Motile nonmuscle cells rapidly remodel their actin cytoskeleton to change shape as they crawl. The mechanisms underlying these dynamic changes in actin-filament concentration, length, and architecture have been the subject of extensive study for nearly three decades. Ameboid movements are complex and multidirectional, making analysis of individual cells difficult. Therefore investigators have tried to simplify their experimental models by (i) purifying individual proteins and analyzing their interactions with actin; (ii) studying the behavior of actin in cytoplasmic extracts. This condition allows the introduction of various inhibitors and stimulators and the addition and subtraction of individual proteins; it also allows the introduction of Listeria monocytogenes and Shigella flexneri, intracellular parasites that form discrete polymerization zones to propel them through the host cell cytoplasm; (iii) microinjecting individual proteins and inhibitory peptides into intact cells or transfecting cells with either sense and antisense cDNA; and (iv) knocking out individual genes. Each approach has advantages and disadvantages that need to be considered in interpreting results and assessing the contribution of individual proteins to the complex orchestration of changes required for actin-based motility. The temptation has been to oversimplify actin-based motility and tout an individual protein as being of primary importance to the exclusion of others. Certain proteins become fashionable and tend to dominate the literature. However, investigators need to maintain an open-minded perspective that is inclusive rather than exclusive.
The controversy over the roles of actin depolymerizing factor (ADF)/cofilin and gelsolin in actin-filament turnover is a case in point. The ADF/cofilin family of proteins and gelsolin were discovered approximately two decades ago (1, 2). Initially, gelsolin received more attention and was considered to be of primary importance in regulating the dynamic actin cytoskeletal changes associated with ameboid movement. However, over the past 5 years, ADF/cofilin has been studied extensively, and many investigators now claim that ADF/cofilin is primarily responsible for remodeling the actin cytoskeleton (3).
To gain a more objective perspective on the contribution of these proteins to actin-filament dynamics, the experimental data derived from each of the approaches outlined above need to be reviewed (see Table 1). First, the investigation of the purified proteins reveals many structural and functional similarities between gelsolin and ADF/cofilin as well as some important functional differences. Examination of tertiary structure indicates that ADF/cofilin shares many structural features with gelsolin (4), and synthetic peptide inhibition experiments suggest that cofilin binds to F-actin similarly to gelsolin (5). Binding of ADF/cofilin causes a reduction in the rotation of the actin filament, and this twist removes the phalloidin-binding site (6). Purified gelsolin also alters the conformation of the actin filament (7) and dissociates phalloidin (8). Recombinant ADF and cofilin increase the off-rate at the pointed (minus) end of actin filaments. Although some controversy continues, the preponderance of data indicate that ADF/cofilin increases the rate of monomer turnover by severing actin filaments, thus increasing the number of filament ends (3) (see Fig. 1A). Recently both native and recombinant Dictyostelium cofilin were shown to sever actin filaments, but to different extents (9). The ADF/cofilin-induced change in actin-filament conformation may also enhance the off rate of the pointed end (6). Gelsolin markedly accelerates actin filament disassembly by severing actin filaments and increasing the number of free pointed ends (see Fig. 1A). As compared with gelsolin (picomolar Kd), the affinity of ADF/cofilin for F-actin is considerably weaker (micromolar Kd) (10). ADF/cofilin function varies with pH (affinity being highest near pH 8.0), with the state of phosphorylation of serine 3 (phosphorylation inactivates actin binding) and with the nucleotide bound to actin (100-fold higher affinity for ADP-actin as compared with ATP- or ADP-Pi-bound actin) (3). Gelsolin also binds to ADP-actin with higher affinity than ATP-actin (11); however, given gelsolin's very tight binding, the physiologic significance of this difference is unclear. The ability of both gelsolin and ADF/cofilin to bind actin is inhibited by the phosphoinositide by product phosphatidylinositol bisphosphate (3, 12). Unlike ADF/cofilin, which is calcium insensitive, gelsolin requires calcium to bind to actin. Calcium binding alters the conformation of the molecule, unmasking the amino-terminal actin-binding sites. Also unlike ADF/cofilin, which does not block monomer exchange at either end of the filament, gelsolin caps the barbed filament end with high affinity (subpicomolar Kd) (12).
