Background: β-Transaminases are promising biocatalysts for the synthesis of β-amino acids.
Results: The first three-dimensional structures were obtained of a native β-transaminase and complexes with a keto acid and two covalently bound β-amino acids.
Conclusion: Dual functionality of the carboxylate- and side chain-binding pockets allows binding of β- and α-amino acids.
Significance: These structures may facilitate the development of improved β-amino acid biocatalysts.
Keywords: Biosynthesis, Crystal Structure, Enzyme Inhibitors, Pyridoxal Phosphate, Site-directed Mutagenesis, Mesorhizobium, Aminotransferase, β-Amino Acid, β-Transaminase, Biocatalysis
Abstract
Chiral β-amino acids occur as constituents of various natural and synthetic compounds with potentially useful bioactivities. The pyridoxal 5′-phosphate (PLP)-dependent S-selective transaminase from Mesorhizobium sp. strain LUK (MesAT) is a fold type I aminotransferase that can be used for the preparation of enantiopure β-Phe and derivatives thereof. Using x-ray crystallography, we solved structures of MesAT in complex with (S)-β-Phe, (R)-3-amino-5-methylhexanoic acid, 2-oxoglutarate, and the inhibitor 2-aminooxyacetic acid, which allowed us to unveil the molecular basis of the amino acid specificity and enantioselectivity of this enzyme. The binding pocket of the side chain of a β-amino acid is located on the 3′-oxygen side of the PLP cofactor. The same binding pocket is utilized by MesAT to bind the α-carboxylate group of an α-amino acid. A β-amino acid thus binds in a reverse orientation in the active site of MesAT compared with an α-amino acid. Such a binding mode has not been reported before for any PLP-dependent aminotransferase and shows that the active site of MesAT has specifically evolved to accommodate both β- and α-amino acids.
Introduction
β-Amino acids occur as precursors of many natural and synthetic compounds that display a wide range of pharmacological activities. Altering and improving the pharmacological properties of these compounds critically depend on the availability of β-amino acids and their derivatives as building blocks. Therefore, several strategies for the synthesis of β-amino acids have been explored over the years, involving either synthetic (1, 2) or combined chemoenzymatic (3) methods. However, fully enzyme-based synthesis methods have clear advantages over synthetic or chemoenzymatic methods (4) and have the potential to increase the feasibility of biocatalytic or fermentative routes toward β-amino compounds.
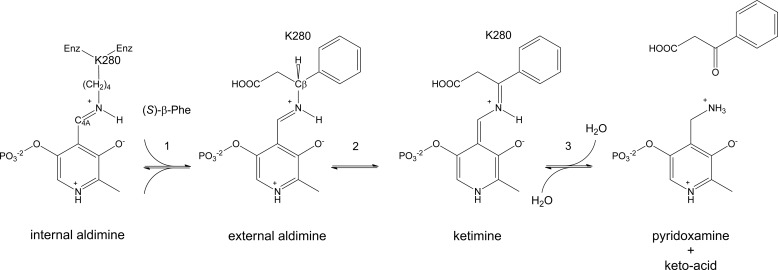
Pyridoxal 5′-phosphate (PLP)3-dependent aminotransferases (also called transaminases) are attractive for the production of amino acids because they have a broad substrate range, can be highly enantioselective, show a high catalytic activity, and are relatively stable (5). Aminotransferases catalyze the transfer of an amino group from an amino compound to a keto acid. In the first half-reaction, the amino group of the amino compound replaces the covalent Schiff base linkage, or imine bond, between the ϵ-amino group of a lysine and the C4A atom of the PLP cofactor, generating an external aldimine (Fig. 1). Lysine-assisted transfer of a proton from the external aldimine to the C4A atom of the cofactor results in a ketimine intermediate, which is hydrolyzed to yield pyridoxamine phosphate (PMP). Subsequently, in the second half-reaction, the amino group of PMP is transferred to a keto acid, which generates a new amino compound (Fig. 1) (6).
FIGURE 1.
Half-reactions catalyzed by MesAT, a β-specific PLP-dependent transaminase. An amino acid ((S)-β-Phe) enters the active site of the enzyme (Enz) (step 1), and an external aldimine is generated. Lys-280, a base, abstracts the Cβ proton (step 2) and transfers the proton to the C4A atom (1,3-prototropic shift; not shown), which results in the ketimine intermediate. Hydrolysis of the ketimine intermediate results in the formation of PMP and a keto acid (3-oxo-3-phenylpropanoic acid) (step 3).
The production of β-amino acids using PLP-dependent aminotransferases has been demonstrated for an ω-transaminase from Polaromonas sp. strain JS666 (7) and a β-transaminase from Mesorhizobium sp. strain LUK (MesAT) (8). The genes of these two enzymes were successfully cloned and expressed in Escherichia coli BL21(DE3) using pET-based expression systems (7, 8), and MesAT has been crystallized (9). MesAT is an enzyme of 445 amino acids with molecular weight of 45 kDa, forms dimers in solution, and accepts β- and α-amino acids (8, 10). The enzyme can convert the aliphatic β-amino acids (R)-3-amino-5-methylhexanoic acid and (R)-3-aminobutyric acid and the aromatic β-amino acid (S)-β-Phe to the corresponding β-keto acids using pyruvate or 2-oxoglutarate as the amino acceptor. In the reverse reaction, MesAT can be used to produce (S)-β-Phe from its β-keto acid ethyl ester in a coupled enzyme reaction containing a lipase to generate the keto acid in situ and rac-3-aminobutyric acid as amino donor (8). Unfortunately, the more stable keto ester is a very poor substrate for aminotransferase-mediated conversion to (S)-β-Phe.
