Abstract
Paclitaxel is a frequently used anticancer drug with considerable inter-individual variability in terms of drug efficiency and toxicity. The reasons for this variability have not been fully explained. The purpose of this study was to evaluate the possible relationship between paclitaxel-induced neurotoxicity and the distribution of genetic variations with reported functional significance in the ABCB1, CYP2C8 and CYP3A4 genes that are all implicated in taxol metabolism. Women (n=36) experiencing paclitaxel-induced neurotoxicity were included in the study, and the ABCB1 G2677A/T and C3435T as well as CYP2C8*3 and CYP3A4*1b allele frequencies were determined using PCR-RFLP and DNA sequence analysis. We showed that the ABCB1 3435T allele, previously reported as a risk allele for neurotoxicity, did not correlate with the occurrence of neurotoxicity in our patient sample (Chi-square test, p=0.61). Furthermore, we showed that neither the CYP2C8*3 nor CYP3A4*1b alleles, that both lead to diminished enzyme activity, correlated with paclitaxel-induced neurotoxicity. The occurrence and variation in severity of neurotoxicity in our Swedish patient sample could therefore not be explained by the reported functional polymorphisms in the ABCB1, CYP2C8 and CYP3A4 genes.
Keywords: paclitaxel, neurotoxicity, ABCB1, CYP2C8, CYP3A4
Introduction
Paclitaxel is a frequently used anticancer drug with a broad spectrum of activity against malignant solid tumors, including breast, ovarian and lung cancer (1). However, the considerable variability in drug efficiency and toxicity of paclitaxel treatment results in unpredictable patient responses. Neurotoxicity is one common adverse effect following treatment with paclitaxel (2). This effect is apparently dose-related, since several studies have shown a trend towards increased neurotoxicity with increased paclitaxel AUC (area under the concentration curve) (3–5). The reasons for this inter-individual variability have not been determined. Many studies are therefore focusing on genetic variation in genes encoding metabolising and drug-transporting proteins. Paclitaxel is mainly metabolised by the CYP2C8 enzyme to its primary metabolite, 6α-hydroxypaclitaxel (6) and to a lesser extent by the CYP3A4 enzyme (7). Furthermore, paclitaxel is a substrate for ABCB1 (MDR1; P-glycoprotein), an important membrane efflux protein (3,8). ABCB1 is expressed in the blood-brain barrier and is thought to protect peripheral nervous tissue, as it transports toxic substances back to the systemic circulation (9,10). Recently, genetic polymorphisms in these enzymes have been described. However, the clinical relevance of these variations has not as yet been fully explored in clinical studies.
In a cohort of 97 Caucasian cancer patients, no significant association was observed between different alleles in the ABCB1, CYP2C8 and CYP3A4 genes and the pharmaco-kinetics of paclitaxel (11). Moreover, a recent study showed no association between polymorphisms in ABCB1 and the pharmacokinetics of paclitaxel (12). These authors conclude that the inter-individual variability in the pharmacokinetics of paclitaxel cannot be attributed to genetic variations in this gene. However, adverse effects such as neurotoxicity might be associated with inherited alleles in these genes through mechanisms unrelated to plasma pharmacokinetics. Genetic variation in ABCB1 has been associated with response to paclitaxel treatment, but the literature has not been conclusive (24).
The purpose of this study was to investigate the association between the occurrence and degree of neurotoxicity following paclitaxel treatment in breast and ovarian cancer and to correlate these clinical findings with the allele frequency of variants in the ABCB1, CYP2C8 and CYP3A4 genes reportedly leading to impaired enzyme activity.
