Background: We examined a key example of regulated transport that involves modulation of cargo binding by a coat component.
Results: We identified a recycling sorting signal recognized by ACAP1 and showed that this binding is regulated by autoinhibition.
Conclusion: The mechanistic understanding of regulated cargo binding has been advanced.
Significance: We elucidated a key regulatory juncture that controls integrin recycling.
Keywords: Adhesion, Akt, G Proteins, Integrin, Receptor Recycling, ACAP1, Integrin, Recycling
Abstract
Coat complexes sort protein cargoes into vesicular transport pathways. An emerging class of coat components has been the GTPase-activating proteins (GAPs) that act on the ADP-ribosylation factor (ARF) family of small GTPases. ACAP1 (ArfGAP with coiled-coil, ankyrin repeat, and PH domains protein 1) is an ARF6 GAP that also acts as a key component of a recently defined clathrin complex for endocytic recycling. Phosphorylation by Akt has been shown to enhance cargo binding by ACAP1 in explaining how integrin recycling is an example of regulated transport. We now shed further mechanistic insights into how this regulation is achieved at the level of cargo binding by ACAP1. We initially defined a critical sequence in the cytoplasmic domain of integrin β1 recognized by ACAP1 and showed that this sequence acts as a recycling sorting signal. We then pursued a combination of structural, modeling, and functional studies, which suggest that phosphorylation of ACAP1 relieves a localized mechanism of autoinhibition in regulating cargo binding. Thus, we have elucidated a key regulatory juncture that controls integrin recycling and also advanced the understanding of how regulated cargo binding can lead to regulated transport.
Introduction
Intracellular transport can be divided into two general types, constitutive versus regulated transport. Studies on constitutive transport events have been at the forefront in advancing a mechanistic understanding of how vesicular transport is achieved. In particular, coat complexes are now appreciated to act as the core machinery in initiating transport. They accomplish this role through two major functions, membrane deformation to achieve vesicle formation and cargo sorting that directs the proper packaging of cargoes into vesicles. A detailed mechanistic understanding of how coat components act in either process has been achieved in recent years (1, 2). In comparison, although regulated transport is appreciated to underlie multiple physiologic events, how this type of transport can be achieved by regulating the function of a coat component has been less clear.
One of the key physiologic examples of regulated transport has been the stimulation-dependent recycling of surface integrins. This process is critical for cell migration because it underlies the dynamic redistribution of surface integrins to the leading edge of cells in achieving directional migration (3–5). As such, there has been intense interest in identifying the signaling components and the transport factors that act in this event. With respect to the transport factors, recent advances have led to the identification of core effectors predicted to mediate the different mechanistic steps of integrin recycling (6–9). Moreover, small GTPases and their catalytic regulators that modulate these core effectors are also being elucidated (10–15).
Distilled to its mechanistic core, regulated transport involves upstream signaling impacting on downstream transport events. Key factors that act at the interface of these two general events have been identified in the case of integrin recycling. Different growth factors have been found to instigate a canonical signaling cascade that results in the protein kinase Akt phosphorylating ACAP1 at Ser-554, which results in the enhanced binding of ACAP1 to integrin β1 at the recycling endosome in stimulating integrin recycling (16).
ACAP1 is a member of the ADP-ribosylation factor (ARF)6 GTPase-activating protein (GAP) family (17). ARF GAPs have been known conventionally as regulators of their cognate small GTPases (18). However, multiple members of this family have been discovered to possess also a novel function as ARF effectors, which involves their roles as coat components (19–23). In particular, ACAP1 is an ARF6 GAP that has been found to function also as a component of a recently defined clathrin complex for endocytic recycling (6). As such, the discovery that regulated cargo binding by ACAP1 underlies how integrin recycling is regulated (16) represents one of the best characterized examples of how regulated transport can be achieved by modulating a coat component. In this study, we advance a further mechanistic understanding of this key example of regulated transport.
EXPERIMENTAL PROCEDURES
Chemicals, Proteins, and Cells
Protein A/G-agarose beads were obtained from Pierce. Glutathione-Sepharose 4B and PreScission protease were from GE Healthcare. GST fusion proteins were purified as described previously (24). His6-tagged full-length ACAP1 has been described previously (24). Recombinant forms of ACAP1 truncations were generated by initially expressing them as GST fusion proteins in Escherichia coli and then purified by binding to glutathione-Sepharose resin followed by PreScission protease cleavage according to the manufacturer's protocol.
A peptide containing 12 residues within the cytoplasmic domain of integrin β1 (H2N-HDRREFAKFEKE-OH) was obtained from CHI Scientific, Inc. (Maynard, MA). Another peptide derived from the cytoplasmic domain of Wbp1 has been described (25). HeLa cells were cultured based on the guidelines of American Type Culture Collection.
Antibodies
The following antibodies have been described previously (11): mouse TS2/16 against integrin β1, mouse 9E10 against the Myc epitope, and rabbit antiserum against ACAP1. Additional antibodies used in this study include mouse 6C5 against GAPDH (Applied Biosystems); mouse SAM1 against human integrin α5 (Santa Cruz Biotechnology); secondary antibodies conjugated to horseradish peroxidase, Cy3, or Cy5 (Jackson ImmunoResearch Laboratories), and Alexa Fluor 546-conjugated transferrin (Invitrogen).
Plasmids, Mutagenesis, and Transfections
The Myc-tagged forms of ACAP1, wild-type and mutant (residues 525–566 deleted, ΔLinker), were subcloned into the SalI and BamHI sites of the mammalian expression vector pcDNA3.1 for transfection studies. The cDNA sequences of different ACAP1 forms were amplified by PCR and then subcloned into the BamHI and EcoRI/SalI sites of the bacterial expression vector pGEX-6P-1 (GE Healthcare) to generate recombinant proteins. To generate different forms of the cytoplasmic domain of integrin β1 fused to GST, the cDNA sequence of β1 was amplified by PCR and then subcloned into the BamHI and EcoRI sites of the pGEX-4T-3 vector (GE Healthcare). Fusion constructs consisting of the β1 cargo peptide (HDRREFAKFEKE) fused to the C terminus of the S554D mutant of the C-terminal portion or the linker mutant were generated by adding an intervening flexible linker (GSSNSGSNSG) using the method of overlap PCR.
All site-directed mutagenesis was carried out using the QuikChange II XL site-directed mutagenesis kit (Stratagene) according to manufacturer's guidelines. Transient transfections were carried out using FuGENE 6 (Roche Applied science).
shRNA and Stable Cell Lines
Nucleotides 224–244 of human integrin β1 were targeted for shRNA (5′-GCCCUCCAGAUGACAUAGAAA; Thermal Scientific). To generate shRNA-resistant forms of human integrin β1, point mutations were introduced within the sequence targeted by shRNA (nucleotides 224–244). The specific mutations are shown underlined in the following sequence: GCCCTCCTGACGATATCGAAA.
For the stable expression of the shRNA sequence in HeLa cells, a lentiviral expression system (Thermal Scientific) was used according to the guidelines provided by the manufacturer. To generate shRNA-resistant forms of human integrin β1, the mutated sequences were subcloned into the BamHI and EcoRI sites of the pENTR plasmid (Invitrogen), followed by recombination with the pLenti6.2 plasmid (Invitrogen). Lentiviral particles that express different shRNA-resistant forms of human integrin β1 were generated using the ViraPower lentiviral expression system (Invitrogen) and blasticidin (Invitrogen) at 10 μg/ml for selection after viral transduction.
In Vivo Assays
Colocalization studies using laser confocal microscopy have been described previously (24). To assess the association of endosomal β1 with ACAP1, a co-precipitation approach was performed as described previously (16).