Information on three-dimensional structures of β-aminotransferases is currently lacking. To enable rational protein engineering approaches for improving the activity of the enzyme for application in the biosynthesis of β-amino acids, we have elucidated the crystal structure of MesAT in the native state, as well as in complex with β-amino acids, a keto acid, and an inhibitor. The structure of the enzyme conforms to a fold type I aminotransferase structure, but the hydrophobic binding pocket is located on the 3′-oxygen side of the PLP cofactor rather than on its phosphate side as found in other aminotransferases. On the other hand, an α-amino acid binds in a normal orientation, with the α-carboxylate on the 3′-oxygen side and the side chain of the amino acid on the phosphate side of the PLP cofactor. The architecture of the active site explains how MesAT can accept β- and α-amino acids (8), whereas the aromatic α-amino acid aminotransferase (AroAT) from Paracoccus denitrificans (11) and most other aminotransferases accept only α-amino acids. The structure also explains the stereo preference of the enzyme for α- and β-amino acids.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The MesAT gene was codon-optimized for E. coli, synthesized by DNA2.0, Inc., and cloned into the expression plasmid pET28b+ with an N-terminal His6 tag using NdeI/HindIII restriction sites. After transformation into E. coli BL21(DE3), cells were grown at 37 °C in 1.4 liters of Terrific broth/sorbitol medium (12) containing 50 μg/ml kanamycin. Protein expression was induced with 0.4 mm isopropyl β-d-thiogalactopyranoside when the absorbance of the culture at 600 nm reached 0.6–0.8, and the temperature was adjusted to 17 °C. The cells were grown for another 48 h; harvested; and resuspended in buffer containing 20 mm Tris-HCl (pH 8.0), 500 mm NaCl, 20 mm imidazole, 1 mg of DNase I (Roche Applied Science), and one cOmplete EDTA-free protease inhibitor tablet (Roche Applied Science). The cells were disrupted at 4 °C by sonication, followed by centrifugation at 31,000 × g for 1 h at 4 °C.
The supernatant was applied to a HisTrap HP affinity chromatography column (GE Healthcare), and after washing, MesAT was eluted with 15 column volumes of a linear gradient of 20–500 mm imidazole in elution buffer (20 mm Tris-HCl (pH 8.0) and 500 mm NaCl). The fractions corresponding to the peak were pooled, and the imidazole was removed by buffer exchange using a HiPrep 26/10 desalting column (GE Healthcare) equilibrated with 25 mm Tris-HCl (pH 8.0) and 0.01% (v/v) β-mercaptoethanol. The resulting protein fractions were applied to a Q-Sepharose HP anion exchange column (GE Healthcare) and eluted with 15 column volumes of a linear gradient of 0–1 m NaCl in 25 mm Tris-HCl (pH 8.0). The fractions containing active enzyme were pooled, concentrated (Ultracel, Mr 30,000 cutoff, Amicon), and applied to a Superdex 200 10/300 GL size exclusion chromatography column (GE Healthcare) equilibrated with 20 mm Tris-HCl (pH 8.0) containing 200 mm NaCl. After elution, the fractions corresponding to the protein peak were pooled, concentrated (Amicon), and dialyzed overnight against buffer containing 20 mm Tris-HCl (pH 7.5). The sample was subsequently concentrated to 10 mg/ml as judged by a protein assay, and the purity of the sample was checked with silver-stained SDS-polyacrylamide gels using a PhastSystem (GE Healthcare).
Mutagenesis
The R412A mutant gene of MesAT (supplemental Table S2) was constructed by site-directed mutagenesis (QuikChange, Stratagene) and transformed into E. coli DH5α ElectroMAX electrocompetent cells (Invitrogen). For overexpression, E. coli BL21(DE3) was used. The mutant construct was confirmed by sequence analysis (GATC Biotech).
Protein Crystallization
A Mosquito crystallization robot (TTP LabTech) was used to search for suitable crystallization conditions. Crystallization experiments were set up at 20 °C. Crystals were found in a JCSG+ Suite (Qiagen) condition containing 0.1 m HEPES (pH 7.5), 8% (v/v) ethylene glycol, and 10% (w/v) PEG 8000. After optimization, it was found that this was also the optimal condition for crystal growth, with crystals reaching sizes of 80 × 50 × 30 μm. Crystals were transferred to a cryoprotection solution consisting of mother liquor with 20% (v/v) ethylene glycol. This was done in four steps of 5 min each, starting with a solution containing 2% (v/v) ethylene glycol, followed by solutions of 5, 10, and finally 20% (v/v) ethylene glycol. Crystals from this last solution were cryo-cooled in liquid nitrogen. For amino acid binding studies, the same steps were followed, but with the cryoprotection solutions supplemented with 2, 5, 10, and 20 mm (S)-β-Phe (PepTech Corp.), (R)-3-amino-5-methylhexanoic acid (Fluorochem), 2-oxoglutarate disodium salt (Fluka), or 2-aminooxyacetic acid (AOA) (Aldrich).