Table 1.
Comparison of the functional and morphological studies of gelsolin and ADF/cofilin
| Experimental method | Gelsolin | ADF/cofilin |
|---|---|---|
| Purified proteins | Ca2+-sensitive, severs actin filaments, caps barbed end. Severing and capping increases free pointed ends, binds actin with very high affinity, and dissociates phalloidin from actin filaments. | pH-sensitive, weakly severs actin filaments, increases pointed end off rate. Severing increases free pointed and barbed ends, binds actin with lower affinity, and dissociates phalloidin from actin filaments. |
| Extracts | Serial platelet extracts demonstrate binding and dissociation of gelsolin from actin immediately after agonist exposure. | Increased depolymerization rate of Listeria actin rocket tails in Xenopus extract, increased dissociation of pointed end in platelet extracts. |
| Whole cells | Localized to Listeria rocket tails, overexpression increases the speed of Listeria and increases fibroblast chemotaxis | Localized to Listeria rocket tails, overexpression in fibroblasts causes rounding of cells, increases chemotaxis in Dictyostelium. |
| Mouse knockout experiments | Decreased chemotaxis of neutrophils, defective platelet function, abnormal stress fibers in fibroblasts, defective retraction of neurites, increased susceptibility to ischemic brain damage, and defective podosome formation in osteoclasts. | Not done. |
See text for references.
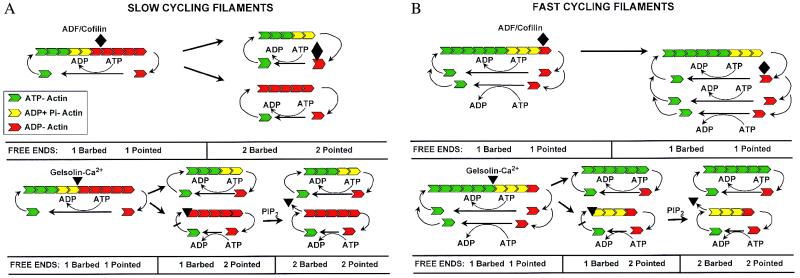
Figure 1.
Schematic diagram of actin-filament cycling before and after addition of ADF/cofilin or gelsolin-Ca2+. (A) Actin filaments have a barbed and pointed end, as demonstrated by electron micrographs of myosin head-decorated actin filaments. The myosin heads bind at a 45° angle, defining a pointed and barbed end. The barbed end has a higher affinity for actin monomers and has a more rapid exchange rate than the pointed end. At steady state (Left), actin monomers come on the filament at the barbed end as ATP actin monomers (solid green). As they enter the filament, the ATP is hydrolyzed, forming an intermediate ADP + Pi (yellow) and then ADP-actin (red). ADP- actin has a much lower affinity for filament ends (Kd of approximately 6 μM) than ATP actin (Kd of 0.03 to 0.1 μM for the barbed end). Once the ADP-actin monomer dissociates from the pointed end, ATP is exchanged for ADP, and the actin monomer can again add to the barbed end [when ADF is bound to ADP-actin, profilin is required for this exchange to occur efficiently (3)]. This process is called treadmilling, because an individual monomer adds to the barbed end, cycles through the filament, and then dissociates from the pointed end. Treadmilling allows remodeling of actin filaments. The rate of remodeling depends on the number of free filament ends. Doubling the number of free ends of the same concentration of filamentous actin would be expected to double the rate of treadmilling (i.e., the identical concentration of actin existing as short filaments would be expected to recycle more rapidly than a population of long filaments). In the slow-cycling filament, significant amounts of ADP-actin exist in the filament; therefore, ADF/cofilin can bind and sever. Each time ADF severs, it doubles the filament ends. Gelsolin-Ca2+ has very high affinity for filaments and rapidly severs them. Because gelsolin also binds and caps the barbed ends, severing and capping doubles the free pointed ends but does not increase the free barbed ends. However, when chemotactic signal transduction pathways increase the concentration of phosphatidylinositol bisphosphate, gelsolin will dissociate from the barbed end, and the number of free barbed ends will double. (B) When actin filaments rapidly cycle, there is reduced time for ATP hydrolysis, and the filament would be expected to have a lower content of ADP-actin. This condition would be expected to reduce the ability of ADF/cofilin to bind to the filament and thus reduce its ability to sever and increase the number of free ends. Therefore, ADF/cofilin would lose its ability to enhance treadmilling. However, gelsolin-calcium, by virtue of its high-affinity binding, would be expected to continue to bind and sever filaments with low ADP content and therefore continue to be effective at increasing the number of filament ends. Space does not permit illustration of exchange of the two actin monomers per filament after gelsolin severing (Bottom Right).