Materials and methods
Patient selection
This study comprised 36 female patients with breast cancer (24 patients) or ovarian cancer (12 patients), all of which developed clinical neurotoxicity grade 1–2 according to the NCI-CTCAE (v. 3.0) during paclitaxel-containing chemotherapy. Information concerning debut of toxicity was obtained from chart-review. Among the breast cancer patients, 4 were treated with paclitaxel as a single drug (2 on a weekly basis), 18 with paclitaxel, epirubicin and capecitabine, while 2 received paclitaxel and herceptin. Among the ovarian cancer patients, 1 received weekly paclitaxel and 11 were administered combination chemotherapy with paclitaxel and carboplatin. Prior chemotherapy had been administered to 15 of the total 36 patients. The patients median age was 58 years (range, 40–80). The patients were diagnosed and treated at the Department of Oncology at Sahlgrenska University Hospital. Informed consent was obtained, and the study was approved by the regional ethics committee. Our control sample consisted of 50 healthy blood donors at Sahlgrenska University Hospital.
Genetic analysis/genotyping
Genomic DNA was isolated from peripheral leucocytes from the 36 blood samples using the Puregene® DNA Isolation Kit (Gentra Systems, Minneapolis, MN, USA). PCR primers were purchased from Invitrogen®.
Genotyping of the CYP2C8*3 allele was performed by a polymerase chain reaction-based restriction fragment length polymorphism (PCR-RFLP) as previously described (6). The analysis was conclusive for 35 samples. Genotyping of the CYP3A4*1b allele was also performed by PCR-RFLP (13). The analysis was conclusive for 31 samples.
Genotyping of the SNP 2677G/T/A of the ABCB1 gene was performed by PCR (14), followed by DNA sequence analysis using the ABI 3100 PRISM Genetic Analyzer and the Big Dye Terminator Cycle Sequencing Ready Reaction Kit (both from Applied Biosystems, Foster City, CA, USA).
Genotyping for the SNP 3435C/T was performed by a PCR-RFLP. The PCR primer sequences 5′TGT TTT CAG CTG CTT GAT GG-3′ and 3′CTC CGG TTG TAT GTA CGG AA-5′ were obtained from Epidauros Biotechnologie AG (Bernried, Germany). The PCR program consisted of 30 cycles at 95°C for 30 sec, 55°C for 30 sec, 72°C for 60 sec and a final elongation step at 72°C for 7 min. The PCR fragment was subjected to cleavage with the DpnII enzyme. The analysis was conclusive for 36 samples.
All products from the PCR-RFLP analysis were separated on a 3% agarose gel.
Statistical evaluation
Statistical analyses were carried out using the SAS system. The differences in allele frequencies among literature control population, blood donor controls and patients were analysed using the Chi-square test. The analysis of association between mean debut dose of paclitaxel and grade of neurotoxicity and 3435C/T SNPs was performed using the Fisher’s exact test.
Results
Patients
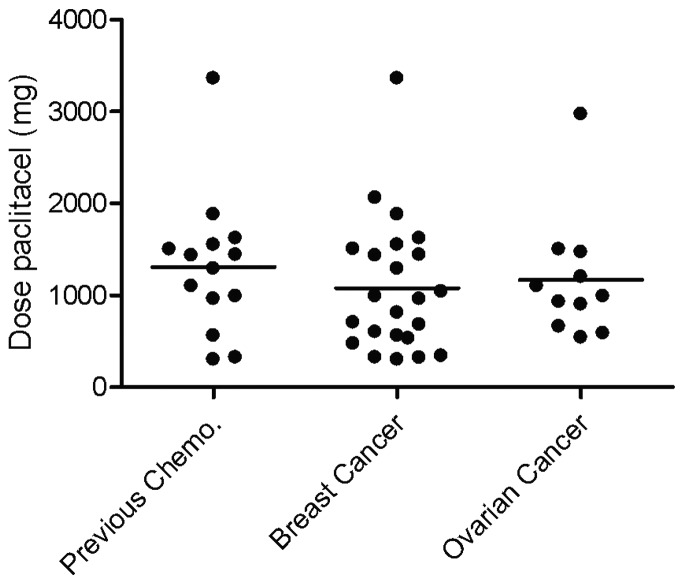
Patients were selected for the study when they experienced neurotoxicity. There was no significant difference in the mean debut dose of paclitaxel for neurotoxicity grade 1 based on diagnosis, or if the patients had received prior chemotherapy. The mean debut dose for neurotoxicity for all analyzed patients was 1129 mg (range, 300–3360) (Fig. 1).