Diffraction Data Collection and Processing
Diffraction data were collected at beamlines ID14-1 and ID14-2 of the European Synchrotron Radiation Facility (ESRF, Grenoble, France) and at beamline X13 of the European Molecular Biology Laboratory Outstation at the Deutsches Elektronen-Synchrotron (DESY, Hamburg, Germany). Reflections were indexed and integrated using XDS (13), and scaling and merging of the data were done with the program SCALA (14) from the CCP4 Software Suite (15). Phaser (16) was used for molecular replacement with a mixed input model generated by the FFAS03 server (17) on the basis of the structures of glutamate-1-semialdehyde 2,1-aminomutase from Thermus thermophilus strain HB8 (Protein Data Bank code 2E7U), d-phenylglycine aminotransferase from Pseudomonas stutzeri strain ST-201 (code 2CY8), and 4-aminobutyrate aminotransferase from pig (code 1OHV (18)). The resulting model was subjected to successive rounds of automatic model building with ARP/wARP (19), followed by manual model building in Coot (20). REFMAC5 was used for refinement of the atomic coordinates and atomic B-factors (21). Data collection and refinement statistics are given in supplemental Table S3. After refinement, the model was validated with MolProbity (22). Stereochemical restraints for the amino acid analogs were generated using the PRODRG2 server (23). Root mean square deviations (r.m.s.d.) were calculated with the RMSDcalc tool of the CaspR server (24), and structural homologs of MesAT were obtained from the Dali server (25). PISA from the CCP4 Software Suite was used for protein interface analysis (26). Simulated annealing composite omit maps where generated with PHENIX (27). Chemical structure drawings were made using the ChemDraw program (CambridgeSoft), and PyMOL (28) was used to generate images of the protein structure.
Enzyme Assay and Analytical Methods
Aminotransferase assays were performed with 10 mm amino donor ((S)-β-Phe), 10 mm amino acceptor (pyruvate), and enzyme at 37 °C in 50 mm MOPS (pH 7.6) containing 50 μm PLP (Acros Organics). Samples were taken at different times and treated according to the following procedure: to 50 μl of sample was added 50 μl of 2 m HCl to quench the reaction. The sample was left on ice for 5 min and neutralized by adding 45 μl of 2 m NaOH, followed by adding 50 μl of demineralized water. In an HPLC autosampler (Jasco), 1-μl sample was mixed with 2 μl of o-phthalaldehyde (Sigma) solution (15 mg of o-phthalaldehyde was dissolved in 50 μl of absolute ethanol, which was then mixed with 4.42 ml of 0.4 m sodium borate (pH 10.4), 15 μl of 30% (w/v) Brij 35 (Fluka), and 11 μl of β-mercaptoethanol) and 5 μl of 0.4 m sodium borate (pH 10.4). The o-phthalaldehyde-derivatized samples were analyzed by HPLC using a Alltech Adsorbosphere C18 column (5 μm, 4.6 × 100 mm) in a Jasco HPLC system. Separation of o-phthalaldehyde-derivatized imines was achieved at room temperature at a flow rate of 1 ml/min using a gradient of eluent A (5% THF in 20 mm sodium acetate (pH 5.5)) and eluent B (99% pure CH3CN) as follows: start with 100:0 eluent A/eluent B for 5 min; change from 100:0 to 80:20 in 7 min; continue with 80:20 for 4 min; change from 80:20 to 40:60 in 8 min; continue with 40:60 for 6 min; change from 40:60 to 100:0 in 2 min; and finally continue for 5 min at 100:0 for re-equilibration of the column. The eluate was analyzed by UV light (338 nm) using a Jasco UV-2075 Plus detector and a Jasco FP-920 fluorescence detector (350 nm excitation and 450 nm emission). Retention times for derivatized l-α-alanine and (S)-β-Phe were 7.7 and 23.2 min, respectively. One unit of enzyme activity is defined as the amount of enzyme that produces 1 μmol/min alanine from 10 mm pyruvate (sodium salt; Fluka) and 10 mm (S)-β-Phe (Acros Organics).
To determine the Vmax and Km values of the R412A mutant of MesAT, initial rate assays were done with varying concentrations of (S)-β-Phe or pyruvate, fixing the non-variant substrate at 10 mm. The reactions were started by adding 680 μg of purified protein and incubating at 37 °C. Inhibition studies were performed by preincubating the reaction mixture (lacking (S)-β-Phe but containing 40 μg of wild-type enzyme) with 5 mm AOA or 5 mm dl-propargylglycine (2-amino-4-pentynoic acid; Sigma-Aldrich) for 5 min, after which (S)-β-Phe was added.
RESULTS
Overexpression, Purification, and Enzyme Activity Measurements of Wild-type and R412A MesAT
Previously, an overexpression system of MesAT was reported that gave a yield of 1.4 mg of pure protein/liter of culture (8). To obtain enhanced expression, we used a codon-optimized synthetic gene that was equipped with an N-terminal His tag, cloned it under the control of the T7 promoter in a pET vector, and cultivated the transformed E. coli BL21(DE3) cells in Terrific broth/sorbitol medium at 17 °C. This resulted in an expression level that allowed the isolation of ∼10 mg of pure enzyme/liter of culture (supplemental Table S1). The specific activity of the purified wild-type enzyme was 1.6 units/mg, which is similar to what was reported previously (8).
Structure Determination of MesAT
The holoenzyme crystallizes in space group C2 with three molecules (chains A, B, and C) per asymmetric unit. The three-dimensional structure of MesAT was elucidated at 2.5 Å resolution by molecular replacement and refined at 1.65 Å resolution. No density is defined for the first 30 N-terminal residues and the C-terminal residue Met-445. The three molecules are very similar to each other, with r.m.s.d. values for the Cα atom positions of ∼0.2–0.25 Å. The three molecules form 1.5 dimer in the asymmetric unit; one dimer consists of chains A and B (supplemental Fig. S1), and the other dimer is made up of chain C and a chain C from a neighboring asymmetric unit related by crystallographic 2-fold symmetry. Both dimers are very similar, with r.m.s.d. values in the order of 0.2 Å (Cα atoms). The two chains in the dimer interact tightly, burying a surface area of ∼4400 Å2, which is one-quarter of their total surface area. The presence of dimers in the crystal is in agreement with the occurrence of MesAT dimers in solution (10).