Cell-extract experiments have emphasized the importance of ADF/cofilin as an enhancer of actin-filament turnover. Xenopus extracts and L. monocytogenes have been used to examine actin turnover in bacterial actin-filament rocket tails after removal and addition of ADF/cofilin and gelsolin. Because ADF/cofilin accelerated the rate of depolymerization and gelsolin did not, it was concluded that ADF/cofilin, but not gelsolin, is essential for depolymerizing actin filaments (13). This conclusion was also supported by experiments by using platelet extracts (14). However, the extracts used for all experiments lacked free calcium (they contained 5–10 mM EGTA); therefore, gelsolin would not be expected to bind actin under these conditions. Because the introduction of calcium into cytoplasmic extracts activates multiple calcium-sensitive proteases, it has not been possible to study gelsolin function in extracts. Furthermore, the regulation of gelsolin function is likely to require oscillations in ionized calcium and phosphatidylinositol bisphosphate concentrations, conditions that cannot be reproduced in vitro. Platelet extracts generated at various time points after agonist stimulation support this supposition and demonstrate that gelsolin rapidly associates and then dissociates from actin within seconds, raising the possibility that gelsolin can enhance actin-filament recycling by severing followed by uncapping (15) (see Fig. 1A).
Investigations of intact cells provide strong evidence for the importance of gelsolin as well as ADF/cofilin in the recycling of actin in vivo. Both gelsolin and ADF/cofilin localize to Listeria actin rocket tails, suggesting that in vivo both proteins are likely to play a role in actin-filament reorganization (13, 16). Microinjection of constitutively active gelsolin accelerates Listeria motility, as does the overexpression of full-length gelsolin (16). Overexpression of gelsolin has also been shown to enhance fibroblast chemotaxis (17), whereas overexpression of Xenopus cofilin (XAC) causes fibroblasts to round up and lose their ability to adhere to substrate (3). Overexpression of cofilin in Dictyostelium discoideum increases the motility of the ameba (18). Mutational analysis of yeast cofilin demonstrates a strong correlation between in vivo and in vitro functional defects in cofilin's ability to disassemble actin, strongly supporting a role for cofilin in the recycling of actin in yeast (19).
The gelsolin knockout mouse provides the strongest evidence in support of gelsolin's role in actin-filament dynamics. Gelsolin-null fibroblasts have increased the numbers of stress fibers that are resistant to depolymerization by cytochalasin B. Furthermore, the chemotactic rates of fibroblasts and neutrophils are reduced and platelets grossly malfunction, causing a marked delay in blood coagulation. Gelsolin-null platelets contain extremely long actin filaments and fail to spread normally in response to agonists (20). Susceptibility to ischemic brain damage is increased (21), and neurite retraction is defective (22). Finally, podosome formation by osteoclasts is blocked, leading to reduced bone resorption (23). To date, viable ADF/cofilin knockout mice have not been generated.
