (36). Thus, alternative and harmless strategies are needed. In that sense, we proposed an example of rational engineering of a meganuclease DNA binding domain that alleviates such sensitivity when methylation occurs at position +2b of a meganuclease target. On the basis of the I-CreI:C1234_Me+2b co-crystal structure, we were able to design a single point mutation (Val-73 to Ala) that increased by about 10-fold the frequency of TM induced by ADCY9m at its endogenous methylated locus. Even though this specific point mutation cannot be applied to all engineered meganucleases, we demonstrated that their sensitivity to methylation could be reduced by determining the methylation pattern of their cognate target and performing minimal point mutations within their DNA binding domain.
ADCY9m and CAPNS1m showed similar nuclease activities toward their unmethylated targets (supplemental Fig. 4). However, they displayed significant difference of activity toward their respective endogenous loci (Fig. 5). We showed that this difference is partly due to the differential methylation status of the CAPNS1 meganuclease target and ADCY9t target; however, we cannot exclude that other epigenetic factors affecting locus accessibility may be involved as recently demonstrated by Daboussi et al. (15). Considering the fact that CpG methylation is linked to the regulation chromatin structure (43–45), it could indirectly modulate the access of meganuclease to its endogenous target. Thus, the activity and specificity of engineered meganucleases are not the only parameters to consider for successful gene editions, and in that sense, we recommend considering the epigenetic pattern of the locus to edit for choosing the optimal meganuclease target.
In summary, our work unravels the intimate relationship between meganuclease activity and the epigenetic status of their cognate target. Such property has been suggested for zinc finger nuclease (46) and has been described recently for TAL DNA binding domains used to assemble TAL effector nucleases (47). Thus, whatever engineered nuclease platform is considered, it becomes clear that epigenetic modifications may represent a bottleneck for the genome engineering applications and especially for those related to stem cells and iPS cells.
Supplementary Material
Acknowledgment
We thank Georges Silva for critical reading of the manuscript.

This article contains supplemental Materials and Methods, Figs. 1–4, and Tables 1–4.
The atomic coordinates and structure factors (codes 4AQX and 4AQU) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CpG
- 5′-cytosine-phosphoguanine
- 5mC
- 5-methylcytosine
- 5-aza-dC
- 5-aza-2-deoxycytidine
- FAM
- 6-carboxyfluorescein
- TM
- targeted modifications
- HGT
- homologous gene targeting
- FA
- fluorescence anisotropy
- ADCY9m
- ADCY9 meganuclease
- ADCY9t
- ADCY9 meganuclease target
- CAPNS1m
- CAPNS1 meganuclease
- PDB
- Protein Data Bank.
REFERENCES
- 1. McMahon M. A., Rahdar M., Porteus M. (2012) Gene editing. Not just for translation anymore. Nat. Methods 9, 28–31 [DOI] [PubMed] [Google Scholar]
- 2. Carroll D. (2011) Genome engineering with zinc-finger nucleases. Genetics 188, 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Christian M., Cermak T., Doyle E. L., Schmidt C., Zhang F., Hummel A., Bogdanove A. J., Voytas D. F. (2010) Targeting DNA double strand breaks with TAL effector nucleases. Genetics 186, 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li T., Huang S., Zhao X., Wright D. A., Carpenter S., Spalding M. H., Weeks D. P., Yang B. (2011) Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 39, 6315–6325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miller J. C., Tan S., Qiao G., Barlow K. A., Wang J., Xia D. F., Meng X., Paschon D. E., Leung E., Hinkley S. J., Dulay G. P., Hua K. L., Ankoudinova I., Cost G. J., Urnov F. D., Zhang H. S., Holmes M. C., Zhang L., Gregory P. D., Rebar E. J. (2011) A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148 [DOI] [PubMed] [Google Scholar]
- 6. Arnould S., Delenda C., Grizot S., Desseaux C., Pâques F., Silva G. H., Smith J. (2011) The I-CreI meganuclease and its engineered derivatives. Applications from cell modification to gene therapy. Protein Eng. Des. Sel. 24, 27–31 [DOI] [PubMed] [Google Scholar]