Structure of the MesAT Monomer
The MesAT monomer has a curved shape and consists of a PLP-binding domain (residues 112–334) and a domain formed by the N and C termini of the polypeptide chain (NC-domain; residues 1–111 and 335–445) (supplemental Fig. S1). These two domains line a cleft into which the PLP cofactor protrudes. The monomer contains 11 α-helices (of at least two or more turns) and 13 β-strands that form a mixed central seven-stranded β-sheet in the large domain and 2 three-stranded antiparallel β-sheets in the NC-domain (supplemental Fig. S1). The overall structure is similar to that of aspartate aminotransferase, the archetypical representative of fold type I aminotransferases (Z-score of 21, r.m.s.d. of 4.2 Å for 309 Cα atoms, and 16% sequence identity; Protein Data Bank code 1BKG) (29, 30), and AroAT from P. denitrificans (Z-score of 21, r.m.s.d. of 4.8 Å for 326 Cα atoms, and 15% sequence identity; code 1AY4) (11, 31), which is specific for l-α-Phe.
PLP Cofactor
Each MesAT monomer contains a PLP cofactor covalently anchored via an imine bond (Schiff base) to the ϵ-amino group of Lys-280. The amide protons of Gly-145, Thr-146, and Thr-314 (from monomer B in the case of the AB dimer) anchor the phosphate group of the PLP cofactor to the protein backbone. The pyridine ring of the PLP cofactor is stacked between Val-255 (at the si-face of the pyridine ring (32)) and Tyr-172 (at the re-face of the pyridine ring). The nitrogen atom of the pyridine ring is in hydrogen bonding distance of Asp-253. These interactions keep the PLP cofactor secured in the active site.
Binding of (S)-β-Phe
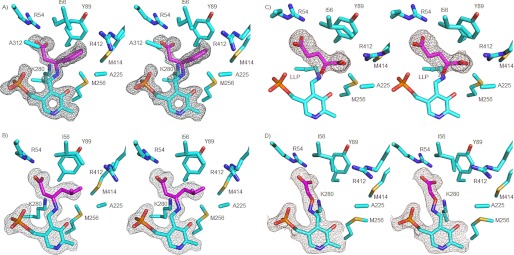
To analyze how (S)-β-Phe binds in the active site, a crystal structure of MesAT with bound (S)-β-Phe was solved at 1.7 Å resolution. (S)-β-Phe binds covalently to the PLP cofactor via its β-amino group, replacing the imine bond between the ϵ-amine of Lys-280 and the C4A atom of the PLP cofactor (Fig. 2A). It binds with an estimated occupancy of ∼80% in the three subunits. The carboxylate group of (S)-β-Phe has a salt bridge interaction with the Nϵ and Nη2 atoms of Arg-54. The aromatic ring of (S)-β-Phe is bound between the side chains of Tyr-89 and Tyr-172 with edge-to-face interactions (Fig. 2A) and also has van der Waals interactions with Ile-56, Ala-225, Met-256, and Met-414 from monomer A and with Ala-312 from monomer B; these residues line a hydrophobic binding pocket that is capped by Arg-412 (monomer A). Thus, residues from both monomers A and B contribute to the binding of the aromatic side chain of (S)-β-Phe. The side chain amino group of Lys-280 is close to the Cβ R-proton of (S)-β-Phe (3.3 Å), in agreement with its role in proton transfer (Fig. 1). The binding of (S)-β-Phe does not induce large-scale conformational changes or domain movements in MesAT. Only local conformational changes have occurred (see “Discussion”).
FIGURE 2.
Stereo figures of simulated annealing composite 2mFo − DFc omit maps contoured at 1σ. A, (S)-β-Phe (magenta) bound to the PLP cofactor. For clarity, Tyr-172 has been omitted here and in B–D. B, (R)-3-amino-5-methylhexanoic acid (magenta). For clarity, Ala-312 has been omitted here as well as in C and D. C, 2-oxoglutarate (magenta) bound in the active site of MesAT. LLP, 2-lysine 3-hydroxy-2-methyl-5-phosphonooxymethylpyridin-4-ylmethane, the internal aldimine. D, AOA (magenta), a β-alanine mimic, bound in the active site of MesAT.
In monomer A, a second (S)-β-Phe molecule is present in a surface pocket, with its aromatic ring stacked between Leu-269 and Leu-368. Its carboxylate and amino groups point into the solvent and do not interact with the protein. However, in the surface pockets of monomers B and C, electron density is present only for the aromatic ring of an (S)-β-Phe molecule, but not for the amino and carboxylate groups. From this, we conclude that (S)-β-Phe binds nonspecifically in this surface pocket.
Binding of (R)-3-Amino-5-methylhexanoic Acid
To investigate how an aliphatic β-amino acid such as (R)-3-amino-5-methylhexanoic acid (8) binds in the active site, a 1.95 Å resolution crystal structure was determined of MesAT in complex with this compound. (R)-3-Amino-5-methylhexanoic acid binds covalently to the PLP cofactor via its β-amino group, replacing the imine bond between the ϵ-amine of Lys-280 and the C4A atom of the PLP cofactor (Fig. 2B). It binds with an estimated occupancy of ∼90% in the three subunits. The carboxylate group of (R)-3-amino-5-methylhexanoic acid has a salt bridge interaction with the Nϵ and Nη2 atoms of Arg-54, and the aliphatic side chain (C4, C5, and C6 atoms) binds in the hydrophobic binding pocket, where also the aromatic side chain of (S)-β-Phe was observed to bind (see above). Binding of (R)-3-amino-5-methylhexanoic acid produces only limited local conformational changes in MesAT.