Figure 1.
Scatter dot plot showing the paclitaxel dose when the patient first experienced neurotoxicity grade 1. The individuals are grouped based on diagnosis. The patients, regardless of diagnosis, that had received previous chemotherapy are also shown. Information about dose is absent for one of the breast cancer patients.
CYP2C8 and CYP3A4
The CYP2C8*3 allele frequency in our study sample (n=35) was determined by PCR-RFLP (data not shown). The allele frequency was 14.3% as compared to the expected frequency of 14% (13–15%) noted in populations of similar Caucasian origin (15).
The allele CYP3A4*1b, with an expected population frequency of 2.0–9.6% (16), was not found in any of our patients (n=31), indicating that this risk allele was not abundant in our patient sample.
ABCB1
The ABCB1 2677G/T/A and 3435C/T SNPs were analysed by DNA sequencing and RFLP after PCR amplification of exon 21 and 26, respectively.
The distribution of alleles at position 3435C/T and 2677G/T/A was compared with the expected population frequencies obtained from the analyses of 200 randomly selected individuals from the southeastern part of Sweden (Table I) (17). The distribution in our patient sample closely resembled the distribution noted in the literature control sample, implying that these two cohorts were not significantly stratified (Chi-square test, p=0.29). This finding contradicts previously published data suggesting that patients carrying the 3435T allele have a higher risk for neurotoxicity.
Table I.
Frequencies of ABCB1 3435C/T alleles in patients, literature controls and blood donor controls.
| Alleles | ||||
|---|---|---|---|---|
|
|
||||
| C/C | C/T | T/T | Total | |
| Patients | 12 | 16 | 8 | 36 |
| Literature controls | 46 | 88 | 66 | 200 |
| Blood donor controls | 11 | 28 | 11 | 50 |
| Total | 58 | 104 | 74 | 286 |
The literature control population sample we used was geographically closely linked to our study population, but to strengthen our data and avoid the possibility of population bias, we also examined the distribution of the 3435T allele in 50 blood donor controls from the same university hospital as our patients. As anticipated, the frequency of each allele did not differ statistically from that of the literature control population sample (Chi-square test, p=0.24). The allele frequencies in the control groups did not differ significantly from published reports including ethnically similar subjects (22,23). Furthermore, there was no statistically significant difference between mean debut dose of paclitaxel, grade of neurotoxicity and 3435C/T SNPs (Chi-square test, p=0.61 or Fisher’s exact test, p=0.69) (Fig. 2).
Figure 2.
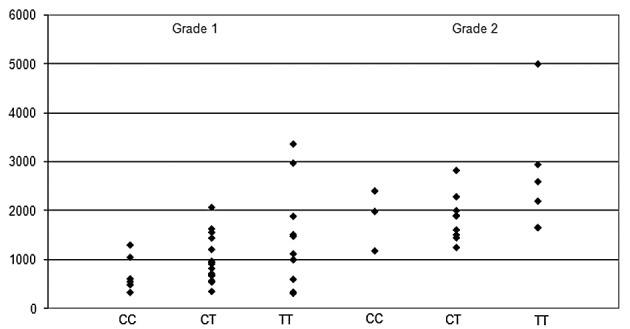
Scatter dot plot showing the paclitaxel dose when the patients first experienced neurotoxicity grade 1 and/or grade 2. Patients are grouped according to the ABCB1 3435C/T alleles. Information about dose is absent for one of the breast cancer patients with the allele T/T.