The endocytic recycling of β1 was assessed using a previously established recycling assay (11, 16). Briefly, the anti-β1 antibody (TS2/16) was bound to the surface of starved HeLa cells at 4 °C for 1 h, followed by incubation for 2 h at 37 °C to allow the accumulation of surface integrin at the recycling endosome. Cells were rinsed three times with ice-cold PBS and then washed twice with an acidic buffer (0.5% glacial acetic acid (pH 3.0) and 0.5 m NaCl) to release the remaining antibody-bound surface β1. Cells were then stimulated for recycling by incubation with prewarmed (37 °C) medium containing 20% FCS. At different time points as indicated, cells were subjected to a second acid wash to release antibody bound to any internalized β1 that had recycled to the cell surface. Cells were then lysed and immunoprecipitated by antibody-bound β1 using protein A/G-agarose beads. Analysis was performed by SDS-PAGE under nonreducing conditions, as the TS2/16 antibody works only for immunoblotting under these conditions.
To examine the recycling of mutant β1 (lacking the linker region) under basal conditions, primaquine was added as described previously (16). Briefly, because the ACAP1 mutant converts β1 recycling from regulated transport to constitutive transport, primaquine (0.3 mm) was added to accumulate endocytic β1 at the recycling endosome. This pool of β1 was then examined for recycling upon the washout of primaquine.
Pulldown Assays
Pulldown assays using GST fusion proteins were carried out as described previously (6). Briefly, GST fusion proteins on glutathione beads were incubated with soluble proteins (4 nm) at 4 °C for 1 h in 0.5 ml of incubation buffer (50 mm HEPES (pH 7.3), 300 mm NaCl, 90 mm KCl, 1 mm EDTA, and 0.5% Nonidet P-40). Beads were then pelleted by centrifugation at 1000 × g for 1 min at 4 °C, followed by two washes with incubation buffer and analysis by SDS-PAGE Western blotting. Coomassie Blue staining was carried out to detect the level of GST fusion proteins on beads. Peptide competition was performed as described previously (25).
Protein Preparation and Crystallization
The different forms of ACAP1 were expressed in E. coli strain BL21(DE3) using vector pGEX-6P-1 induced at 16 °C for 18 h with 0.2 mm isopropyl β-d-thiogalactopyranoside. The harvested cells were broken by sonication in PBS containing 140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 1.8 mm KH2PO4 (pH 7.0). After centrifugation for 30 min at 14,000 rpm, the supernatant was incubated with glutathione-Sepharose 4B at 4 °C. After removing the GST tag, the proteins were further purified by anion exchange chromatography (Resource Q, GE Healthcare) with a 0–1000 mm gradient of NaCl in 20 mm Tris (pH 8.5) and 10% glycerol and by size exclusion chromatography (Superdex 200 10/300 GL, GE Healthcare). Recombinant proteins were then concentrated to 3 mg/ml in 20 mm Tris (pH 8.5). Preliminary conditions were screened with an Index kit (Hampton Research) and then optimized. Crystals were grown in reservoir solution containing 0.2 m ammonium sulfate, 0.1 m sodium citrate (pH 5.0–5.1), and 14–16% PEG 3350 at 16 °C using the hanging drop vapor diffusion method.
Data Collection and Structure Determination
Crystals of the wild-type C-terminal portion were flash-frozen in reservoir solution and 25% glycerol. Single-wavelength anomalous diffraction data of the ACAP1 C-terminal domain were collected to 2.2 Å at beamline BL17A (Quantum-270 CCD detector) of the Photon Factory in Japan. Crystals of the C-terminal portion with the S554D point mutation and also the fusion construct (S554D-β1) were soaked in 0.2 m ammonium sulfate, 0.1 m sodium citrate (pH 5.1), and 20% PEG 8000 for 1 h and were frozen in liquid nitrogen in the same cryoprotecting solution as the wild type. Data were collected at beamline BL17U of the Shanghai Synchrotron Radiation Facility in China. All data were processed with the HKL2000 program suite (26).
The internal zinc ion positions were determined using the program SHELXD (27). The identified heavy atoms were refined, and initial phases were generated using the program SOLVE (28) with the single-wavelength anomalous diffraction experimental phasing module. Phase improvement, density modification, and a crude model were traced automatically using the program RESOLVE (29). OASIS (30), DM (31), RESOLVE, and OASIS-DM-ARP/wARP (32) iterative methods were used to further improve the phases and to complete the model. Loops that were not built automatically were then manually built and refined in Coot (33). Structure refinement was done using Refmac (34) or CNS (35). The initial phase of the C-terminal portion with the S554D point mutation and also of the fusion construct was calculated by molecular replacement using Phaser (36) based on the wild-type form of the C-terminal portion as the model. The data processing and structure refinement statistics are listed in Table 1. Surface and ribbon representations of molecular structures were generated using PyMOL.
TABLE 1.
Data collection and refinement statistics
| Data processing statistics | |||
| Parameters | C-terminal portion (WT) | C-terminal portion (S554D) | S554D-β1 fusion |
| Space group | P21212 | P21212 | P21212 |
| Cell parameters | a = 107.8, b = 163.5, and c = 41.1 Å; α = β = γ = 90° | a = 107.6, b = 163.5, and c = 41.2 Å; α = β = γ = 90° | a = 108.3, b = 164.9, and c = 41.7 Å; α = β = γ = 90° |
| Wavelength (Å) | 1.2800 | 1.2000 | 1.0000 |
| Resolution range (Å) | 48.6–2.20 (2.28–2.20)a | 50.0–2.30 (2.38–2.30) | 50.0–2.20 (2.24–2.20) |
| Completeness (%) | 97.3 (81.8) | 97.4 (91.9) | 97.3 (83.8) |
| Redundancy | 5.6 (3.3) | 11.5 (4.6) | 11.5 (7.5) |
| Average I/σ(I) | 19.9 (1.6) | 17.5 (2.0) | 37.3 (4.5) |
| Unique reflections | 36,997 | 32,470 | 37,309 |
| Rmerge (%)b | 6.7 (35.1) | 9.5 (39.3) | 9.9 (41.0) |
| Refinement statistics | |||
| r.m.s.d. (Å)c | 45–2.30 | 50–2.30 | 40–2.30 |
| Bond lengths (Å) | 0.011 | 0.008 | 0.012 |
| Bond angles | 1.272° | 1.121° | 1.351° |
| Rwork (%)d | 20.2 | 21.4 | 18.9 |
| Rfree (%)d | 21.9 | 24.1 | 22.5 |
a Corresponding parameters for the highest resolution shell are shown in parentheses.
b Rmerge = ΣhΣi|Iih − 〈Ih〉 |/ΣhΣi〈Ih〉, where 〈Ih〉 is the mean intensity of the observation Iih reflection h.
c r.m.s.d., root mean standard deviation.
d Rwork = Σ(‖Fp(obs)| − |Fp(calc)‖)/Σ|Fp(obs)|; Rfree = R factor for a selected subset (5%) of the reflections that was not included in prior refinement calculations.
Molecular Modeling of the Linker Region
The linker region (residues 525–568) was modeled using the crystal structure of the fusion construct as the template. This region was divided into multiple five-residue segments, which were then sequentially elongated toward the N- or C-terminal direction. The initial conformation of each segment was obtained by searching for homologous sequences in the Protein Data Bank. Possible positions of the each segment were then searched using the flexible docking program AutoDock4 (37) and the protein structure modeling program Modeler (38). The most closely related model was retained. After each elongation, energy minimization was performed to optimize the position of the newly added segment. These steps were repeated until the entire region was built. The final models were then optimized using further energy minimization and molecular dynamics simulations.