Binding of 2-Oxoglutarate
To analyze how a keto acid binds in the active site of MesAT, we determined the crystal structure of the enzyme with bound 2-oxoglutarate at 1.85 Å resolution (Fig. 2C). The compound binds noncovalently in the active site. Its α-carboxylate has a bidentate salt bridge interaction with the Nη1 and Nη2 atoms of Arg-412. The γ-carboxylate binds with one of its oxygen atoms to the Nϵ and Nη2 atoms of Arg-54; the other oxygen atom is not involved in hydrogen bond formation. The keto oxygen atom of 2-oxoglutarate is located at 3.0 Å from the nitrogen atom of the Lys-280 side chain, which is covalently bound to the C4A atom of the PLP cofactor. The binding of 2-oxoglutarate thus leaves the internal aldimine intact. The orientation of 2-oxoglutarate in the active site of MesAT suggests that amino group transfer from PMP results in the synthesis of l-glutamate, in agreement with chiral HPLC analysis of the product of a reaction with (S)-β-Phe as amino donor and 2-oxoglutarate as amino acceptor (data not shown). Binding of 2-oxoglutarate does not result in large-scale conformational changes in MesAT. However, Arg-412 reorients its side chain such that its Nη1 and Nη2 atoms can make a salt bridge interaction with the α-carboxylate of 2-oxoglutarate. This reorientation of the arginine side chain in response to the binding of an amino acid in the active site of an aminotransferase is referred to as the “arginine switch” (33).
The importance of Arg-412 for activity was confirmed by mutagenesis. The protein yield of the R412A variant was similar to that of the wild-type enzyme. The kcat values of the R412A mutant were significantly lower than those of the wild-type enzyme (Table 1). Furthermore, the Km value for pyruvate had increased by a factor of 28, whereas that for (S)-β-Phe had decreased by a factor of 4. The relationship between the reaction rate of the R412A mutant and the substrate concentration indicated substrate inhibition, as observed in the wild-type enzyme (Table 1) (34).
TABLE 1.
Specific activity at 37 °C of wild-type MesAT and the R412A mutant using (S)-β-Phe as amino donor and pyruvate as amino acceptor
| MesAT | Activity |
(S)-β-Phe |
Pyruvate |
Source | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Specific | Relative | Km | kcat | kcat/Km | Ki | Km | kcat | kcat/Km | Ki | ||
| units/mg | % | mm | s−1 | s−1 mm−1 | mm | mm | s−1 | s−1 mm−1 | mm | ||
| WT | 1.6 | 100 | 1.2a | 1.3 | 1.1 | 3.2a | 3.9a | 1.3 | 0.33 | 177a | Ref. 8 |
| R412A | 6 × 10−3 | 0.4 | 0.29 | 5 × 10−3 | 2 × 10−2 | 47 | 109 | 4.1 × 10−2 | 3.8 × 10−4 | 2500 | This study |
a Adopted from Kim et al. (8).
Binding of AOA
AOA (35) and 2-amino-4-pentynoic acid (propargylglycine) (36, 37) are known inhibitors of aminotransferase activity. Preincubation of MesAT with these inhibitors at 5 mm resulted in activity decreases of 10% upon treatment with dl-propargylglycine and of 96% upon treatment with AOA. Analysis of the 1.9 Å resolution crystal structure of MesAT with bound AOA (Fig. 2D) shows that the amino group of AOA binds covalently to the C4A atom of PLP, as has also been observed for the interaction of AOA with aspartate aminotransferase (38). The ether oxygen atom (Oχ1) is close to Lys-280 (2.9 Å), and the carboxylate group binds via a salt bridge to the Nϵ and Nη2 atoms of Arg-54. The binding of AOA to the PLP cofactor is irreversible; the amine–Oχ1 bond cannot be weakened by Lys-280. Binding of AOA to the PLP cofactor thus prevents PMP formation and thereby inhibits aminotransferase activity. No significant conformational changes are observed upon AOA binding.
DISCUSSION
Structure of MesAT
The structure of MesAT presented here is the first structure of a transaminase with specificity toward β-amino acids as well as α-amino acids. The structure of the enzyme is similar to that of aspartate aminotransferase (39), the archetypical fold type I aminotransferase. The enzyme assembles into a homodimer, in which residues from both monomers A and B contribute to the binding of amino acids and their respective oxo acid substrates. In contrast to aspartate aminotransferase, the binding of ligands does not induce large domain movements. Structural changes that do occur in MesAT upon substrate/inhibitor binding are localized to the active site and consist of a 16° rotation of the pyridine ring of the PLP cofactor upon formation of the external aldimine. This rotation liberates Lys-280 and allows it to function as the proton-transferring lysine. Another structural change is the rearrangement of the Arg-412 side chain, also known as the arginine switch (33), upon binding of 2-oxoglutarate (mentioned above).