- 7. Stoddard B. L. (2011) Homing endonucleases. From microbial genetic invaders to reagents for targeted DNA modification. Structure 19, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stoddard B. L. (2005) Homing endonuclease structure and function. Q. Rev. Biophys. 38, 49–95 [DOI] [PubMed] [Google Scholar]
- 9. Arnould S., Chames P., Perez C., Lacroix E., Duclert A., Epinat J. C., Stricher F., Petit A. S., Patin A., Guillier S., Rolland S., Prieto J., Blanco F. J., Bravo J., Montoya G., Serrano L., Duchateau P., Pâques F. (2006) Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 355, 443–458 [DOI] [PubMed] [Google Scholar]
- 10. Smith J., Grizot S., Arnould S., Duclert A., Epinat J. C., Chames P., Prieto J., Redondo P., Blanco F. J., Bravo J., Montoya G., Pâques F., Duchateau P. (2006) A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 34, e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rosen L. E., Morrison H. A., Masri S., Brown M. J., Springstubb B., Sussman D., Stoddard B. L., Seligman L. M. (2006) Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res. 34, 4791–4800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arnould S., Perez C., Cabaniols J. P., Smith J., Gouble A., Grizot S., Epinat J. C., Duclert A., Duchateau P., Pâques F. (2007) Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J. Mol. Biol. 371, 49–65 [DOI] [PubMed] [Google Scholar]
- 13. Grizot S., Smith J., Daboussi F., Prieto J., Redondo P., Merino N., Villate M., Thomas S., Lemaire L., Montoya G., Blanco F. J., Pâques F., Duchateau P. (2009) Efficient targeting of a SCID gene by an engineered single-chain homing endonuclease. Nucleic Acids Res. 37, 5405–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Redondo P., Prieto J., Muñoz I. G., Alibés A., Stricher F., Serrano L., Cabaniols J. P., Daboussi F., Arnould S., Perez C., Duchateau P., Pâques F., Blanco F. J., Montoya G. (2008) Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature 456, 107–111 [DOI] [PubMed] [Google Scholar]
- 15. Daboussi F., Zaslavskiy M., Poirot L., Loperfido M., Gouble A., Guyot V., Leduc S., Galetto R., Grizot S., Oficjalska D., Perez C., Delacote F., Dupuy A., Chion-Sotinel I., Le Clerre D., Lebuhotel C., Danos O., Lemaire F., Oussedik K., Cedrone F., Epinat J. C., Smith J., Dickson G., Popplewell L., Koo T., Vandendriessche T., Chuah M. K., Duclert A., Duchateau P., Paques F. (2012) Chromosomal context and epigenetic mechanisms control the efficacy of genome editing by rare-cutting designer endonucleases. Nucleic Acids Res. 40, 6367–6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chapdelaine P., Pichavant C., Rousseau J., Pâques F., Tremblay J. P. (2010) Meganucleases can restore the reading frame of a mutated dystrophin. Gene Ther. 17, 846–858 [DOI] [PubMed] [Google Scholar]
- 17. Gao H., Smith J., Yang M., Jones S., Djukanovic V., Nicholson M. G., West A., Bidney D., Falco S. C., Jantz D., Lyznik L. A. (2010) Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 61, 176–187 [DOI] [PubMed] [Google Scholar]
- 18. Jaenisch R., Bird A. (2003) Epigenetic regulation of gene expression. How the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254 [DOI] [PubMed] [Google Scholar]
- 19. Bryk M., Quirk S. M., Mueller J. E., Loizos N., Lawrence C., Belfort M. (1993) The td intron endonuclease I-TevI makes extensive sequence-tolerant contacts across the minor groove of its DNA target. EMBO J. 12, 4040–4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ellison E. L., Vogt V. M. (1993) Interaction of the intron-encoded mobility endonuclease I-PpoI with its target site. Mol. Cell. Biol. 13, 7531–7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Werner E., Wende W., Pingoud A., Heinemann U. (2002) High resolution crystal structure of domain I of the Saccharomyces cerevisiae homing endonuclease PI-SceI. Nucleic Acids Res. 30, 3962–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bolduc J. M., Spiegel P. C., Chatterjee P., Brady K. L., Downing M. E., Caprara M. G., Waring R. B., Stoddard B. L. (2003) Structural and biochemical analyses of DNA and RNA binding by a bifunctional homing endonuclease and group I intron splicing factor. Genes Dev. 17, 2875–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang J., Kim H. H., Yuan X., Herrin D. L. (1997) Purification, biochemical characterization, and protein-DNA interactions of the I-CreI endonuclease produced in Escherichia coli. Nucleic Acids Res. 25, 3767–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arosio D., Costantini S., Kong Y., Vindigni A. (2004) Fluorescence anisotropy studies on the Ku-DNA interaction. anion and cation effects. J. Biol. Chem. 279, 42826–42835 [DOI] [PubMed] [Google Scholar]