All energy minimization and molecular dynamics simulations were performed with NAMD (39) and the CHARMM all-atom force field for protein (40) with CMAP corrections. For the molecular dynamics simulations, the final models were solvated with TIP3P water boxes, and the systems were then neutralized with Na+ and Cl− ions. At the periodic boundary condition, a 12-Å cutoff was used for van der Waals interactions, and particle mesh Ewald summation was used to calculate the electrostatic interactions in all simulations. Temperature was controlled at 310 K using Langevin dynamics with the damping coefficient 1/ps, and pressure was controlled at 1 atm by the Langevin piston method. The backbone atoms observed in the crystal structures were fixed during the simulations. For each model, 20 ns of simulation was carried out, and the final snapshot was taken for analysis and comparison.
Accession Codes
The coordinates of the C-terminal portion (wild-type and S554D) and the S554D-β1 fusion construct have been deposited in the Protein Data Bank with accession codes 3JUE, 4F1P, and 3T9K, respectively.
RESULTS
ACAP1 Binds to a Sequence in β1 That Acts as a Recycling Sorting Signal
Experimentally, cargo sorting requires the demonstration that a coat component binds to a specific sequence in the cargo and also that this sequence is required for targeting the cargo into a particular intracellular pathway. In this regard, although we had previously found that ACAP1 binds directly to the cytoplasmic domain of integrin β1, the precise sequence in β1 recognized by ACAP1 and whether this sequence acts functionally as a recycling sorting signal had not been determined. Thus, we first sought to address these questions.
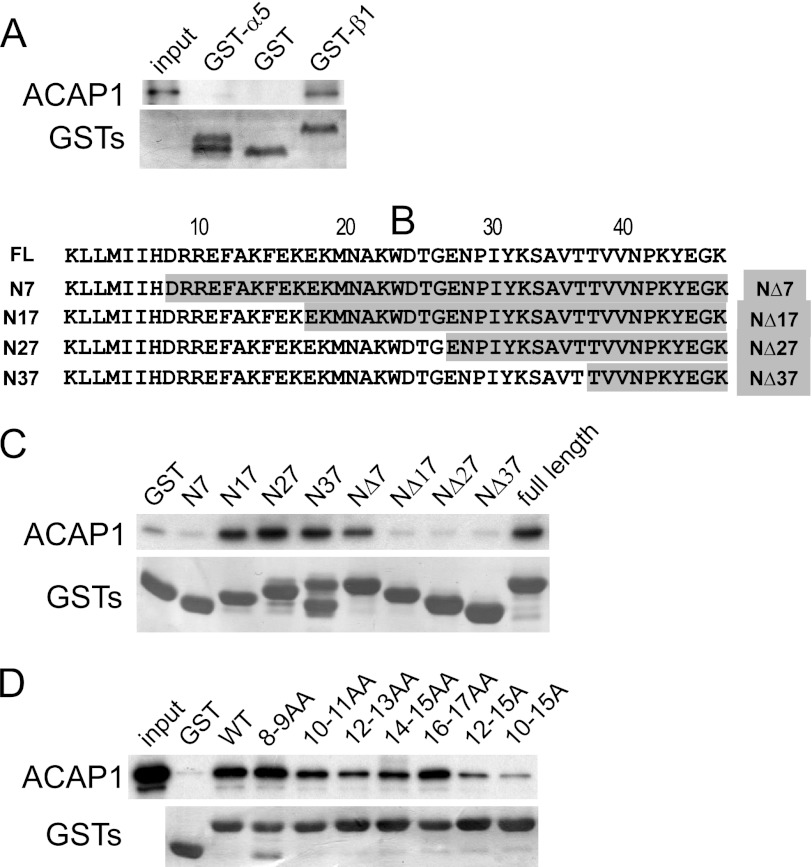
Previous studies have examined HeLa cells to elucidate how ACAP1 acts in integrin recycling (6, 16). In these cells, β1 pairs mainly with α5 in forming functional integrin heterodimers that recognize fibronectin as the ligand. Thus, we first confirmed that ACAP1 interacts directly with the cytoplasmic domain of β1, but not with the cytoplasmic domain of α5 (Fig. 1A). We next generated progressive truncations for the cytoplasmic domain of β1 from either its membrane-proximal or C-terminal end (Fig. 1B). These constructs were then analyzed as GST fusion proteins for binding to ACAP1 (Fig. 1C), which suggested that a region in β1 encompassed by residues 8–17 is critical for its direct binding to ACAP1. To further pinpoint the critical residues, we next generated a series of alanine-scanning mutants in the context of the entire cytoplasmic domain of integrin β1. Focusing on the region encompassed by residues 8–17, we systematically replaced paired native residues with paired alanines. With further analysis, we defined a region in β1 encompassed by residues 10–15 as being the most critical in mediating the direct binding of the β1 cargo by ACAP1 (Fig. 1D).
FIGURE 1.
Identifying a sequence in integrin β1 critical for its direct binding to ACAP1. A, ACAP1 binds directly to the cytoplasmic domain of integrin β1. The cytoplasmic domain of either α5 or β1 was fused to GST, and the resulting fusion proteins were bound to beads for incubation with soluble ACAP1 in pulldown experiments. B, truncation mutants of the cytoplasmic domain of integrin β1. Residues are numbered from the membrane-proximal end. FL, full-length ACAP1. C, identifying a region in the cytoplasmic domain of β1 responsible for its direct binding to ACAP1. Different truncations of β1 as GST fusion proteins were bound to beads for incubation with full-length ACAP1 as soluble recombinant protein in pulldown experiments. Beads were immunoblotted for ACAP1 and Coomassie Blue-stained for GST fusion proteins. D, alanine-scanning mutagenesis identifies specific residues within the cytoplasmic domain of β1 responsible for its direct binding to ACAP1. Residues within the cytoplasmic domain of β1 were mutated to alanines as indicated. The mutants as GST fusion proteins were then bound to beads for incubation with soluble ACAP1 in pulldown experiments. Beads were immunoblotted for ACAP1 and Coomassie Blue-stained for GST.
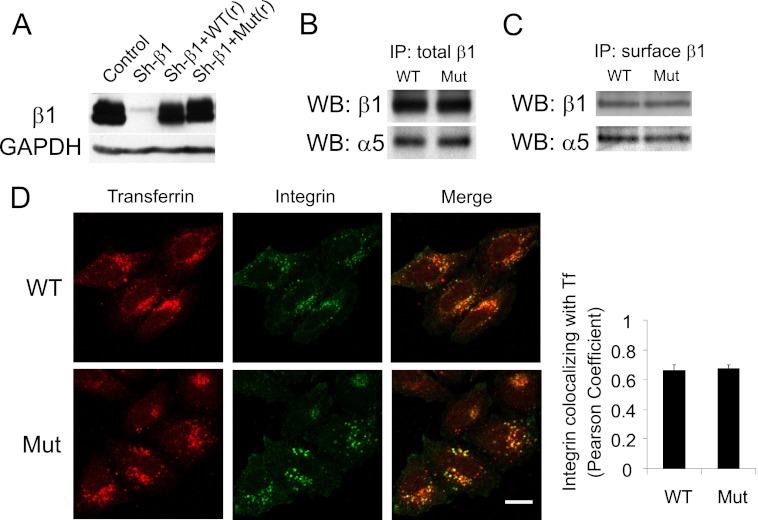
We then examined whether these residues in β1 act functionally as a recycling sorting signal. Stable cell lines were generated in which endogenous β1 was replaced with transfected forms (Fig. 2A). This was accomplished using a lentiviral system that expressed shRNA directed against endogenous β1 and that also stably expressed either wild-type or mutant (with residues 10–15 in the cytoplasmic domain mutated to alanines) β1 that was siRNA-resistant. Co-precipitation analysis confirmed that the β1 mutant could still pair with α5 in forming heterodimers (Fig. 2B). Moreover, the mutant heterodimers had levels at the cell surface that were similar to that of the wild type (Fig. 2C). Additionally, similar to the wild type, a surface pool of the mutant integrin could be tracked to accumulate at the recycling endosome under basal conditions (Fig. 2D).