Different Binding Modes of (S)-β-Phe, (R)-3-Amino-5-methylhexanoic Acid, and 2-Oxoglutarate
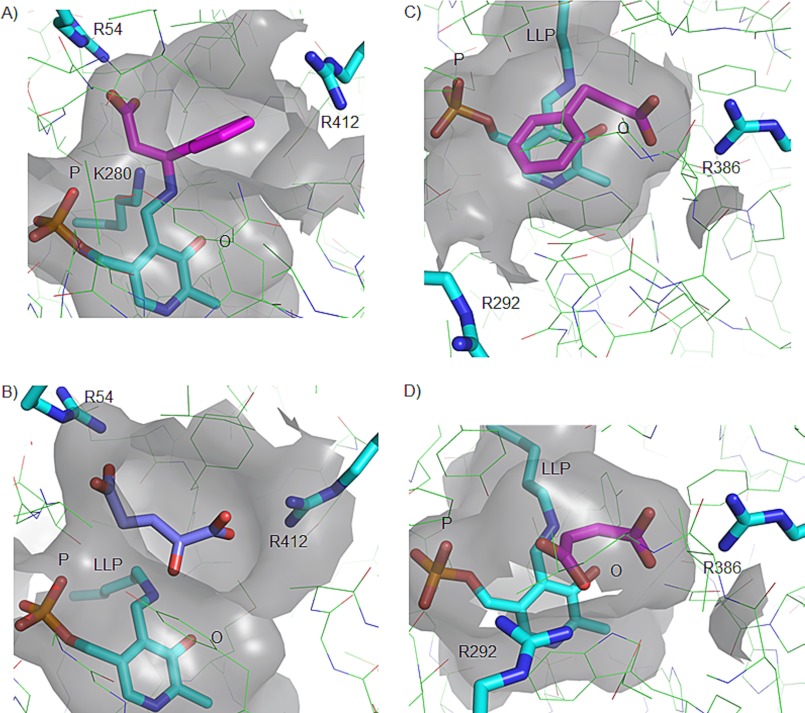
The way in which 2-oxoglutarate binds in the active site of MesAT and the manner in which (S)-β-Phe and (R)-3-amino-5-methylhexanoic acid bind are very different. The side chains of (S)-β-Phe (Fig. 2A) and (R)-3-amino-5-methylhexanoic acid (Fig. 2B) bind in a pocket on the 3′-oxygen side of the PLP cofactor, which we denote the O-pocket. However, the side chain of the α-keto acid 2-oxoglutarate, a model for the α-amino acid l-glutamate, binds with its γ-carboxylate group on the phosphate side of the PLP cofactor (Fig. 2C), in the P-pocket. Thus, MesAT has two distinct side chain-binding pockets, one for the side chains of β-amino acids (O-pocket) (Fig. 3) and one, on the other end of the active site, for the side chains of α-keto acids and presumably also α-amino acids (P-pocket) (Fig. 3). The enzyme has also two carboxylate-binding pockets; one, involving Arg-54 in the P-pocket, binds the α-carboxylate group of β-amino acids and AOA, a β-alanine mimic (Fig. 2D), whereas the other, which contains Arg-412 in the O-pocket, binds the α-carboxylate of α-keto acids such as 2-oxoglutarate (Fig. 2C). As a consequence, an α-amino acid and a β-amino acid have very different binding modes in MesAT (Fig. 3, A and B).
FIGURE 3.
Comparison of the active site architectures of the β-aminotransferase MesAT (A and B) and the α-aminotransferase AroAT (C and D). A and B, surface renditions of the active site of MesAT bound to (S)-β-Phe (magenta) and to 2-oxoglutarate (magenta), respectively. C and D, surface renditions of the active site of AroAT bound to 3-phenylpropionate (magenta) based on Protein Data Bank code 1AY8 (31) and to maleate (magenta) based on code 1AY5 (31), respectively. The positions of the PLP cofactor and Arg-412 and Arg-386 in the active sites of MesAT and AroAT, respectively, are similar. Arg-412 represents the arginine switch of MesAT, and Arg-292 represents the arginine switch of AroAT. LLP, 2-lysine 3-hydroxy-2-methyl-5-phosphonooxymethylpyridin-4-ylmethane, the internal aldimine. See “Discussion” for explanation of the P-pocket (P) and O-pocket (O).
Intriguingly, the O-pocket that binds the aliphatic and hydrophobic side chain of β-amino acids also binds the α-carboxylate group of α-keto acids and presumably also of α-amino acids. This dual functionality is made possible by a switch in position of the Arg-412 side chain, the arginine switch residue (33). When a β-amino acid binds, the Arg-412 side chain is oriented away from the active site, providing space in the O-pocket for the hydrophobic side chain of the β-amino acid (Fig. 2, A and B). This orientation of Arg-412 is stabilized by a hydrogen bond of its side chain Nη atom to the carbonyl oxygen atom of Ala-225 (monomer B) (data not shown). In contrast, upon 2-oxoglutarate binding, the side chain of Arg-412 switches back toward the hydrophobic O-pocket (Fig. 2C); the hydrogen bond with Ala-225 is broken, and Arg-412 now has a salt bridge interaction with the α-carboxylate of 2-oxoglutarate. In this way, the Arg-412 side chain allows the enzyme to accept both α-amino/α-keto acids and aliphatic or aromatic β-amino/β-keto acids in the same active site. Because of the dual functionality of the active site pockets of MesAT, we prefer to use O- and P-pocket (rather than L- and S-pocket) nomenclature, which reflects the presumed size of these pockets (40).
For aminotransferase activity, Arg-412 is virtually essential, as reflected by the 870- and 55-fold reduction of the catalytic efficiencies for pyruvate and (S)-β-Phe, respectively, upon mutation of this residue to Ala (Table 1). Whereas the decreased apparent Km for (S)-β-Phe can be due to the reduced catalytic rate at 10 mm pyruvate, the increased Km for pyruvate strongly supports that Arg-412 is important for binding pyruvate through electrostatic interactions with its carboxylate.