Discussion
With the aim to study the inter-individual variation of paclitaxel-induced neurotoxicity in relation to genetic variation, relevant gene polymorphisms in the three metabolising enzymes, CYP2C8, CYP3A4 and ABCB1, were analysed. CYP2C8 is the principle enzyme metabolising paclitaxel. CYP2C8*3, which is found primarily in Caucasians, has shown only 15% of the enzymatic turnover number of wild-type CYP2C8 for paclitaxel in vitro (6). Lower enzyme activity for CYP2C8*3 compared with the wild-type has also been shown in human liver microsomes (18). In an attempt to verify in vivo whether the suggested decrease in enzyme activity for this allele correlated to neurotoxicity, we analysed the CYP2C8*3 allele frequency in patients experiencing at least grade 1–3 neurotoxicity after treatment with paclitaxel. We reported an allele frequency of 14.3% as compared with the expected frequency of 14% (13–15%) (15). We concluded that neurotoxicity as a side effect of paclitaxel treatment is not associated with an increased incidence of CYP2C8*3 allele frequency. The suggested decrease in enzyme activity shown for this allele by the in vitro data has therefore not been confirmed in this patient group.
CYP3A4 is the most abundant P-450 enzyme in the human liver and it metabolises more than 50% of prescription drugs. The CYP3A4*1b allele has been associated with prostate cancer and estrogen receptor-negative breast cancer (25). Moreover, CYP3A4*1b has been shown to influence enzyme activity (26).
The allele frequency of the 3A4*1b allele differs between populations, and the expected frequency in our Swedish sample was not previously determined. However, as this allele was not detected in our patient sample, we conclude that it is probably of minor importance in the pathogenesis of neurotoxicity.
P-glycoprotein, encoded by ABCB1, is an ATP-driven drug export pump. It is expressed in many tissues such as the intestine and the blood-brain barrier. Furthermore, high expression in ovarian tumour cells has been shown to correlate with a poor response to treatment with paclitaxel (19). Several SNPs in the ABCB1 gene have been described and two of them (2677G/T/A and 3435C/T) have been correlated to p-glycoprotein expression in the Caucasian population. As a result, individuals homozygous for the 3435T alleles were shown to express lower levels of p-glycoprotein in the intestine and subsequently higher plasma levels of the drug (20). Patients with low p-glycoprotein expression would be more likely to suffer from neurotoxicity, since this would indicate a higher systemic concentration of the drug. Recently, the ABCB1 alleles G2677T/A were shown to correlate with the response to paclitaxel treatment in ovarian cancer in a Swedish patient sample (17). In this study by Gréen et al, the frequency of the T allele was higher in patients experiencing a favorable response. However, the 3435C/T SNP was not found to correlate with treatment outcome. Moreover, no significant difference in SNP frequencies could be found between the reference population and the patients with ovarian cancer. There are several previous studies showing an association between the 3435C allele and poor response. Consequently, Kafka et al showed that the alleles T/T at position 3435 correlated with a complete clinical response to chemotherapy in locally advanced breast cancer (21).
The association of the 2677G/T/A and 3435C/T SNPs with paclitaxel-induced neurotoxicity and neutropenia was recently evaluated in a German study by Sissung et al (12). In this study, comprising 22 patients experiencing peripheral neuropathy, none of the patients carrying the 3435C/C alleles developed peripheral neuropathy after paclitaxel treatment. In addition, there was a trend towards an increased risk of neurotoxicity for individuals carrying 3435C/T or 3435T/T. This result was not reproduced in our Swedish patient sample. Given the close proximity of the study populations in these two studies, we conclude that the 3435T allele does not increase the risk for paclitaxel-induced neurotoxicity.
The German study also showed that 3435 T/T in combination with 2667T/T conferred a risk of neutropenia. Linkage disequilibrium between these two SNPs has been proposed (22) and it was also evident in our data.
In conclusion, by analysing the CYP2C8, CYP3A4 and ABCB1 genes implicated in paclitaxel pharmacogenetics, we showed no association between these alleles and paclitaxel-induced neurotoxicity. Our data contradict the previously reported association between the 3435T allele (12) and neurotoxicity. This implies that other genetic and/or environmental mechanisms regulate the individual difference in susceptibility to paclitaxel-induced neurotoxicity.