FIGURE 2.
Replacing endogenous β1 with a β1 mutant that cannot bind efficiently to ACAP1. A, cell lines stably expressing different forms of integrin β1. The lentiviral system was used to replace endogenous β1 with transfected forms, which involved stably expressing shRNA against integrin β1 (Sh-β1), followed by stable expression of wild-type or mutant β1, which was resistant to shRNA through the introduced silent mutations. Expression was assessed by immunoblotting of whole cell lysates, with the level of GAPDH serving as a loading control. B, mutant β1 (Mut) assembles with α5 similarly as wild-type β1. Stable cell lines as described above were lysed, followed by immunoprecipitation (IP) for β1 and then immunoblotting for proteins as indicated. WB, Western blot. C, integrin heterodimers with mutant β1 are expressed at similar levels on the cell surface as those with wild-type β1. Stable cell lines as described above were bound with anti-β1 antibody at the cell surface. This pool of surface β1 was then immunoprecipitated, followed by immunoblotting for proteins as indicated. D, the mutant and wild-type integrins accumulate similarly at the recycling endosome under basal conditions. After allowing the surface pools of integrin and transferrin (Tf) to internalize for 2 h under basal conditions, cells were assessed by confocal microscopy comparing the distribution of integrin (green) and transferrin (red). Scale bar = 15 μm. The graph shows quantitative colocalization analysis, which reveals that wild-type and mutant integrins accumulate to a similar degree with internalized transferrin. The mean ± S.E. from three experiments is shown.
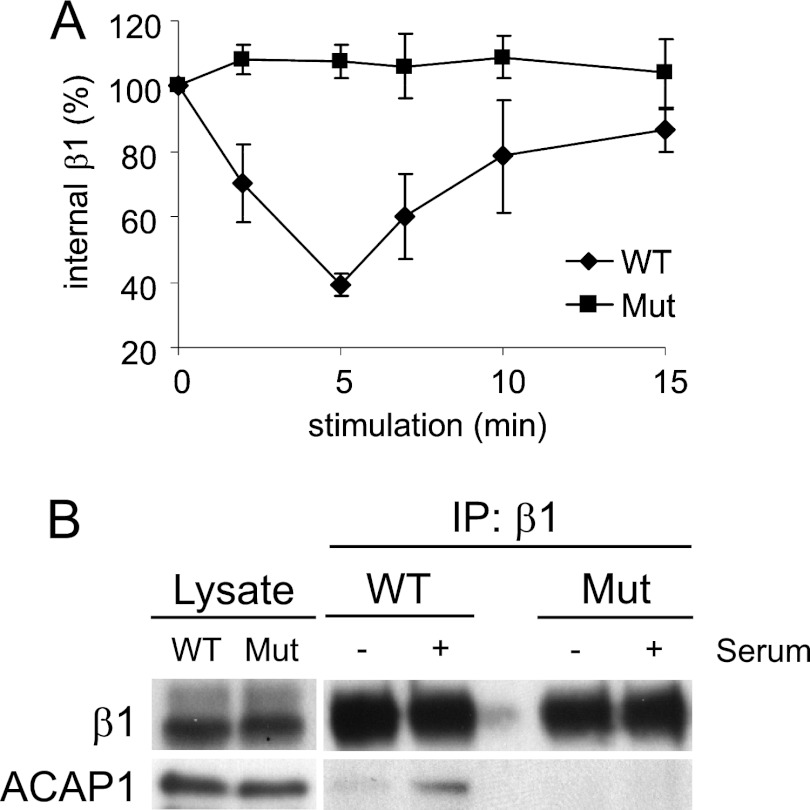
We then examined the recycling efficiency of the mutant from this internal compartment using a previously established integrin recycling assay (11, 16). Briefly, an antibody was used to bind to the surface pool of integrin β1. Under basal conditions, this surface pool underwent internalization and then accumulated the recycling endosome. Upon stimulation by serum (or by specific growth factors), this pool of β1 was induced to recycle to the cell surface. Using this assay, we found that the β1 mutant exhibited a reduced ability to recycle (Fig. 3A). We also confirmed that the β1 mutant exhibited defective binding to ACAP1 in vivo, which involved a co-precipitation approach to detect ACAP1 binding to the endosomal pool of β1 (Fig. 3B). Thus, we concluded that the residues in the cytoplasmic domain of β1 critical for direct binding to ACAP1 also function as a recycling sorting signal.
FIGURE 3.
Mutant β1 that cannot bind efficiently to ACAP1 also cannot recycle efficiently. A, integrin β1 with a mutation (Mut) of residues critical for its direct binding to ACAP1 shows a reduced ability to recycle. A lentiviral system was used to deplete endogenous β1, followed by stable expression of transfected forms as indicated. Surface β1 integrins (tracked by antibody binding) were then allowed to accumulate at the recycling endosome under basal conditions, followed by stimulation at the times indicated for their recycling. The mean ± S. E. from three experiments is shown. B, stimulation-dependent association of endosomal β1 with ACAP1 is reduced by mutations in β1 that reduce its binding to ACAP1. Endosomal β1, tracked as described above, was assessed for association with ACAP1 by co-precipitation. IP, immunoprecipitation.
C-terminal Portion of ACAP1 Recapitulates Regulated Cargo Binding
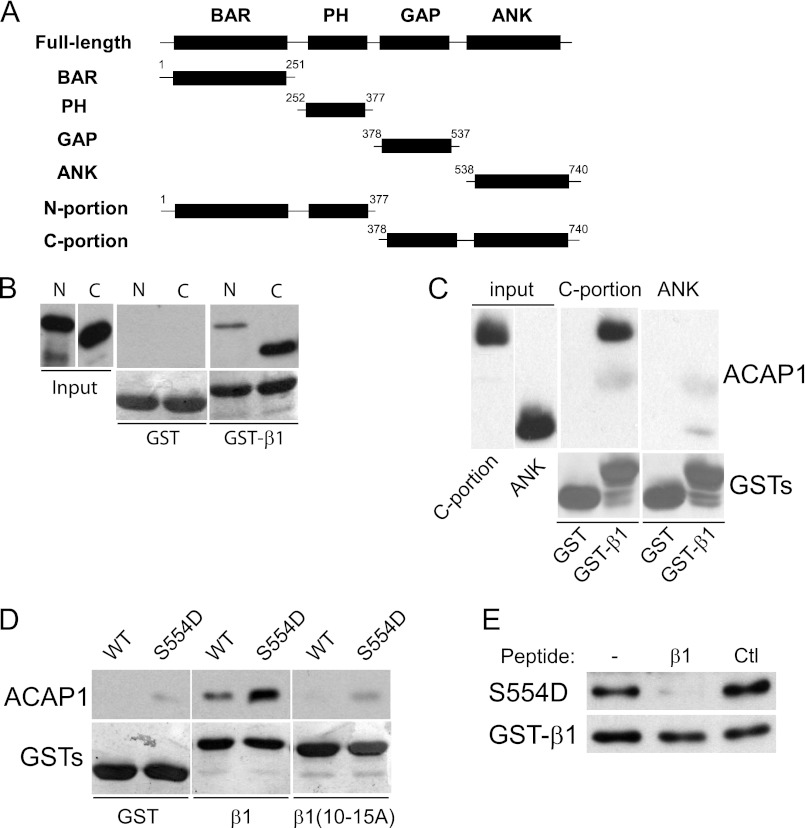
We next sought to define the region in ACAP1 responsible for its direct binding to the β1 cargo. ACAP1 is predicted to possess four domains (Fig. 4A), the BAR (Bin/amphiphysin/Rvs), PH (pleckstrin homology), GAP, and ANK (ankyrin repeat) domains (17). We initially sought to generate each domain separately. However, the GAP domain was found to be unstable. Pursuing an explanation, we noted that the domain organization of ACAP1 was predicted to be similar to that of ASAP2 (previously known as Pap-β) and ASAP3 (17). The structure of the GAP and ANK domains in these ARF GAPs had been solved (41, 42), suggesting that the ANK domain acts as a scaffold in stabilizing the folding of the GAP domain. Thus, we next generated the GAP-ANK domains of ACAP1 (referred to as the C-terminal portion of ACAP1), which was successfully purified as a recombinant protein. We also generated the remaining portions of ACAP1, consisting of its BAR and PH domains (referred to as the N-terminal portion). Both portions of ACAP1 were then assessed for their ability to bind to the β1 cargo in a pulldown experiment. This analysis revealed that the C-terminal portion bound to the β1 cargo significantly better than the N-terminal portion (Fig. 4B). We also found that the ANK domain alone showed a markedly reduced ability to bind β1 (Fig. 4C). Thus, the results led us to focus on the C-terminal portion construct for further studies on cargo binding.