Comparison of MesAT and AroAT
To investigate how MesAT differs from an aminotransferase that accepts only α-amino acids, the MesAT structures were compared with AroAT from P. denitrificans (11). In both enzymes, the PLP cofactor has a similar position and orientation. In contrast to MesAT, AroAT has a single α-carboxylate-binding pocket, and charged as well as uncharged α-amino acids bind with their α-carboxylate groups to Arg-386 in the O-pocket (Fig. 3, C and D). In MesAT, the α-carboxylate group of (S)-β-Phe binds to Arg-54 in the P-pocket, whereas the side chain of (S)-β-Phe is bound in the O-pocket (Fig. 3A). As a consequence, in MesAT, β-amino acids bind in a reverse orientation in comparison with the substrates of AroAT (Fig. 3, A, C, and D). Moreover, the arginine switch of MesAT (Arg-412) is located in the O-pocket of the enzyme, where it binds the α-carboxylate group of an α-amino acid, whereas in AroAT, the arginine switch (Arg-292) is located in the P-pocket of the enzyme and binds the side chain carboxylate group of α-amino/α-keto acids (Fig. 3, A–D). These differences between MesAT and AroAT show that although these enzymes share the same fold, the architectures of their active sites are very different. In MesAT, the active site architecture has evolved to accommodate β-amino acids while retaining the ability to accommodate α-amino/α-keto acids.
Covalent Adducts in the Active Site of MesAT Represent External Aldimine Intermediates
Structures obtained from crystals soaked with (S)-β-Phe and (R)-3-amino-5-methylhexanoic acid show that covalent PLP–β-amino acid adducts have formed in the active site of MesAT (Fig. 2, A and B). These adducts represent external aldimine intermediates because the Nβ, Cα, Cβ, and Cγ atoms are not coplanar, as would occur in the ketimine intermediate (Fig. 1). The non-coplanarity of the Nβ, Cα, Cβ, and Cγ atoms suggests that the reaction has stopped before abstraction of the Cβ proton.
Different explanations for the trapping of the external aldimine may be considered. Proton abstraction is most efficient if, in the transition state, the bond to be broken is oriented perpendicular to the plane of the PLP ring system (41). The crystal structures indicate that the Cβ proton is indeed nearly perpendicular to the PLP plane, deviating 20–30° from the perpendicular position. Such a deviation is probably not sufficient to fully prevent proton abstraction.
Another explanation may be related to the observation that in none of the external aldimine intermediate-bound structures density is present for a hydrolytic water molecule near the Cβ atom that could convert the ketimine intermediate into the pyridoxamine intermediate (Fig. 1). The equilibrium of the aminotransferase reaction in the crystal structure of MesAT may thus lie toward the external aldimine intermediate rather than the ketimine intermediate, which could explain why the external aldimine intermediates of (S)-β-Phe and (R)-3-amino-5-methylhexanoic acid are trapped.
Enantioselectivity of MesAT
MesAT is enantioselective toward the β-amino acids (S)-β-Phe, (R)-3-amino-5-methylhexanoic acid, and (R)-3-aminobutyric acid (8). These preferred enantiomers have the same stereo configuration of functional groups on the Cβ atom as (S)-β-Phe. The preference for these enantiomers can be fully explained by the architecture of the active site, which forces these substrates to bind in an orientation in which the carboxylate group binds to Arg-54 in the P-pocket and the side chain in the O-pocket, followed by addition of an amino group at the si-face of the β-carbon of the β-keto acid. The insights obtained from the three-dimensional structure of MesAT upon its interaction with α- and β-amino/keto acids may facilitate structure-based protein engineering efforts to enhance the biocatalytic potential of β-transaminases for the production of β-amino acids of pharmacological interest.
Supplementary Material
Acknowledgments
We acknowledge the ESRF and the EMBL Outstation at DESY for provision of synchrotron radiation facilities. We thank the ID14-1 and ID14-2 (ESRF) and X13 (DESY) beamline staff for assistance.
This work was supported by the Netherlands Ministry of Economic Affairs and the B-Basic Partner Organizations through B-Basic, a public-private Nederlandse Organisatie voor Wetenschappelijk Onderzoek-Advanced Chemical Technologies for Sustainability (NWO-ACTS) Program.

This article contains supplemental Fig. S1 and Tables S1–S3.
- PLP
- pyridoxal 5′-phosphate
- PMP
- pyridoxamine phosphate
- MesAT
- Mesorhizobium sp. strain LUK aminotransferase
- AroAT
- aromatic α-amino acid aminotransferase
- AOA
- 2-aminooxyacetic acid
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Cole D. C. (1994) Recent stereoselective synthetic approaches to β-amino acids. Tetrahedron 50, 9517–9582 [Google Scholar]
- 2. Weiner B., Szymański W., Janssen D. B., Minnaard A. J., Feringa B. L. (2010) Recent advances in the catalytic asymmetric synthesis of β-amino acids. Chem. Soc. Rev. 39, 1656–1691 [DOI] [PubMed] [Google Scholar]
- 3. Liljeblad A., Kanerva L. T. (2006) Biocatalysis as a profound tool in the preparation of highly enantiopure β-amino acids. Tetrahedron 62, 5831–5854 [Google Scholar]
- 4. Koeller K. M., Wong C. H. (2001) Enzymes for chemical synthesis. Nature 409, 232–240 [DOI] [PubMed] [Google Scholar]
- 5. Taylor P. P., Pantaleone D. P., Senkpeil R. F., Fotheringham I. G. (1998) Novel biosynthetic approaches to the production of unnatural amino acids using transaminases. Trends Biotechnol. 16, 412–418 [DOI] [PubMed] [Google Scholar]
- 6. Braunstein A. E. (1964) Binding and reactions of the vitamin B6 coenzyme in the catalytic center of aspartate transaminase. Vitam. Horm. 22, 451–484 [DOI] [PubMed] [Google Scholar]