The data presented here will be re-analysed in a large, randomised, phase III study comprising more than 300 cases of breast cancer. In this study, polymorphisms in genes related to paclitaxel pharmacogenomics will be analysed with the aim of obtaining an insight into predictive or prognostic factors relating to paclitaxel efficiency and toxicity.
In addition to SNPs, inter-individual genetic variation is also composed of small insertions/deletions in the genome, VNTRs, CNVs as well as epigenetic effects such as DNA methylation, which also opens possibilities for further studies on genetic variation and paclitaxel-induced adverse events.
Acknowledgements
This work was supported by grants from the Assar Gabrielsson Foundation.
References
- 1.Socinski MA. Cytotoxic chemotherapy in advanced non-small cell lung cancer: a review of standard treatment paradigms. Clin Cancer Res. 2004;10:4210–4214. doi: 10.1158/1078-0432.CCR-040009. [DOI] [PubMed] [Google Scholar]
- 2.Fellner S, Bauer B, Miller DS, et al. Transport of paclitaxel (Taxol) across the blood-brain barrier in vitro and in vivo. J Clin Invest. 2002;110:1309–1318. doi: 10.1172/JCI15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walle UK, Walle T. Taxol transport by human intestinal epithelial Caco-2 cells. Drug Metab Dispos. 1998;26:343–346. [PubMed] [Google Scholar]
- 4.Huizing MT, Giaccone G, van Warmerdam LJ, et al. Pharmacokinetics of paclitaxel and carboplatin in a dose-escalating and dose-sequencing study in patients with non-small cell lung cancer. The European Cancer Centre. J Clin Oncol. 1997;15:317–329. doi: 10.1200/JCO.1997.15.1.317. [DOI] [PubMed] [Google Scholar]
- 5.Sonnichsen DS, Hurwitz CA, Pratt CB, Shuster JJ, Relling MV. Saturable pharmacokinetics and paclitaxel pharmacodynamics in children with solid tumors. J Clin Oncol. 1994;12:532–538. doi: 10.1200/JCO.1994.12.3.532. [DOI] [PubMed] [Google Scholar]
- 6.Dai D, Zeldin DC, Blaisdell JA, Chanas B, Coulter SJ, Ghanayem BI, Goldstein JA. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics. 2001;11:597–607. doi: 10.1097/00008571-200110000-00006. [DOI] [PubMed] [Google Scholar]
- 7.Harris JW, Rahman A, Kim BR, Guengerich FP, Collins JM. Metabolism of taxol by human hepatic microsomes and liver slices: participation of cytochrome P450 3A4 and an unknown P450 enzyme. Cancer Res. 1994;54:4026–4035. [PubMed] [Google Scholar]
- 8.Malingré MM, Beijnen JH, Rosing H. Co-administration of GF120918 significantly increases the systemic exposure to oral paclitaxel in cancer patients. Br J Cancer. 2001;84:42–47. doi: 10.1054/bjoc.2000.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordon-Cardo C, O’Brien JP, Boccia J, et al. Multidrug-resistance gene (P-glycoprotein) is expressed by endothelial cells at blood-brain barrier sites. Proc Natl Acad Sci USA. 1989;86:695–698. doi: 10.1073/pnas.86.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schinkel AH, Smit JJ, van Tellingen O. Disruption of the mouse mdr1a P-glycoprotein gene leads to a deficiency in the blood-brain barrier and to increased sensitivity to drugs. Cell. 1994;77:491–502. doi: 10.1016/0092-8674(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 11.Henningsson A, Marsh S, Loos WJ. Association of CYP2C8, CYP3A4, CYP3A5 and ABCB1 polymorphisms with the pharmacokinetics of paclitaxel. Clin Cancer Res. 2005;11:8097–8104. doi: 10.1158/1078-0432.CCR-05-1152. [DOI] [PubMed] [Google Scholar]