FIGURE 4.
C-terminal portion of ACAP1 reproduces regulated cargo binding. A, schematic showing different domain constructs of ACAP1 generated as recombinant proteins. B, binding to the cytoplasmic domain of integrin β1 by different portions of ACAP1. The cytoplasmic domain of β1 as GST fusions bound to beads was incubated with the N- or C-terminal portion of ACAP1 in pulldown experiments. C, the ANK domain has reduced ability in binding to the β1 cargo. Recombinant forms of ACAP1 as indicated were incubated with GST-β1 on beads in pulldown experiments. D, regulation of cargo binding by Ser-554 is reproduced by the C-terminal portion. Different GST fusions on beads as indicated were incubated with the C-terminal portion of ACAP1 (WT or mutant S554D) in pulldown experiments. WB, Western blot. E, the recycling sorting signal in β1 as a free peptide competes for binding of the mutant (S554D) C-terminal portion to GST-β1 on beads. The pulldown experiment was performed. A free peptide containing an irrelevant sequence of similar length (derived from the cytoplasmic domain of Wbp1) was used as control (Ctl).
We had found previously that the mutation of Ser-554 in ACAP1 to aspartate (S554D), which mimicked constitutive phosphorylation, enhanced cargo binding (16). When this mutation was introduced into the C-terminal portion, we found that binding to β1 was also enhanced (Fig. 4D). In further support of the functional relevance of this binding, we found that mutating the recycling sorting signal in β1 reduced the ability of the S554D mutation to enhance cargo binding by the C-terminal portion (Fig. 4D). We also performed a competition experiment and confirmed that a peptide encompassing the recycling sorting signal in β1 eliminated enhanced cargo binding of the C-terminal portion induced by the S554D mutation (Fig. 4E). Thus, as the C-terminal portion recapitulated how cargo binding is achieved by ACAP1, we next pursued structural analysis of this construct.
Structural Analysis of the C-terminal Portion
We initially attempted to solve the crystal structure of the C-terminal portion using the molecular replacement method, reasoning that the GAP-ANK domains of ASAP2 and ASAP3 had been solved previously and thus could be used as models (41, 42). However, this approach failed, predicting that the corresponding domains in ACAP1 possessed more differences than anticipated. Next, pursuing anomalous diffraction based on the zinc ion coordinated in the GAP domain, we solved the crystal structure of the C-terminal portion to 2.2 Å resolution (Table 1). Of the 363 residues in the C-terminal portion (residues 378–740), those that resided in the GAP or ANK domain could be clearly traced (250 residues). Residues that could not be traced resided in a region N-terminal to the GAP domain (residues 378–403), a region C-terminal to the ANK domain (residues 698–740), and a region between the GAP and ANK domains (residues 525–568).
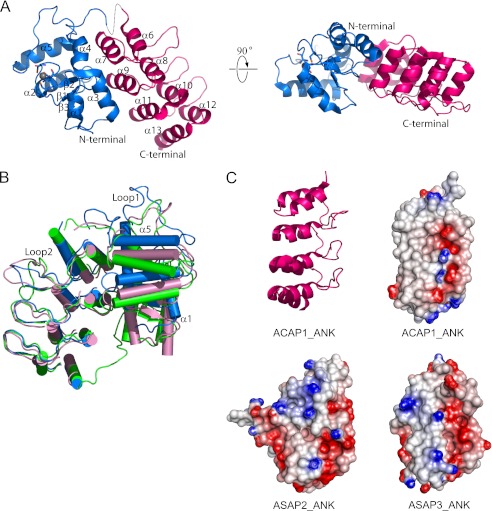
The general features of the GAP and ANK domains were similar to those of the corresponding domains in ASAP2 and ASAP3 (41, 42). The GAP domain of ACAP1 exhibits a three-stranded β-sheet flanked by five α-helices on three sides (Fig. 5A). A zinc ion is well coordinated by four cysteines (Cys-420, Cys-423, Cys-440, and Cys-443), which reside within a conserved GATA-like motif (Cys-X2-Cys-X16-Cys-X2-Cys). The ANK domain contains four ankyrin repeats (Fig. 5A), forming an elongated structure with approximate dimensions of 45 × 25 × 20 Å3. Each repeat consists of the ubiquitous β-hairpin-helix-loop-helix motif, and their adjacent long loops form the β-hairpin motif and are stabilized by hydrogen bonds. The helices proximal to the β-hairpin motif are shorter than the helices on the distal side. As a result, a concave surface facing the tips of the β-hairpin is formed.
FIGURE 5.
Structure of the C-terminal portion of ACAP1. A, the structure of the C-terminal portion of ACAP1 shown at two different orientations. The GAP domain is colored blue. The ANK domain is colored red. The zinc ion is shown as a gray sphere. The secondary structural elements (α-helices and β-sheets) are also labeled. B, comparing the GAP domain of ACAP1 with the corresponding domains in ASAP2 and ASAP3. The three structures are shown in schematic representation and superimposed based on their ANK domains. ACAP1 is colored blue, ASAP2 is colored green, and ASAP3 is colored pink. C, comparison of the predicted electrostatic surfaces of the different ANK domains. The ribbon representation of the ANK domain of ACAP1 is shown for orientation guidance. The electrostatic surfaces of the different ANK domains are shown, with negatively charged regions colored red and positively charged regions colored blue.
The C-terminal portion also possesses features that could explain why the molecular replacement method could not be used to solve its structure based on the corresponding domains of ASAP2/3. The root mean square deviation of the GAP domains between ACAP1 and ASAP2 is 1.6 Å for 118 Cα atoms. When ACAP1, ASAP2, and ASAP3 were superimposed with respect to their ANK domains, the GAP domain of ACAP1 exhibited significant displacement. As a result, the angle spanned by the ANK and GAP domains of ACAP1 was increased by ∼20° compared with those of ASAP2 and ASAP3 (Fig. 5B). Also, loop 1, which connects helices 4 and 5 in the GAP domain of ACAP1, differed significantly from the corresponding regions of ASAP2 and ASAP3 (Fig. 5B). Furthermore, although the ANK domains of all three ARF GAPs exhibited similar general features, their predicted electrostatic surfaces showed obvious differences (Fig. 5C).