Despite strong evidence pointing to an integral role for gelsolin in regulating actin-filament turnover and actin-based motility, two recent reviews claim that the enhanced turnover of actin filaments observed in vivo is primarily because of the action of the ADF/cofilin family (3, 24). In a recent issue of PNAS, McGrath et al. have addressed this issue in living cells by using photoactivation of fluorescence and fluorescence recovery after photobleaching, combined with measurement of the number of free barbed and pointed ends in permeabilized cells (25). Their studies represent the most thorough analysis to date of actin-filament turnover rates, actin-filament length, and filament number in different populations of motile cells. They find that cells with faster motility have shortened actin-filament half lives and a reduced fraction of polymerized actin as compared with cells with slow motility. The turnover rate of actin filaments positively correlates, whereas the fraction of polymerized actin inversely correlates with cell speed. Gelsolin-null fibroblasts move more slowly, have very prolonged filament half lives, and contain a greater fraction of polymerized actin as compared with wild-type fibroblasts. By permeabilizing similar populations of cells and measuring the nucleation rate of pyrene-labeled actin in the presence and absence of the barbed-end capping agent cytochalasin B, they have also estimated the number of free barbed and pointed filament ends. These values in turn allow an estimate of average filament length and number, as well as the percentage of free barbed ends. The number of filaments as well as the number of free barbed ends increases, and the mean actin-filament length decreases in more rapidly moving cells, consistent with an increase in actin-filament severing and uncapping. Of course, nucleation of new actin filaments by the Arp2/3 complex may also contribute to the increased number of free barbed ends. However, because this complex caps the pointed ends of actin filaments, ARP2/3 nucleation cannot explain the marked increase in pointed ends observed in rapidly moving cells. It is also unlikely that the increase in filament number is caused by an increase in cofilin severing activity. Measurement of cofilin in the Triton-soluble (not associated with the actin cytoskeleton) and Triton-insoluble (associated with the actin cytoskeleton) fractions demonstrates a 50% decrease in cofilin binding to actin filaments in highly motile cells as compared with more stationary cells. The reduced half lives of filaments in motile cells reduce the concentration of ADP in actin filaments, and this condition reduces the affinity of ADF/cofilin for actin. This is a curious paradox. A number of investigators have claimed that cofilin is required for the rapid recycling of actin filaments at the leading edge, yet increased turnover depletes the filament of ADP-actin and reduces ADF/cofilin activity (see Fig. 1B). This may explain why the leading edge of actively migrating keratocytes is depleted of ADF/cofilin (26).
The authors are very modest in their conclusions and point out that gelsolin-null cells continue to move, albeit at slower speeds; they attribute this behavior to ADF/cofilin. What should the general reader conclude from these findings? The wisest conclusion is that both ADF/cofilin and gelsolin are likely to play important roles in the dynamic cycling of actin filaments. Future work should be directed at determining how these two proteins may work together in the cell to coordinate remodeling of the actin cytoskeleton. Perhaps ADF/cofilin is responsible for the recycling of actin in stationary cells, whereas gelsolin is responsible for recycling filaments at the leading edge of motile cells. The ADF/cofilin vs. gelsolin controversy reminds us that oversimplification and exclusivity will only interfere with progress toward our ultimate goal, to understand how the cell truly regulates the actin cytoskeleton to achieve actin-based motility.
Footnotes
See companion article on page 6532 in issue 12 of volume 97.