- 25. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leslie A. G. (2006) The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 27. Vagin A., Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 28. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 30. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lakowicz, (2006) in Principles of Fluorescence Spectroscopy (Lakowicz J. R., ed) pp. 366–374, Springer Science and Business Media, LLC, New York [Google Scholar]
- 33. Chevalier B., Sussman D., Otis C., Noël A. J., Turmel M., Lemieux C., Stephens K., Monnat R. J., Jr., Stoddard B. L. (2004) Metal-dependent DNA cleavage mechanism of the I-CreI LAGLIDADG homing endonuclease. Biochemistry 43, 14015–14026 [DOI] [PubMed] [Google Scholar]
- 34. Chevalier B. S., Monnat R. J., Jr., Stoddard B. L. (2001) The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites. Nat. Struct. Biol. 8, 312–316 [DOI] [PubMed] [Google Scholar]
- 35. Silva G., Poirot L., Galetto R., Smith J., Montoya G., Duchateau P., Pâques F. (2011) Meganucleases and other tools for targeted genome engineering. Perspectives and challenges for gene therapy. Curr. Gene Ther. 11, 11–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Palii S. S., Van Emburgh B. O., Sankpal U. T., Brown K. D., Robertson K. D. (2008) DNA methylation inhibitor 5-aza-2′-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol. Cell. Biol. 28, 752–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hudson K., Luo S., Hagemann N., Preuss D. (2011) Changes in global gene expression in response to chemical and genetic perturbation of chromatin structure. PLoS One 6, e20587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Molina R., Redondo P., Stella S., Marenchino M., D'Abramo M., Gervasio F. L., Charles Epinat J., Valton J., Grizot S., Duchateau P., Prieto J., Montoya G. (2012) Nonspecific protein-DNA interactions control I-CreI target binding and cleavage. Nucleic Acids Res. doi: 10.1093/nar/gks320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jurica M. S., Monnat R. J., Jr., Stoddard B. L. (1998) DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI. Mol. Cell 2, 469–476 [DOI] [PubMed] [Google Scholar]
- 40. Argast G. M., Stephens K. M., Emond M. J., Monnat R. J., Jr. (1998) I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J. Mol. Biol. 280, 345–353 [DOI] [PubMed] [Google Scholar]
- 41. Feng S., Cokus S. J., Zhang X., Chen P. Y., Bostick M., Goll M. G., Hetzel J., Jain J., Strauss S. H., Halpern M. E., Ukomadu C., Sadler K. C., Pradhan S., Pellegrini M., Jacobsen S. E. (2010) Conservation and divergence of methylation patterning in plants and animals. Proc. Natl. Acad. Sci. U.S.A. 107, 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lucas P., Otis C., Mercier J. P., Turmel M., Lemieux C. (2001) Rapid evolution of the DNA-binding site in LAGLIDADG homing endonucleases. Nucleic Acids Res. 29, 960–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Choy J. S., Wei S., Lee J. Y., Tan S., Chu S., Lee T. H. (2010) DNA methylation increases nucleosome compaction and rigidity. J. Am. Chem. Soc. 132, 1782–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Karymov M. A., Tomschik M., Leuba S. H., Caiafa P., Zlatanova J. (2001) DNA methylation-dependent chromatin fiber compaction in vivo and in vitro. Requirement for linker histone. FASEB J. 15, 2631–2641 [DOI] [PubMed] [Google Scholar]
- 45. Gilbert N., Thomson I., Boyle S., Allan J., Ramsahoye B., Bickmore W. A. (2007) DNA methylation affects nuclear organization, histone modifications, and linker histone binding but not chromatin compaction. J. Cell Biol. 177, 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hoshaw J. P., Unger-Wallace E., Zhang F., Voytas D. F. (2010) A transient assay for monitoring zinc finger nuclease activity at endogenous plant gene targets. Methods Mol. Biol. 649, 299–313 [DOI] [PubMed] [Google Scholar]
- 47. Bultmann S., Morbitzer R., Schmidt C. S., Thanisch K., Spada F., Elsaesser J., Lahaye T., Leonhardt H. (2012) Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res. 40, 5368–5377 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.