Linker Region between the GAP and ANK Domains Regulates Cargo Binding
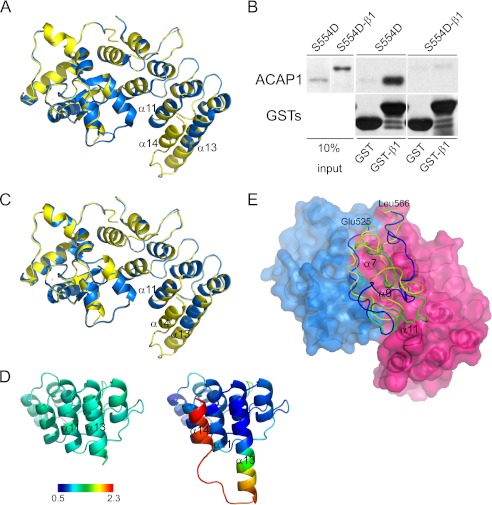
We next sought insight into how cargo binding is accomplished by ACAP1 by attempting to solve a co-crystal structure consisting of the C-terminal portion bound to the β1 cargo peptide. However, repeated trials using different lengths of the β1 peptide, introduction of the S554D mutation to enhance β1 binding, and also changing crystallization conditions failed to achieve this goal. A general approach to overcome such an obstacle has been to fuse the interacting partners covalently, which further enhances their interaction. Thus, we next pursued this approach by fusing the β1 peptide to the C terminus of the C-terminal portion and also introducing the S554D mutation to further enhance cargo binding (S554D-β1). We then sought to crystallize this fusion construct. In this effort, we had to overcome the tendency of the β1 peptide to become cleaved during the prolonged incubation needed for crystallization. Eventually, we solved the fusion construct to 2.2 Å resolution (Table 1). It was found to superimpose quite well with the wild-type counterpart (Fig. 6A), with a root mean square deviation of 0.46 Å for 249 Cα atoms (residues 405–524 and 569–697). However, we still could not visualize the β1 cargo peptide in this structure.
FIGURE 6.
Linker region between the GAP and ANK domains predicted to mediate regulation of cargo binding by Ser-554. A, structure of the S554D-β1 fusion construct and comparison with the wild-type C-terminal portion. The wild-type form is colored blue, and the fusion construct is colored yellow. B, fusion of the β1 cargo peptide to the S554D form of the C-terminal portion prevents the resulting fusion construct from binding intermolecularly to the same cargo peptide on beads in a pulldown assay. The fusion construct was compared with the non-fusion counterpart in binding to GST-β1 in a pulldown experiment. Input shows proteins stained with Coomassie Blue, whereas pulldown results were immunoblotted for the proteins indicated. C, structure of the C-terminal portion with the S554D mutation and comparison with the S554D-β1 fusion construct. The S554D mutant is colored blue, and the fusion construct is colored yellow. D, temperature factor distribution of C-terminal helices in the ANK domain. The temperature factor (B factor) distribution was compared between S554D (left) and the fusion construct (right). The average B factor value was defined to 1.0, and the relative B factor value was derived from the ratio of every B factor value of Cα to the average value. The color bar shows the different values along a gradient, from 0.5 Å2 (dark blue) to 2.3 Å2 (dark red). A lower B factor predicts more stability. E, molecular modeling of the linker region. The GAP domain is colored blue, and the ANK domain is colored red. The linker region is modeled onto the surface representation of these domains, with the wild-type form shown in green, S554D shown in yellow, and S554A shown in blue. The regions of the ANK domain predicted to be covered by the linker are also labeled.
We confirmed by a functional assay that the fusion construct resulted in the C-terminal portion binding to the fused β1 peptide. Whereas the S554D mutant alone could bind GST-β1 on beads in a pulldown assay, the fusion construct showed marked reduced ability (Fig. 6B). We pursued two additional approaches, which also suggested that the fusion construct was active. First, by solving the S554D mutation without the fused β1 peptide (Table 1) and by comparing with the fusion construct, we found that they were virtually superimposable (Fig. 6C). Thus, as the S554D mutant form of the C-terminal portion is active in cargo binding (see Fig. 4), the fusion construct is also likely active. Second, we noted that an additional helix (α14) became detectable in the fusion construct compared with the S554D construct without the fused β1 peptide (Fig. 6C). This finding suggested that the fused β1 peptide interacted with the C-terminal portion, resulting in enhanced stability at the C terminus of the ANK domain, where the fusion occurred. Consistent with this explanation, we performed temperature factor analysis, which also predicted enhanced stability at the C terminus of the ANK domain (Fig. 6D). Thus, the results are consistent with the fusion construct leading to enhanced cargo binding of the β1 peptide by the C-terminal portion.
A general explanation for why a portion of a crystal structure cannot be visualized is that it is too flexible, predicting that additional component(s) help to stabilize cargo binding by the C-terminal portion. Thus, future studies will be needed to identify such predicted component(s). In the meantime, we sought further mechanistic insight into how cargo binding could be regulated by Ser-554 by considering that the S554D mutation did not induce a major conformational change within the C-terminal portion in explaining how this mutation enhanced cargo binding. We further noted that the linker region between the GAP and ANK domains, which contains Ser-554, could not be traced in any of the solved constructs. Thus, we next pursued molecular modeling to gain insight into whether this linker region could be involved in the regulation of cargo binding by Ser-554.
Initially, smaller segments of the linker region were searched for homologous templates in the Protein Data Bank. These segments were then modeled into longer segments using protein structure and flexible docking programs while also considering energy minimization and taking into consideration a physiologic aqueous environment. From these analyses, the linker region was modeled to cover an adjoining region contributed mainly by helices α7, α9, and α11 of the ANK domain (Fig. 6E). In comparison, the S554D mutation was predicted to induce the linker region to tilt away from the ANK domain near helix α11 (Fig. 6E). Thus, molecular simulation suggested the possibility that the linker region could be acting as a “flap” that covered the cargo binding site, with the S554D mutation inducing this flap to “uncover” from the binding site.
We also examined the effect of the S554A mutation, which has been shown previously to inhibit cargo binding (16). Compared with either the wild-type or S554D form, molecular simulation predicted the greatest difference imposed by the S554A mutation would occur in the linker region covering the ANK domain near helix α7 (Fig. 6E). Future studies will be needed to elucidate how coverage in this region of the ANK domain would result in a dominant-negative inhibition of cargo binding by the S554A mutation. In the meantime, as the wild-type form of Ser-554 is more likely to represent the physiologic unphosphorylated state of Ser-554 than S554A, we considered that the comparison between the wild-type form and S554D is more likely to reflect how phosphorylation at Ser-554 affects the role of the linker region in modulating cargo binding.