References
- 1.Bamburg J E, Harris H E, Weeds A G. FEBS Lett. 1980;121:178–182. doi: 10.1016/0014-5793(80)81292-0. [DOI] [PubMed] [Google Scholar]
- 2.Yin H L, Stossel T P. Nature (London) 1979;281:583–586. doi: 10.1038/281583a0. [DOI] [PubMed] [Google Scholar]
- 3.Bamburg J. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 4.Hatanaka H, Ogura K, Moriyama K, Ichikawa S, Yahara I, Inagaki F. Cell. 1996;85:1047–1055. doi: 10.1016/s0092-8674(00)81305-7. [DOI] [PubMed] [Google Scholar]
- 5.Van Troys M, Dewitte D, Verschelde J L, Goethals M, Vandekerckhove J, Ampe C. J Biol Chem. 1997;272:32750–32758. doi: 10.1074/jbc.272.52.32750. [DOI] [PubMed] [Google Scholar]
- 6.McGough A, Pope B, Chiu W, Weeds A. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGough A, Chiu W, Way M. Biophys J. 1998;74:764–772. doi: 10.1016/S0006-3495(98)74001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen P G, Janmey P A. Biochemistry. 1994;269:32916–32923. [PubMed] [Google Scholar]
- 9.Itchekovkin I, Han J, Pang K M, Knecht D A, Condeelis J S. Cell Motil Cytoskeleton. 2000;45:293–306. doi: 10.1002/(SICI)1097-0169(200004)45:4<293::AID-CM5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 10.Ressad F, Didry D, Xia G-X, Hong Y, Chua N-H, Pantaloni D, Carlier M-F. J Biol Chem. 1998;273:20894–20902. doi: 10.1074/jbc.273.33.20894. [DOI] [PubMed] [Google Scholar]
- 11.Laham L E, Way M, Yin H L, Jamney P A. Eur J Biochem. 1995;234:1–7. doi: 10.1111/j.1432-1033.1995.001_c.x. [DOI] [PubMed] [Google Scholar]
- 12.Sun H Q, Yamamoto M, Mejillano M, Yin H L. J Biol Chem. 1999;274:33179–33182. doi: 10.1074/jbc.274.47.33179. [DOI] [PubMed] [Google Scholar]
- 13.Rosenblatt J, Agnew B J, Abe H, Bamburg J R, Mitchison T J. J Cell Biol. 1997;136:1323–1332. doi: 10.1083/jcb.136.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlier M-F, Laurent V, Santolini J, Melki R, Didry D, Xia G-X, Hong Y, Chua N-H, Pantaloni D. J Cell Biol. 1997;136:1307–1323. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lind S E, Janmey P A, Chaponnier C, Herbert T J, Stossel T P. J Cell Biol. 1987;105:833–842. doi: 10.1083/jcb.105.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laine R O, Phaneuf K L, Cunningham C C, Kwiatkowksi D, Azuma T, Southwick F S. Infect Immun. 1998;66:3775–3782. doi: 10.1128/iai.66.8.3775-3782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham C C, Stossel T P, Kwiatkowski D J. Science. 1991;251:1233–1236. doi: 10.1126/science.1848726. [DOI] [PubMed] [Google Scholar]
- 18.Aizawa H, Sutoh K, Yahara J. J Cell Biol. 1996;132:335–344. doi: 10.1083/jcb.132.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lappalainen P, Drubin D G. Nature (London) 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- 20.Witke W, Sharpe A H, Hartwig J H, Azuma T, Stossel T P, Kwiatkowski D J. Cell. 1995;81:41–51. doi: 10.1016/0092-8674(95)90369-0. [DOI] [PubMed] [Google Scholar]
- 21.Endres M, Fink K, Zhu J, Stagliano N E, Bondada V, Geddes J W, Azuma T, Mattson M P, Kwiatkowski D J, Moskowitz M A. J Clin Invest. 1999;103:347–354. doi: 10.1172/JCI4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu M, Witke W, Kwiatkowski D J, Kosik K S. J Cell Biol. 1997;138:1279–1287. doi: 10.1083/jcb.138.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chellaiah M, Kizer N, Silva M, Alvarez U, Kwiatkowski D, Hruska K A. J Cell Biol. 2000;148:665–678. doi: 10.1083/jcb.148.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlier M-F, Ressad F, Pantaloni D. J Biol Chem. 1999;274:33827–33830. doi: 10.1074/jbc.274.48.33827. [DOI] [PubMed] [Google Scholar]
- 25.McGrath J L, Osborn E A, Tardy Y S, Dewey C F, Jr, Hartwig J H. Proc Natl Acad Sci USA. 2000;97:6532–6537. doi: 10.1073/pnas.100023397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svitkina T M, Borisy G G. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]