- 12.Sissung TM, Mross K, Steinberg SM, Behringer D, Figg WD, Sparreboom A, Mielke S. Association of ABCB1 alleles with paclitaxel-mediated peripheral neuropathy and neutropenia. Eur J Cancer. 2006;42:2893–2896. doi: 10.1016/j.ejca.2006.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavalli SA, Hirata MH, Hirata RD. Detection of MboII polymorphism at the 5′ promoter region of CYP3A4. Clin Chem. 2001;47:348–351. [PubMed] [Google Scholar]
- 14.Kim DH, Park JY, Sohn SK, et al. Multidrug resistance-1 gene polymorphisms associated with treatment outcomes in de novo acute myeloid leukemia. Int J Cancer. 2006;118:2195–2201. doi: 10.1002/ijc.21666. [DOI] [PubMed] [Google Scholar]
- 15.Spratlin J, Sawyer MB. Pharmacogenetics of paclitaxel metabolism. Crit Rev Oncol Hematol. 2007;61:222–229. doi: 10.1016/j.critrevonc.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 16.Lamba JK, Lin YS, Schuetz EG, Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54:1271–1294. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- 17.Gréen H, Söderkvist P, Rosenberg P, Horvath G, Peterson C. Mdr-1 single nucleotide polymorphisms in ovarian cancer tissue: G2677T/A correlates with response to paclitaxel chemotherapy. Clin Cancer Res. 2006;12:854–859. doi: 10.1158/1078-0432.CCR-05-0950. [DOI] [PubMed] [Google Scholar]
- 18.Bahadur N, Leathart JB, Mutch E, et al. CYP2C8 polymorphisms in Caucasians and their relationship with paclitaxel 6alpha-hydroxylase activity in human liver microsomes. Biochem Pharmacol. 2002;64:1579–1589. doi: 10.1016/s0006-2952(02)01354-0. [DOI] [PubMed] [Google Scholar]
- 19.Szakács G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci USA. 2000;97:3473–3478. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kafka A, Sauer G, Jaeger C, Grundmann R, Kreienberg R, Zeillinger R, Deissler H. Polymorphism C3435T of the MDR-1 gene predicts response to preoperative chemotherapy in locally advanced breast cancer. Int J Oncol. 2003;22:1117–1121. [PubMed] [Google Scholar]
- 22.Siddiqui A, Kerb R, Weale ME, et al. Association of multidrug resistance in epilepsy with a polymorphism in the drug-transporter gene ABCB1. N Engl J Med. 2003;348:1442–1448. doi: 10.1056/NEJMoa021986. [DOI] [PubMed] [Google Scholar]
- 23.Cascorbi I, Gerloff T, Johne A, et al. Frequency of single nucleotide polymorphisms in the P-glycoprotein drug transporter MDR1 gene in white subjects. Clin Pharmacol Ther. 2001;69:169–174. doi: 10.1067/mcp.2001.114164. [DOI] [PubMed] [Google Scholar]
- 24.Green H. Pharmacogenomics of importance for paclitaxel chemotherapy. Pharmacogenomics. 2008;9:671–674. doi: 10.2217/14622416.9.6.671. [DOI] [PubMed] [Google Scholar]
- 25.Keshava C, McCanlies EC, Weston A. CYP3A4 poly-morphisms – potential risk factors for breast and prostate cancer: a HuGE review. Am J Epidemiol. 2004;160:825–841. doi: 10.1093/aje/kwh294. [DOI] [PubMed] [Google Scholar]
- 26.Rodríguez-Antona C, Sayi JG, Gustafsson LL, Bertilsson L, Ingelman-Sundberg M. Phenotype-genotype variability in the human CYP3A locus as assessed by the probe drug quinine and analyses of variant CYP3A4 alleles. Biochem Biophys Res Commun. 2005;338:299–305. doi: 10.1016/j.bbrc.2005.09.020. [DOI] [PubMed] [Google Scholar]