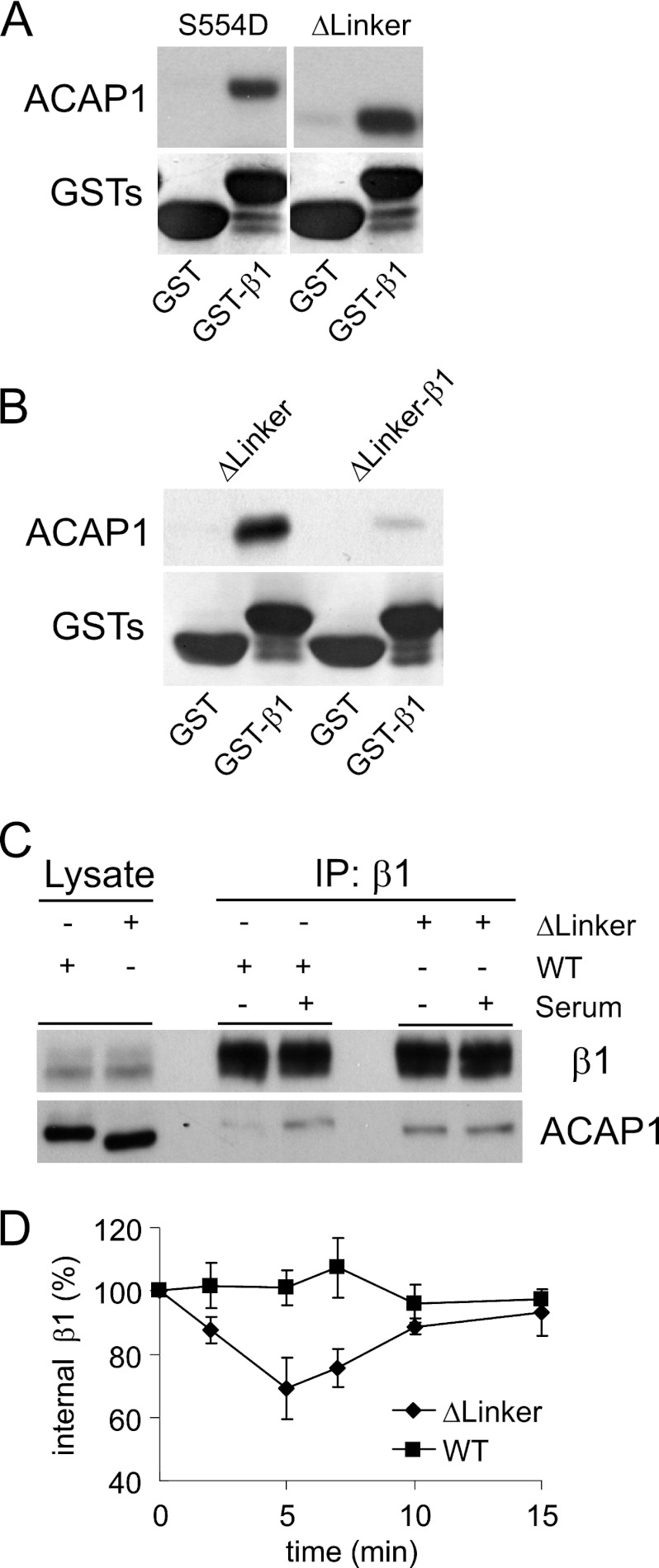
In particular, as comparison between the wild-type and S554D forms suggested that the linker region could be acting to inhibit cargo binding, we next sought functional support by excising this region and then assessing for cargo binding by the resulting mutant. Notably, cargo binding was enhanced, similar to that seen for the S554D mutant. This enhancement could be observed either by assessing the binding of the linker mutant to GST-β1 on beads in pulldown assay (Fig. 7A) or in the context of a competition experiment in which the ability of a fusion construct (composed of the linker mutant fused to the β1 peptide) was assessed for intermolecular binding to GST-β1 (Fig. 7B). We also sought in vivo confirmation for the effect of excising the linker region. We generated a full-length form of ACAP1 with the linker region excised. After transfection into cells, the ability of the ACAP1 mutant to bind endosomal β1 and its effect on β1 recycling were assessed. Similar to the previously observed effects of the S554D mutation (16), excision of the linker region also resulted in enhanced binding to endosomal β1 under basal conditions (Fig. 7C). Moreover, the expression of this mutant in cells converted β1 recycling from a regulated to a constitutive (stimulation-independent) process (Fig. 7D), similar to that seen previously for the effect of the S554D mutant (16). Thus, the results support the role of the linker region acting to inhibit cargo binding.
FIGURE 7.
Linker region inhibits cargo binding. A, excision of the linker region results in enhanced cargo binding by ACAP1 in vitro. The different forms of ACAP1 were incubated with GST-β1 in a pulldown experiment. B, fusion of the β1 peptide to the linker mutant prevents the resulting fusion construct from binding intermolecularly to the same cargo peptide on beads in a pulldown assay. The fusion construct was compared with the non-fusion counterpart in binding to GST-β1 in a pulldown experiment. The input shows proteins stained with Coomassie Blue, whereas pulldown results were immunoblotted for the proteins indicated. C, excision of the linker regions results in enhanced cargo binding by ACAP1 in vivo. ACAP1 (either wild-type or with the linker region excised) was expressed in HeLa cells. The association of endosomal β1 with either form of ACAP1 was then assessed through co-precipitation (IP). D, excision of the linker region induces β1 recycling under basal (no stimulation) conditions. The integrin recycling assay was performed under basal conditions on HeLa cells that stably expressed either wild-type or mutant (with the linker region deleted) ACAP1. The mean ± S.E. from three experiments is shown.
DISCUSSION
We have advanced the mechanistic understanding of how ACAP1 achieves regulated cargo sorting. Cargo sorting is defined operationally by a coat component binding to a particular sequence in the cargo and by demonstrating that this sequence acts functionally as a sorting signal. Thus, we initially identified a minimal region in the β1 cargo recognized by ACAP1 and showed that this binding represents cargo sorting because this minimal region acts functionally as a recycling sorting signal. We next identified a minimal region in ACAP1 that reproduces regulation of cargo binding by Ser-554, which involves the C-terminal portion (composed of the GAP and ANK domains).
We then pursued structural studies on this C-terminal portion. However, whereas this portion of ACAP1 could be largely solved, cargo binding of the β1 peptide could not be visualized, despite efforts to enhance this binding through the use of a fusion construct. Importantly, a linker region between the GAP and ANK domains containing Ser-554 also could not be solved. Thus, to pursue further insight into how Ser-554 regulates cargo binding, we pursued molecular modeling of the linker region. This approach suggested the possibility that the linker region could act to prevent cargo binding and that phosphorylation at Ser-554 could relieve this inhibition. We confirmed this autoinhibitory mechanism by excising the linker region and showing that the resulting mutant behaved similar to the S554D mutant in promoting cargo binding and integrin recycling.
Autoinhibition has emerged as a common mechanism of regulating the function of a protein. In most cases, however, autoinhibition has been shown to involve large conformational changes within the protein. Key transport factors that act in this manner have included the cytohesin class of ARF guanine nucleotide exchange factors, which undergo large conformational changes upon phosphorylation so that their catalytic domain can contact cognate ARFs as substrates (43). In contrast, our results with ACAP1 suggest a more localized mechanism of autoinhibition, which involves a flexible linker region between the GAP and ANK domains that acts to obstruct cargo binding, with a key residue within this region (Ser-554) regulating this obstruction. Thus, our findings expand an appreciation for the mechanistic spectrum by which autoinhibition can occur in modulating the function of a transport factor.
Our findings also advance a mechanistic understanding of a key regulatory juncture that controls integrin recycling. As integrin recycling is critical for cell migration, there has been great interest in identifying both upstream signaling components and downstream transport effectors of this recycling event (3–5). From this effort, there has also been an elucidation of how a signaling component (Akt) impacts on a transport factor (ACAP1) in explaining how integrin recycling involves regulated transport (16). We have now further advanced a mechanistic understanding of this key regulatory juncture in integrin recycling.
Acknowledgment
We thank the Supercomputing Center of the Chinese Academy of Sciences for computational resources.
This work was supported, in whole or in part, by National Institutes of Health Grant GM073016. This work was also supported by National Science Foundation of China Grants 31000635 and 31021062 and Chinese Ministry of Science and Technology 973 Project Grants 2011CB910301 and 2011CB910900.
The atomic coordinates and structure factors (codes 3JUE, 4F1P, and 3T9K) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ARF
- ADP-ribosylation factor
- GAP
- GTPase-activating protein.