- 7. Bea H. S., Park H. J., Lee S. H., Yun H. (2011) Kinetic resolution of aromatic β-amino acids by ω-transaminase. Chem. Commun. 47, 5894–5896 [DOI] [PubMed] [Google Scholar]
- 8. Kim J., Kyung D., Yun H., Cho B. K., Seo J. H., Cha M., Kim B. G. (2007) Cloning and characterization of a novel β-transaminase from Mesorhizobium sp. strain LUK: a new biocatalyst for the synthesis of enantiomerically pure β-amino acids. Appl. Environ. Microbiol. 73, 1772–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim B., Park O. K., Bae J. Y., Jang T. H., Yoon J. H., Do K. H., Kim B. G., Yun H., Park H. H. (2011) Crystallization and preliminary x-ray crystallographic studies of β-transaminase from Mesorhizobium sp. strain LUK. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 231–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim J., Kyung D., Yun H., Cho B. K., Kim B. G. (2006) Screening and purification of a novel transaminase catalyzing the transamination of aryl β-amino acid from Mesorhizobium sp. LUK. J. Microbiol. Biotechnol. 16, 1832–1836 [Google Scholar]
- 11. Oue S., Okamoto A., Nakai Y., Nakahira M., Shibatani T., Hayashi H., Kagamiyama H. (1997) Paracoccus denitrificans aromatic amino acid aminotransferase: a model enzyme for the study of dual substrate recognition mechanism. J. Biochem. 121, 161–171 [DOI] [PubMed] [Google Scholar]
- 12. Barth S., Huhn M., Matthey B., Klimka A., Galinski E. A., Engert A. (2000) Compatible-solute-supported periplasmic expression of functional recombinant proteins under stress conditions. Appl. Environ. Microbiol. 66, 1572–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 14. Kabsch W. (2010) Integration, scaling, space group assignment, and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 16. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jaroszewski L., Rychlewski L., Li Z., Li W., Godzik A. (2005) FFAS03: a server for profile-profile sequence alignments. Nucleic Acids Res. 33, W284–W288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Storici P., De Biase D., Bossa F., Bruno S., Mozzarelli A., Peneff C., Silverman R. B., Schirmer T. (2004) Structures of γ-aminobutyric acid (GABA) aminotransferase, a pyridoxal 5′-phosphate, and [2Fe-2S] cluster-containing enzyme, complexed with γ-ethynyl-GABA and with the antiepilepsy drug vigabatrin. J. Biol. Chem. 279, 363–373 [DOI] [PubMed] [Google Scholar]
- 19. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Automated macromolecular model building for x-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 22. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schuttelkopf A. W., van Aalten D. M. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 24. Claude J. B., Suhre K., Notredame C., Claverie J. M., Abergel C. (2004) CaspR: a web server for automated molecular replacement using homology modeling. Nucleic Acids Res. 32, W606–W609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holm L., Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 27. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. DeLano W. L. (2002) The PyMOL Molecular Graphics System, San Carlos, CA [Google Scholar]
- 29. Grishin N. V., Phillips M. A., Goldsmith E. J. (1995) Modeling of the spatial structure of eukaryotic ornithine decarboxylases. Protein Sci. 4, 1291–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schneider G., Käck H., Lindqvist Y. (2000) The manifold of vitamin B6-dependent enzymes. Structure 8, R1–R6 [DOI] [PubMed] [Google Scholar]
- 31. Okamoto A., Nakai Y., Hayashi H., Hirotsu K., Kagamiyama H. (1998) Crystal structures of Paracoccus denitrificans aromatic amino acid aminotransferase: a substrate recognition site constructed by rearrangement of hydrogen bond network. J. Mol. Biol. 280, 443–461 [DOI] [PubMed] [Google Scholar]
- 32. Soda K., Yoshimura T., Esaki N. (2001) Stereospecificity for the hydrogen transfer of pyridoxal enzyme reactions. Chem. Rec. 1, 373–384 [DOI] [PubMed] [Google Scholar]
- 33. Eliot A. C., Kirsch J. F. (2004) Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem. 73, 383–415 [DOI] [PubMed] [Google Scholar]
- 34. Tipton K. F. (1996) in Enzymology LabFax (Engel P. C., ed) pp. 115–174, Academic Press, San Diego, CA [Google Scholar]
- 35. John R. A., Charteris A. (1978) The reaction of amino oxyacetate with pyridoxal phosphate-dependent enzymes. Biochem. J. 171, 771–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marcotte P., Walsh C. (1975) Active site-directed inactivation of cystathionine γ-synthetase and glutamic pyruvic transaminase by propargylglycine. Biochem. Biophys. Res. Commun. 62, 677–682 [DOI] [PubMed] [Google Scholar]
- 37. Tanase S., Morino Y. (1976) Irreversible inactivation of aspartate aminotransferases during transamination with l-propargylglycine. Biochem. Biophys. Res. Commun. 68, 1301–1308 [DOI] [PubMed] [Google Scholar]
- 38. Marković-Housley Z., Schirmer T., Hohenester E., Khomutov A. R., Khomutov R. M., Karpeisky M. Y., Sandmeier E., Christen P., Jansonius J. N. (1996) Crystal structures and solution studies of oxime adducts of mitochondrial aspartate aminotransferase. Eur. J. Biochem. 236, 1025–1032 [DOI] [PubMed] [Google Scholar]
- 39. Kirsch J. F., Eichele G., Ford G. C., Vincent M. G., Jansonius J. N., Gehring H., Christen P. (1984) Mechanism of action of aspartate aminotransferase proposed on the basis of its spatial structure. J. Mol. Biol. 174, 497–525 [DOI] [PubMed] [Google Scholar]
- 40. Shin J. S., Kim B. G. (2002) Exploring the active site of amine:pyruvate aminotransferase on the basis of the substrate structure-reactivity relationship: how the enzyme controls substrate specificity and stereoselectivity. J. Org. Chem. 67, 2848–2853 [DOI] [PubMed] [Google Scholar]
- 41. Toney M. D. (2011) Controlling reaction specificity in pyridoxal phosphate enzymes. Biochim. Biophys. Acta 1814, 1407–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.