REFERENCES
- 1. Bonifacino J. S., Glick B. S. (2004) The mechanisms of vesicle budding and fusion. Cell 116, 153–166 [DOI] [PubMed] [Google Scholar]
- 2. Pucadyil T. J., Schmid S. L. (2009) Conserved functions of membrane active GTPases in coated vesicle formation. Science 325, 1217–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caswell P., Norman J. (2008) Endocytic transport of integrins during cell migration and invasion. Trends Cell Biol. 18, 257–263 [DOI] [PubMed] [Google Scholar]
- 4. Pellinen T., Ivaska J. (2006) Integrin traffic. J. Cell Sci. 119, 3723–3731 [DOI] [PubMed] [Google Scholar]
- 5. Margadant C., Monsuur H. N., Norman J. C., Sonnenberg A. (2011) Mechanisms of integrin activation and trafficking. Curr. Opin. Cell Biol. 23, 607–614 [DOI] [PubMed] [Google Scholar]
- 6. Li J., Peters P. J., Bai M., Dai J., Bos E., Kirchhausen T., Kandror K. V., Hsu V. W. (2007) An ACAP1-containing clathrin coat complex for endocytic recycling. J. Cell Biol. 178, 453–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caswell P. T., Chan M., Lindsay A. J., McCaffrey M. W., Boettiger D., Norman J. C. (2008) Rab-coupling protein coordinates recycling of α5β1 integrin and EGFR1 to promote cell migration in three-dimensional microenvironments. J. Cell Biol. 183, 143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jović M., Naslavsky N., Rapaport D., Horowitz M., Caplan S. (2007) EHD1 regulates β1 integrin endosomal transport: effects on focal adhesions, cell spreading, and migration. J. Cell Sci. 120, 802–814 [DOI] [PubMed] [Google Scholar]
- 9. Hasan N., Hu C. (2010) Vesicle-associated membrane protein 2 mediates trafficking of α5β1 integrin to the plasma membrane. Exp. Cell Res. 316, 12–23 [DOI] [PubMed] [Google Scholar]
- 10. Roberts M., Barry S., Woods A., van der Sluijs P., Norman J. (2001) PDGF-regulated Rab4-dependent recycling of αvβ3 integrin from early endosomes is necessary for cell adhesion and spreading. Curr. Biol. 11, 1392–1402 [DOI] [PubMed] [Google Scholar]
- 11. Powelka A. M., Sun J., Li J., Gao M., Shaw L. M., Sonnenberg A., Hsu V. W. (2004) Stimulation-dependent recycling of integrin β1 regulated by ARF6 and Rab11. Traffic 5, 20–36 [DOI] [PubMed] [Google Scholar]
- 12. Caswell P. T., Spence H. J., Parsons M., White D. P., Clark K., Cheng K. W., Mills G. B., Humphries M. J., Messent A. J., Anderson K. I., McCaffrey M. W., Ozanne B. W., Norman J. C. (2007) Rab25 associates with α5β1 integrin to promote invasive migration in three-dimensional microenvironments. Dev. Cell 13, 496–510 [DOI] [PubMed] [Google Scholar]
- 13. Pellinen T., Arjonen A., Vuoriluoto K., Kallio K., Fransen J. A., Ivaska J. (2006) Small GTPase Rab21 regulates cell adhesion and controls endosomal traffic of β1 integrins. J. Cell Biol. 173, 767–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oh S. J., Santy L. C. (2010) Differential effects of cytohesins 2 and 3 on β1 integrin recycling. J. Biol. Chem. 285, 14610–14616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mai A., Veltel S., Pellinen T., Padzik A., Coffey E., Marjomäki V., Ivaska J. (2011) Competitive binding of Rab21 and p120RasGAP to integrins regulates receptor traffic and migration. J. Cell Biol. 194, 291–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li J., Ballif B. A., Powelka A. M., Dai J., Gygi S. P., Hsu V. W. (2005) Phosphorylation of ACAP1 by Akt regulates the stimulation-dependent recycling of integrin β1 to control cell migration. Dev. Cell 9, 663–673 [DOI] [PubMed] [Google Scholar]
- 17. Nie Z., Randazzo P. A. (2006) ArfGAPs and membrane traffic. J. Cell Sci. 119, 1203–1211 [DOI] [PubMed] [Google Scholar]
- 18. D'Souza-Schorey C., Chavrier P. (2006) ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 19. Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M. F., Ravazzola M., Amherdt M., Schekman R. (1994) COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77, 895–907 [DOI] [PubMed] [Google Scholar]
- 20. Yang J. S., Lee S. Y., Gao M., Bourgoin S., Randazzo P. A., Premont R. T., Hsu V. W. (2002) ARFGAP1 promotes the formation of COPI vesicles, suggesting function as a component of the coat. J. Cell Biol. 159, 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mazelova J., Astuto-Gribble L., Inoue H., Tam B. M., Schonteich E., Prekeris R., Moritz O. L., Randazzo P. A., Deretic D. (2009) Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. EMBO J. 28, 183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bai M., Gad H., Turacchio G., Cocucci E., Yang J. S., Li J., Beznoussenko G. V., Nie Z., Luo R., Fu L., Collawn J. F., Kirchhausen T., Luini A., Hsu V. W. (2011) ARFGAP1 promotes AP-2-dependent endocytosis. Nat. Cell Biol. 13, 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pryor P. R., Jackson L., Gray S. R., Edeling M. A., Thompson A., Sanderson C. M., Evans P. R., Owen D. J., Luzio J. P. (2008) Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell 134, 817–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dai J., Li J., Bos E., Porcionatto M., Premont R. T., Bourgoin S., Peters P. J., Hsu V. W. (2004) ACAP1 promotes endocytic recycling by recognizing recycling sorting signals. Dev. Cell 7, 771–776 [DOI] [PubMed] [Google Scholar]
- 25. Lee S. Y., Yang J. S., Hong W., Premont R. T., Hsu V. W. (2005) ARFGAP1 plays a central role in coupling COPI cargo sorting with vesicle formation. J. Cell Biol. 168, 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 27. Schneider T. R., Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 28. Terwilliger T. C., Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Terwilliger T. C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hao Q., Gu Y. X., Zheng C. D., Fan H. F. (2000) OASIS: a computer program for breaking phase ambiguity in one-wavelength anomalous scattering or single isomorphous substitution (replacement) data. J. Appl. Crystallogr. 33, 980–981 [Google Scholar]
- 31. Cowtan K. D. (1994) Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography, Vol. 31, pages 34–38, Daresbury Laboratory, Warrington, United Kingdom [Google Scholar]
- 32. Perrakis A., Morris R., Lamzin V. S. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 33. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 35. Brunger A. T. (2007) Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 36. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., Olson A. J. (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sali A., Blundell T. L. (1993) Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 39. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. MacKerell A. D., Jr., Bashford D., Bellott M., Dunbrack R. L., Jr., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., 3rd, Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiórkiewicz-Kuczera J., Yin D., Karplus M. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 41. Mandiyan V., Andreev J., Schlessinger J., Hubbard S. R. (1999) Crystal structure of the ARF GAP domain and ankyrin repeats of PYK2-associated protein β. EMBO J. 18, 6890–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ismail S. A., Vetter I. R., Sot B., Wittinghofer A. (2010) The structure of an Arf-ArfGAP complex reveals a Ca2+ regulatory mechanism. Cell 141, 812–821 [DOI] [PubMed] [Google Scholar]
- 43. DiNitto J. P., Delprato A., Gabe Lee M. T., Cronin T. C., Huang S., Guilherme A., Czech M. P., Lambright D. G. (2007) Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Mol. Cell 28, 569–583 [DOI] [PMC free article] [PubMed] [Google Scholar]